Abstract
Huntington disease (HD) is the most common inherited neurodegenerative disorder. It has no cure. The protein huntingtin causes HD, and mutations to it confer toxic functions to the protein that lead to neurodegeneration. Thus, identifying modifiers of mutant huntingtin-mediated neurotoxicity might be a therapeutic strategy for HD. Sphingosine kinases 1 (SK1) and 2 (SK2) synthesize sphingosine-1-phosphate (S1P), a bioactive lipid messenger critically involved in many vital cellular processes, such as cell survival. In the nucleus, SK2 binds to and inhibits histone deacetylases 1 and 2 (HDAC1/2). Inhibiting both HDACs has been suggested as a potential therapy in HD. Here, we found that SK2 is nuclear in primary neurons and, unexpectedly, overexpressed SK2 is neurotoxic in a dose-dependent manner. SK2 promotes DNA double-strand breaks in cultured primary neurons. We also found that SK2 is hyperphosphorylated in the brain samples from a model of HD, the BACHD mice. These data suggest that the SK2 pathway may be a part of a pathogenic pathway in HD. ABC294640, an inhibitor of SK2, reduces DNA damage in neurons and increases survival in two neuron models of HD. Our results identify a novel regulator of mutant huntingtin-mediated neurotoxicity and provide a new target for developing therapies for HD.
Introduction
Huntington disease (HD) is an incurable disease and is the most common inherited neurodegenerative disorder. HD is characterized by involuntary movements, personality changes, and dementia, and results in death 10–20 years after the appearance of symptoms. HD is caused by the N-terminal polyglutamine (polyQ) expansions in the protein huntingtin (Htt). The polyQ expansion primarily leads to degeneration of neurons in the striatum. The cortex and hippocampus are also affected as the disease progresses (1).
The polyQ expansion confers toxic functions to the protein, and identifying modifiers of mutant Htt (mHtt)-mediated neurotoxicity has been proposed as a therapeutic strategy for HD. Over the past decade, therapeutic efforts to reduce neurotoxicity focused on histone deacetylases (HDACs) (2,3). HDACs deacetylate a variety of nuclear, cytoplasmic, and mitochondrial proteins, and, therefore, regulate numerous cellular processes including cell survival. Although both neuroprotective and neurotoxic roles have been described for the nuclear HDACs, pharmacological inhibitors of nuclear HDACs generally promote survival in models of neurodegenerative disorders (4,5).
Sphingosine kinases (SK) catalyze the phosphorylation of sphingosine to form sphingosine-1-phosphate (S1P). Mammalian cells contain two sphingosine kinases, cytoplasmic SK1 and nuclear/mitochondrial SK2. A double knock-out of these kinases is embryonically lethal, which illustrates their importance. S1P is a second messenger involved in a number of extracellular, nuclear, and cytosolic signaling pathways (6). In the central nervous system, astrocytes secrete S1P that binds to the surface G-protein-coupled receptors (S1PR1-5) (7), four of which are found in neurons (7). These receptors regulate migration and synapse formation (7). In the cytosol of neuronal cells, SK1 regulates autophagy (8,9). In the nucleus, SK2 and S1P form a complex with HDAC1/2 that inhibits their deacetylase activity (10).
The S1P pathway has been proposed as a therapeutic target for multiple diseases (11,12). We recently demonstrated that cytoplasmic SK1 regulates the degradation of mHtt (8). Here, we hypothesized that nuclear SK2 may also modify mHtt-associated neurotoxicity. Unexpectedly, we discovered that SK2 is toxic for primary neurons and induces the formation of DNA double-strand breaks (DSBs). We also found that SK2 is hyperphosphorylated in brain samples from a mouse model of HD, the BACHD mice. Remarkably, an inhibitor of SK2, ABC294640, protects against degeneration in two neuron models of HD. This suggests that targeting the SK2 pathway in HD is an attractive therapeutic strategy.
