SUMMARY
While retrograde cargo selection in the Golgi is known to depend on specific signals, it is unknown whether anterograde cargo is sorted and anterograde signals have not been identified. We suggest here that S-palmitoylation of anterograde cargo at the Golgi membrane interface is an anterograde signal, and that it results in concentration in curved regions at the Golgi rims by simple physical chemistry. The rate of transport across the Golgi of two S-palmitoylated membrane proteins is controlled by Spalmitoylation. The bulk of S-palmitoylated proteins in the Golgi behave analogously, as revealed by click chemistry-based fluorescence and electron microscopy. These palmitoylated cargo concentrate in the most highly curved regions of the Golgi membranes, including the fenestrated perimeters of cisternae and associated vesicles. A palmitoylated transmembrane domain behaves similarly in model systems.
eTOC blurb:
Examples exist of secretory pathway receptor-mediated protein sorting for retrograde cargo but not for anterograde. Ernst et al. uncover an anterograde cargo sorting mechanism: Spalmitoylation at the Golgi acts as a biophysical switch that induces “self-sorting” of membrane cargo to the cisternal rim, enabling its efficient transport through the Golgi.
Graphical Abstract
INTRODUCTION
What are the basic principles of cargo selection for anterograde transport in the Golgi stack? Thirty years ago, we first reported that palmitoyl (C16)-Coenzyme A stimulated intra-Golgi transport in a cell-free extract (Glick and Rothman, 1987). Removing palmitoyl-CoA from the extract revealed a strong requirement for acyl-CoA which was met only by palmitoyl-CoA and not C14 or C18 acyl-CoAs. Transport was inhibited by adding a non-hydrolyzable analogue of palmitoyl-CoA, confirming that acyl chain transfer to an acceptor was required. Remarkably, the assembly of COPI coated vesicles from the Golgi cisternal rims, containing the cargo protein VSV G, was revealed to be strongly dependent on this palmitoylation event (Pfanner et al., 1989). The molecular basis of this striking and fundamental finding has remained obscure since then.
Now, as a result of important progress in understanding the enzymes that mediate palmitoylation (Fukata et al., 2004; Lobo et al., 2002; Roth et al., 2002; Politis et al., 2005) and advances allowing sites of palmitoylation to be visualized by light microscopy (Kolb et al., 2001), as well as broad advances in molecular cell biology, this investigation is now in a position to be reopened. A large variety of transmembrane and soluble proteins are modified with palmitate by its attachment onto cysteine residues of proteins via a thioester linkage, which strikingly can alter the biophysical properties of the acylated proteins (Smotrys and Linder, 2004). In most cases, these sites of acylation are retained, while in other cases, particularly for soluble proteins involved in signaling cascades, a highly dynamic turnover is observed (Fukata and Fukata, 2010, Linder and Deschenes, 2007, Demers et al., 2014, El-Husseini et al., 2002, Salaun et al., 2010). Furthermore, S-palmitoylation was shown to induce wide ranging regulatory effects on proteins including: membrane targeting, protein-protein interactions, protein folding and stability, sorting of soluble palmitoylated proteins to the plasma membrane (Salaun et al., 2010, Linder and Deschenes, 2007), and recently, modulation of membrane protein spontaneous curvature (Chlanda et al., 2017). Currently, proteomic analysis has identified more than 500 proteins as S-palmitoylated which includes both integral and peripheral membrane proteins (Blanc et al. 2015). Given the ubiquity of S-palmitoylation and its involvement in key physiological processes, it is no surprise that palmitoylation has been linked to a number of human diseases (Greaves and Chamberlain, 2011).
In mammals, S-palmitoylation is catalyzed by a family of 23 acyltransferase enzymes (Fukata et al., 2004, Lobo et al., 2002, Roth et al., 2002). These enzymes share a conserved membrane topology with 4–6 transmembrane domains, as well as a prominent DHHC tetrapeptide positioned in a cysteine-rich domain (CRD) that is crucial for acylation activity (Politis et al., 2005). The DHHC-CRD domain resides on a cytosolic loop, thereby allowing it to access substrates close to the membrane. Palmitoylation by DHHC enzymes is thought to occur via a two-step mechanism in which the DHHC enzymes first form an acyl-enzyme intermediate on cysteine residues (autoacylation), and then subsequently transfer the palmitate to a cysteine residue on the target protein (Jennings and Linder, 2012, Mitchell et al., 2010). Recently, the structure of DHHCs 15 and 20 were solved in a lipidic cubic phase, suggesting the orientation of the active site close to the membrane (Rana et al., 2018). Early biochemical and localization studies have concluded that the majority of the DHHC enzymes reside on the endoplasmic reticulum and the Golgi apparatus (Ohno et al., 2006).
In the present study, we revisit the requirement for palmitoyl-CoA on the sorting and trafficking of protein cargo within the Golgi. We mapped the subcellular localization of the 23 DHHC isoforms and discovered that the majority of Golgi-localized DHHCs are concentrated in the cis-Golgi. By employing a metabolic labeling strategy paired with click chemistry and super-resolution fluorescence and electron microscopy, we confirmed that the bulk of palmitate was incorporated into anterograde cargo into the cis-Golgi. Unexpectedly, we discovered that palmitoylation in the cis-Golgi accelerates the transport of these proteins through the Golgi. Strikingly, the palmitoylated proteins are concentrated in the highly curved rims of the cis-Golgi, which we suggest may explain the observed acceleration in rate.
RESULTS
Anterograde transported cargo is S-palmitoylated in the cis-Golgi, and Golgi-localized DHHCs are concentrated in the cis.
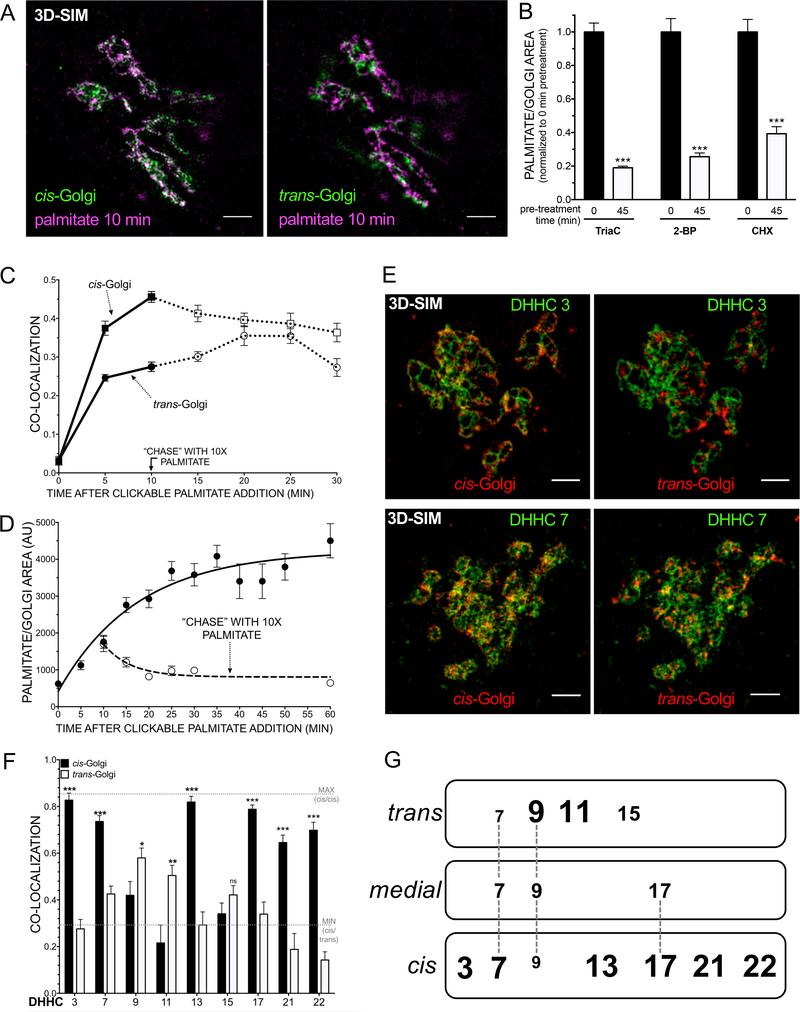
In order to identify the main site of bulk S-acylation in HeLa cells, a metabolic labeling strategy was combined with a click chemistry-based approach (Kolb et al., 2001). HeLa cells were labeled with different molecular species of fatty acid analogues, fixed, and subjected to a copper-dependent azide-alkyne cycloaddition (CuAAC) to azide-Alexa 647. Only the C16 and C18 probes efficiently labeled the Golgi, but not the C14 analogue, while lipid-droplet like structures were observed in each case (Fig. 1A, Fig.S1A). Importantly, labeling of the Golgi with the C16 probe required activation with CoA (sensitive to Triacsin C (TriaC), inhibitor of the long chain fatty acid CoA synthetase, Omura et al., 1986), catalysis by DHHC palmitoyltransferases (sensitive to 2-bromopalmitate (2-BP), inhibitor of DHHCs, Coleman et al., 1992), and the presence of biosynthetic cargo at the Golgi (sensitive to cycloheximide (CHX) -induced purge of the Golgi, (Taylor et al., 1984; Todorow et al., 2000)), altogether supporting that the majority of signal observed stemmed from S-palmitoylated proteins (Fig. 1B, Fig. S1B-D). Furthermore, the use of C16 and C18 probes coincided with the appearance of distinct protein bands after SDS-PAGE and in-gel fluorescence which, as expected of S-palmitoylated proteins, were hydroxylamine-sensitive (Fig.S1E), cleaving S-linked acyl bonds rather than O-linked acyl bonds (Magee et al., 1984). We next quantified the colocalization of the (C16) palmitate probe at different times of labeling from confocal Zstacks with regard to endogenous cis and trans Golgi markers, the ER and lipid droplets (Fig. S1F). The majority of the observed signal over the course of the pulse originated from the Golgi and lipid droplets. 3D structured illumination super-resolution microscopy (SIM) revealed that within the Golgi, the main site of incorporation was in the ciscompartment (Fig. 1A,C). Within the next 10 minutes of a “chase” experiment, in which an excess of natural palmitic acid was added to largely replace the analogue from that time onwards, co-localization with the cis-Golgi declined while increasing in the trans- Golgi (with no appearance of the probe in the ER, Fig. 1C, Fig. S1G). This would be expected if the bulk of labeled proteins were anterograde-directed cargo. Consistent with this finding, the bulk of the protein at the Golgi exited within 20 minutes, as revealed by quantifying several hundreds of cells in a “chase” experiment (Fig. 1D, Fig. S1H).
Figure 1. Anterograde transported cargo is S-palmitoylated in the cis-Golgi, and Golgi-localized DHHCs are concentrated in the cis.
A HeLa cells were metabolically labeled with 50 μM alkyne-palmitate for 10 min, fixed, permeabilized, clicked to azideAF647, immunostained for endogenous Golgi markers (cis-Golgi: GPP130, trans-Golgi: p230), and subjected to structured illumination super-resolution microscopy (3D-SIM, max. intensity Z-projections; scale bars: 2 μm). B Labeling of the Golgi with alkynepalmitate requires CoA-activation, DHHC enzyme function, and the presence of biosynthetic cargo. Pretreatment of cells with Triacsin C (TriaC, 100 μM, inhibitor of the long chain fatty acyl-CoA synthetase), 2-bromopalmitate (2-BP, 100 μM, competitive DHHC inhibitor), or cycloheximide (CHX, 100 μg/ml, inhibitor of protein biosynthesis) for 45 min prior to a 10 min pulse of alkyne-palmitate prevents Golgi labeling. The average integrated intensity of alkyne-palmitate signal at the Golgi in presence of inhibitors was quantified and compared to mock-treated controls (n>60 cells/timepoint and condition, mean and SEM, two-tailed unpaired t-tests (***: p<0.001)). C 3D-SIM of alkynepalmitate with cis- and trans-Golgi markers for a 5 or 10 min pulse (50 μM), followed by a chase with natural palmitate at 10x molar excess after 10 min. Co-localization (Pearson´s R) was determined and subjected to two-tailed, unpaired t-tests (***: p<0.001; n>10 cells/timepoint, mean and SEM). D Hela cells were constantly labeled with 50 μM alkyne-palmitate, or pulsed for 10 min followed by a chase with natural palmitate at 10x molar excess (500 μM). After click chemistry, the integrated fluorescence intensity of the Golgi area was quantified (n>20 cells/timepoint and condition, mean and SEM). E Co-localization analysis of human DHHC 3 and 7 (3DSIM). SNAP-DHHC isoforms were transfected for 24 h. After fixation, endogenous Golgi markers were immunolabeled (cis-Golgi: GPP130, trans-Golgi: p230), and SIM images of the Golgi area were obtained (max. intensity Z-projections are given, scale bars: 2 μm). F Localization of human DHHC candidates within the Golgi. SNAP-DHHC fusions of 9 Golgi candidates were scored for co-localization as described in E, and subjected to two-tailed, unpaired t-tests (ns: not significant; *: p<0.05; **: p<0.01; ***: p<0.001; n>20 cells/timepoint, mean and SEM). G Schematic of the localization of human DHHC palmitoyltransferases within the Golgi. See also Fig. S1.
