Abstract
Friedreich's ataxia (FRDA) is a neurodegenerative disease caused by inherited deficiency of the mitochondrial protein Frataxin (FXN), which has no approved therapy and is an area in which biomarkers are needed for clinical development. Here, we investigated the consequences of FXN deficiency in patient-derived FRDA fibroblast cell models, the FRDA mouse model KIKO, and in whole blood collected from patients with FRDA. We observed decreased mitochondrial copy number in all the three FRDA models tested: cells, mice and patient blood. In addition, we observed 40% residual mitochondrial gene expression in FRDA patient blood. These deficiencies of mitochondrial biogenesis in FRDA cells and patient blood are significantly correlated with FXN expression, consistent with the idea that the decreased mitochondrial biogenesis is a consequence of FXN deficiency. The observations appear relevant to the FRDA pathophysiological mechanism, as FXN-dependent deficiency in mitochondrial biogenesis and consequent mitochondrial bioenergetic defect could contribute to the neurodegenerative process. The observations may also have translational potential, as mitochondrial biogenesis could now be followed as a clinical biomarker of FRDA as a correlate of disease severity, progression, and therapeutic effect. Also, mitochondrial copy number in blood is objective, scalar and more investigator-independent than clinical-neurological patient rating scales. Thus, FXN deficiency causes mitochondrial deficiency in FRDA cells, the KIKO mouse model, and in whole blood of patients with FRDA, and this deficiency could potentially be used in clinical trial design.
Introduction
Frataxin (FXN) is a nuclear-encoded mitochondrial protein whose function in mitochondria supports the formation of iron–sulfur clusters, protection from oxidative stress, and maintenance of iron homeostasis (1,2). Decreased FXN expression as a result of expanded GAA repeats, in the first intron of FXN gene, leads to a progressive, neuro-muscular disease called Friedreich’s ataxia (FRDA) (3). It is characterized by impaired proprioception, muscle weakness, hypertrophic cardiomyopathy and diabetes (4). FRDA is most common autosomal recessive disorder affecting 1 in 20,000 Caucasians (5). Currently, there is no cure or effective treatment for this devastating disease (6).
Decreased FXN in FRDA increases mitochondrial and nuclear DNA damage in yeast, mice and humans (7–10). FXN deficiency also results in enhanced oxidative stress and diminished mitochondrial function (2,11). Despite of evidence, that decrease in FXN is associated with enhanced mitochondrial DNA damage and diminished mitochondrial function in FRDA, effect of FXN deficiency on overall mitochondrial biogenesis is unclear.
Here, we investigated the effect of decreased FXN on mitochondrial biogenesis in FRDA patient fibroblast cells, KIKO mice and whole blood of patients with FRDA. We observe a significant decrease in mitochondrial copy number in all the three models tested. In addition, FXN knockdown decreases mitochondrial copy number in healthy fibroblast and its overexpression increases mitochondrial protein expression in FRDA patient fibroblast, suggesting FXN-dependent mitochondrial biogenesis defect in FRDA. Further, we observe significant positive correlation between FXN mRNA expression to mitochondrial copy number in FRDA fibroblasts and whole blood. There is also decrease in mitochondrial subunit gene expression in FRDA whole blood. The measurement of mitochondrial copy number in blood is objective, rapid and appears to relate to FXN expression, which relates to disease severity, age of onset, and progression. Thus, we show that mitochondrial copy number could potentially be used as a biomarker for determining disease severity and in support of effective therapeutic evaluation.
Results
There is a decrease in mitochondrial mass in FRDA fibroblasts
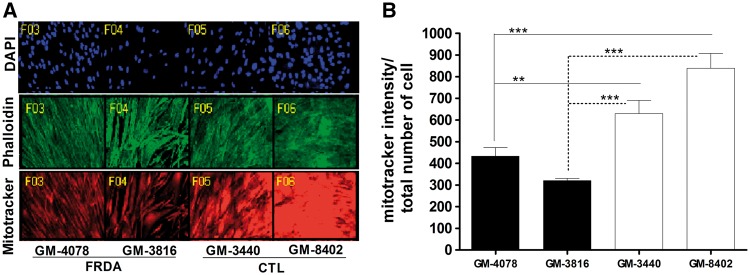
FXN functions in mitochondrial iron-sulfur cluster biogenesis. Iron-sulfur clusters are required in mitochondrial enzyme complexes I, II and III and other mitochondrial enzymes (1). Therefore, a decrease in FXN might impact mitochondrial function and number. Healthy and FRDA fibroblast derived from patients were quantified for mitochondrial mass by staining with MitoTracker® Deep Red FM, which stains active mitochondria and is retained after fixation. Its intensity per cell was measured with MetaXpress® image acquisition and analysis software. FRDA fibroblasts, GM4078 had 68% (P < 0.017, n = 8) and a 52% (P < 1.6×10−4, n = 8) residual mitochondrial mass compared to healthy fibroblasts, GM-3440 and GM-8402, respectively (Fig. 1). Similarly, FRDA fibroblast GM3816 had 51% (P < 2×10−4, n = 8) and 38% (P < 3.5×10−6, n = 8) residual mitochondrial mass compared to GM-3440 and GM-8402, respectively (Fig. 1). Thus, we observe a significant decrease in mitochondrial mass in FRDA fibroblasts compared to healthy fibroblasts.