Results
SK2 is nuclear in primary cultured cortical and striatal neurons
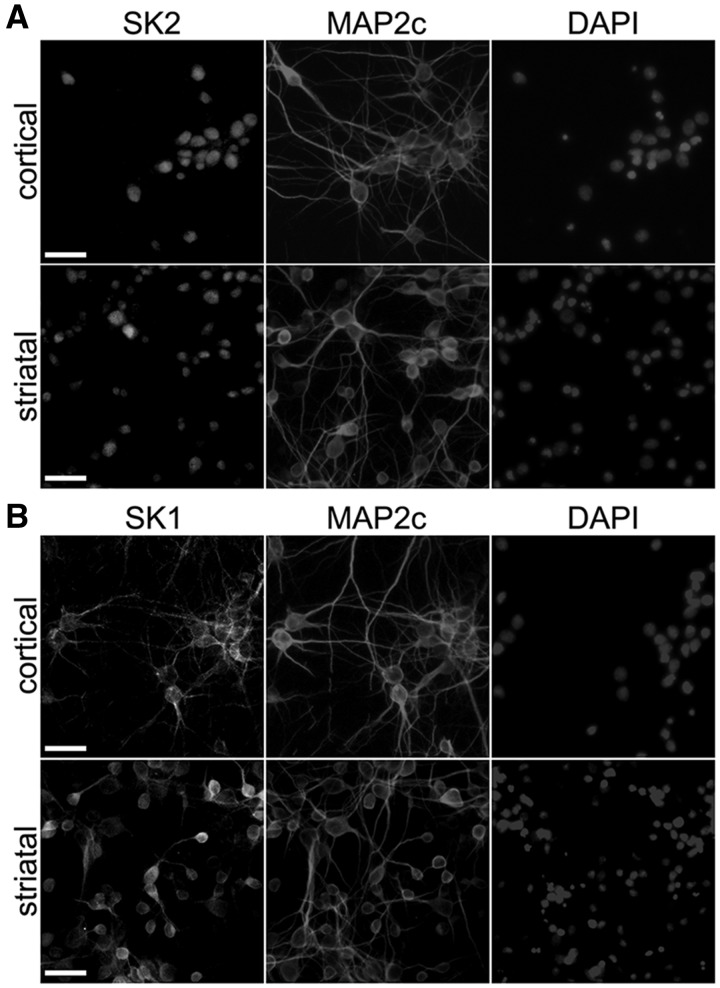
In non-neuronal cells, SK2 is localized to the nucleus and mitochondria (10,11,13). In neurons, the localization of SK2 is less clear. Several studies demonstrated that SK2 is nuclear, at least in cancerous neuron-like SH-SY5Y cells (12), but yet another study showed the involvement of SK2 in autophagy, a strictly cytoplasmic process (14). We, therefore, decided to determine the localization of SK2 in cultured cortical and striatal neurons, which are the most affected neuronal types in HD. As a control, we assessed the localization of SK1. Neurons were cultured from the embryonic rat cortices and striata. Neurons were fixed and stained with an antibody against SK2 or SK1. An antibody against microtubule-associated protein MAP2c was used to visualize the cytosol and dendrites, and Hoechst dye to visualize the nuclei. The SK2 staining was specific to the nucleus and the localization of SK1 was cytosolic and prominently dendritic (Fig. 1A and B).
Figure 1.
SK2 is nuclear in primary cultured cortical and striatal neurons. (A) Primary cortical and striatal cultures at 14 days in vitro (DIV) were fixed and stained with antibodies against SK2 and MAP2c, and with the nuclear Hoechst dye (DAPI). Scale bar is 30 μm. (B) Primary cortical and striatal cultures at 14 DIV were fixed and stained with antibodies against SK1 and MAP2c, and with DAPI. Scale bar is 30 μm.
SK2 promotes neurotoxicity in cortical and striatal neurons
Raising levels of nuclear S1P in neurons were proposed to be therapeutic in psychiatric conditions (12). Yet another study showed that S1P produced by SK2 was toxic to neurons (15). Therefore, we determined if the expression of SK2 contributes positively or negatively to neuronal health and fate in our neuronal model. A novel microscopy system, automated imaging and longitudinal analysis, enable us to track large cohorts of individual neurons over their lifetimes (16–19). We can then apply statistical approaches used in clinical medicine to the survival data, and measure neurodegeneration or neuroprotection with unprecedented specificity (17–19). The single-cell analysis proved to be 100–1,000-fold more sensitive than conventional approaches for measuring neuronal survival (8,17–20).
