Abstract
SHORT VEGETATIVE PHASE (SVP) genes are members of the well-known MADS-box gene family that regulates vital developmental processes in plants. In Arabidopsis, there are two SVP paralogs, SVP/AGAMOUS-LIKE22 (SVP/AGL22) and AGL24. SVP protein suppresses the flowering process, whereas AGL24 acts as a flowering activator. Phylogenetic analysis of SVP genes representing most of the sequenced eudicot species showed that the SVP gene family could be divided into three major clades in eudicots (SVP1, SVP2, and SVP3), most likely resulting from an ancient whole-genome triplication in core eudicots. Among them, the SVP1 (SVP) and SVP2 (AGL24) clades are retained in nearly all species, whereas the SVP3 clade has been lost in Brassicaceae, Myrtaceae, and some species in other families. Reflecting lineage-specific tandem duplication and whole-genome duplication, SVP gene copy numbers ranged from 3 to 11 in the analyzed species. Sequence analysis showed that SVP3 proteins have obvious differences with SVP1 and SVP2 in the C-terminal (C) domain and intervening (I) domain. Positive selection analysis also showed that the ω (dN/dS) value was highest in the SVP3 clade, with 17 positive selection sites detected in the SVP3 clade. Promoter analysis for cis-regulatory elements showed that some genes in the SVP2 and SVP3 clades may be regulated by abscisic acid, ethylene, and gibberellin. RNA-seq data from grape, poplar, and apple revealed that genes in SVP3 group are highly expressed in vegetative organs such as buds, leaves, cotyledons, and dormant buds in particular, indicating the involvement of genes belong to SVP3 group in the dormancy process. Overall, the findings underscore the functional diversity of the SVP genes in eudicots.
Keywords: SVP, core eudicots, evolutionary history, gene duplication, positive selection, functional diversity
Introduction
The MADS-box gene family encodes transcription factors with crucial roles in floral organ identity regulation and plant development (Ng and Yanofsky 2001). MADS-box genes can be classified into the following 14 clades: StMADS11, AGL17, AGL12, TM3, FLOWERING LOCUS C (FLC), AGL6, AGL2, SQUA, AG, TM8, OsMADS32, DEF/GLO, GGM13, and AGL15 (Gramzow and Theissen 2013; Chen et al. 2017). Some of these clades include floral organ identity genes that can be subdivided into four functional classes: A, B, C, and E genes. These genes are responsible for five different homeotic functions, with A specifying sepals, A + B + E for petals, B + C + E for stamens, C + E for carpels, and D (sister of C genes) for ovules (Pelaz et al. 2000; Honma and Goto 2001). In contrast, other MADS-box genes, such as SHORT VEGETATIVE PHASE (SVP), SOC1, and GMADS, are generally not regarded as core genes involved in floral organ identity (Gregis et al. 2009; Tao et al. 2012).
In Arabidopsis, the two SVP paralogs, SVP/AGAMOUS-LIKE22 (SVP/AGL22) and AGL24, are implicated in floral transition and development. SVP is expressed broadly during vegetative development in leaves and shoot apices, and it acts together with FLC to negatively regulate SOC1 and FT to suppress flowering (Willmann and Poethig 2011). In contrast, AGL24 acts as a flowering activator and promotes SOC1 expression during inflorescence development (Gregis et al. 2009). svp and agl24 mutants are phenotypically distinct in terms of flowering time but have similar leaf-like sepals (Hartmann et al. 2000; Gregis et al. 2009). In the early stage of flower development, SVP and AGL24 act together with AP1 to maintain floral meristem identity. In temperature-dependent flowering regulation, the SVP-FLM-β complex is predominately formed at low temperatures and prevents precocious flowering (Lee, Ryu, et al. 2013; Marin-Gonzalez et al. 2015). Additionally, SVP can reduce gibberellin biosynthesis at the shoot apex to regulate the floral transition (Andres et al. 2014).
Homologs of Arabidopsis SVP and AGL24 with similar functions have been identified and characterized in many species including potato (Garcia-Maroto et al. 2000), Brassica plants (Lee et al. 2007), barley (Trevaskis et al. 2007), rice (Fornara et al. 2008), and the woody perennials Paulownia kawakamii (Prakash and Kumar 2002) and eucalyptus (Brill and Watson 2004). However, SVP homologs with distinct functions have also been identified in other species. For instance, overexpression of MtSVP1 (SVP1 of Medicago truncatula) caused floral defects and delayed flowering in Arabidopsis but only affected floral development in M. truncatula, indicating that MtSVP might have undergone subfunctionalization (Jaudal et al. 2014). Of the four SVP homologous genes in kiwifruit, only SVP1 and SVP3 are able to complement the svp mutant, whereas the others have distinct roles during bud dormancy (Wu et al. 2012; Voogd et al. 2015). SVP genes from Narcissus tazetta, the herbaceous perennial Gentiana triflora, and the basal eudicot Epimedium sagittatum are also associated with dormancy transition (Li et al. 2015, 2016; Yamagishi et al. 2016).
In this study, we performed systematic phylogenetic analyses of the SVP subfamily in eudicots and found that an ancient whole-genome triplication (WGT, γ) event generated three groups of SVPs. Besides the well-known clades of SVP and AGL24, there is a third group of SVP genes (SVP3), some of which have been lost in Brassicaceae and other species. In some species, tandem duplication and lineage-specific duplication contributed to SVP gene family expansion. We calculated the rates of synonymous and nonsynonymous substitution, performed promoter region analysis, and investigated the expression patterns of duplicated gene pairs in the three clades. The present findings indicate the functional diversity of SVP genes in eudicots.
Materials and Methods
Identification of SVP Homologous Genes in Eudicots
All eudicot and Amborella whole-genome data were downloaded from the Phytozome v12.0 database (Goodstein et al. 2012) (https://phytozome.jgi.doe.gov/pz/portal.html) and the genome databases listed in supplementary table S1, Supplementary Material online. The water lily (Nymphaea colorata) genome was recently sequenced by our laboratory and stored in the database: www.angiosperms.org (unpublished data). SVP homologs were obtained by running a local BLASTp search (Altschul et al. 1990) using the Arabidopsis SVP sequence (AT2G22540) as a query against all protein sequences in each genome with an E-value cutoff of 10−5. All hits were analyzed for the MADS-box (PF00319) and K-box (PF01486) domains using HMMER software (Finn et al. 2011). Multiple sequences were aligned using the accurate alignment software MAFFT with default parameters (Katoh and Standley 2013), and a phylogenetic tree was generated. The maximum likelihood (ML) tree was constructed with FastTree software using the JTT+CAT model (Price et al. 2009).