Figure 1.
There is decrease in mitochondrial mass in FRDA fibroblast cell model. Human fibroblast cells were cultured in 96 well plate and stained with DAPI, phalloidin and mitotracker deep red FM. (A) Representative Image obtained from MetaXpress® Hi-content image acquisition, Molecular Devices. (B) Amount of mitochondrial mass measured as mitotracker intensity per cell. Bars represent averages ± standard deviations (n = 8, *P < 0.05, **P < 0.01, ***P < 0.001).
FXN expression correlates with mitochondrial biogenesis in fibroblast cells
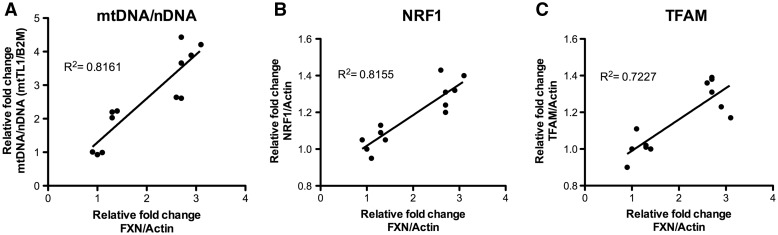
To investigate the relationship of FXN to mitochondrial biogenesis, we measured mitochondrial copy number as ratio of mitochondrial DNA over nuclear DNA (mt/nDNA) by quantitative PCR (qPCR). In addition, the relationship of mitochondrial biogenesis replication factors—Transcription Factor A-Mitochondrial (TFAM) and Nuclear Respiratory Factory 1 (NRF1) to FXN level was quantified, by qRT-PCR and linear regressions was carried out (Fig. 2). A significant and strong positive correlation is observed between FXN mRNA expression and mitochondrial biogenesis markers—mt/nDNA, TFAM and NRF1, with r2 correlation coefficients of 0.816 (P < 0.0001), 0.722 (P < 0.0001) and 0.815 (P < 0.0005), respectively. This clearly indicates that FXN expression is significantly and positively correlated with mitochondrial biogenesis in fibroblast cells.
Figure 2.
Frataxin expression is correlated to mitochondrial biogenesis marker in human fibroblast cell model. Linear regression analysis of FXN mRNA expression with (A) mitochondrial DNA to nuclear DNA ratio (mtTL1/B2M) (B) NRF1 mRNA expression and (C) TFAM mRNA expression.
FXN knockdown in healthy fibroblasts decreases their mitochondrial copy number
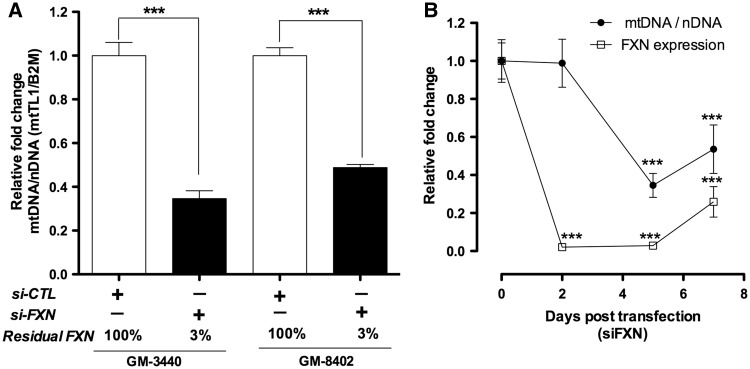
To further understand FXN-dependent effect of mitochondrial biogenesis, FXN expression was knocked down using FXN siRNA (siFXN) in two different healthy fibroblasts GM-3440 and GM-8402. Scrambled siRNA (siCTL) was used as a control to determine any non-specific target effect of RNAi knockdown. As expected, we see no significant difference in FXN expression and mitochondrial copy number when comparing siCTL-treated cells to untreated healthy fibroblast, confirming absence of any non-specific targeting effect of RNAi knockdown (data not shown). siFXN treatment significantly decreased FXN expression with residual FXN of 3% (P < 3.6×10−4, n = 3) in both GM3440 and GM8402 (Fig. 3A). There was also decrease in mt/nDNA ratio with residual mitochondrial copy number of 35% (P < 7.4×10−4, n = 3) and 47% (P < 1.9×10−4, n = 3) in GM3440 and GM8402, respectively; 5 days post transfection (Fig. 3A). Thus, a long-term FXN knockdown significantly decreases mitochondrial biogenesis.
Figure 3.