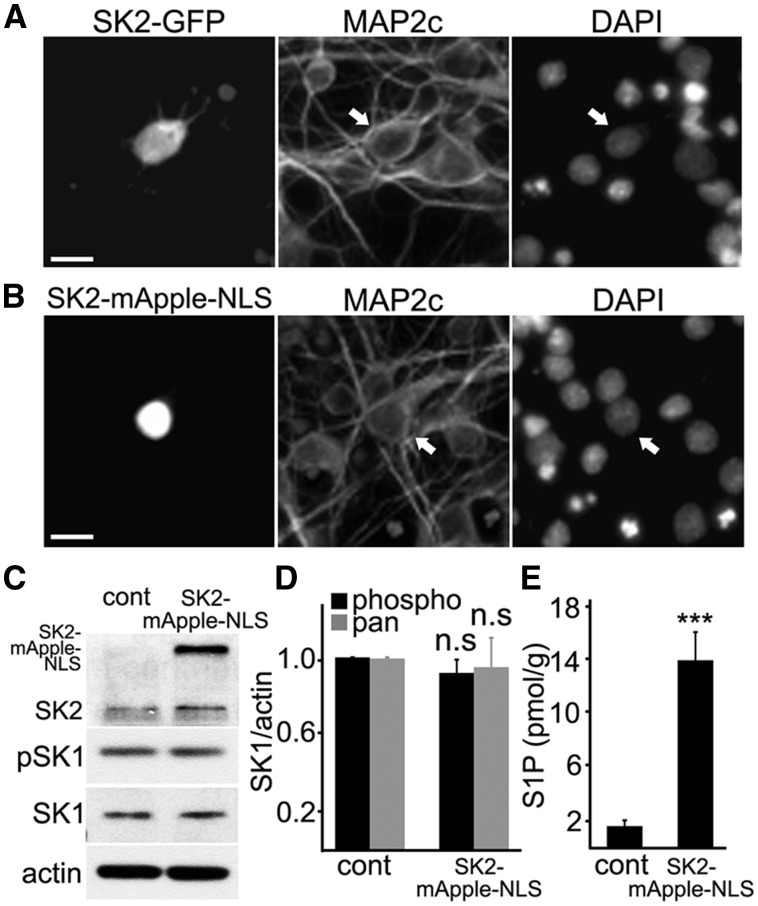
To start, we cloned a fluorescently tagged SK2 construct (SK2-GFP) and transfected primary neurons with it. Interestingly, we found that transfected neurons exhibited a significant cytosolic localization of this construct (Fig. 2A), unlike the endogenous SK2 (Fig. 1A), which might be an artifact of overexpression. To exclude any cytosolic SK2 contribution to affecting neuronal homeostasis, we decided to clone fluorescently tagged SK2 that would be specifically targeted to the nucleus due to the SV40 nuclear localization signal located on the C-terminus of a fluorescent protein mApple (SK2-mApple-NLS). Most neurons transfected with SK2-mApple-NLS exhibited nuclear localization of the mApple fluorescence, as expected (Fig. 2B).
Figure 2.
Targeting ectopically expressed SK2 to the neuronal nucleus. (A) Previous studies used the SK2 constructs tagged with a fluorescent protein to SK2's C-terminus. Primary cortical neurons were transfected with SK2-GFP, fixed, and stained with an antibody against MAP2c and with DAPI. Scale bar is 10 μm. Arrow depicts a transfected neuron. Transfected neurons exhibit a significant cytosolic localization of the SK2-GFP construct, unlike the endogenous SK2. (B) To target SK2 specifically to the neuronal nucleus, SK2 was fused to mApple (a red fluorescent protein), which contains the SV40 nuclear localization signal (NLS) on its C-terminus. Primary cortical neurons were transfected with SK2-mApple-NLS, fixed, and stained with an antibody against MAP2c and with DAPI. Neurons transfected with SK2-mApple-NLS exhibit nuclear localization of the construct. Scale bar is 10 μm. Arrow depicts a transfected neuron. (C) Primary cortical neurons were nucleofected either with mApple (cont) or SK2-mApple-NLS, plated, and maintained for 3 days. Neuronal lysates were then analyzed by western blotting with antibodies against pan-SK2 (SK2), pan-SK1 (SK1) and phosphorylated SK1 (pSK1). Actin was used as a loading control. (D) Quantification of SK1 and pSK1 band intensities from (C) normalized to actin. n.s., not significant; P = 0.79 and P = 0.37 (t-test) for pSK1 and SK1, respectively. Results were pooled from three independent experiments. (E) Primary cortical neurons were nucleofected with mApple (cont) or SK2-mApple-NLS plated and maintained for 3 days. The levels of S1P were measured by liquid chromatography and mass spectrometry from the nuclear neuronal fraction. ***P = 0.0012 (t-test). Results were pooled from two independent experiments with triplicates in each one.