Palmitoylation of Cysteine residues in membrane proteins occurs at juxta-membrane positions, with more than 500 proteins previously identified via proteomic analyses (Blanc et al. 2015). Several DHHC palmitoyltransferases are localized within the Golgi (Ohno et al., 2006). These enzymes generally accept C16 (palmitate) and to a lower extent C18 (stearate), but not shorter (C14, myristate) fatty acids as Coenzyme A- activated substrates (Politis et al., 2005; Jennings and Linder, 2012; Mitchell et al., 2010). S-palmitoylation in the cis- prior to the trans-Golgi suggested the presence of distinct DHHCs in the cis-Golgi. In order to identify DHHC isoform candidates that exclusively localize to the Golgi, the subcellular localization of a mouse orthologue library of all 23 DHHCs was mapped (Fukata et al., 2004). Indeed, 17 of the 23 DHHCs exhibited a partial Golgi localization, with 11 predominantly localizing to the Golgi (Fig.S2A-C). The corresponding human orthologues were cloned as aminoterminal fusion proteins to the SNAP-tag protein and the intra-Golgi localization of the human DHHC candidates was then probed with endogenous cis- and trans-Golgi markers using confocal microscopy (Fig. 1F, S2D). While the human DHHC 16 and 18 localized to the ER and PM respectively, the remaining 9 candidates exhibited an asymmetrical distribution across the cis-to-trans Golgi axis: DHHCs 3, 7, 13, 17, 21, and 22 specifically localized to the cis. zDHHCs 9, 11, and 15 localized partially to the trans Golgi, and to differing extents, to post-Golgi structures (Fig. 1G, Fig. S2D).
DHHCs 3 and 7 account for the majority of S-palmitoylated cargo proteins at the Golgi and catalyze transport of bulk S-palmitoylated anterograde cargo to the plasma membrane.
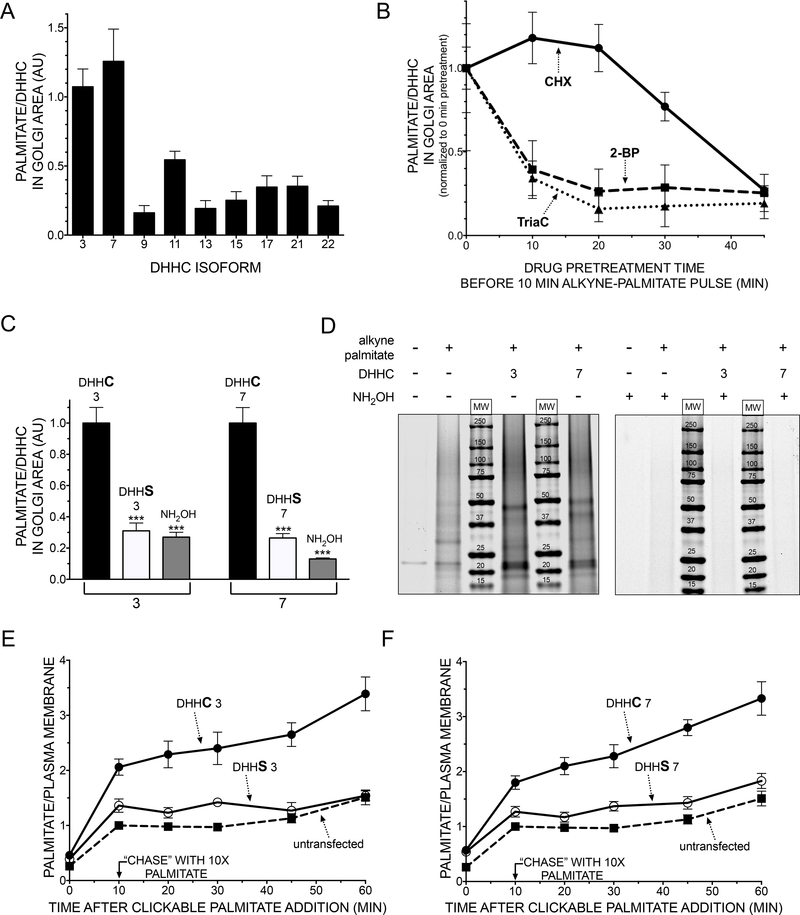
To determine which Golgi-localized S-palmitoyltransferases contribute to the observed incorporation of palmitate analogue into anterograde cargo in the cis-Golgi (Fig. 1C), each Golgi candidate (human) DHHC enzyme was overexpressed (Fig. 2A, Fig. S2E). DHHC3 and DHHC7 dramatically increased both the rate of incorporation as well as the rate of Golgi exit when chased with natural palmitate (Fig. S3A,B). Analogous to untransfected cells, DHHC-catalyzed S-palmitoylation at the Golgi required activation with CoA, DHHC enzyme function, and the presence of biosynthetic cargo (Fig. 2B, Fig. S1B-D). Furthermore, the increased incorporation of palmitate in the Golgi area resulted from acyl transfer since enzymatically inactive versions of DHHC3 and DHHC7 (DHHS) did not increase the incorporation (Fig. 2C, Fig. S3C,D). As expected, the palmitate analogue incorporated by both DHHC3 and DHHC7 was released from fixed, permeabilized cells upon exposure to hydroxylamine (Fig. 2C, Fig. S1B).
Figure 2. DHHCs 3 and 7 account for the majority of S-palmitoylated cargo proteins at the Golgi and catalyze their transport to the plasma membrane.
A Human SNAP-tagged DHHC candidates were transiently transfected into HeLa cells and metabolically labeled with azide-palmitate for 20 minutes. The ratio of the integrated intensity of palmitate and the respective DHHC was quantified (n>10, mean and SEM). B HeLa cells overexpressing SNAP-DHHC3 were pretreated with Triacsin C (TriaC, 100 μM), 2-bromopalmitate (2-BP, 100 μM), or cycloheximide (CHX, 100 μg/ml) prior to a 10 min pulse with alkyne-palmitate. Confocal Z-stacks were obtained, and the ratio of palmitate to DHHC fluorescence was quantified as in A (n>30 cells per timepoint and condition, mean and SEM). C Cells transfected with DHHC isoforms 3 and 7, or their enzymatically inactive (DHHS) variants were labeled with alkyne-palmitate for 20 min. To estimate the fraction of thioester-bound palmitate in the Golgi area, the samples were incubated with 1.5M neutral hydroxylamine for 2h followed by an extensive washout. The ratio of palmitate and DHHC fluorescence was quantified, normalized to the respective wt construct, and subjected to two-tailed unpaired t-tests (***: p<0.001, n>50 cells per condition, mean and SEM). D Overexpressed DHHCs 3 and 7 catalyze S-palmitoylation of numerous protein substrates. Cells expressing SNAP-DHHC3, 7, or mock-transfected cells were labeled for 20 min with alkyne-palmitate, clicked to azideAF647, and subjected to in-gel fluorescence analysis. E-F Cells were transfected with DHHC isoforms 3 (E), 7 (F), or their inactive (DHHS) variants, metabolically labeled with alkyne-palmitate for 10 min, and subjected to a chase with natural palmitate at 10x molar excess for the indicated time. After fixation, permeabilization, and labeling, the samples were subjected to TIRF microscopy (n>20 cells per construct, timepoint, and condition, mean and SEM). See also Fig. S2–4.
Consistent with the initial concentration of basal alkyne palmitate into the cis-Golgi (Fig. 1A,C), DHHC3 and DHHC7 are localized to cis- but not trans-Golgi regions when colocalized with endogenous markers using 3D-SIM (Fig. 1E, Fig. S3E). DHHC7 was recently mapped to the trans-Golgi, but by employing overexpressed Golgi markers (Du et al., 2017). In fact, we noticed that strong DHHC overexpression results in the disappearance of endogenous Golgi markers (Fig. S3F), and conclude that the localization of these DHHCs within the stack can only be accurately mapped in the presence of these endogenous markers (at low DHHC expression levels). The closely related DHHCs 3 and 7 are ubiquitously expressed (Ohno et al., 2006), and various studies revealed that overexpression of the corresponding mouse orthologues efficiently catalyzed S-palmitoylation of a broad variety of substrates (Gottlieb et al., 2015; Greaves et al., 2017). DHHC3 and DHHC7 protein substrates consist of numerous distinct proteins visible in SDS-PAGE that are sensitive to hydroxylamine cleavage (Fig. 2D).
As the rapid Golgi exit of S-palmitoylated proteins (with lack of rising palmitate levels in the ER) implied that the majority of DHHC substrates are anterograde cargo, putatively plasma membrane residents, bioinformatical predictions on compartment-specific datasets were performed (ER, Golgi, PM). We compared i) the total fraction of proteins containing high-confidence S-palmitoylation sites, ii) the average number of predicted sites per protein, and iii) the fraction of proteins containing signal sequences, cytoplasmic Cys residues, and high-confidence S-palmitoylation sites (Fig. S3G,H), supporting that indeed anterograde cargo are a major class of DHHC substrates, as all values peak in the PM dataset.
We therefore metabolically labeled cells expressing either DHHC3, 7, or their enzymatically inactive variants (DHHS) with alkyne palmitate for 10 min, chased the probe for different times, and after fixation and permeabilization performed total internal reflection fluorescence (TIRF) microscopy. Strikingly, over-expressing either DHHC3 or DHHC7 (Fig. 2E,F, Fig. S4A,B) markedly accelerated the rate and extent of appearance of S-palmitoylated proteins at the cell surface (3-fold after 60 min). Furthermore, this acceleration required active DHHC forms of the enzymes, with no acceleration observed with DHHS point mutants. To confirm transit of the anterograde palmitoylated cargo, a strategy was employed that would allow for transmission electron microscopy of palmitoylated proteins. HeLa cells were stably transfected with SNAP-DHHC3 to catalyze the incorporation of alkyne-palmitate into the Golgi (Fig. S4C). Our previous approach was adapted by employing azide-biotin as a ligand for alkyne-palmitate, which was then detected using a fluorogold-labeled (containing 1.4nm nanogold and Alexa488) Streptavidin, allowing the use of light and electron microscopy (Fig. S4D). We again performed a time-course experiment to monitor the distribution of the probe within intracellular membranes over time, and its enrichment was quantified in the Golgi. In agreement with the partitioning of alkyne-palmitate observed through the Golgi on the light level (Fig. 1–2), an extensive labeling of Golgi membranes after 30 min of pulse was confirmed (Fig. S4E). After 60 minutes, a marked labeling of the plasma membrane concurrent with a reduction of membrane labeling within the Golgi was observed (Fig. S4E,F).
Palmitoylation of membrane proteins accelerates their rate of intra-Golgi protein transport
The marked increase in the amount of S-palmitoylated protein detected at the plasma membrane upon overexpression of DHHC3 or 7 (in the cis-Golgi) prompted the hypothesis that palmitoylation accelerates trafficking of anterograde cargo at the level of the Golgi. There is some precedent as palmitoylation at the Golgi had been established as an important step in the vectorial transport of multiple soluble proteins, e.g. H-Ras, CSP, and SNAP25 (Gonzalo and Linder, 1998; Greaves et al., 2008; Apolloni et al., 2000). For such soluble proteins, palmitoylation acts as a post-ER membrane targeting signal, restricting peripheral protein function to the Golgi and PM, while enabling their release via thioesterases. Further, a plethora of integral membrane proteins have been identified as being S-palmitoylated, including most viral spike proteins, plasma membrane channels, and receptors (Schmidt and Schlesinger, 1979; Chien et al., 1996; Charollais and Van Der Goot, 2009). Would palmitoylation of individual anterograde (integral) membrane protein cargo impact their trafficking, as we observed for the bulk of palmitoylated proteins?