SVP Sequence Alignment and Phylogenetic Tree Construction
All SVP amino acid sequences were aligned using MAFFT then used to construct the phylogenetic tree. Both Bayes and ML phylogenetic analyses were performed using CIPRES v3.3 (Miller et al. 2010) (https://www.phylo.org/portal2). For Bayes, we used MrBayes v3.2 (Ronquist et al. 2012) set to 8,000,000 generations and 4 Markov chains, and the first 25% of the trees from all runs were discarded. For ML, we used RAxML v8.2.10 (Stamatakis 2014) and assumed a general time of nucleotide evolution. Default parameter settings were used to construct SVP phylogenetic trees in FastTree v2.1.10 (Price et al. 2009).
Cis-Regulatory Element Prediction in the 3-kb Upstream Regions of SVP Genes
For genes in all three SVP clades, the promoter sequences comprising at least 3 kb of the upstream regions were downloaded from the genome databases (Phytozome v12.0) listed in supplementary table S1, Supplementary Material online. The PlantCARE (Lescot 2002) and PLACE (Higo et al. 1999) databases were used to predict the conserved cis-regulatory elements in the promoter regions.
Analysis of Positive Selection
The ratios of synonymous substitution (dS) and nonsynonymous substitution (dN) in or between different clades were calculated with the Ka/Ks calculator (Zhang et al. 2006). In this study, we used phylogenetic analysis by ML (PAML) (Yang 2007) to evaluate positive selection in the SVP family. To explore how selection occurred in SVP1, SVP2, and SVP3, a branch model was used to evaluate the ω value for each clade. A likelihood ratio test (LRT) was performed to accept or reject the hypothesis of model 0 (only one ω value in the tree) and model 2 (allow ω value to vary among foreground and background branches) in the three clades. The significance of the LRT was calculated with the assumption that it was twice the difference in the log of maximum-likelihoods and was distributed normally as a χ2 distribution, with the degrees of freedom (df) calculated by the difference in the number of parameters in the models. Positive selection was also detected using the branch site model. The test was based on the comparison between the following two models: A model (MA) that allowed positive selection on one or more branches and another model (MA1) that did not allow positive selection. Each of the three clades was set as foreground for testing positive selection sites. The LRT was used to accept one of the models, but in this case, the P-value obtained for the χ2 distribution of 2LRT was divided by two df. When the LRT suggested the action of positive selection, Bayes Empirical Bayes (BEB) analysis was used to evaluate the posterior probability (pp) that each codon belonged to the site class of positive selection on the foreground branch.
Homology Modeling and Evolutionary Conservation Analysis across Residues
Motif analysis was conducted using MEME v5.0 software (Bailey et al. 2009). Sequence logos were generated from the web-logo website (http://meme-suite.org/). Using gene IDs from different SVP groups, we determined the collinearity of genomic segments from the same species and different species using the PGDD website (http://chibba.agtec.uga.edu/duplication/) (Lee, Tang, et al. 2013). A synteny plot for the SVP family was constructed using PLAZA v3.0 (Proost et al. 2015). The structures of all SVP proteins were generated de novo using the I-TASSER Server (Yang et al. 2015). Top-scoring models were chosen, and all structures in the figures were visualized using PyMOL v2.0 software (Alexander et al. 2011). The evolutionary conservation scores across amino acid residues were calculated in the Consurf server (Ashkenazy et al. 2016) using a sequence alignment that included three clades of SVP members. All images were modified and represented using PyMOL v2.0 software (Alexander et al. 2011).
Gene Expression Analysis
RNA expression data were downloaded from previous studies of soybean (Shen et al. 2014), grape (Khalil-Ur-Rehman et al. 2017), poplar (SRR6031364-95 in NCBI-SRA database), and apple (Kumar et al. 2017). Using Hisat2 v2.0.1 (Kim et al. 2015), reads were mapped to the soybean genome (Glycine max Wm82.a2.v1), grape genome (Vitis vinifera Genoscope.12X), poplar genome (Populus trichocarpa v3.0), and apple genome (Malus_x_domestica-genome_GDDH13_v1.1). The reads were assembled and quantified using StringTie v1.2.2 software (Pertea et al. 2015). Gene expression levels were calculated as fragments per kilobase of exon per million fragments mapped (FPKM). Differentially expressed genes were analyzed with the R package of Ballgown (Frazee et al. 2015). Transcriptome analysis was performed according to a previously described transcriptome protocol (Pertea et al. 2016).
Results
Identification of SVP Genes in Eudicots
Using the Arabidopsis SVP (AT2G22540) protein sequence as a query, 10,275 hits were obtained in 2 basal angiosperms and 81 eudicots (fig. 1). MADS-box genes in angiosperms can be divided into 14 subclades, and the SVP subclade represents only 1 group among them (Gramzow and Theissen 2013). Therefore, the BLAST hits were not limited to SVP genes, so a phylogenetic tree was subsequently constructed using all of the obtained sequences. The approximate ML tree generated using FastTree software (supplementary fig. S1, Supplementary Material online) placed all of the candidate genes into 12 subgroups (AP1, FLC, SEP, SOC1, AG, GMADS, AGL15, ANR1, AGL32, AP3/PI, SVP, and AGL12). The SVP group contained some genes that were experimentally characterized as SVP homologs, including Arabidopsis SVP (AT2G22540) and AGL24 (AT4G24540). Based on this analysis, 310 full-length sequences from the SVP group were retained for subsequent analyses (supplementary table S1, Supplementary Material online).