To confirm that SK1 is not downregulated or upregulated when SK2 is overexpressed, neurons were nucleofected with either mApple (control) or the SK2-mApple-NLS construct. Unlike the lipid-based transfection techniques, nucleofection provides significant transfection efficiency, which allows neuronal extracts to be analyzed by biochemical methods (Fig. 2C). Cultured neurons were lysed, and the levels of phospho-SK1 and pan-SK1 were assessed (Fig. 2C and D). As expected, the levels of both were unchanged.
We localized SK2 to the nucleus, but does ectopically expressed SK2 produce S1P in the neuronal nuclei? Cortical neurons were again nucleofected with mApple or SK2-mApple-NLS constructs. At 48 h after nucleofection, nuclei were isolated, and the S1P levels were analyzed by liquid chromatography and mass spectrometry. We observed a sevenfold increase in S1P in neurons that overexpressed the SK2-mApple-NLS construct in comparison with mApple-expressing neurons (Fig. 2E).
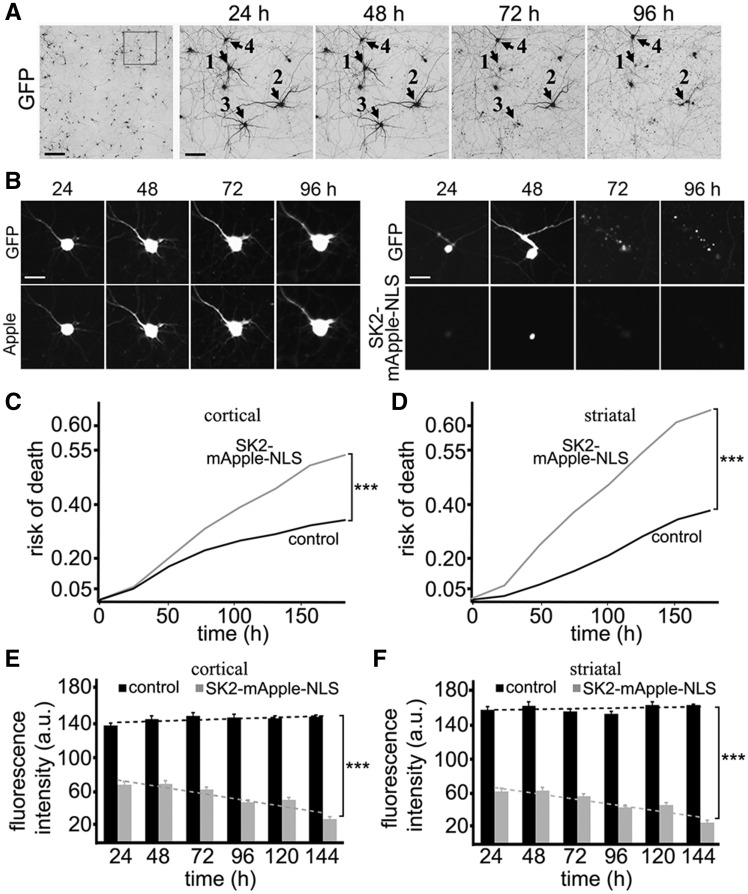
Then we tested how SK2 affects neuronal survival. Primary neurons were transfected either with GFP (a marker of viability and morphology) and mApple or with GFP and SK2-mApple-NLS (Fig. 3A–D). Loss of the green fluorescence is a sensitive marker of neuronal death (18–21). Therefore, by analyzing when each neuron lost its fluorescence, we can measure neuronal survival with cumulative hazard statistics (Fig. 3A–D). Surprisingly, the SK2 construct was toxic for primary cortical and striatal neurons, in comparison with control mApple-expressing neurons (Fig. 3C and D).
Figure 3.