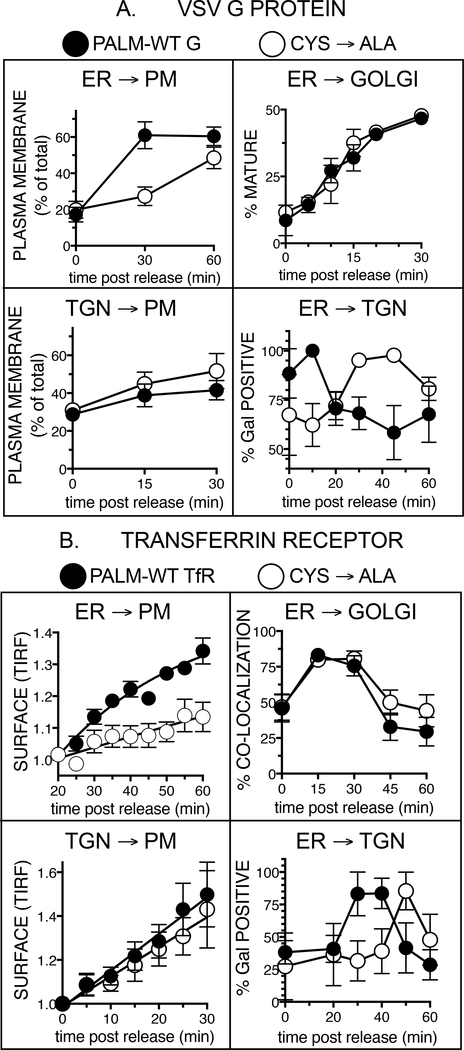
To investigate this in more detail, we compared the rate of anterograde transport of two well characterized model palmitoylated cargo proteins (VSV G protein and transferrin receptor) compared to non-palmitoylated mutants in which the attachment site cysteines were mutated to alanines (VSVG-C490A, TfR-C62AC67A). S-palmitoylation for both model proteins depended on the presence of Cys and their release from the ER (Fig. S5A,H). For VSV G, a wave of anterograde cargo can be generated by employing a temperature-sensitive mutant (tsO45, Bergman, 1989) that under the non-permissive temperature (40.5°C) is blocked in the ER, and can be released at 32°C. Employing a surface biotinylation strategy, we observed a marked difference in the amount detected on the cell surface 30 min post release from the ER: wt VSV G exhibited 50% increased PM levels, while similar levels of Cys→Ala VSVG were only observed after at 60 min (Fig. 3A, upper left panel, Fig. S5B).
Figure 3. Palmitoylation of membrane proteins accelerates their rate of intra-Golgi protein transport.
A HeLa cells transfected with wt VSVG-GFP or its non-palmitoylated variant VSV G-C490-GFP were shifted from non-permissive (40.5°C) to permissive temperature (32°C) to generate an anterograde wave of cargo. A Upper left panel: Surface arrival of S-palmitoylated VSV G is accelerated. Cells transfected with VSV G or C490A were released from the ER, and subjected to surface biotinylation with sulfoNHS-biotin at the indicated timepoint. A ratio of surface to total was quantified for n=3 independent experiments (mean and SEM). A Upper right panel: S-palmitoylation of VSV G does not impact the rate of entry into the Golgi. Constructs were released from the ER, and the rate of acquisition of EndoH-resistant N-glycans was monitored via Western blot. Quantification of n=4 independent experiments (mean and SEM). A Lower left panel: Surface arrival does not differ after release from the trans Golgi/TGN. Cargo was arrested in the trans-Golgi via a 20°C bl ock for 2h prior to releasing the temperature block for the indicated times. Quantification of n=3 independent experiments (mean and SEM). A Lower right panel: Pull-down experiments employing the Galactose-specific lectin Jacalin. Quantification of n=3 independent experiments, normalized to wt G peak at 10 min (mean and SEM). B HeLa cells transfected with transferrin receptor (TfR-FM4-HALO) or its non-palmitoylated variant (TfR-C62AC67AFM4-HALO) were released from the ER using a cell-permeable small molecule solubilizer (D/D) to generate an anterograde wave of cargo. B Upper left panel: the rate of surface arrival was quantified employing live-cell TIRF imaging. The integrated intensity per cell over time from n=4 independent experiments per construct and condition was quantified, and normalized to initial fluorescence intensity (mean and SEM). B Upper right panel: Co-localization with a cis Golgi marker (HPL) after release from the ER for the indicated times. Quantification of n>20 cells per timepoint and construct (mean and SEM). B Lower left panel: Surface arrival of TfR constructs does not differ when released from trans Golgi/TGN. Live TIRF imaging of cells transfected with TfR constructs was performed after a 20°C bloc k for 2h in presence of D/D solubilizer. Quantification of n>5 cells per construct (mean and SEM). B Lower right panel: Acquisition of Galactose in the trans Golgi is accelerated for palmitoylated TfR. Acquisition of Galactose in the trans Golgi was monitored as for VSV G in A by employing Jacalin. Quantification of n=3 independent experiments (mean and SEM). See also Fig. S5.
Where did this acceleration occur? Over 30 years ago Rose and colleagues had compared the rate of acquisition of EndoH resistance carbohydrates between wt and the same Cys→Ala mutant VSV G protein and did not observe any significant differences in these rates (Rose et al., 1984). We revisited these experiments, and indeed an identical rate of entry of wt and variant G into the Golgi was observed (Fig. 3A, upper right panel, Fig. S5C). EndoH-resistance is achieved upon arrival of the cargo in the cis-to-medial Golgi, where the concentrations of alpha-Mannosidases I/II peak, hence monitoring the rate of transport from the ER to the Golgi, but not beyond this point. We therefore speculated that any contribution of S-palmitoylation to ER→PM trafficking was not at the ER→Golgi, but either within the Golgi or Golgi/TGN→PM. When performing surface biotinylation experiments in conjunction with a 20°C block (to accumulate cargo in the TGN prior to its release), no differences were observed in surface arrival between the constructs (Fig. 3A, lower left panel, Fig. S5D), supporting a palmitoylation-dependent acceleration of G protein from the cis- to the trans-Golgi.
To test this possibility, the rate of acquisition of Galactose was measured in the trans Golgi by using the terminal Galactose residue-specific lectin Jacalin. While wt G rapidly became positive for Galactose 10 minutes post release and was fully processed within the following 10 min, in which further extension of the N-glycans masks detection by Jacalin, Cys→Ala G protein revealed a significantly delayed peak at 30 min and elevated levels even at 60 min (Fig. 3A, lower right panel, Fig. S5E). An accelerated arrival of wt G in the trans-Golgi was confirmed by confocal co-localization with endogenous Golgi markers (Fig. S5F), excluding the possibility of an altered access to the glycosyltransferase within the trans-Golgi.
To assess whether these results are retained for a different palmitoylated membrane cargo independent of the temperature shift, we next employed a modified version of the transferrin receptor (TfR-FM4-HALO) containing FM domain repeats, leading to aggregation of the model cargo in the ER and allowing for a triggered release using a solubilizing drug (Bottanelli et al., 2016). Importantly, similar to VSV G, TfR has been established as palmitoylated protein at two juxtamembrane cysteine residues (C62 and C67; Alvarez et al., 1990). A Cys→Ala variant (C62A-C67A) was generated and live cell TIRF microscopy of single cells transfected with either construct was performed. In agreement with the impact of palmitoylation of G protein, surface arrival of palmitoylated TfR (Fig. 3B, upper left panel, Fig. S5I) was significantly delayed, with similar mutant surface levels observed only 90 min post release (Fig. S5J,K). Again, this acceleration did not stem from different rates of ER→Golgi trafficking, which was concluded from time-course experiments in which wt or Cys→Ala TfR from the ER was released and the extent of co-localization was monitored with a cis-Golgi marker (Fig. 3B, upper right panel, Fig. S5L). Employing a 20°C temperature bloc k resulted in similar rates of surface arrival when measured by live cell TIRF microscopy (Fig. 3B, lower left panel), while employing a 16°C block (cis-Golgi) resulted i n an accelerated surface arrival for wt TfR (Fig. S5M). In agreement with VSV G, these data strongly suggest an acceleration of intra-Golgi transport for palmitoylated TfR. The acquisition rate of Galactose in the trans-Golgi was again monitored. Peaks for wt TfR were observed at 30 min, while the non-palmitoylated mutant exhibited a 20-minute delay peaking at 50 min post release (Fig. 3B, lower right panel, Fig. S5N).
In summary, for both model palmitoylated anterograde cargo proteins, the acceleration of transport only occurred within the Golgi, indicated by the observation that neither the kinetics of transport from ER→Golgi, nor TGN→PM, were affected by palmitoylation. Confirming this result, the rate of transit from ER→TGN was accelerated for both cargo proteins by palmitoylation, strongly supporting the finding that central regulation of anterograde traffic occurs by palmitoylation at the cis-Golgi.
Modulation of DHHC levels and their activity impacts transport of both membrane and secretory proteins.
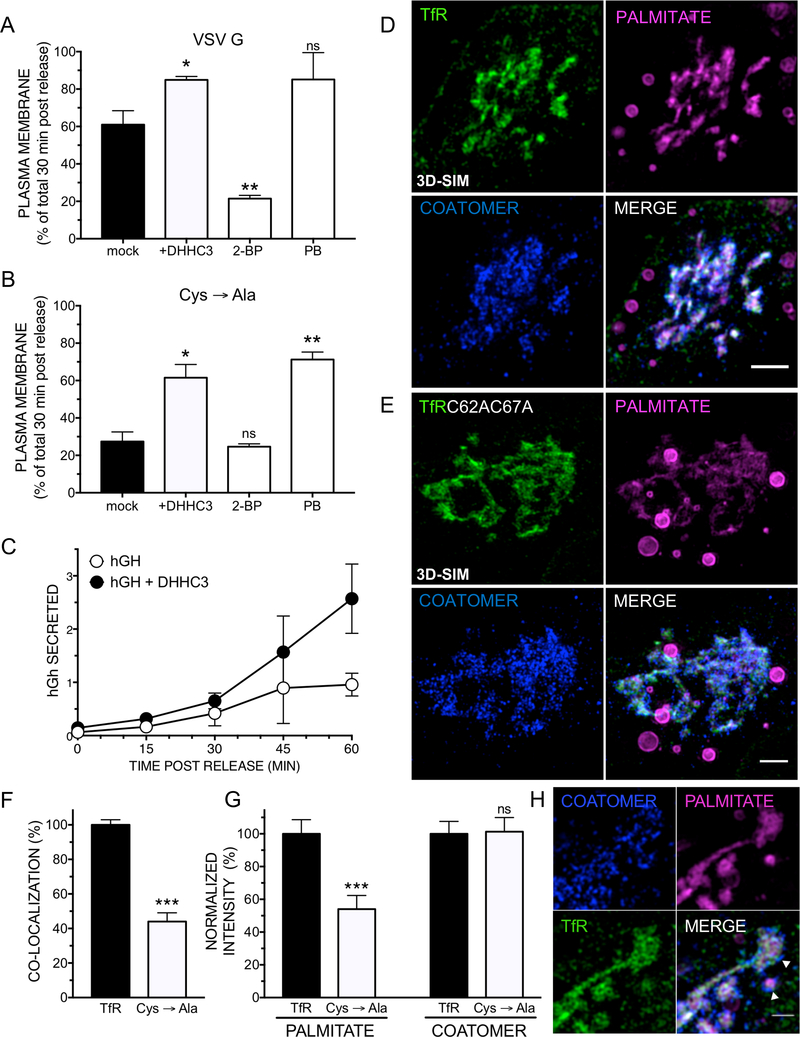
Overexpression of DHHC3 and 7 had significantly catalyzed partitioning of S-palmitoylated proteins as a class to the plasma membrane (Fig. 2E-F, Fig. S4A). We therefore set out to test the effect of modulation of S-palmitoylation on the trafficking of two reporter proteins, VSV G and TfR, from the ER to the plasma membrane. The anterograde trafficking was examined in three conditions: i) upon overexpression of DHHC3; ii) inhibition of global S-palmitoylation with 2-bromopalmitate (2-BP); and iii) inhibition of de-palmitoylation with palmostatin B (PB). We employed surface biotinylation experiments to monitor bulk arrival of VSV G at the plasma membrane. While in untreated cells a 2- to 3-fold increase in surface palmitoylated G was observed, no significant difference was observed in 2-BP-pretreated cells (Fig. 4A-B, Fig. S5G). Interestingly, either overexpression of DHHC3 or inhibition of de-palmitoylation with palmostatin B markedly accelerated surface arrival of wt and C490 variants. A corresponding acceleration in surface arrival was also detected for the TfR reporter: PM levels increased 3-fold after 60 min, whereas cells pretreated with 2-BP exhibited a marked reduction in TfR at the plasma membrane (Fig. S5O,P).