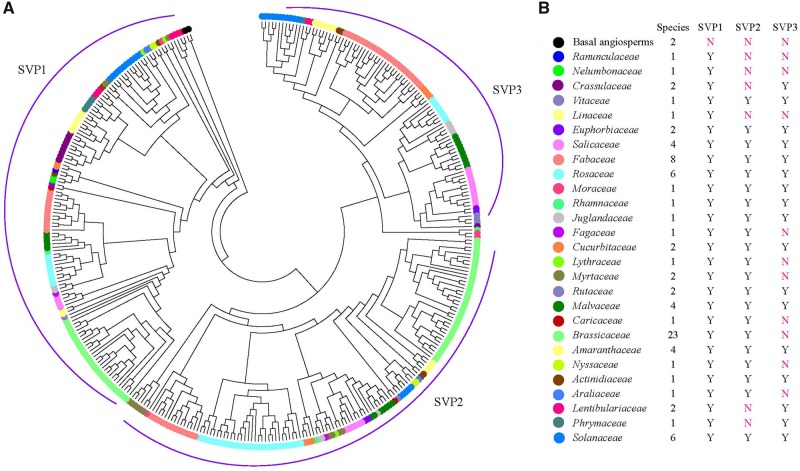
Fig. 1.
—Phylogeny of all SVP genes in eudicots and two basal angiosperms. (A) Using basal angiosperm SVP genes (from Amborella trichopoda and water lily) as the outgroup, the SVP members from eudicots were classified into three clades (SVP1, SVP2, and SVP3) based on their phylogeny. (B) The left panel lists all the eudicot families and the number of species included in this study. The right panel indicates which SVP clades were present in each eudicot family. Y, present; N, absent.
Phylogenetic Analysis Revealed SVP Gene Family Expansion by Whole-Genome Duplication and Tandem Duplication
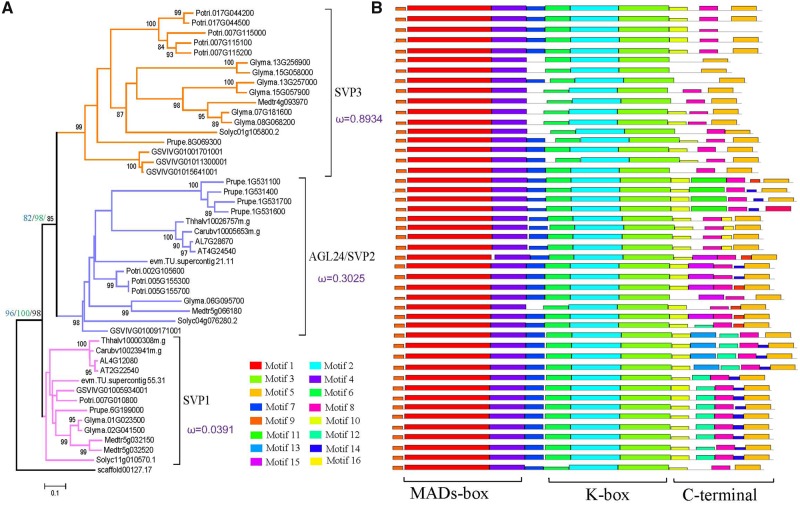
To investigate the evolutionary history of the plant SVP gene family, we constructed ML and Bayes trees using the 310 full-length SVP group protein sequences and 2 basal angiosperm (Amborella trichopoda and N.colorata) SVPs as an outgroup. Based on the topology and pp >80, SVP genes in eudicots were divided into three clades (figs. 1 and 2). The SVP1 clade contained genes that were previously experimentally characterized as SVP genes, including those from Arabidopsis thaliana, P.trichocarpa, and V.vinifera. The AT4G24540 sequence in the SVP2 clade was previously identified and characterized as AGL24. SVP1 clade genes were present in all of the analyzed eudicot species. SVP2 clade genes were absent in the Ranunculaceae, Nelumbonaceae, Crassulaceae, Linaceae, Lentibulariaceae, and Phrymaceae. The SVP3 clade clustered with SVP2 as a sister group, and SVP3 genes were absent in the Ranunculaceae, Nelumbonaceae, Linaceae, Fagaceae, Lythraceae, Myrtaceae, Rutaceae, Caricaceae, Myssaceae, Araliaceae, and Brassicaceae (fig. 1 and supplementary table S1, Supplementary Material online). Only one SVP gene was identified in the basal angiosperms A. trichopoda and N. colorata, but many copies were identified in eudicots. The analysis suggested that there were at least 3 copies of SVP in each eudicot species, but there were many species with at least 6 copies, such as Gossypium hirsutum with 11 copies, indicating expansion of the SVP genes in some species. Examples of a high copy number for a specific clade include Utricularia gibba and Kalanchoe laxiflora, each with six copies in the SVP1 clade; Fragaria vesca with five copies and Prunus mume with six copies in the SVP2 clade; and G.max with six copies and G. hirsutum with five copies in the SVP3 clade.