SK2 is neurotoxic to cortical and striatal neurons in a dose-dependent manner. (A) An example of survival analysis. Primary cortical neurons were transfected with GFP (a morphology and viability marker) and tracked with an automated microscope. Images collected after 24 h demonstrate the ability to return to the same field of neurons and to follow them over time. Each image is a montage of non-overlapping images captured in one well of a 24-well plate. Scale bar is 200 μm. A region from the original images at different time points is zoomed in to demonstrate longitudinal single-cell tracking (right panels). Black arrows depict three neurons that degenerate by 72 h (neurons 1 and 3) and by 96 h (neuron 2) after transfection. Neuron 4 survived the entire experiment. Scale bar is 50 μm. (B) Longitudinal imaging of a neuron expressing GFP and mApple (left panel) and a neuron expressing GFP and SK2-mApple-NLS (right panel). The left neuron remains alive throughout the experiment. The right neuron died by 72 h. Scale bar is 10 μm. (C) Primary cortical neurons were transfected either with GFP + mApple (control) or with GFP + SK2-mApple-NLS. Transfected neurons were tracked with an automated microscope. Cumulative risk of death was calculated from Kaplan–Meier curves (JMP software). Cumulative risk of death curves demonstrates that the SK2-mApple-NLS construct is neurotoxic. ***P < 0.001 (log-rank test). Two hundred neurons were analyzed. Results were pooled from three independent experiments. (D) Primary striatal neurons were transfected either with GFP + mApple (control) or with GFP + SK2-mApple-NLS. Transfected neurons were tracked with an automated microscope. Cumulative risk of death was calculated from Kaplan–Meier curves. Cumulative risk of death curves demonstrate that the SK2-mApple-NLS construct is also neurotoxic to striatal neurons. ***P < 0.001 (log-rank test). Two hundred neurons were analyzed. Results were pooled from three independent experiments. (E) The average mApple-fluorescence and SK2-mApple-NLS intensities and single neuron survival to determine the dose-dependent toxicity in cortical primary neurons that express the mApple or SK2-mApple-NLS construct. Neuronal survival is not affected by mApple expression. However, higher SK2-mApple-NLS intensity lead to higher risk of death. m(control) = 0.1487; m(SK2-mApple-NLS) = −0.4246. ***P < 0.0001 (t-test). One hundred fifty neurons were analyzed. Results were pooled from three independent experiments. (F) The average mApple-fluorescence and SK2-mApple-NLS fluorescence intensities and single neuron survival to determine the dose-dependent toxicity in striatal primary neurons that express the mApple or SK2-mApple-NLS construct. Note that neuronal survival is affected by SK2-mApple-NLS expression. m(control) = 0.1005; m(SK2-mApple-NLS) = -0.4476. ***P < 0.0001 (t-test). One hundred fifty neurons were analyzed. Results were pooled from three independent experiments.
Our data indicate that overexpressed SK2 is neurotoxic. At least one study also demonstrated that artificially high levels of S1P produced by SK2 were toxic to neurons (22). However, others showed that increased SK2 activity was neuroprotective or, at least, not toxic to neurons (12,23). We, therefore, wondered if the toxicity of high expression levels of SK2-mApple-NLS could "mask" protective properties of the SK2 construct at lower expression levels in our survival analyses. To analyze this further, we took advantage of the fact that the fluorescence intensity of an expressed protein (e.g., mApple) is directly proportional to the amount of the expressed protein (20,21,24). By analyzing the levels of SK2-mApple-NLS, we can determine an effect of the dose-dependent toxicity in neuronal survival. In control cortical and striatal neurons, the levels of mApple did not correlate with a time when a neuron would die (Fig. 3E and F). Expectedly, the levels of SK2-mApple-NLS correlated with neuronal death, with higher doses of the SK2 construct being more toxic (Fig. 3E and F). Therefore, we conclude that SK2 is toxic to primary neurons in our model.