Frataxin knockdown in healthy fibroblasts decreases mitochondrial DNA copy number. Human fibroblast cells are treated with FXN siRNA and CTL siRNA. (A) qPCR analysis of mt/nDNA ratio 5 days post transfection in GM3440 and GM8402. (B) qPCR analysis of mt/nDNA ratio at different time points in GM3440. Bars represent averages ± standard deviations (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
We then investigated the effect of FXN knockdown on mt/nDNA ratio at different time points in GM3440 (Fig. 3B). We observe that 2 days post-siFXN transfection, although FXN was significantly knocked down with 2% residual FXN (P < 5.8×10−5, n = 3), there was no significant difference in mt/nDNA copy number. After 5 days of siFXN transfection, FXN remains significantly knocked down with 3% residual FXN (P < 3.6×10−4, n = 3) and mt/nDNA copy number significantly decreases with 35% residual mitochondria (P < 7.4×10−6, n = 3). Thus, a prolonged FXN-deficiency appears to be required for a decline in mitochondrial biogenesis. We previously demonstrated that there is a lag of 4.5 days for depletion of FXN protein in mitochondria, after knockdown of FXN transcript in the nucleus (30). The delay in decline of mitochondrial biogenesis by mtDNA/nDNA ratio observed at 5 days post-siFXN in Figure 3B is likely the result of mitochondrial FXN protein turnover. Also consistent with our hypothesis, at 7 days post-transfection as the siRNA wears off, residual FXN increases by 25% (P < 4.1×10−4, n = 3) and simultaneously residual mt/nDNA copy number also increases by 53% (P < 2.5×10−4, n = 3). Overall, FXN deficiency produces a decline in mitochondrial biogenesis, whereas FXN increase produces a rise in mitochondrial biogenesis.
FXN overexpression in FRDA patient fibroblast increases mitochondrial biogenesis
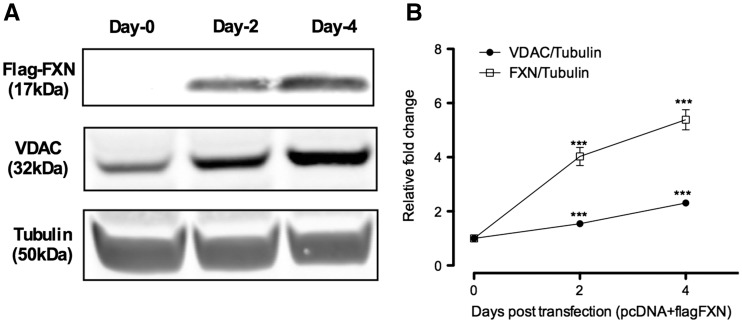
We also investigated the consequences of FXN overexpression above the low levels observed in FRDA patient fibroblast on mitochondrial biogenesis as visualized by Voltage- Dependent Anion Channel (VDAC) expression, a commonly used marker of mitochondrial biogenesis. Patient fibroblast GM4078 was transfected, with a vector (pcDNA3.1-frataxin-flag) (12) which expressed FXN protein with a C-terminal flag epitope tag. Fibroblast transfected with vector (pcDNA3.1), which did not have any insert, was used as a negative control. The construct (pcDNA3.1-frataxin-flag) drove high FXN expression in fibroblast cell as detected by anti-FLAG antibody (Fig. 4A). There was increase in FXN expression by 4 fold (P < 8.9×10−4, n = 3) and 5.3 fold (P < 3.2×10−4, n = 3), respectively, 2 days and 4 days post-transfection (Fig. 4B). VDAC expression increased by 1.5 fold (P < 5.8×10−5, n = 3) and 2.3 fold (P < 4.2×10−5, n = 3) (Fig. 4B), respectively, 2 days and 4 days post-transfection. This confirms FXN-dependent effect on mitochondrial biogenesis.
Figure 4.
Frataxin over-expression in patient fibroblast increases mitochondrial protein expression. Human fibroblast cells are treated with (pcDNA3.1-frataxin-flag) or (pcDNA3.1). (A, B) Western blot analysis of FXN and VDAC normalized to tubulin. Bars represent averages ± standard deviations (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
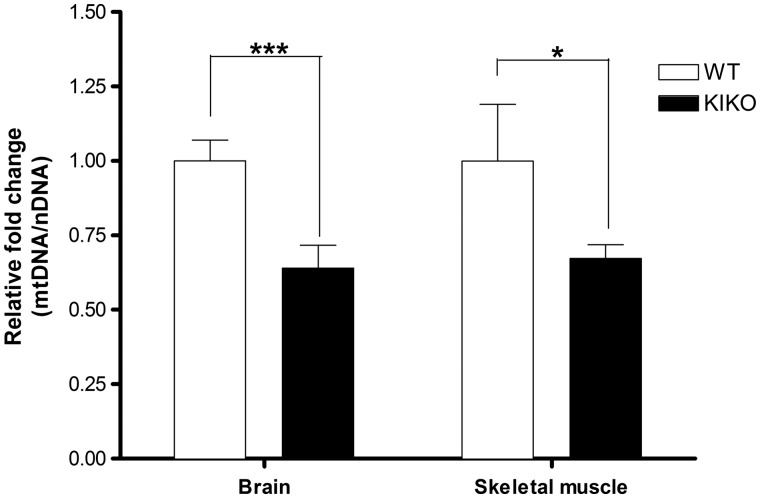
Mitochondrial copy defect in brain and muscle of KIKO mouse model of FRDA
Since stable and transient FXN deficiency decreased mitochondrial biogenesis in cell models, we investigated mitochondrial biogenesis in the FRDA KIKO mouse model that has a ∼40% residual FXN level (13). Brain and skeletal muscle tissues were collected from 10 Knock-in Knock- out (KIKO) mice and 7 age-matched wildtype (WT) mice. qPCR was used to analyze mitochondrial DNA copy number as ratio of mt-ND1 (mitochondrial gene) to Cftr (nuclear gene). We observed significant decrease, with 63% (P < 0.004, n = 7) residual mt/nDNA in the brain, and 66% (P < 0.03, n = 7) residual mt/nDNA in skeletal muscle tissue, respectively, compared to wild type mice (Fig. 5).