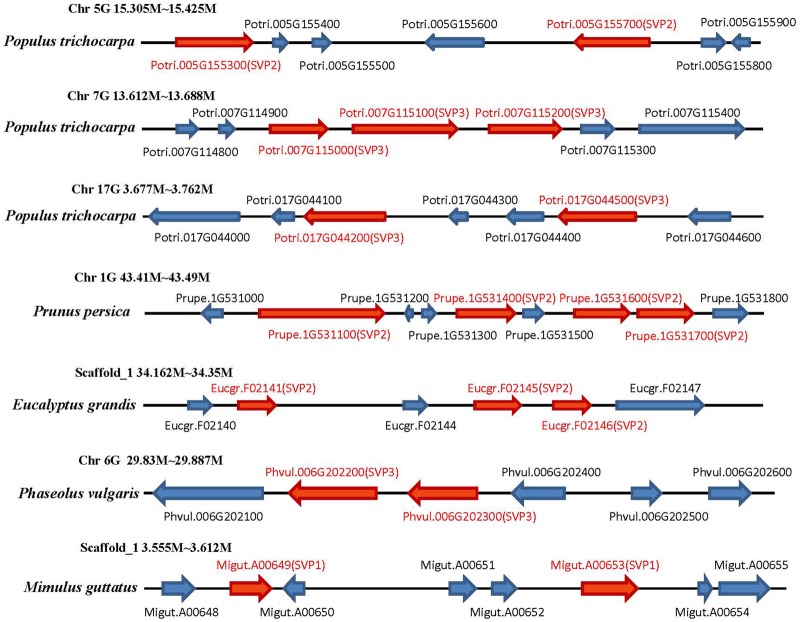
Whole-genome duplications have a major impact on gene copy number in plants (Maere and De Peer 2010). Synteny analyses showed clear collinearity relationships in SVP1 and SVP3 and in SVP2 and SVP3, such as in P. trichocarpa, G. max, and P. vulgaris (supplementary fig. S2, Supplementary Material online). We used the synonymous substitution rates (Ks) as a proxy value for time to compare the date of gene duplications. Most of the Ks values were >1 among the three different clade gene pairs (supplementary table S2, Supplementary Material online). The three SVP clades probably resulted from an ancestral polyploidy event shared by all core eudicots, with some species having many copies belonging to specific clades. For example, P. persica had four copies and poplar had three copies of genes in the SVP2 clade. These genes were located in adjacent regions in collinear genomic segments (such as in P. trichocarpa, G. max, and P. persica; fig. 3). Most Ks values of the SVP gene pairs were <0.5 (supplementary table S3, Supplementary Material online), suggesting expansion of the SVP gene family by tandem duplication. There was also evidence of lineage-specific duplication events contributing to the expansion of the SVP gene family in poplar and soybean, resulting in five and six copies, respectively (fig. 2A: SVP3 clade). However, the tandem duplication events differed between poplar and soybean. In G. max, tandem duplication occurred prior to a whole-genome duplication event, whereas the opposite process appears to have occurred in poplar. In collinear genomic segments (including ten genes adjacent to SVP genes), seven genes adjacent to the SVP genes belonged to corresponding gene families only in three Brassicaceae species (SVP1 group), and four adjacent genes belonged to the corresponding gene families in poplar and soybean species (SVP3 group) (supplementary fig. S4, Supplementary Material online).
Fig. 3.
—Structure of SVP tandem duplication gene clusters in specific species. Conserved regions with variable numbers of SVP genes in different orientations located in syntenic regions of the genome. Chr and Scaffold indicate the chromosome and scaffold number for the individual species. SVP genes are marked in red, and the orientations of these genes are indicated by the arrows.
Fig. 2.
—Phylogenetic tree of representative SVP genes in eudicots. (A) The phylogenetic tree was constructed using ML and Bayes method, and the A. trichopoda SVP gene was used as the outgroup. Group SVP1, magenta; AGL24/SVP2, blue; SVP3, yellow. The ω (dN:dS) values calculated by PAML software are marked in each clade. (B) Motif analysis showing structural differences in the three SVP clades. SVP genes include the following three domains: MADS-box, K-box, and C-terminal region.
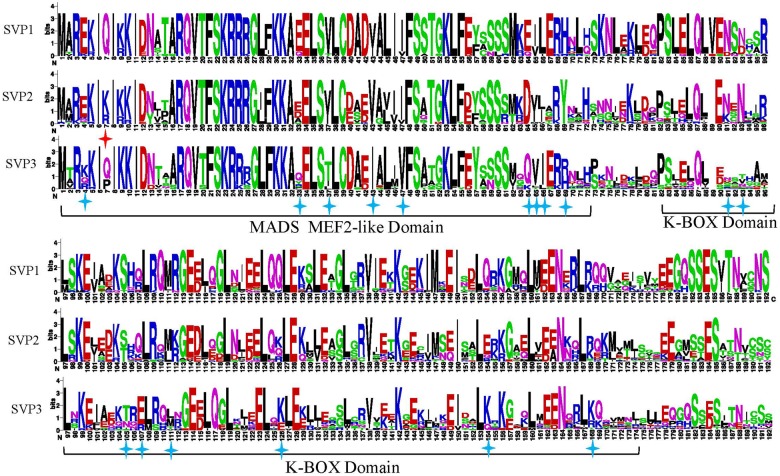
SVP3 Clade under Positive Selection
A possible functional divergence of SVP genes was found among the three clades in eudicots, and we hypothesized that the different SVP clades had undergone adaptive evolution and that some amino acid residues might correlate with functional diversification. To test this hypothesis, we investigated selection models for each clade using Codeml (Yang 2007). An initial examination of selection patterns among SVP1, SVP2, and SVP3 using branch model analysis showed a significant difference between the null model and two-branch model (P = 0.0262). The results indicated low dN:dS (ω) values for SVP1 (0.0391) and SVP2 (0.3025) but a relatively higher ω value for SVP3 (0.8934) (table 1 and fig. 2). The branch site model was subsequently used to test the three groups, with each individual group as foreground and all three groups as background. BEB was used to calculate the pp of sites resulting from the site class with ω > 1. Seventeen positive selection sites were detected in the SVP3 clade, one site was identified in the SVP2 clade, and no sites were identified in the SVP1 clade. Two sites in the SVP3 clade with pp >0.95, V37 (BEB =0.993*) and E64 (BEB =0.959*) were identified as having potential roles in functional divergence. These two sites were both located in the MADS-box domain. The other 15 sites under positive selection were located in the MADS-box and K-box domains (tables 2 and 3 and fig. 4).
Table 1.
LRT of Branch Model in the SVP Gene Family in Eudicots
| Model | ln L | Parameter Estimates ω |
P-Value | ||
|---|---|---|---|---|---|
| SVP1 | SVP2 | SVP3 | |||
| Model 2 | −10309.033696 | 0.0391 | 0.3025 | 0.8934 | 0.0262* |
| Model 0 | −10313.657443 | 0.1925 | |||
Table 2.