Figure 4. Modulation of DHHC levels and their activity impacts transport of both membrane and soluble proteins.
A-B Modulation of VSV G surface arrival by overexpression of SNAP-DHHC3, the competitive DHHC inhibitor 2-bromopalmitate (2BP), or the de-palmitoylation inhibitor palmostatin B (PB). HeLa cells were transfected with VSV G (A) or C490A (B) for 24 h and incubated at 40.5°C. If indicated, e xpression of DHHC3 was induced in the stably transfected cells for 5h. Un-induced, VSV Gtransfected parallel setups were incubated for 1 h with 2-BP (100 μM) or PB (25 μM). VSVG was released from the ER for 30 min prior to biotinylation of surface-exposed proteins. Quantification of n=3 independent experiments, mean and SEM. The results of two-tailed, unpaired t-tests are given (ns: not significant; *: p<0.05; **: p<0.01; ***: p<0.001). C Overexpression of DHHC3 modulates the secretion of soluble cargo. The model protein hGh-FM4-GFP was transfected into dox-SNAP-DHHC3 cells. Expression was induced for 5h, prior to releasing hGh from the ER for the indicated times using D/D solubilizer. Cell contents were compared to hGh in the medium after chloroformmethanol precipitation (quantification of n=3 independent experiments, mean and SEM). Upon entry into the Golgi, S-palmitoylated TfR segregates with palmitate into areas enriched with coatomer. D-E 3D-SIM of TfR-FM4-SNAP (D) or TfRC62AC67AFM4-SNAP (E) 15 min post release from the ER with a simultaneous metabolic labeling using alkyne-palmitate. After fixation, S-palmitoylated proteins were subjected to CuAAC to azide-AF647, and SNAP-tagged TfR constructs (AF488) and endogenous coatomer (CM1, AF568) were immunolabeled. F Quantification of co-localization (Pearson’s R) of TfR constructs with palmitate in the cis Golgi (15 min post release from the ER, n=30 cells/construct, SIM max. intensity Z-projections, normalized to wt TfR). G Quantification of the ratio (integrated intensity) of palmitate and coatomer per TfR construct and for the Golgi area as in F. H Zoom into Golgi area indicating enlarged cisternal rims (15 min post release of TfR from the ER). White arrowheads: putative carriers positive for TfR, palmitate, and encompassed with coatomer (lower arrow), TfRenriched rim area (upper arrow). Scale bar: 1 μm.
The acceleration in transport observed for G and TfR model Cys →Ala variants upon overexpression of DHHC 3 (Fig. 4B, Fig. S5O) prompted us to test whether overexpression of DHHC and the resultant increase in the rate of transport of Spalmitoylated proteins through the Golgi could globally impact secretion. We thus probed an FM-domain-containing model secretory protein, the human growth hormone (hGH-FM4-GFP), and monitored the fraction of hGH secreted in presence or absence of overexpressed DHHC3 (Fig. 4C, Fig. S5Q). Strikingly, levels of hGH increased 3-fold in the medium of cells with a concomitant reduction of intracellular levels 60 min post release from the ER.
Palmitoylated membrane cargo concentrates in highly curved regions at the cisternal rims of the Golgi
Altogether, the data thus far supports that S-palmitoylation of membrane proteins results in higher rates of anterograde transport from the cis- to trans-Golgi. These results implied that S-palmitoylated proteins are sorted differentially upon entry into the Golgi, which prompted us to perform 3D-SIM of wt and Cys→Ala TfR constructs upon entry into the Golgi. Strikingly, wt and mutant TfR exhibited a marked differential distribution: wt appeared clustered, while mutant TfR appeared dispersed across cisGolgi membranes (Fig. 4D,E). Furthermore, labeling of the Golgi area with palmitate increased significantly in presence of wt TfR, with the increase in staining stemming from areas that co-localized with the protein and that were juxtaposed by coatomer (Fig. 4D-H).
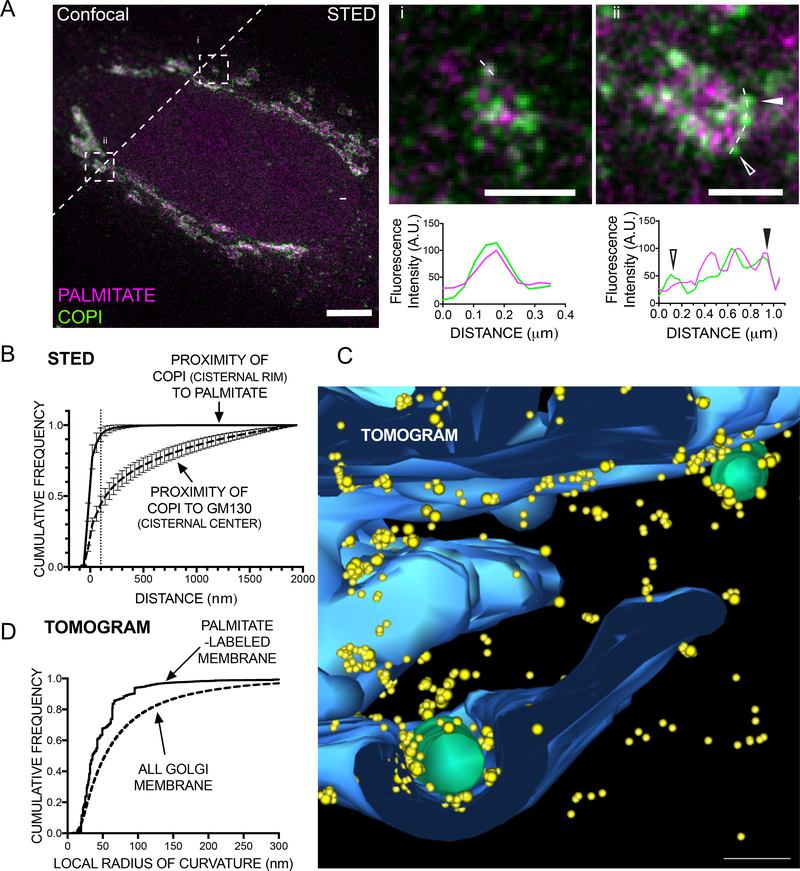
The differential distribution of TfR constructs in the Golgi prompted us to localize Spalmitoylated proteins as a class and at high resolution employing STED nanoscopy. Cells were pulsed with alkyne-palmitate, and probed for a nanoscale co-localization with coatomer (marker of the highly-curved cisternal rim) or a marker for the flat centers of cis-Golgi cisternae (GM130). Strikingly, coatomer-positive clusters representing putative COPI buds/vesicles at the Golgi were frequently observed that were enriched in palmitate (Fig. 5A). We next set out to quantify the average proximity of COPI to S-palmitoylated proteins, finding that the majority (~93%) of COPI structures were positioned within 100 nm of S-palmitoylated proteins. On the other hand, GM130 appeared segregated from COPI with only ~44% of coatomer signal within 100 nm (dashed line) from the cisternal center (Fig. 5B).
Figure 5. S-Palmitoylated membrane cargo concentrates in highly curved regions at the cisternal rims of the Golgi.
A STED nanoscopy of S-palmitoylated proteins at the Golgi. doxSNAP-DHHC3 cells were induced for 5 h and subsequently metabolically labeled with alkyne-palmitate for 20 min, followed by a 10 min chase in delipidated medium. After fixation and permeablilization, palmitoylated proteins were detected with azide-ATTO594, and coatomer or GM130 were immunolabeled with ATTO647N. Left: overview of Golgi area labeled for STED microscopy. Upper right: zoom into individual cisternal elements (dashes boxes i & ii). Lower right panel: line profiles obtained from the magnified images (dashed lines, arrowheads highlight the position of distinct COPIclusters). B Quantification of STED images (cumulative frequency distribution of the proximity of pixels) obtained from Golgi areas (dashed line: 100nm; n=10 Golgi areas per pair, mean and SD). C Electron tomography of S-palmitoylated proteins at the Golgi. Expression of DHHC3 and labeling of the Golgi with alkyne-palmitate was performed as in A, and followed by fixation and permeabilization of the cells. After click chemistry to azide-biotin, S-palmitoylated proteins were detected using streptavidin-fluoronanogold and subjected to double tilt electron tomography. 3D-modelling of the tomogram data was performed (Golgi membranes) and the loci of 3575 gold-labeled palmitoylated proteins in the tomogram were annotated. Magnification of a cisternal rim area of palmitate-labeled Golgi membranes (3D tomogram model). Yellow: gold clusters (=palmitoylated proteins), blue: rendered Golgi membranes, green: vesicular/tubular structures; scale bar: 50 nm. Full model: Fig. S6E. D Local curvature was assessed at each point along the annotated membrane curves, and the cumulative frequency of total Golgi membrane curvature (dashed line) is compared to the underlying curvature observed in loci positive for S-palmitoylated proteins (solid line). See also Fig. S6.
To obtain further clues as to the underlying mechanisms, we obtained double-tilt electron tomograms of cells pulsed with alkyne-palmitate and generated 3D models of the position of S-palmitoylated proteins with the underlying Golgi membranes (Fig. S6AE). First, we systematically quantified the local curvature encountered in the entire Golgi (Fig. S6F) before and after a pulse of palmitate. Strikingly, the distribution drastically shifted towards the presence of highly-curved membranes as palmitate was incorporated into the Golgi (Fig. S6G). It was evident upon inspection that the goldlabeled palmitoylated proteins were concentrated in the highly curved tubulo-vesicular rims of the stack (Fig. 5C, Fig. S6A-E). To quantify this effect, we measured the underlying local curvature at all positions within the tomogram exhibiting palmitate labeling, and compared it to the local curvature distribution of Golgi membranes as a whole (Fig. 5D, Fig. S6F). Strikingly, palmitoylated cargo exhibited a strong preference for membranes whose local radius of curvature was < 50 nm, about the size of a COPI transport vesicle and the tubular-vesicular network encountered at the rim of Golgi cisternae (“non-compact region”, Ladinsky et al., 1999). These palmitoylated cargo could either be acylated within the curved regions or concentrate there after being palmitoylated elsewhere, depending on where the DHHC enzymes are localized. In fact, DHHCs are located separately from their palmitoylated products, co-localizing instead at the light level with markers of the stacked, flat regions of the Golgi cisternae (Fig. S6H), suggesting that the palmitoylated cargo migrates from its site of synthesis in the stacked regions of the cis Golgi to concentrate at the tubule-vesicular rims.
Curvature preference may be an intrinsic property of palmitoylated anterograde cargo
These results suggest that palmitoylated cargo – on the basis of the acyl chain – concentrate in highly curved Golgi rims and when they do so in large numbers actually increase the proportion of membrane in such curved regions, thereby putatively resulting in higher rates of overall anterograde transport. The simplest basis for these effects of the acyl chain would be physical-chemical. In other words, an increase in the intrinsic preference of the cargo membrane protein for curved membranes due to the physical chemistry of the fatty acid chain.