Figure 5.
Mitochondrial copy number decreases in brain and skeletal muscle tissue of KIKO mice model of FRDA. DNA was extracted from brain and skeletal muscle tissues of KIKO and WT mice followed by qPCR analysis of mitochondrial DNA copy number over nuclear DNA copy number (mt-ND1/Cftr). Bars represent averages ± standard deviations (n = 7, *P < 0.05, **P < 0.01, ***P < 0.001).
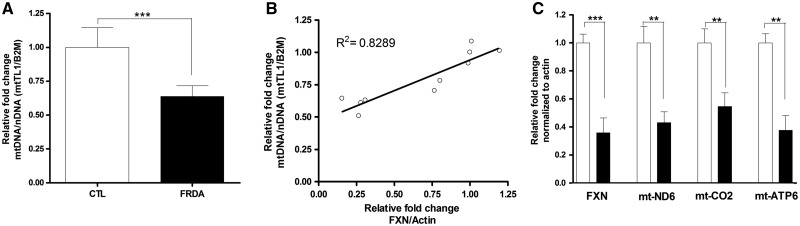
Mitochondrial biogenesis is deficient in FRDA patient blood
Given the consistent deficiency in mitochondrial biogenesis in patient cells, FXN knockdown cells and KIKO mouse model, the question arose whether the mitochondrial biogenesis defect could be observed in patient blood, and could be used as a minimally invasive biomarker. We used whole blood from five affected individuals and five age-matched controls to determine the effect of decreased FXN on mitochondrial biogenesis. Consistent with our other results, there was a mitochondrial biogenesis defect in FRDA patient blood with 60% residual (P < 3.2×10−4, n = 5) mt/nDNA compared to age matched controls (Fig. 6A). In addition, there is strong and significant positive correlation of FXN mRNA expression to mitochondrial copy number with correlation coefficient 0.8289 (n = 5, P < 0.0003) (Fig. 6B). Trends toward deficient mitochondrial complex-I gene expression in FRDA has been shown (14). We further confirm defective expression of multiple mitochondrial complex subunit genes by measuring expression-mtND6 (complex-I), mtCO2 (complex-II) and mtATP6 (complex-V) in patients with FRDA by using qRT-PCR (Fig. 6C). We see about 40% (n = 5, P < 0.002), residual mitochondrial gene expression in patients with FRDA compared to healthy controls in 3/3 mitochondrial genes. Thus, there is defective mitochondrial gene expression as well as mitochondrial biogenesis as measured by mt/nDNA ratio in blood of patients with FRDA.
Figure 6.
Mitochondrial biogenesis defect can be determined in whole blood of patients with FRDA. DNA and RNA was extracted from whole blood of patients with FRDA and age-matched controls. (A) qPCR analysis of mitochondrial DNA copy number over nuclear DNA copy number (mtTL1/B2M). (B) Linear regression analysis of mitochondrial copy number to FXN mRNA expression. (C) qRT-PCR analysis of mitochondrial subunit gene expression-mtND6 (complex-I), mtCO2 (complex-II) and mtATP6 (complex-V). Bars represent averages ± standard deviations (n = 7, *P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
FXN-deficiency causes a mitochondrial biogenesis defect
FXN deficiency in FRDA impairs mitochondrial respiratory chain function and enzymes involved in antioxidant defense mechanism (2,11). FXN deficiency also increases mitochondrial and nuclear DNA damage in multiple FRDA models (7–10). Although diminished mitochondrial function and enhanced mitochondrial DNA damage have been reported in FRDA, its effect on overall mitochondrial biogenesis is unclear. Decreased expression of PGC1-alpha, a transcriptional regulator of mitochondrial biogenesis, has been reported in FRDA patient fibroblasts (15). Another study shows increase in PGC1alpha and signals upstream in FRDA patient fibroblast due to ATP deficiency and ROS production. However, its effect on mitochondrial content is not clear (16).