LRT of Branch Site Model for the SVP Gene Family in Eudicots
| Clade | Model | ln L | Parameter Estimates | Positive Selection Sites | P-Value | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVP1 | Model A | −10171.89 | Site class | 0 | 1 | 2a | 2b | Not found | 1 |
| f | 0.7944 | 0.2055 | 0.00000 | 0.00000 | |||||
| ω0 | 0.1585 | 1.0000 | 0.15856 | 1.00000 | |||||
| ω1 | 0.1585 | 1.0000 | 1.00000 | 1.00000 | |||||
| Model A null | −10171.89 | 1 | |||||||
| SVP2 | Model A | −10171.81 | Site class | 0 | 1 | 2a | 2b | 7 Q 0.572 | 1 |
| f | 0.6698 | 0.1736 | 0.12430 | 0.03223 | |||||
| ω0 | 0.1581 | 1.0000 | 0.15818 | 1.00000 | |||||
| ω1 | 0.1581 | 1.0000 | 1.00000 | 1.00000 | |||||
| Model A null | −10171.81 | 1 | |||||||
| SVP3 | Model A | −10165.50 | Site class | 0 | 1 | 2a | 2b | Table 3 | 0.7910 |
| f | 0.5675 | 0.1473 | 0.22641 | 0.05877 | |||||
| ω0 | 0.1544 | 1.0000 | 0.15441 | 1.00000 | |||||
| ω1 | 0.1544 | 1.0000 | 3.43907 | 3.43907 | |||||
| Model A null | −10165.54 | 1 | |||||||
Table 3.
Sites Predicted to be Under Positive Selection in the SVP3 Gene
| Amino Acid Position | SVP Amino Acid (AtSVP Site) | BEB Value |
|---|---|---|
| 4 | 4 E | 0.747 |
| 33 | 33E | 0.826 |
| 37 | 37 V | 0.993** |
| 43 | 43 V | 0.840 |
| 47 | 47 I | 0.853 |
| 63 | 64 E | 0.959* |
| 64 | 65 V | 0.857 |
| 65 | 66 L | 0.804 |
| 67 | 69 H | 0.602 |
| 81 | 91 N | 0.513 |
| 83 | 93 D | 0.837 |
| 90 | 105 S | 0.517 |
| 92 | 107 R | 0.839 |
| 96 | 111 M | 0.797 |
| 111 | 126 Q | 0.811 |
| 128 | 154 Q | 0.776 |
| 141 | 168 R | 0.565 |
Fig. 4.
—Logos indicating sequence conservation and positive selection sites in the three SVP clades. Positive selection sites in the SVP2 and SVP3 clades are marked with red and blue stars, respectively. One of the SVP2 positive selection sites was located in the MADS-box domain. Nine and eight SVP3 positive selection sites were located in the MADS-box and K-box domains, respectively.
In some species, such as P. persica, P. trichocarpa, and K. laxiflora, a high number of SVP genes belonged to a specific clade. To test whether there was a significant difference in the selection pressure among the three SVP clades, we performed a branch model test in each species. Free ratio model tests were used, which allows an independent ω value to be calculated for each branch. These analyses indicated three branches in K. laxiflora, four branches in P. trichocarpa, and one branch in peach with a ω ratio >1 (supplementary fig. S3, Supplementary Material online). Among these branches, two branches of K. laxiflora existed in the SVP1 clade, and two branches of strawberry and four branches of apple in the SVP2 clade were subjected to selection. These findings indicated that selection constraints occurred not only in the SVP3 clade but also in SVP1 and SVP2.
Promoter Cis-Regulatory Element Prediction
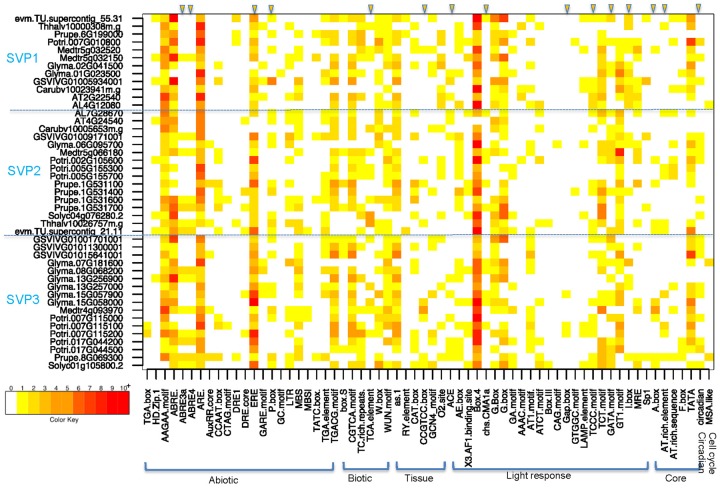
To investigate the possible role of cis-regulatory elements in the promoter regions of SVP genes, we analyzed the 3 kb region upstream of the translation start site of 45 SVP genes. Based on their functional differences, the identified cis-regulatory elements were classified into the following seven groups: Abiotic, biotic, tissue-specific, core promoter element, light responsive, circadian, and cell cycle. Cis-Regulatory elements related to abiotic stress response, such as ABRE3a and ABRE4 involved in the abscisic acid response, were present in six SVP2 genes, seven SVP3 genes, and only one SVP1 gene. The ethylene response-related ERE element was more abundant in the SVP3 group than in SVP1 and SVP2. The P-box motif associated with gibberellin responsiveness was identified in nine SVP2 and eight SVP3 genes (fig. 5 and supplementary tables S4 and S5, Supplementary Material online). The biotic stress-related TCA-element, which is involved in salicylic acid responsiveness, was found in three SVP1, seven SVP2, and five SVP3 genes (fig. 5 and supplementary tables S4 and S5, Supplementary Material online). The CCGTCC-box is a regulatory element related to meristem-specific activation and was present in one SVP1, three SVP2, and five SVP3 genes (fig. 5 and supplementary tables S4 and S5, Supplementary Material online). There were large differences in the number of genes containing the following light response elements, and the numbers in parentheses indicate the number of analyzed genes in SVP1, SVP2, and SVP3 groups containing the indicated element: ACE (3, 3, 6); chs-CMA1a (1, 5, 5); GATA-motif (4, 8, 11); I-box (10, 3, 4); and A-box (1, 4, 5). Additionally, the GAP-box was present in only three SVP3 genes (fig. 5 and supplementary tables S4 and S5, Supplementary Material online). The circadian responsive element (Circadian) was found in three SVP1, six SVP2, and nine SVP3 genes (fig. 5 and supplementary tables S4 and S5, Supplementary Material online).