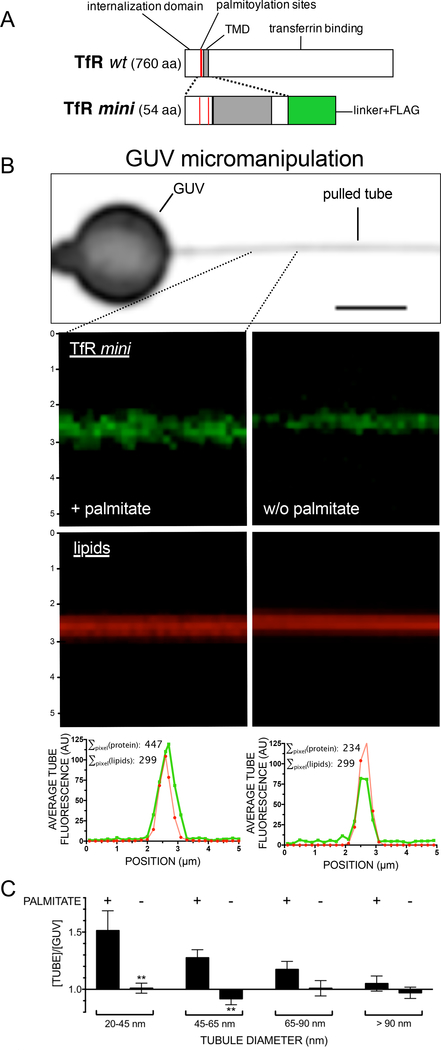
To test this concept, we utilized a well-characterized system that allows intrinsic curvature preference to be directly measured when presented with a choice between a large vesicle and a small highly curved and contiguous tubule (Bo and Waugh, 1989). We expressed and purified from mammalian suspension cells a 54-amino acid fragment of transferrin receptor consisting of the transmembrane helical domain flanked by juxtamembrane sequences at both ends (Fig. 6A, Fig. S7A,B). The peptide (termed “TfR mini”) was prepared in both palmitoylated and de-palmitoylated (hydroxylaminecleaved) forms (Fig. S7C,D) and then reconstituted into giant unilamellar vesicles (GUVs). Tubules (Fig. 6B) ranging from 20– 150 nm diameter can be pulled out from GUVs following adhesion of an applied micro-pipette tip (Bo and Waugh, 1989). TfRmini initially in the GUV can then distribute at equilibrium between the curved tubule and the continuous, essentially planar GUV bilayer. The concentration of the TfR-mini in the tubule relative to the concentration remaining in the GUV can then be determined using established methods as can the tubule diameter (Bo and Waugh, 1989; Hochmuth et al., 1982; Sorre et al., 2009).
Figure 6. Curvature preference may be an intrinsic property of palmitoylated anterograde cargo.
A Schematic representation of a minimal construct of the transferrin receptor, TfRmini. Red: Cys residues 62 and 67, grey: transmembrane domain, green: linker and FLAG epitope. B Micromanipulation of giant unilamellar vesicles (GUVs). GUVs containing biotinylated and fluorescently-labeled lipids were formed, and recombinant proteins were reconstituted into GUVs. Next, two micropipettes were employed, one containing a streptavidin bead, one to exert suction on the GUV while pulling a tube out of the membrane due to the biotin-streptavidin interaction (upper panel). The ratio of the fluorescence of the recombinant protein in the tubule versus the GUV donor membrane divided by the intensity of membrane lipids in tubule versus the donor membrane was calculated (representative individual channels for S-palmitoylated or non-palmitoylated TfRmini: middle panels). The model tubes exhibit identical sum-of-lipid pixel intensities in the lipid channel and thus allow the direct comparison of sum of protein pixel intensities (lower panels; tube/GUV enrichment: +50% (S-palmitoylated TfRmini); −20% (de-palmitoylated TfRmini)). C S-palmitoylated TfRmini partitions into tubules with high curvature. TfRmini-containing GUVs were micromanipulated to generate different classes of tube diameters, and the relative excess of recombinant protein in the tube was plotted against increasing tube diameter bins for palmitoylated and de-palmitoylated (hydroxylamine-cleaved) TfRmini. See also Fig. S7.
We observed that the transferrin receptor transmembrane domain was strongly concentrated in a palmitoylation-dependent manner into the tubules (Fig. 6B,C). Enrichment increased progressively as the tubule diameter decreased (i.e. curvature increased). The greatest concentration occurred for diameters of curvature below 65 nm, and removing the attached palmitate groups abolished all curvature preference.
DISCUSSION
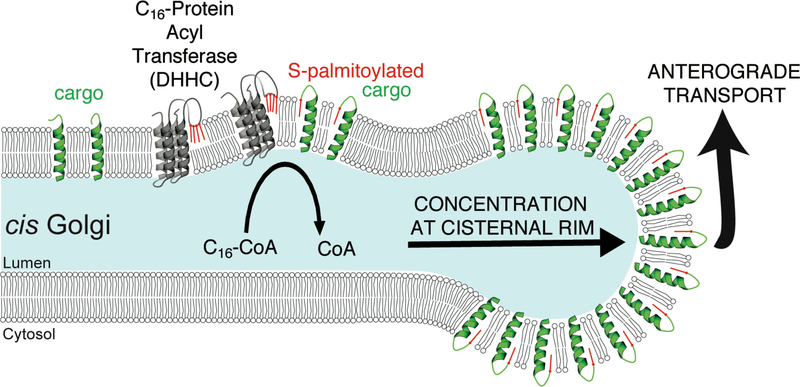
Mammalian Golgi cisternae are stacked due to the action of GRASP proteins, and it has been established that vesicle transport machinery (ARF1, COPI) preferentially accumulate at the highly-curved rims of cisternae (Farquhar, 1985; Orci et al., 1986). Early electron microscopic studies of the distribution of VSV G within Golgi cisterna revealed a still-unexplained increase in surface density of VSV G in the curved rims as compared to the flatter centers of the cisternae (Orci et al., 1986). The data presented here introduce a new concept to cell biology, and in doing so, bear the potential to resolve a number of long-standing puzzles, including our own still unexplained results from three decades ago, that originally pointed to a palmitoyl transfer reaction from Coenzyme A as an important determinant of COPI vesicle budding from Golgi cisterna. The new idea is that membrane proteins – by virtue of acylation – acquire preference for curved regions of the lipid bilayer, and thus are spontaneously sorted towards the cisternal rim (Fig. 7). In so doing, the palmitoylated proteins increase the rate of vesicle/tubule budding from these same regions by lowering the energy needed to further bend the local membrane to yield pinched-off carriers. The resultant diffusive flux (the gradient generated by removal of S-palmitoylated proteins from the rim) carries both non-acylated membrane as well as secretory cargo in the center of the cisternae toward the rim. This is supported by our observation that anterograde trafficking of the model cargos G and TfR is markedly accelerated across the Golgi (Fig. 3), and that partitioning of bulk anterograde cargo is enhanced by maximizing the rate of palmitoylation through over-expression of DHHCs (Fig. 4A-C).
Figure 7. S-palmitoylation acts as a sorting signal for anterograde membrane cargo at the cis-Golgi.
Upon interaction with DHHCs in the cis-Golgi, anterograde membrane cargo is rapidly S-palmitoylated. This leads to an increased affinity for the highly positively curved cisternal rim, from which budding occurs, and an extraction of cargo from stacked (flat) Golgi membranes. Partitioning into in curved regions at the rims of the Golgi cisternae increases the incorporation of S-palmitoylated proteins into coatomer-positive carriers, resulting in an acceleration of anterograde transport across the Golgi.
Deforming the membrane into a vesicle, or tubule, requires energy, typically provided by an externally-applied shaping device such as a spherical, or helical, coat. This needed energy may also be provided from within the membrane if its constituent proteins intrinsically prefer high curvature. When such curvature-seeking proteins transit into regions of higher curvature, they lower the free energy of the system (applied device plus membrane), providing energy to drive the budding process. By reducing the activation energy for curving the membrane, curvature-seeking proteins will not only increase the rate of budding (vesicles or tubules) above the basal rate resulting from the external device alone, but will themselves further concentrate into the budding membrane during the process. From a rate perspective, the optimal reaction will occur when the curvature preference of the membrane proteins considerably exceeds the physical curvature of the external membrane-shaping device. This is in agreement with our observation that the palmitoylated TfR transmembrane domain has a preference for bilayers with a diameter of curvature below 65 nm but lacks a curvature preference when it is not palmitoylated (Fig. 6C). In comparison, the diameter of the vesicle enclosed in a COPI coat is about 70–80 nm (Orci et al., 1986) and a peripheral Golgi tubule has a diameter of 50 nm (Ladinsky et al., 1999).
A classical target sequence for palmitoylation of membrane proteins by DHHC family palmitoyltransferases simply appears to consist of one or more Cys residues close to the cytoplasmic end of the transmembrane domain (Aicart-Ramos et al., 2011). Based on our results, this simple motif might be considered as a sorting signal marking the cargo bearing it for rapid anterograde transport, that is recognized by DHHCs in the cisGolgi (Fig. 1F,G) and actuated by attaching a fatty acid chain(s).
Although further detailed studies will be required, it appears that these straightforward concepts, originating from detailed analysis of the TfR and G protein, are likely to apply broadly to the anterograde-directed cargo in the Golgi. Notably, the bulk of palmitoyl acceptors in the Golgi membranes are cargo molecules (Fig. 1B, Fig. 2B,D) that transit the stack in the anterograde direction (Fig. 1C,D, Fig. 2E,F, Fig. S4E). As a class, these palmitoylated cargo demonstrate accelerated anterograde transport upon increasing the rate of their palmitoylation (Fig. 2E,F, Fig. S3A,B) and they localize to the curved rims of the Golgi (Fig. 5B,C), strongly concentrating in regions whose local curvature is less than 50 nm (Fig. 5D). As may be generalized from the physics governing the behavior of model S-palmitoylated transmembrane peptides (Fig. 6), not only do palmitoylated anterograde cargo as a class partition into curved regions at the rims of the Golgi cisternae, but producing more of them increases the proportion of the total Golgi surface that is in the very highly curved regions from which budding occurs (Fig. S6G). In support of this view, a significant fraction of anterograde-targeted proteins (e.g. plasma membrane residents) are predicted by bioinformatics to be substrates for palmitoylation (Fig. S3G,H). However, while we show that S-palmitoylation is a key new mechanism to promote anterograde traffic within the Golgi based on a membrane-intrinsic, physical-chemical process, this does not exclude other mechanisms which may augment sorting of cargo to the cisternal rim, e.g. heterooligomerization of S-palmitoylated and non-palmitoylated cargo, protein folds that result in spontaneous curvature of the hydrophobic moiety (polytopic membrane proteins), or lectin-mediated interactions of S-palmitoylated membrane cargo with cargo in the lumen of the Golgi.
Why might palmitoylation of certain membrane proteins impart curvature preference? Acceptor Cys residues are frequently observed within a few amino acids of the cytoplasmic end of a transmembrane domain. This enables the fatty acid to insert into the cytoplasmic leaflet, adding surface area locally and changing the overall shape in the membrane (the diameter of a fatty acid in a liquid disordered phase is about 0.4 nm as compared to an a-helix of about 1.2 nm), thus being larger on the cytoplasmic than on the lumenal side. Anchoring a previously flexible local amino acid sequence to the cytoplasmic leaflet, would convert it into a restricted loop-like extension of the transmembrane domain, further adding to the cone-like shape. This change in the shape (spontaneous curvature) now modulates the properties of the cargo so that it prefers highly curved regions of membrane, which in the Golgi stack are mainly present at the rims of the cisternae, thus concentrating the anterograde cargo in the regions where transport occurs. That intrinsic curvature preference causes partitioning to curved membranes is an established principle in membrane biochemistry (Frolov et al., 2011; Sheetz and Singer, 1974), and that S-palmitoylation can impact the spontaneous curvature of a membrane protein was recently demonstrated (Chlanda et al., 2017), supporting our proposed mechanism.