Here, we clearly show that FXN-deficiency causes a mitochondrial biogenesis defect in multiple situations, including FRDA patient fibroblast cells, healthy fibroblast cells in which FXN has been knocked down, tissues of an FRDA mouse model, and in blood of patients with FRDA (Figs 1–4,6). In addition, we show FXN overexpression in FRDA patient fibroblast cells increases mitochondrial protein expression (Fig. 5). After the siRNA knockdown ‘wears off ’ at 7 days, mtDNA/nDNA rises immediately (Fig. 3B). Also, mitochondrial biogenesis is correlated to FXN expression (Figs 2, 6B). These observations support the idea that FXN deficiency causes defects in mitochondrial biogenesis, and that FXN rescues mitochondrial biogenesis. This also supports the idea that mitochondrial biogenesis can be measured to estimate some downstream consequence of FXN deficiency, such as disease severity, progression and therapeutic effectiveness.
Potential mechanism for FXN-dependent mitobiogenesis defect
Since we see defective mitochondrial biogenesis in multiple FRDA models, we tested the possibility of FXN knockdown in healthy cells and its effect on mitochondrial biogenesis. As expected, FXN knockdown in healthy human fibroblast showed significant decrease in FXN expression and mitochondrial copy number by 97% and 60%, respectively; 5 days post-siFXN transfection (Fig. 3) However, 2 days post-siFXN transfection; there was no change in mitochondrial copy number despite of significant reduction in FXN expression by 97% (Fig. 3B). Our previous experiments demonstrate that even after FXN transcript loss in the nucleus, it takes about 4.5 days for FXN protein to decrease in the mitochondria, which was consistent with the time at which mitochondrial biogenesis decreases in FXN knocked down healthy fibroblast (Fig. 3B).
There could be multiple potential mechanisms that link FXN deficiency to mitochondrial biogenesis defect. Since FXN's functions include the formation of iron–sulfur clusters (17) and iron–sulfur clusters are involved in mitochondrial electron transport and antioxidant defense (18), a defect in mitochondrial iron–sulfur cluster proteins could lead to deficiencies in a critical metabolite required for mitochondrial biogenesis. Defects in such a metabolite could lead to selective mitophagy and reduced mitobiogenesis (19). Alternatively, multiple DNA replication and repair proteins contain iron-sulfur clusters, and a decrease in expression or activity of such enzymes could inhibit mitochondrial replication and or repair (20,21). Thus, better understanding of cellular and molecular events involved in downstream of frataxin’s iron-sulfur function and upstream of mitochondrial biogenesis could perhaps identify novel therapeutic targets.
Mitochondrial biogenesis defect and FRDA pathophysiology
Since mitochondrial biogenesis was observed to decrease in multiple tissues of KIKO mice, one question is what consequence this would have for mitochondrial pathophysiology. There has been agreement that decreased mitochondrial function can contribute to multiple neurodegenerative disorders including Huntington's disease, Parkinson’s disease, Charcot-Marie-Tooth disease and FRDA (22,23). In addition, FXN is highly expressed in tissues with high energy demand, including large sensory neurons of dorsal root ganglion, skeletal muscle, heart and pancreas, which are also the major tissues affected in FRDA (24). A defect in energy generation due to deficient mitochondrial biogenesis (as shown here) appears to drive cellular apoptosis in FRDA (21,25) and could potentially contribute to the progressive degenerative nature of the disease.
Summary and prospects
Defective mitochondrial biogenesis has been indirectly proposed in FRDA based on diminished iron-sulfur cluster enzyme activity and decrease in mitochondrial DNA in cell circulating free nucleic acid in plasma of human blood (17,26–28). The results presented here clearly shows that FXN deficiency in vitro and in vivo leads to mitochondrial biogenesis defects in cell and animal models of FRDA, and in blood of patients with FRDA. Thus, FXN deficiency causes defects in mitochondrial biogenesis, which could be relevant to understanding the progressive neurodegenerative pathomechanism of FRDA.
Since there is no approved therapy for FRDA and there is a dearth of blood-based biomarkers, deficient mt/nDNA ratio could serve as biomarkers of disease severity, disease progression, and deficient mitochondrial gene expression and therapeutic effect. Thus, a significant positive correlation of FXN expression and deficient mitochondrial biogenesis in patient blood (Fig. 6) suggests the prospect that mitochondrial biogenesis could become a reliable and minimally invasive blood-based biomarker for determining disease progression and finding effective therapy in FRDA. Since we show the mitobiogenesis defect responds to frataxin increase, drugs that increase frataxin would be expected to increase mitochondrial biogenesis in blood of FRDA patients.
Materials and Methods
Cell culture
Human control (GM-8402 and GM-3440) and FRDA patient fibroblasts (GM-3816 and GM-4078) were obtained from the Coriell Institute for Medical Research repository. They were maintained at 37 °C in a humidified atmosphere with 5% CO2. DMEM (Corning, Inc., Corning, NY, USA) supplemented with 10% fetal bovine serum (JR-Scientific, Woodland, CA, USA), 1x Penicillin-Streptomycin Solution (Corning, Inc., Corning, NY, USA) was used as growth media. Media was changed every two days.