Fig. 5.
—Cis-Regulatory elements identified in the three SVP clades. The promoter regions (3 kb upstream of the transcription start site) of SVP genes in each of the three clades were scanned for cis-regulatory elements. Seven groups of cis-regulatory elements (abiotic, biotic, tissue, light response, core promoter element, circadian, and cell cycle) are listed in the X axis, and gene IDs are listed in the Y axis. Yellow triangles with a blue outline denote a difference in the three SVP clades for the indicated cis-regulatory element. The color key from white to red indicates the number of cis-regulatory elements in the promoter region from 0 to 10+.
Conservation and Variation in Sequence Composition, Motifs, and Spatial Structure
To identify conserved and variant features of SVP sequences, sequence logos were generated to visualize their components (fig. 4). Among the three clades, SVP3 exhibited the most site variation, and many of these sites were in the K-box domain. At the motif level, most SVP genes in all species were highly conserved and had similar structures, including the typical MADS-box domain, K-box domain, and C-terminal region. Eight motifs (1, 2, 3, 4, 5, 6, 8, and 9) were conserved in all SVP sequences, and some of these were highly conserved sequences in the MADS-box and K-box domains (fig. 2B). Motifs 5 and 8 were the most highly conserved in SVP genes. Motif 7 in the intervening (I) region was highly conserved in the SVP1 and SVP2 clades but was lost in some species in the SVP3 clade. Much greater differences were observed among the three SVP groups for the C-terminal region. Motifs 12 and 14 were specific to the SVP1 clade, and motif 13 was unique to Brassicaceae species. Motifs 11, 15, and 16 existed in some species in the SVP2 clade. Motifs 11–16 were all absent or lost in the SVP3 group. Using the multiple sequence alignment of all SVP genes, we calculated the conservation score for each position using the ConSurf web server (http://consurf.tau.ac.il/2016//credits.php). We found that the MADS-box and K-box domains formed a groove, which could potentially interact with DNA regions (fig. 6). The MADS-box domain was highly conserved in all SVP genes compared with the K-box domain and the C-terminal region.
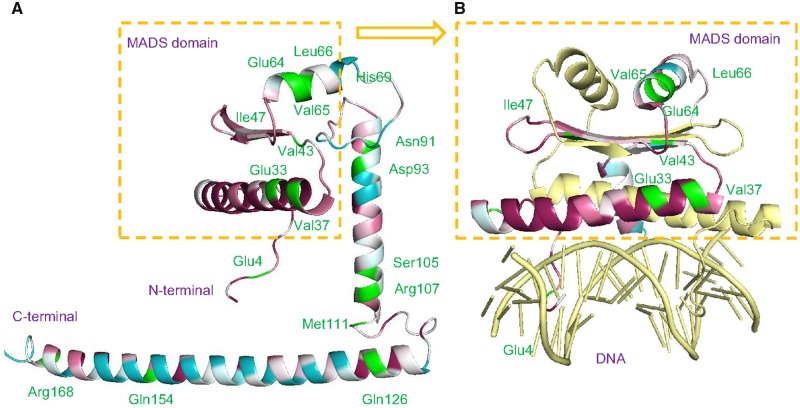
Fig. 6.
—Positive selection sites in the three-dimensional structure of the SVP protein. Residues are marked with different colors according to the degree of conservation. Conserved sites, magenta; variable sites, blue; and positive selection sites, green. (A) Positive selection sites shown in a variation of the three-dimensional structure of the SVP protein. (B) The MADS-box domain binding to DNA.
Expression Profiles of SVP Genes in Eudicot Plants
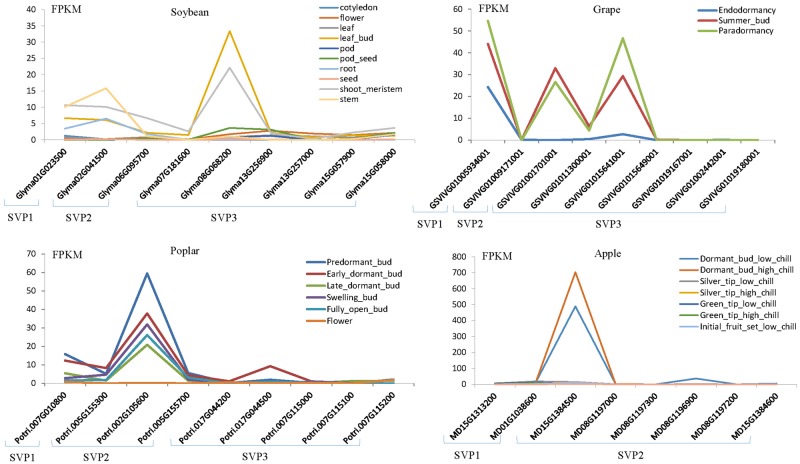
To investigate the possible functional roles of different SVP homologs, we analyzed SVP expression patterns from G. max (Shen et al. 2014), V. vinifera (Khalil-Ur-Rehman et al. 2017), P. trichocarpa (SRR6031364-95 NCBI-SRA database), and M. domestica (Kumar et al. 2017) based on published RNA-seq data (fig. 7). In soybean (fig. 7A), SVP expression profiles were analyzed for cotyledons, flowers, leaves, axillary buds, pods, seeds, roots, shoot meristems, and stems. The SVP2 gene Glyma06G095700 was not expressed in any of the tissues, whereas the SVP2 gene Glyma02G041500 was highly expressed in axillary buds and apical meristems (16 and 10 FPKM [fragments per kilobase of exon per million fragments mapped], respectively). The soybean SVP1 gene was expressed in roots, stems, axillary buds, and shoot meristems. SVP3 genes were highly expressed in axillary buds and shoot meristems (33 and 22 FPKM, respectively), indicating their involvement in bud development. To assess the relationship between bud dormancy and SVP genes, we analyzed different tissues during bud dormancy (fig. 7B–D). In grape, the SVP1 gene GSVIVG01005934001 was expressed during the endormancy, summer bud, and paradormancy phases (24, 44, and 54 FPKM, respectively), and two SVP3 genes (GSVIVG01001701001 and GSVIVG0101564100) were highly expressed in summer buds and during paradormancy (33, 27 and 30, 47 FPKM, respectively). In poplar, one SVP2 gene (Potri.002G105600) exhibited the highest expression in predormant buds (59 FPKM), but its levels gradually decreased in early dormant buds, swelling buds, fully open buds, and late dormant buds (38, 32, 26, and 21 FPKM, respectively). The poplar SVP1 gene Potri.007G010800 was expressed in all stages of bud development. In apple, only one SVP2 gene (MD15G1384500) was highly expressed in dormant buds in low and high chill temperatures (489 and 703 FPKM), and its expression level was reduced with warmer temperatures.