Our newly proposed physical-chemical way of thinking about anterograde transport in the Golgi might help explain many puzzling disparate observations. For example, we observed that VSV G protein appeared concentrated in the Golgi rims at every level of the stack (Orci et al., 1986). Second, over-expressing VSV G leads to an increase in the rate of transport accompanied by a dramatically increased prevalence of tubulation (i.e. curvature) at the rims (Trucco et al., 2004). Third, palmitoyl-CoA was known to dramatically accelerate the rate of budding of VSV G-containing COPI vesicles (and transport) in cell-free systems (Glick and Rothman, 1987; Pfanner et al., 1989; Ostermann et al., 1993).
In conclusion, we can envisage S-palmitoylation as the first example of a physicalchemical, membrane-intrinsic sorting process by which anterograde cargo spontaneously clusters within highly-curved regions of the cisternal rim and to the exclusion of retrograde cargo and their receptors (Fig. 7), thereby enabling their uptake into highly-curved tubular (Trucco et al., 2004; CDC42-mediated: Park et al., 2015; ARF1-mediated: Bottanelli et al., 2017) or vesicular (COPI: Orci et al., 1986; Pellett et al., 2013; Rothman, 2014) carriers, and thus resulting in an efficient net anterograde cargo transport.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Andreas Ernst (andreas.ernst@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HeLa cells were maintained at 37°C in 5 % CO 2 in DMEM (Gibco, Grand Island, NY,
United States) supplemented with 10 % FBS (Gibco). EXPI293F cells (Thermo Fisher
Scientific) were maintained at 37°C in 8 % CO 2 in Expi293 expression medium (Thermo Fisher Scientific). A stably transfected, doxycycline-inducible SNAP-hDHHC3 HeLa cell line was generated employing pCW57.1 (a gift from David Root, Addgene plasmid #41393) and SNAP-DHHC3 plasmid.
METHOD DETAILS
Cell Culture and reagents
HeLa cells were maintained at 37°C in 5 % CO 2 in DMEM (Gibco, Grand Island, NY, United States) supplemented with 10 % FBS (Gibco). Plasmid transfections were performed with Fugene HD (Promega, Madison, WI, USA) as recommended by the manufacturer. Release of FM4-domain-containing constructs from the ER was performed with the disaggregating drug D/D solubilizer (Clontech, Mountain View, CA, USA) at a final concentration of 1.5 μM. HA-tagged mouse zDHHC constructs were gift from Y. Fukata and M. Fukata (Fukata et al., 2004). Human DHHC cDNA clones isoforms 3, 7, 9, 11, 13, 15, 17, 21, and 22 were obtained from Origene (Rockville, MD, USA), and subcloned to pSNAPf plasmids (NEB, Ipswich, MA, USA). (tsO45)VSV G-3GFP and TfR-FM4-HALO were a gift from Derek Toomre (Yale). SNAP-tag ligands (benzylguanine-conjugates) were purchased from NEB. Live-cell imaging and temperature blocks were performed in in HEPES-buffered imaging medium supplemented with 20 mM glucose (Molecular Probes, Eugene, OR, U.S.A) and in temperature-controlled incubators. EndoH assay kit was purchased from Promega and used according to the instructions of the manufacturer. Jacalin-agarose was purchased from Thermo Fisher.
Cycloheximide (final C: 100 μg/ml), 2-bromopalmitate (final C: 100 μM), and Triacsin C (final C: 100 μM) were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A), palmostatin B from Merck Millipore (Darmstadt, Germany).
Immunofluorescence, ligand labeling, microscopy
For imaging experiments, 100,000 HeLa cells were seeded on glass-bottom dishes (MatTek, Ashland, MA, USA) 24 h prior to transfection. Live-cell labeling of SNAP- and HALO-tagged proteins was performed for 1h at a final concentration of 5 μM, followed by three washes in medium and a subsequent additional incubation in medium for 30 min. The cells were either imaged live (in HEPES-buffered imaging solution (Gibco, Grand Island, NY, United States) supplemented with 0.1% FBS), or chemically fixed in 4 % paraformaldehyde in PBS for 10 min at RT as indicated. Cells were then permeabilized in permeabilization buffer (0.3 % NP-40, 0.05 % Triton X-100, 0.1 % BSA (IgG free), 1X PBS) for 2 min at RT, washed three times in wash buffer (0.05 % NP-40, 0.05 % Triton X-100, 0.2 % BSA (IgG free), 1X PBS), and subsequently blocked in blocking buffer (0.05 % NP-40, 0.05 % Triton X-100, 5 % goat serum, 1X PBS) for 1h. Primary antibodies were diluted at 1:500 in blocking buffer, and added to cells for 1h at RT. Next, the cells were washed three times in wash buffer (0.05 % NP-40, 0.05 % Triton X-100, 0.2 % BSA (IgG free), 1X PBS), incubated with a dilution of a fluorophorelabeled secondary antibody (1:1000) in blocking buffer for 1 h at RT, washed three times with wash buffer, and twice in PBS. The cells were mounted with ProLong Gold antifade reagent (Life Technologies, Carlsbad, CA) and 1 mm thick precleaned microscope slides (Thermo Fisher Scientific, Waltham, MA). Antibodies: anti-GPP130 (BioLegend, San Diego, CA, USA), anti-p230 (BD Biosciences, San Jose, CA, USA), anti-betaCOP (CM1, Palmer et al., 1993 and BetaEAGE, Duden et al., 1991), GM130 BD Transduction Laboratories, Franklin Lakes, NJ, U.S.A). All secondary antibodies conjugated to fluorescent dyes were purchased from Thermo Fisher (Waltham, MA, USA). Confocal imaging was performed on a Zeiss LSM710duo microscope with a 63x oil objective. TIRF imaging was performed on a DeltaVision OMX (Little Chalfont, UK). 3D-structured illumination (SIM) imaging was performed on a DeltaVision OMX (Little Chalfont, UK). STED imaging was carried out on a commercial Leica TCS SP8 STED 3X equipped with a white light excitation source. Excitation wavelengths: 594 and 650 nm. For detection HyD 1 and HyD 2 were set to 604- to 644- and 665- to 705-nm windows. The 775-nm depletion laser was used for both ATTO594 and ATTO647N. The two colors were imaged sequentially line by line. Image quantifications (Pearson´s R, integrated intensity, line profiles) were performed in ImageJ and Volocity software (PerkinElmer, Waltham, MA, U.S.A).
Metabolic labeling, click chemistry
Alkyne-functionalized fatty acid analogues were obtained from Cayman (Ann Arbor, MI, USA), azide-palmitate was obtained from Thermo Fisher, and lipid stocks were kept at 10 mM in ethanol and at −20°C. For metabolic labeli ng reactions, the lipid analogues were added to delipidated medium (DMEM + 10% charcoal-stripped FBS, Thermo Fisher), sonicated, incubated at 37°C for 10 min, a nd added at a final concentration of 50 μM to 70%-confluent HeLa cells. CuAAC (Copper(I)-catalyzed Azide-Alkyne Cycloaddition) was performed using the Cell reaction kit from Thermo Fisher (Waltham, MA, USA) according to the manufacturer. Azide- and alkyne- conjugated dyes were obtained from Thermo Fisher and used at a final concentration of 5 μM. After CuAAC, unreacted dye was removed with 2% (w/v) of delipidated BSA in PBS for 1h. For in-gel fluorescence, HeLa cells were labeled with the functionalized lipid analogues for 20 minutes in delipidated medium. The cells were scraped, sonicated on a microtip sonicator for 10 s, and subjected to CuAAC to azide-AF647. The samples were lysed in 50 mM HEPES/KOH, 150 mM NaCl, 1% (v/v) TX-100, sonicated, subjected to chloroform-methanol precipitation and dried in a speed-vac. Pellets were either treated with neutral hydroxylamine or 50mM HEPES/KOH, 150mM NaCl, and re-precipitated. Finally, the pellets were resuspended in 6x protein samples buffer (16% SDS), subjected to SDS-PAGE, and scanned on an infrared imager (LI-COR Odyssey; Lincoln, NE, USA). Hydroxylamine-cleavage of thioesters in microscopy samples was performed at 1.5 M in PBS (pH 7.2) for 2 h, and extensively washed with PBS containing 0.05 % Triton X-100 prior to imaging.
Bioinformatics
All reviewed human proteins annotated as membrane proteins were downloaded from Uniprot, and three datasets that correspond to different subcellular compartments (ER, Golgi, plasma membrane) were generated, and only proteins annotated as having experimentally verified subcellular locations were used in the study. The css-palm sites were predicted using CSS-Palm 2.0 (Ren et al., 2008). In order to find the inner loops of the proteins we used scampi 2.0 together with SignalP4.0 (Peters et al. 2015; Nielsen 2017).
Surface biotinylation assay
HeLa cells grown in 6-well plates to 50% confluency were transfected using 1 μg of the respective plasmid DNA and 3 μl of Fugene HD transfection reagent (Promega) for 24 h. Cells were washed twice in PBS Ca/Mg (PBS with 1 mM MgCl2, 0.1 mM CaCl2) and subsequently incubated in 600 μl/well of a 500 μg/ml sulfo-NHS-biotin solution (Pierce, buffer: 150 mM NaCl, 10 mM triethanolamine pH 9.0, 2 mM CaCl2, freshly prepared) for 30 min on ice. The cells were then washed in quenching buffer (PBS Ca/Mg, 100 mM glycine), and incubated in 600 μl/well of quenching buffer for 20 min on ice. The cells were washed twice with PBS (without Mg/Ca) and incubated for 10 min with 300 μl of lysis buffer (50 mM HEPES–NaOH, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholate, and protease inhibitor cocktail (Roche)). Cells were scraped, transferred to Eppendorf tubes and sonicated for 3 min. Next, the tubes were incubated for 15 min with constant agitation at RT. Finally, the lysate was centrifuged for 10 min at 13 000 rpm and 4 °C, and the supernatant was retained. 40 μl of Neutravidincoupled beads (Thermo Fisher) were washed twice with 300 μl of lysis buffer. Centrifugation steps with beads were performed for 1 min at 3000×g and 4 °C. The retained supernatant was then added to the beads and placed on a rotor wheel for 1 h at RT. Samples were eluted with 6× SDS-sample buffer (16% SDS) for 10 min at 95 °C. Eluates were obtained after brief centrifugation at 16 000×g and RT. Input (lysate, 2.5% of total) and eluates (25% of total) were subjected to SDS–PAGE (Bis–Tris 4–12% gradient gels, Invitrogen) in MES buffer (Invitrogen), and transferred to PVDF membranes via wet blot at 100 V for 1 h. The blots were decorated with anti-rabbitIRdye680 coupled antibodies (Li-Cor) and analyzed on a LI-COR Odyssey device. Signal intensities were quantified using Li-Cor software.
Lectin assay
HeLa cells grown in 10 cm dishes were grown to 50% confluency were transfected using 3 μg of the respective plasmid DNA and 9 μl of Fugene HD transfection reagent (Promega) for 24 h. After release of the cargos from the ER (temperature shift or D/D solubilizer) for the indicated times, the cells were scraped in 300 μl PBS containing protease inhibitors (Roche), pelleted at 16,000xg and resuspended 300 μl of lysis buffer (50 mM HEPES–NaOH, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholate, and protease inhibitor cocktail (Roche)), and sonicated with a microtip sonicator for 10s, amplitude 15. The lysate was centrifuged for 8 min at 3,000xg and 4 °C, and the supernatant was retained. 50 μl of Jacalin-coupled agarose was used per lysate. After 3 washes in lysis buffer, lysates were added to the beads and placed on a rotor wheel for 1 h at RT. After 3 washes in lysis buffer, the samples were eluted with 6× SDS-sample buffer (16% SDS) for 10 min at 95 °C. Eluates were obtained after brief centrifugation at 16 000×g and RT and subjected to Western blot.