Mice model and tissue isolation
10 knock in-knock-out (KIKO) mice (Jackson Laboratories #012329 B6.Cg-Fxn(tm1.1Pand) Fxn(tm1Mkn/J)) and 7 littermate wild type (WT) mice were used for this study. They were 9 months old and individually housed in a vivarium maintained at 22–24 °C and 40–60% relative humidity with a 12-h light/12-h dark cycle. The University of California Institutional Animal Care and Use Committee approved all the experimental procedures. The mice were euthanized with brief isoflurane vaporizer followed by cervical dislocation and tissues were immediately removed and stored in RNA later at −20 °C until utilized for experiments.
Patient consent and blood collection
The human clinical study was reviewed and approved by the Investigational Review Board of the University of California, Los Angeles. All patients were provided written informed consent prior to initiation of the study. Study included five patients with FRDA and five age-matched control. Blood was collected in PAXgene blood RNA tube for RNA extraction and BD Vacutainer Lavender K2-EDTA Blood Collection Tubes for DNA extraction.
DNA and RNA isolation and quantification
Total DNA was extracted from fibroblast cells, mouse tissues and whole blood using DNeasy plus mini kit, DNeasy blood & tissue kit and QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) respectively, following manufacturer’s instruction. DNA was quantified by a NanoDrop 2000c Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Total RNA was extracted from human fibroblast cells and whole blood using RNeasy plus mini kit (Qiagen, Valencia, CA) and PAXgene Blood RNA Kit (PreAnalytiX, Switzerland), respectively, following manufacturer’s instruction. RNA quantity and quality were measured by a NanoDrop 2000c Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Quantitative PCR (qPCR)
cDNA was synthesized from mRNA with iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) per manufacturer’s instruction in a C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). qPCR was performed using SensiFAST SYBR No-ROX Kit (Bioline, Taunton, MA, USA) in Roche Lightcycler 480 (Roche Diagnostics, Indianapolis, IN, USA). The second derivative of the amplification curve was used to determine the cycle threshold, and the data were analyzed by delta CT calculation. Primer sets used in qPCR are listed in Table 1.
Table 1.
qPCR primer list
| Species | Gene | Sequence (5' → 3') |
|---|---|---|
| Human | mt-TL1 (DNA) Forward | CACCCAAGAACAGGGTTTGT |
| mt-TL1 (DNA) Reverse | TGGCCATGGGTATGTTGTTA | |
| Human | B2M (DNA) Forward | TGCTGTCTCCATGTTTGATGTATCT |
| B2M (DNA) Reverse | TCTCTGCTCCCCACCTCTAAGT | |
| Human | TFAM Forward | GTGATTCACCGCAGGAAAAGC |
| TFAM Reverse | GTGCGACGTAGAAGATCCTTTC | |
| Human | NRF1 Forward | AGGAACACGGAGTGACCCAA |
| NRF1 Reverse | TATGCTCGGTGTAAGTAGCCA | |
| Human | mt-ND6 Forward | GTAGGATTGGTGCTGTGG |
| mt-ND6 Reverse | GGATCCTCCCGAATCAAC | |
| Human | mt-CO2 Forward | ACCTTTCATGATCACGCCCT |
| mt-CO2 Reverse | GGGCAGGATAGTTCAGACGG | |
| Human | mt-ATP6 Forward | GAAGCGCCACCCTAGCAATA |
| mt-ATP6 Reverse | GCTTGGATTAAGGCGACAGC | |
| Human | β-ACTB Forward | GCCAACACAGTGCTGTCTGG |
| β-ACTB Reverse | CTGCTTGCTGATCCACATCTGC | |
| Human | FXN Forward | ATCTTCTCCATCCAGTGGACCT |
| FXN Reverse | GCTGGGCATCAAGCATCTTTT | |
| Mouse | mt-Nd1 (DNA) Forward | TCCGAGCATCTTATCCACGC |
| mt-Nd1 (DNA) Reverse | GTATGGTGGTACTCCCGCTG | |
| Mouse | Cftr (DNA) Forward | ATGGTCCACAATGAGCCCAG |
| Cftr (DNA) Reverse | GAACGAATGACTCTGCCCCT |
Protein extraction and western blot
Fibroblast cells were homogenized with 1× cell lysis buffer (Cell Signaling Technologies, Danvers, MA, USA) containing 1× Halt phosphatase and protease inhibitor cocktail (Thermo-Fisher, Waltham, MA, USA) and 1% PMSF (Sigma-Aldrich, St. Louis, MO, USA). 50µg of lysates were loaded per lane into 4–12% Bis–Tris gels (Invitrogen, Waltham, MA, USA). Electrophoresis was carried out according to the manufacturer’s recommendations. Following electrophoresis, the proteins were transferred to nitrocellulose membranes by the iBlot device (Invitrogen, Waltham, MA, USA) and blocked with an Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE, USA) for 1 h. Membranes were incubated overnight with the following primary antibodies in blocking buffer: 1:1000 dilution VDAC (D73D12, Cell Signaling Technologies, Danvers, MA, USA) and 1:2000 dilution tubulin (T9026, Sigma-Aldrich, St. Louis, MO, USA). Subsequently, the membranes were incubated with a corresponding pair of IRDye 680CW and IRDye 800CW-coupled secondary antibodies (LI-COR Biosciences, Lincoln, NE, USA) at 1:20,000 dilution. Proteins were visualized with the Odyssey infrared imager and software (LI-COR Biosciences, Lincoln, NE, USA) according the manufacturer’s instruction.