Fig. 7.
—RNA-seq expression patterns of different SVP genes. (A) Heat map showing different expression levels of Glycine max (soybean) SVP genes in cotyledons, flowers, leaves, leaf buds, pods, pod seeds, stems, roots, seeds, and shoot meristems. (B) Heat map showing different expression levels of grape SVP genes in endodormancy buds, summer buds, and paradormancy buds. (C) Heat map showing different expression levels of poplar SVP genes in predormant buds, early dormant buds, late dormant buds, swelling buds, fully open buds, and flowers. (D) Heat map showing different expression levels of apple SVP genes in dormant buds, silver tips, green tips, and initial fruits under low and high chill treatment.
Discussion
The SVP genes in Arabidopsis, SVP and AGL24, are well studied and are known to have opposing functions in terms of flowering time regulation. In contrast, our knowledge about SVP genes in other species is still limited. SVP genes have different roles apart from flowering time regulation in some species, including the regulation of bud dormancy in kiwifruit (Wu et al. 2012) and reproductive organ development in gymnosperms (Chen et al. 2017). Investigating the evolutionary history of the SVP genes is an important first step for advancing our understanding of their functional diversity. The present findings provide a broad overview of SVP genes in eudicots based on analyses of their evolution and expansion history, structural variation, promoter cis-regulatory elements, adaptive selection, and functional diversification.
Evolution, Conservation, and Expansion of SVP Genes in Eudicots
SVP genes are highly conserved in all plants, especially in angiosperms (Kaufmann et al. 2005). However, a previous study about MADS-box gene evolution in gymnosperms revealed that the SVP gene family expanded in gymnosperms and that disrupting their function could affect reproductive organ development (Chen et al. 2017). Compared with gymnosperms, the SVP gene family did not expand much more in ortholog lineages, despite the existence of at least one gene in each subclade in most species. Our phylogenetic and collinearity analyses indicated that at least one ancient genome duplication event may have occurred in the molecular evolution of the SVP subfamily in eudicots, resulting in three clades (SVP1, SVP2, and SVP3). The triplication (γ) took place ∼140 Ma after the monocot–dicot split and before the separation of the asterid and rosid clades (Bowers et al. 2003; Jaillon et al. 2007). We therefore infer that the three SVP clades probably derived from an ancient WGT (γ) event (table 1 and supplementary fig. S1, Supplementary Material online).
Half-life can be used to estimate the longevity of genes resulting from an ancient duplication event (Lynch and Conery 2000). A previous study showed that the half-life of γ duplication is 31.3 Myr, with 4.4% of duplicates still retained (Maere et al. 2005). In our study, the SVP1 and SVP2 clades were highly conserved and present in nearly all eudicots (fig. 1). Although all perennial plants retained SVP3 genes, they were not present in annual plants such as Brassicaceae. We hypothesize that the lifespan of SVP3 genes has been influenced by their biological function as well as selection processes. A similar evolutionary pattern was reported for the MADS-box gene AGL6, which was lost in Brassicaceae species and other core eudicots (Viaene et al. 2010). Some species were found to have multiple SVP3 members, such as P. trichocarpa (5) and G. max (6), resulting from tandem duplication and lineage-specific whole-genome duplication. In Arabidopsis, the most recent α WGD took place ∼50–60 Ma, and the half-life of the α WGD was estimated to be 17.3 Myr (Lynch and Conery 2003). In soybean and poplar, the respective whole-genome duplication events are estimated to have occurred ∼13 Ma (Tuskan et al. 2006; Schmutz et al. 2010), which may explain the retention of SVP3 genes in the soybean and poplar genomes. However, the timing of tandem duplication events differed between soybean and poplar. In soybean, tandem duplication occurred before whole-genome duplication, whereas the opposite appears to have occurred in poplar. Tandem duplication of SVP genes also occurred in U.gibba (Lan et al. 2017), and the authors of that study proposed that under substantial purifying selection, the individualized genomic architecture of a plant may play an important role in adapting to special environmental conditions (Lan et al. 2017).
The gene balance or gene dosage hypothesis is one model describing the maintenance of duplicate genes (Birchler and Veitia 2010), with the dosage change in quantity affecting the function of the whole complex (Birchler and Veitia 2012). In our study, there were many examples of SVP gene family expansion resulting from tandem duplication events, such as in soybean, grape, and P. trichocarpa. The increased number of SVP genes in such species may have been beneficial for flowering time regulation, especially in variable environments. Thus, dosage selection may have contributed to the retention of tandemly duplicated genes and functional divergence in the SVP subfamily.
Diverse Functions of SVP Genes in Eudicots
Gene duplication is regarded as the main driving force for acquiring new genes and creating genetic novelty in organisms, including neofunctionalization and subfunctionalization (Taylor and Raes 2004). Gene duplication allows a gene to be free from certain selection pressure and eventually accumulate mutations that can lead to a new function or complete loss of function (Prince and Pickett 2002). A previous study showed that selection on a new function can also contribute to duplicate retention (Panchy et al. 2016). In our study, the SVP3 clade had the highest dN:dS values among the three SVP clades and was under positive selection.