Electron microscopy of S-palmitoylated proteins
doxSNAP-DHHC3 HeLa cells were plated in MatTek dishes and grown to 80% confluency. Expression of DHHC3 was induced by adding a final concentration of 2.5μg/ml doxycycline (Sigma-Aldrich) for 5 h. Next, the cells were metabolically labeled with 50 μM alkyne-palmitate for the indicated times, washed with delipidated medium, and fixed with 2% gluteraldehyde/0.1M sodium cacodylate buffer for 1 h RT. The samples were rinsed in cacodylate buffer for 30 minutes, and subsequently PBS containing 5% charcoal stripped serum for 4 hours with repeated exchanges to remove residual glutaraldehyde. Next, the cells were permeabilized for 1 h in 0.1% saponincontaining blocking buffer (see immunofluorescence, without TX100 and NP40). Next, CuAAC to azide-PEG4-biotin was performed for 30 min RT, and the cells were repeatedly rinsed with PBS containing 5% charcoal stripped serum for 1 h, RT. Finally, the cells were incubated with 10ug/ml streptavidin-fluronanogold 488 (Nanoprobes, Yaphank, NY, USA) in blocking buffer o/n, and rinsed for 1 hour in blocking buffer prior to a second round of fixation with 2% gluteraldehyde/0.1M cacodylate buffer for 30 minutes, and a final rinse in cacodylate buffer for 30 minutes. Next, the cells were rinsed in distilled water, and rinsed in 0.02 M sodium citrate for 30 minutes (to quench aldehydes). Nanogold in the samples was developed using a SilverEnhancer kit (Nanoprobes) for 5 min, and stopped using distilled water. Next, the cells were postfixed with 0.1 % osmium-tetroxide for 30 minutes, then rinsed and left in 0.5% aqueous uranyl acetate overnight, then rinsed for 30 minutes in distilled water.
Samples were dehydrated in an ethanol series 50%, 70%, 95% and 100%, 10 minutes for each step. The cells were infiltrated with the epoxy resin Embed 812 (Electron microscopy Sciences, Hatfield, PA, USA) gel capsules inverted over the cells and hardened overnight at 60°C. Sample blocks were cut using a Leica UltraCut UC7. 60nm sections were collected on formvar/carbon coated grids and contrast stained using 2% uranyl acetate and lead citrate. Grids were viewed on a FEI Tencai Biotwin TEM at 80Kv. Images were taken using Morada CCD and iTEM (Olympus) software. Automated quantification of gold in Golgi membranes was performed with a custom script in MatLab (https://bitbucket.org/omarzaki123/golgi-gold-counter/src). The 250 nm sections for tomography were viewed using a FEI Tecnai TF20 at 200 kV with tilt angles from 60o to −60o. Data was collected using a FEI Eagle 4K × 4K digital camera. The volume reconstruction was done using IMOD and segmentation done using 3Dmod (Boulder Laboratory for 3-Dimensional Electron Microscopy of Cells, University of Colorado).
Estimation of membrane curvature on tomogram data
Membrane outlines, and where appropriate the locations of palmitate clusters, were manually annotated in EM tomograms using IMOD. These annotations, consisting of either 2D curves (in serial sections) representing Golgi membrane, or individual 3D points representing the location of palmitate clusters were loaded into python using the PyIMOD library. Local curvature was assessed at each point along the annotated membrane curves by fitting a segment of a circle to a small neighborhood (14 control points spanning ~30nm) around each point. We chose to estimate curvature from the 2D manually annotated curves rather than attempting to reconstruct a full 3D surface as the complicated Golgi morphology made linking contours between sections both time consuming and error prone. Membrane associated with palmitate was identified by performing nearest neighbor analysis between the palmitate locations and the annotated membrane. As curvature estimation was performed in 2D rather than 3D out of plane curvature is neglected and both total curvature and the palmitate curvature preference are likely to be under-estimated. Assuming a random 3D orientation, trends should nonetheless be visible.
Estimation of proximities in STED images
The proximity of either COPI to palmitate or COPI to a Golgi cisternal marker was assessed as follows: A threshold was chosen (20%) to identify Golgi areas labelled by palmitate and GM130. A euclidian distance transform was performed on this mask, giving the distance of every pixel in the image to the edge of the mask. Using this distance image, a cumulative histogram of COPI labelling was computed as a function of distance. This can be interpreted as the fraction of the total labelling that could be found within a given distance of the edge of palmitate or Golgi labelling. Negative distances indicate pixels which are within the Golgi mask. The code is implemented in python using the scipy toolkit and is available as part of the python-microscopy package (www.python-microscopy.org).
Purification of recombinant proteins from EXPI293 cells
Plasmid encoding for FLAG-tagged TfR-mini was transfected into EXPI293 cells according to the manufacturer (Thermo Fisher), and incubated at 37°C, 8% CO 2 for 48h. Cells were pelleted at 1,000xg for 10 min, washed in PBS, and re-pelleted. Pellets from 50 ml of cell culture were resuspended in 8ml of 50 mM HEPES/KOH pH 7.3, 175 mM NaCl, 5 mM EDTA, 1mM PMSF, 1mM TCEP, protease inhibitor cocktail, and 8% (v/v) TX-100. Next, the pellets were sonicated at amplitude 30 for 1 min, and centrifuged at 800xg for 2 min before repeating the sonication. Next, the lysate was placed rotating at 4°C for 3h, then centrifuged at 20,000xg rpm for 25 min at 4°C. The supernatant was transferred to a conical tube containing FLAG-affinity resin equilibrated in lysis buffer, and rotated for 3h at 4°C. The resin was added to a column, settled, and washed with 25 ml HEPES buffer (without TX-100). AF647-NHS ester was diluted in HEPES buffer to a final concentration of 50 μM, and added to the resin, and incubated for 1 h on ice. After two additional washes, the column was washed twice with 4 ml HEPES buffer containing 50 mM OG for 10 min on ice. If indicated, thioester-bound palmitate was cleaved in 1 ml of 1.5 M neutral hydroxylamine in HEPES, and washed twice in HEPES/OG. Finally, the proteins were eluted in HEPES/OG containing 125 ng/ml FLAG peptide (Sigma-Aldrich) in 300 μl aliquots, 30 mins per elution, for a total of 1.5 ml. Elutions were analyzed on 4–20% Bis-Tris gradient gels and stained with Coomassie, or analyzed for in-gel fluorescence on a LI-COR Odyssey infrared scanner.
Identification of S-palmitoylated proteins
For acyl-PEG and acyl-biotin-exchange assays (APE, ABE) assays, cells were lysed with TEA buffer (pH 7.3, 150 mM NaCl, 50 mM TEA, 1x Protease Inhibitor Mixture (Roche)) containing 4% SDS w/v. Typically, 200 ug of total protein in 95.5 μl lysis buffer was treated with 2 μl of 0.5 M TCEP for a final 10 mM TCEP with 30 min nutation. 25 mM of N-ethylmaleimide (NEM, Sigma) was freshly made and 2.5 μl was added to the sample, followed by a 2h incubation at room temperature. The alkylation step was then terminated by a methanol-chloroform-water (MCW) precipitation, with addition of prechilled methanol (400 μl), chloroform (150 μl), and distilled water (300 μl). The reactions were then centrifuged at 20,000g for 5 min at 4 C. The aqueous layer was removed and 1 mL of MeOH was added before another centrifugation step at 20,000g for 3 min. Once again, the supernatant was decanted, 800 μl of MeOH were added, and the sample was centrifuged again before being dried in a speed-vacuum. The samples were resuspended in 100 μl of TEA buffer, warmed to 37 C and sonicated for 5s, before undergoing two additional rounds of MCW precipitation to ensure complete removal of NEM. The samples were then resuspended in 30 μl of TEA buffer containing 4% SDS and 4 mM EDTA, and they were treated with 90 μl of 2M neutralized NH2OH (Sigma) dissolved in TEA buffer (pH 7.3) and containing 0.2% Triton X-100. The final concentration of NH2OH was 1.5 M. The control samples were not treated with NH2OH but rather TEA containing 0.2% Triton. The samples were incubated at room temperature for 1 h to overnight with nutation. Afterwards, another MCW precipitation was done, and the samples were resuspended in 30 μl TEA buffer containing 4% SDS, 4 mM EDTA. For APE assays, the sample was warmed to 37 C and sonicated for 5 s, before being treated with 90 μl TEA buffer containing 0.2% Triton X-100 and 1.33 mM mPEG-Mal (10 kDa). For ABE assays, HPDP-biotin was employed. The final concentration of the mPEG-Mal/HPDP-biotin was 1 mM. Samples were then incubated for 2h at room temp before a final MCW precipitation. For APE assays, the protein pellet was then resuspended in SDS-sample buffer (16% SDS), heated to 95 C for 5 min, and then loaded on a NuPAGE 4–12% Bis-Tris Gel, separated by SDS PAGE, and analyzed by Western Blot. For ABE-assays, samples were lysed in 500 μl lysis buffer (50 mM HEPES–NaOH, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholate, and protease inhibitor cocktail (Roche)), and concentrated with Neutravidin-agarose (Pierce, Appleton, WI, U.S.A) prior to SDS-PAGE and Western blot.
Identification of S-palmitoylated proteins was performed employing 50 μM alkyne-palmitate for 30 min in cells overexpressing the protein of interest. The cells were then scraped and clicked to 5 μM biotin-azide (ThermoFisher, Waltham, MA, U.S.A) in a waterbath sonicator for 10 min. After 20 min additional agitation of the samples, the cells were lysed as described above, concentrated with Neutravidin-agarose and subjected to SDS-PAGE and Western blot.
Micromanipulation of proteo-giant unilamellar vesicles
Proteo-GUVs reconstitution: Giant unilamellar vesicles (GUVs) were made in a 295 mOsm sucrose solution following the classical electroformation protocol (Mathivet et al., 1996) and contained a mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC from Avanti Polar Lipids), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE from Avanti Polar Lipids), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N[biotinyl(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG-biot from Avanti Polar Lipids) and Atto 488 labeled DOPE (DOPE-Atto from Atto-Tec) at a ratio 87:10:1:0.2 mol/mol.
Prior to the experiment, 20 μl GUVs were added to 180 μl of a solution containing 15 mM Hepes/KOH pH 7.3 and glucose with a final osmolarity of 330 mOsm (HG buffer). The larger osmolarity slightly deflated the GUVs which facilitated micromanipulation and the formation of a membrane protrusion inside the micropipette. 3 μl of Alexa-647 labeled protein at 3 μM were added to this GUV solution. OG was then 50 times below it CMC, GUVs remained intact and proteins spontaneously inserted into the GUV membrane. These proteo-GUVs were used for micromanipulation.
Micromanipulation experiments: Micromanipulations were performed in a glass bottom dish (Mattek, part No. P35G-0–20-C) preincubated for one hour with a 10% bovine Serum Albumin Solution and extensively rinsed with HG buffer. In the dish, 10 μl of the proteo-GUVs solution and 0.1 μl of 5μm biotinylated silica beads (from Bangs Labs) were added to 400 μl of HG buffer. Because of their density, beads and proteo-GUVs settled down at the bottom of the dish where they could be seized by two micropipettes under a confocal microscope (Leica SP8). All observations were performed with a 20x air objective. When the bead was brought in contact with the proteo-GUV membrane, streptavidin-biotin bonds formed leading to a strong adhesion between the membrane and the bead. Upon separation of the bead, a tube was pulled from the membrane as previously described (Bo and Waugh, 1989). Because the surface and volume of the proteo-GUV remained constant during the experiment (Hochmuth, 1982), the radius of the tube, rt, was obtained from the displacement of the protrusion in the pipette, ΔLp, upon displacement of the bead, ΔLb, through the relation: rt=(1-rp/Rv)rpDLv/DLt, where rp is the micropipette radius and Rv the vesicle radius.
Relative concentration of the protein in the membranes: The fluorescence intensities of the protein and DOPE-Atto were measured in both membranes: IGUV/prot, IGUV/Lip in the proteo-GUV respectively and Itube/prot and Itube/Lip in the tube respectively. The relative density of the protein in the tubes and in the proteo-GUV, R, was then obtained, as previously proposed (Sorre et al., 2009), by: R= [Itube/prot/IGUV/prot]/[Itube/Lip/IGUV/Lip].
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA) for unpaired, two-tailed t-tests. Differences were considered significant if P-value <0.05(*), <0.01(**), or <0.001(***). Automated quantification of gold particles in Golgi membranes was performed with a custom script in MatLab (MathWorks, Natick, MA, USA): https://bitbucket.org/omarzaki123/golgi-gold-counter/src. Pearson´s correlation coefficient was calculated using the “co-localization threshold” plugin in ImageJ (confocal sections), or Volocity software (Z-stacks; Quorum Technologies, Lewes, UK).