Plasmid and siRNA transfection
Fibroblast cells were seeded in six-well plates at 0.2×106 cells per well and transfected with negative control siRNA (cat. 12935300, Thermo-Fisher, Waltham, MA, USA), FXN siRNA (siRNA sequence= CCAAACAAGCAAAUCUGGCUAUCUU), pcDNA3.1-frataxin-flag (12) or negative control vector pcDNA3.1(addgene). Lipofectamine RNAiMAX (Thermo Scientific, Waltham, MA, USA) was used for siRNA transfection and Lipofectamine™ 3000 (Thermo Scientific, Waltham, MA, USA) was used for plasmid transfection, following manufacturer’s instruction.
Measuring mitochondrial mass by Hi content screening
Fibroblast cells were seeded in 96-well plates at density 1×105 cells per well. They were then treated with 200 nM MitoTracker® Deep Red FM for 30 min at 37 °C in 5% CO2, fixed in 4% paraformaldehyde for 20 min, permeablized with 0.2% Triton-X (Sigma) for 5 min and stained with DAPI (1:1000, Kirkegaard & Perry Laboratories, Gaithersburg, MD) and Alexa Fluor 488 phalloidin (1:1000, Thermo Scientific, Waltham, MA, USA) for 30 min. Cells were imaged with ImageXpress Micro System (Molecular Devices) at 20X magnification, one site per well, with binning of 1 and gain of 2 using laser-based focusing. Images were captured using DAPI filter, Cy5 filter and GFP filter for DAPI, Mitotracker and Alexa Fluor respectively (29). Mitotracker intensity per cell was measured using MetaXpress® image acquisition and analysis software.
Conflict of Interest statement. None declared.
Funding
RO1NS07777, RO1EY12245, PO1AG025532 to G.A.C.
References
- 1. Koutnikova H., Campuzano V., Foury F., Dollé P., Cazzalini O., Koenig M. (1997) Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]
- 2. Bradley J., Blake J., Chamberlain S., Thomas P., Cooper J., Schapira A. (2000) Clinical, biochemical and molecular genetic correlations in Friedreich’s ataxia. Hum. Mol. Genet., 9, 275–282. [DOI] [PubMed] [Google Scholar]
- 3. Campuzano V., Montermini L., Moltò M.D., Pianese L. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423. [DOI] [PubMed] [Google Scholar]
- 4. Parkinson M.H., Boesch S., Nachbauer W., Mariotti C., Giunti P. (2013) Clinical features of Friedreich's ataxia: classical and atypical phenotypes. J. Neurochem., 126, 103–117. [DOI] [PubMed] [Google Scholar]
- 5. Vankan P. (2013) Prevalence gradients of Friedreich's Ataxia and R1b haplotype in Europe co‐localize, suggesting a common Palaeolithic origin in the Franco‐Cantabrian ice age refuge. J. Neurochem., 126, 11–20. [DOI] [PubMed] [Google Scholar]
- 6. Pandolfo M. (2013) Treatment of Friedreich's ataxia. Expert Opin. Orphan Drugs, 1, 221–234. [Google Scholar]
- 7. Heidari M.M., Houshmand M., Hosseinkhani S., Nafissi S., Scheiber-Mojdehkar B., Khatami M. (2009) A novel mitochondrial heteroplasmic C13806A point mutation associated with Iranian Friedreich’s ataxia. Cell. Mol. Neurobiol., 29, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houshmand M., Panahi M.S.S., Nafisi S., Soltanzadeh A., Alkandari F.M. (2006) Identification and sizing of GAA trinucleotide repeat expansion, investigation for D-loop variations and mitochondrial deletions in Iranian patients with Friedreich's ataxia. Mitochondrion, 6, 87–93. [DOI] [PubMed] [Google Scholar]
- 9. Singh I., Faruq M., Padma M.V., Goyal V., Behari M., Grover A., Mukerji M., Srivastava A.K. (2015) Investigation of mitochondrial DNA variations among Indian Friedreich's ataxia (FRDA) patients. Mitochondrion, 25, 1–5. [DOI] [PubMed] [Google Scholar]
- 10. Haugen A.C., Di Prospero N.A., Parker J.S., Fannin R.D., Chou J., Meyer J.N., Halweg C., Collins J.B., Durr A., Fischbeck K. (2010) Altered gene expression and DNA damage in peripheral blood cells from Friedreich's ataxia patients: cellular model of pathology. PLoS Genet., 6, e1000812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calabrese V., Lodi R., Tonon C., D'Agata V., Sapienza M., Scapagnini G., Mangiameli A., Pennisi G., Stella A.M.G., Butterfield D.A. (2005) Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J. Neurol. Sci., 233, 145–162. [DOI] [PubMed] [Google Scholar]