SVP homologs in different species have been found to have different functions. In Arabidopsis, SVP and AGL24 have opposite functions during the floral transition (Hartmann et al. 2000; Yu et al. 2002). SVP homologs in the SVP2 and SVP3 clades may function as suppressors during bud dormancy in kiwifruit, to prevent the growth of flowers and premature development during unfavorable winter periods (Wu et al. 2012). PpDAM5 and PpDAM6 of the SVP3 clade are involved in lateral bud endodormancy in peach (Yamane et al. 2011). In our analysis of published RNA-seq data, we found that one of the SVP3 genes in soybean was highly expressed in axillary buds and shoot meristems, and two grape SVP3 genes were highly expressed during endodormancy and paradormancy. Two SVP2 genes in poplar and apple exhibited high expression patterns in all stages of bud development and may therefore be involved in bud dormancy. Promoter cis-regulatory element analysis revealed a greater abundance of ABRE (abscisic acid response), ERE (ethylene response), and P-box (gibberellin responsiveness) elements in the SVP3 clade than in SVP1 and SVP2. It is well established that abscisic acid, ethylene, and gibberellin greatly affect bud dormancy (Horvath et al. 2004). Twenty-three light response elements were identified in the promoter regions of SVP genes, and further analysis will be required to understand their contribution to the functions of SVP genes. Altogether, these examples underscore the diverse functions of SVP genes in eudicots.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81502437) and a startup fund from Fujian Agriculture and Forestry University to L.Z.
Literature Cited
- Alexander N, Woetzel N, Meiler J.. 2011. Bcl:: cluster: a method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int Conf Comput Adv Bio Med Sci. 2011:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andres F, et al. 2014. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci U S A. 111:E2760–E2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, et al. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44(W1):W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2010. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 109(37):14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH.. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. [DOI] [PubMed] [Google Scholar]
- Brill EM, Watson JM.. 2004. Ectopic expression of a Eucalyptus grandis SVP orthologue alters the flowering time of Arabidopsis thaliana. Funct Plant Biol. 31(3):217–224. [DOI] [PubMed] [Google Scholar]
- Chen F, Zhang XT, Liu X, Zhang LS.. 2017. Evolutionary analysis of MIKCc-type MADS-Box genes in gymnosperms and angiosperms. Front Plant Sci. 8:895.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(Suppl):W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M.. 2008. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. J Exp Bot. 59:2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, et al. 2015. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 33:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maroto F, Ortega N, Lozano R, Carmona MJ.. 2000. Characterization of the potato MADS-box gene STMADS16 and expression analysis in tobacco transgenic plants. Plant Mol Biol. 42:499–513. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Theissen G.. 2013. Phylogenomics of MADS-Box genes in plants—two opposing life styles in one gene family. Biology (Basel) 2:1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM.. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60(4):626–637. [DOI] [PubMed] [Google Scholar]
- Hartmann U, et al. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21:351–360. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T.. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27:297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T, Goto K.. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409:525–529. [DOI] [PubMed] [Google Scholar]
- Horvath DR, Anderson JV, Chao WS, Foley ME.. 2004. Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci. 9:12–12. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467. [DOI] [PubMed] [Google Scholar]
- Jaudal M, et al. 2014. Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J Exp Bot. 65:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G.. 2005. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347:183–198. [DOI] [PubMed] [Google Scholar]
- Khalil-Ur-Rehman M, et al. 2017. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 17:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G, et al. 2017. Chilling affects phytohormone and post-embryonic development pathways during bud break and fruit set in apple (Malus domestica Borkh.). Sci Rep. 7:42593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, et al. 2017. Long-read sequencing uncovers the adaptive topography of a carnivorous plant genome. Proc Natl Acad Sci U S A. 114:E4435–E4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SH, Lee JS, Ahn JH.. 2007. A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochim Biophys Acta. 1769:455–461. [DOI] [PubMed] [Google Scholar]
- Lee JH, Ryu K, et al. 2013. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342(6158):628–632. [DOI] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH.. 2013. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41:D1152–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30(1):325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XF, et al. 2015. Narcissus tazetta SVP-like gene NSVP1 affects flower development in Arabidopsis. J Plant Physiol. 173:89–96. [DOI] [PubMed] [Google Scholar]
- Li Z, Zeng S, Li Y, Li M, Souer E.. 2016. Leaf-like sepals induced by ectopic expression of a SHORT VEGETATIVE PHASE (SVP)-Like MADS-box gene from the basal eudicot Epimedium sagittatum. Front Plant Sci. 7:1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The evolutionary demography of duplicate genes. J Struct Funct Genomics 3:35–44. [PubMed] [Google Scholar]
- Maere S, et al. 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 102:5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Peer YV.. 2010. Duplicate retention after small- and large-scale duplications In: Evolution after Gene Duplication. p. 31–56. [Google Scholar]
- Marin-Gonzalez E, et al. 2015. SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant Physiol. 169:1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). p. 1–8.
- Ng M, Yanofsky MF.. 2001. Function and evolution of the plant MADS-box gene family. Nat Rev Genet. 2:186–195. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171:2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF.. 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405(6783):200–203. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL.. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, et al. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash AP, Kumar PP.. 2002. PkMADS1 is a novel MADS box gene regulating adventitious shoot induction and vegetative shoot development in Paulownia kawakamii. Plant J. 29:141–151. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB.. 2002. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 3:827–837. [DOI] [PubMed] [Google Scholar]
- Proost S, et al. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. [DOI] [PubMed] [Google Scholar]
- Shen Y, et al. 2014. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26:996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, et al. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70:549–561. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J.. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 38:615–643. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, et al. 2007. Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 143:225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa. Science 313:1596–1604. [DOI] [PubMed] [Google Scholar]
- Viaene T, Vekemans D, Becker A, Melzer S, Geuten K.. 2010. Expression divergence of the AGL6 MADS domain transcription factor lineage after a core eudicot duplication suggests functional diversification. BMC Plant Biol. 10:148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd C, Wang TC, Varkonyi-Gasic E.. 2015. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J Exp Bot. 66(15):4699–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann MR, Poethig RS.. 2011. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 138:677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RM, et al. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J Exp Bot. 63:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, et al. 2016. Identification and functional analysis of SVP ortholog in herbaceous perennial plant Gentiana triflora: implication for its multifunctional roles. Plant Sci. 248:1–7. [DOI] [PubMed] [Google Scholar]
- Yamane H, et al. 2011. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J Exp Bot. 62:3481–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP.. 2002. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci U S A. 99:16336–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.