Supplementary Material
Highlights:
DHHC S-palmitoyltransferases are enriched in the cis-Golgi
S-palmitoylation induces concentration of membrane cargo at the cisternal rim
The rate of anterograde transport across the Golgi is controlled by S-palmitoylation
ACKNOWLEDGEMENTS
This work was supported by the G. Harold and Leila Y. Mathers foundation ( to J.E.R.), a NIH MIRA grant (GM118084; J.E.R.), and a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft to A.M.E. We thank Al Mennone at the Yale Center for Cellular and Molecular Imaging Facility (CCMI, NIH grant S10OD020142).
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I (2011). Protein palmitoylation and subcellular trafficking. Biochem Biophys Acta, 1808(12): 2981–94. [DOI] [PubMed] [Google Scholar]
- Alvarez E, Girones N and Davis RJ (1990). Inhibition of the receptor-mediated endocytosis of diferric transferrin is associated with the covalent modification of the transferrin receptor with palmitic acid. Journal of Biological Chemistry, 265(27), 16644–16655. [PubMed] [Google Scholar]
- Apolloni A, Prior IA, Lindsay M, Parton RG and Hancock JF (2000). H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Molecular and cellular biology, 20(7), 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo L, and Waugh RE (1989). Determination of bilayer membrane bending stiffness by tether formation from giant, thin-walled vesicles. Biophys J, 55, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JE (1989). Using temperature-sensitive mutants of VSV to study membrane protein biogenesis. Methods in cell biology, 32, 85–110. [DOI] [PubMed] [Google Scholar]
- Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, van der Goot FG (2015). SwissPalm: Protein Palmitoylation database. F1000Res, 4:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottanelli F, Kromann EB, Allgeyer ES, Erdmann RS, Wood Baguley S, Sirinakis G, Schepartz A, Baddeley D, Toomre DK, Rothman JE, Bewersdorf J (2016). Two-colour live-cell nanoscale imaging of intracellular targets. Nat Commun, 7:10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottanelli F, Kilian N, Ernst AM, Rivera-Molina F, Schroeder LK, Kromann EB, Lessard MD, Erdmann RS, Schepartz A, Baddeley D, Bewersdorf J, Toomre D, Rothman JE (2017). A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Mol Biol Cell, 28(12): 1676–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais J and Van Der Goot FG (2009). Palmitoylation of membrane proteins (Review). Molecular membrane biology, 26(1–2), 55–66. [DOI] [PubMed] [Google Scholar]
- Chien AJ, Carr KM, Shirokov RE, Rios E and Hosey MM (1996). Identification of palmitoylation sites within the L-type calcium channel β2a subunit and effects on channel function. Journal of Biological Chemistry, 271(43), 26465–26468. [DOI] [PubMed] [Google Scholar]
- Chlanda P, Mekhedov E, Waters H, Sodt A, Schwartz C, Nair V, Blank PS, Zimmerberg J (2017). Palmitoylation contributes to membrane curvature in influenza A virus assembly and hemagglutinin-mediated membrane fusion. J Virol, 91(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Rao P, Fogelsong RJ, Bardes ESG (1992). 2-bromopalmitoyl-CoA and 2-bromopalmitate: promiscuous inhibitors of membrane-bound enzymes. Biochem Biophys Acta, 1125(2): 203–9. [DOI] [PubMed] [Google Scholar]
- Demers A, Ran Z, Deng Q, Wang D, Edman B, Lu W and Li F, 2014. Palmitoylation is required for intracellular trafficking of influenza B virus NB protein and efficient influenza B virus growth in vitro. Journal of General Virology, 95(6), pp.1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Murakami S, Sun Y, Kilpatrick CL, and Luscher B (2017). DHHC7 Palmitoylates Glucose Transporter 4 (Glut4) and Regulates Glut4 Membrane Translocation. Journal of Biological Chemistry, 292(7), 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE (1991). Beta-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to beta-adaptin. Cell, 64(3): 649–65. [DOI] [PubMed] [Google Scholar]
- El-Husseini AED, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA and Bredt DS, 2002. Synaptic strength regulated by palmitate cycling on PSD-95. Cell, 108(6), pp.849–863. [DOI] [PubMed] [Google Scholar]
- Farquhar MG (1985). Progress in unraveling pathways of Golgi traffic. Annual review of cell biology, 1(1), 447–488. [DOI] [PubMed] [Google Scholar]
- Frolov VA, Shnyrova AV, and Zimmerberg J (2011). Lipid Polymorphisms and Membrane Shape. Cold Spring Harbor Perspectives in Biology, 3(11), a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA and Bredt DS (2004). Identification of PSD-95 palmitoylating enzymes. Neuron, 44(6), 987–996. [DOI] [PubMed] [Google Scholar]
- Fukata Y and Fukata M, 2010. Protein palmitoylation in neuronal development and synaptic plasticity. Nature Reviews Neuroscience, 11(3), pp.161–175. [DOI] [PubMed] [Google Scholar]
- Glick BS and Rothman JE (1987). Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature, 326(6110), 309–312. [DOI] [PubMed] [Google Scholar]
- Gonzalo S and Linder ME (1998). SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Molecular Biology of the Cell, 9(3), 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb CD, Zhang S, and Linder ME (2015). The Cysteine-rich Domain of the DHHC3 Palmitoyltransferase Is Palmitoylated and Contains Tightly Bound Zinc. The Journal of Biological Chemistry, 290(49), 29259–29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J and Chamberlain LH, 2011. DHHC palmitoyl transferases: substrate interactions and (patho) physiology. Trends in biochemical sciences, 36(5), pp.245–253. [DOI] [PubMed] [Google Scholar]
- Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NC and Chamberlain LH (2017). Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proceedings of the National Academy of Sciences, 114(8), 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Salaun C, Fukata Y, Fukata M and Chamberlain LH (2008). Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. Journal of Biological Chemistry, 283(36), 25014–25026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmuth RM, Wiles HC, Evans EA, and McCown JT (1982). Extensional flow of erythrocyte membrane from cell body to elastic tether. II. Experiment. Biophysical Journal, 39(1): 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BC and Linder ME (2012). DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. Journal of Biological Chemistry, 287(10), 7236–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB (2001). Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 40(11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE and Staehelin LA (1999). Golgi structure in three dimensions: functional insights from the normal rat kidney cell. The Journal of cell biology, 144(6), 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G, Zheng H, and Rothman JE (2013). Stapled Golgi cisternae remain in place as cargo passes through the stack. eLife, 2, e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G, Dunlop MH, Lerich A, Zheng H, Bottanelli F, Rothman JE (2014). The Golgi ribbon structure facilitates anterograde transport of large cargoes. Mol Biol Cell. 25(19): 3028–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME and Deschenes RJ, 2007. Palmitoylation: policing protein stability and traffic. Nature reviews Molecular cell biology, 8(1), pp.74–84. [DOI] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME and Deschenes RJ (2002). Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. Journal of Biological Chemistry, 277(43), 41268–41273. [DOI] [PubMed] [Google Scholar]
- Magee AI, Koyama AH, Malfer C, Wen D, and Schlesinger MJ (1984). Release of fatty acids from virus glycoproteins by hydroxylamine. Biochimica et Biophysica Acta (BBA) - General Subjects, 798(2), 156–166. [DOI] [PubMed] [Google Scholar]
- Martin BR, Wang C, Adibekian A, Tully SE and Cravatt BF (2012). Global profiling of dynamic protein palmitoylation. Nature methods, 9(1), 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivet L, Cribier S, and Devaux PF (1996). Shape change and physical properties of giant phospholipid vesicles prepared in the presence of an AC electric field. Biophys J, 70, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Mitchell G, Ling Y, Budde C and Deschenes RJ (2010). Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. Journal of Biological Chemistry, 285(49), 38104–38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H (2017). Predicting Secretory Proteins with SignalP. Methods Mol Biol, 1611: 59–73. [DOI] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T and Igarashi Y (2006). Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain- containing proteins. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1761(4), 474–483. [DOI] [PubMed] [Google Scholar]
- Omura S, Tomoda H, Xu QM, Takahasi Y, Iwai Y (1986). Triacsins, new inhibitors of acyl-CoA synthetase produced by Steptomyces sp. J Antibiot (Tokyo), 39(9):1211–8. [DOI] [PubMed] [Google Scholar]
- Orci L, Glick BS and Rothman JE (1986). A new type of coated vesicular carrier that appears not to contain clathrin: Its possible role in protein transport within the Golgi stack. Cell, 46(2), 171–184. [DOI] [PubMed] [Google Scholar]
- Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Söllner TH, and Rothman JE (1997). Bidirectional Transport by Distinct Populations of COPICoated Vesicles. Cell, 90(2), 335–349. [DOI] [PubMed] [Google Scholar]
- Ostermann J, Orci L, Tani K, Amherdt M, Ravazzola M, Elazar Z and Rothman JE, (1993). Stepwise assembly of functionally active transport vesicles. Cell, 75(5), 1015–1025. [DOI] [PubMed] [Google Scholar]
- Palmer DJ, Helms JB, Beckers CJ, Orci L, Rothman JE (1993). Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J Biol Chem. 268(16): 12083–9. [PubMed] [Google Scholar]
- Park SY, Yang JS, Schmider AB, Soberman RJ, Hsu VW (2015). Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature, 521(7553): 529–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PA, Dietrich F, Bewersdorf J, Rothman JE, and Lavieu G (2013). Inter- Golgi transport mediated by COPI-containing vesicles carrying small cargoes. eLife, 2, e01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS and Hang HC (2016). Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proceedings of the National Academy of Sciences, 113(16), 4302–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Tsirigos KD, Shu N, Elofsson A (2016). Improved topology predictions using the first and last hydrophobic helix rule. Bioinformatics, 32(8): 1158–62. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Orci L, Glick BS, Amherdt M, Arden SR, Malhotra V and Rothman JE, (1989). Fatty acyl-coenzyme A is required for budding of transport vesicles from Golgi cisternae. Cell, 59(1), 95–102. [DOI] [PubMed] [Google Scholar]
- Politis EG, Roth AF and Davis NG (2005). Transmembrane topology of the protein palmitoyl transferase Akr1. Journal of Biological Chemistry, 280(11), 1015610163. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, and Lippincott- Schwartz J (1997). ER-to-Golgi transport visualized in living cells. Nature, 389(6646):81–5. [DOI] [PubMed] [Google Scholar]
- Rana MS, Kumar P, Lee CJ, Verardi R, Rajashankar KR, Banerjee A (2018). Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science, 359(6372). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, and Yao X (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Engineering, Design and Selection, 21(11), 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Adams GA and Gallione CJ (1984). The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proceedings of the National Academy of Sciences, 81(7), 2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L and Davis NG (2002). The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. The Journal of cell biology, 159(1), 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE (2014). The Principle of Membrane Fusion in the Cell (Nobel Lecture). Angew. Chem. Int. Ed, 53, 12676–12694. [DOI] [PubMed] [Google Scholar]
- Salaun C, Greaves J and Chamberlain LH, 2010. The intracellular dynamic of protein palmitoylation. The Journal of cell biology, 191(7), pp.1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MF and Schlesinger MJ (1979). Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell, 17(4), 813–819. [DOI] [PubMed] [Google Scholar]
- Sheetz MP and Singer SJ (1974). Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proceedings of the National Academy of Sciences, 71(11), 4457–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys JE and Linder ME, 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annual review of biochemistry, 73(1), pp.559–587. [DOI] [PubMed] [Google Scholar]
- Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, Goud B, and Bassereau P (2009). Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc Natl Acad Sci U S A, 106, 56225626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Limbrick AR, Allan D, and Judah JD (1984). Isolation of highly purified Golgi membranes from rat liver use of cycloheximide in vivo to remove Golgi contents. Biochim Biophys Acta. 769(1), 171–178. [DOI] [PubMed] [Google Scholar]
- Todorow Z, Spang A, Carmack E, Yates J, and Schekman R (2000). Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America, 97(25), 13643–13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco A, Polishchuk RS, Martella O, Pentima AD, Fusella A, Giandomenico DD, and Luini A (2004). Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat Cell Biol, 6(11), 1071–1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.