- 12. Shan Y., Napoli E., Cortopassi G. (2007) Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet., 16, 929–941. [DOI] [PubMed] [Google Scholar]
- 13. Perdomini M., Hick A., Puccio H., Pook M.A. (2013) Animal and cellular models of Friedreich ataxia. J. Neurochem., 126, 65–79. [DOI] [PubMed] [Google Scholar]
- 14. Salehi M.H., Kamalidehghan B., Houshmand M., Meng G.Y., Sadeghizadeh M., Aryani O., Nafissi S. (2014) Gene expression profiling of mitochondrial oxidative phosphorylation (OXPHOS) Complex I in Friedreich Ataxia (FRDA) patients. PloS One, 9, e94069.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marmolino D., Manto M., Acquaviva F., Vergara P., Ravella A., Monticelli A., Pandolfo M. (2010) PGC-1alpha down-regulation affects the antioxidant response in Friedreich's ataxia. Plos One, 5, e10025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Giménez J.L., Gimeno A., Gonzalez-Cabo P., Dasí F., Bolinches-Amorós A., Mollá B., Palau F., Pallardó F.V. (2011) Differential expression of PGC-1α and metabolic sensors suggest age-dependent induction of mitochondrial biogenesis in Friedreich ataxia fibroblasts. PloS One, 6, e20666.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rotig A., Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. (1997) Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet., 17, 215–217. [DOI] [PubMed] [Google Scholar]
- 18. Shan Y., Schoenfeld R.A., Hayashi G., Napoli E., Akiyama T., Iodi Carstens M., Carstens E.E., Pook M.A., Cortopassi G.A. (2013) Frataxin deficiency leads to defects in expression of antioxidants and Nrf2 expression in dorsal root ganglia of the Friedreich's ataxia YG8R mouse model. Antioxid. Redox Signal., 19, 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Priault M., Salin B., Schaeffer J., Vallette F., Di Rago J., Martinou J. (2005) Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ., 12, 1613–1621. [DOI] [PubMed] [Google Scholar]
- 20. Bhalla A.D., Khodadadi‐Jamayran A., Li Y., Lynch D.R., Napierala M. (2016) Deep sequencing of mitochondrial genomes reveals increased mutation load in Friedreich's ataxia. Ann. Clin. Transl. Neurol., 3, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen Y., McMackin M.Z., Shan Y., Raetz A., David S., Cortopassi G. (2016) Frataxin Deficiency Promotes Excess Microglial DNA Damage and Inflammation that Is Rescued by PJ34. PloS One, 11, e0151026.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin M.T., Beal M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443, 787–795. [DOI] [PubMed] [Google Scholar]
- 23. Schapira A.H. (2008) Mitochondrial dysfunction in neurodegenerative diseases. Neurochem. Res., 33, 2502–2509. [DOI] [PubMed] [Google Scholar]
- 24. González‐Cabo P., Palau F. (2013) Mitochondrial pathophysiology in Friedreich's ataxia. J. Neurochem., 126, 53–64. [DOI] [PubMed] [Google Scholar]
- 25. Yang J., Cavadini P., Gellera C., Lonnerdal B., Taroni F., Cortopassi G. (1999) The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum. Mol. Genet., 8, 425–430. [DOI] [PubMed] [Google Scholar]
- 26. Dantham S., Srivastava A.K., Gulati S., Rajeswari M.R. (2016) Plasma circulating cell-free mitochondrial DNA in the assessment of Friedreich's ataxia. J. Neurol. Sci., 365, 82–88. [DOI] [PubMed] [Google Scholar]
- 27. Lodi R., Cooper J.M., Bradley J.L., Manners D., Styles P., Taylor D.J., Schapira A.H.V. (1999) Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc.Natl Aca. Sci., 96, 11492–11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puccio H., Simon D., Cossée M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. (2001) Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet., 27, 181–186. [DOI] [PubMed] [Google Scholar]
- 29. Kitami T., Logan D.J., Negri J., Hasaka T., Tolliday N.J., Carpenter A.E., Spiegelman B.M., Mootha V.K. (2012) A chemical screen probing the relationship between mitochondrial content and cell size. PloS One, 7, e33755.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu C., Cortopassi G. (2007) Frataxin knockdown causes loss of cytoplasmic iron–sulfur cluster functions, redox alterations and induction of heme transcripts. Arch. Biochem. Biophys., 457, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]