SK2 induces DNA double-strand breaks
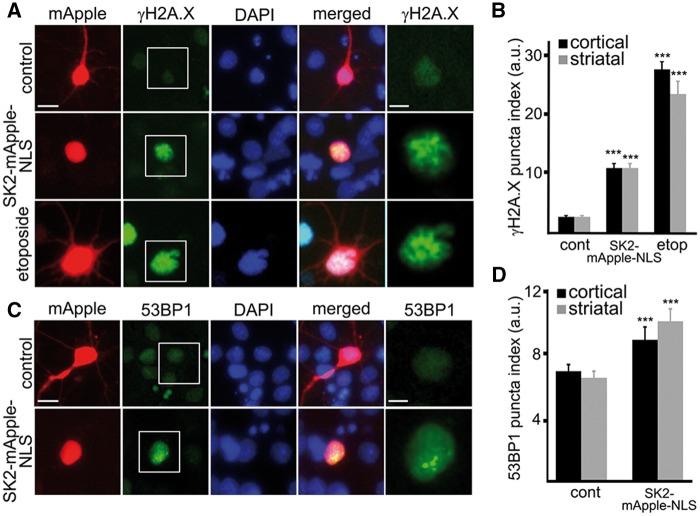
The toxicity of SK2 localized to the nucleus was unexpected. Although the toxicity can be caused by a wide variety of SK2-dependent mechanisms (25–27), we hypothesized that overexpressed SK2 promotes DNA DSBs. Indeed, SK2 inhibits HDAC1/2 (10), and pharmacological inhibition or knockdown of HDAC1 results in the formation of DSBs and neurotoxicity (4,28). To test that, primary cortical and striatal neurons were transfected either with mApple or SK2-mApple-NLS, fixed, and stained with an antibody against γH2A.X, a marker of DNA DSBs, phosphorylated histone H2A variant X (Fig. 4A and B). In a positive control, neurons were treated with etoposide, a drug commonly used for inducing DNA damage in cells (29). We observed that DNA DSBs were very rare in non-transfected and mApple-transfected neurons (Fig. 4A). Interestingly, almost 80% (77% ± 10) of cortical and striatal neurons transfected with the SK2-mApple-NLS construct were positive for γH2A.X and had γH2A.X puncta index increased in the nuclei (Fig. 4A and B).
Figure 4.
SK2 promotes formation of DNA DSBs in primary neurons. (A) Cortical neurons were transfected with mApple (control) or with SK2-mApple-NLS. At 24 h after transfection, cells were fixed, stained with an antibody against γH2A.X (a marker of DSBs) and with the nuclear Hoechst dye (DAPI), and imaged. As a positive control, neurons were treated with the DNA damaging drug etoposide. Note the γH2A.X puncta index in cells treated with etoposide and in SK2-mApple-NLS-expressing neurons is significantly greater than non-treated mApple-expressing neurons. Scale bar is 5 μm. See right panel for higher magnification image of γH2A.X. Scale bar is 2 μm. (B) Quantification of the γH2A.X puncta index in cortical and striatal neurons transfected with mApple (cont, control) or with SK2-mApple-NLS or transfected with mApple and treated with 5 µM etoposide (etop). ***P < 0.0001 (t-test). A.u., Arbitrary units. Three hundred neurons were analyzed from three independent experiments. (C) Cortical neurons were transfected with mApple or with SK2-mApple-NLS. At 24 h after transfection, cells were fixed, stained with an antibody against 53BP1 (a DNA damage repair protein) and with DAPI, and imaged. 53BP1 was diffuse in mApple-expressing neurons. In SK2-mApple-NLS-expressing neurons, 53BP1 formed puncta. Scale bar is 5 μm. See right panel for higher magnification image of 53BP1. Scale bar is 2 μm. (D) Quantification of the 53BP1 puncta index in cortical and striatal neurons transfected with mApple (cont) or with SK2-mApple-NLS. ***P = 0.0008 and ***P < 0.0001 (t-test) for cortical striatal neurons, respectively. A.u., arbitrary units. Three hundred neurons were analyzed from three independent experiments.
We recently published a paper on DNA damage in neurons induced by doxorubicin, a commonly used chemotherapy drug (30). In that study, we confirmed doxorubicin-associated DNA damage with an antibody against p53-binding protein 1 (53BP1), which acts as a scaffold that recruits additional proteins to DNA break sites. Therefore, we determined if ectopically overexpressed SK2 promotes the formation of 53BP1 puncta in neuronal nuclei. Neurons were transfected with either mApple or SK2-mApple-NLS, fixed, and stained for 53BP1. Neurons that overexpressed SK2 had 53BP1 localized to puncta. In control mApple-expressing or non-transfected neurons, 53BP1 was diffuse, as expected (Fig. 4C and D). Based on these data, we conclude that overexpressed SK2 induces the accumulation of DNA DSBs in post-mitotic neurons, which may lead to neurotoxicity.
Neurodegeneration and DNA DSBs promoted by SK2 are reduced by an SK2 inhibitor
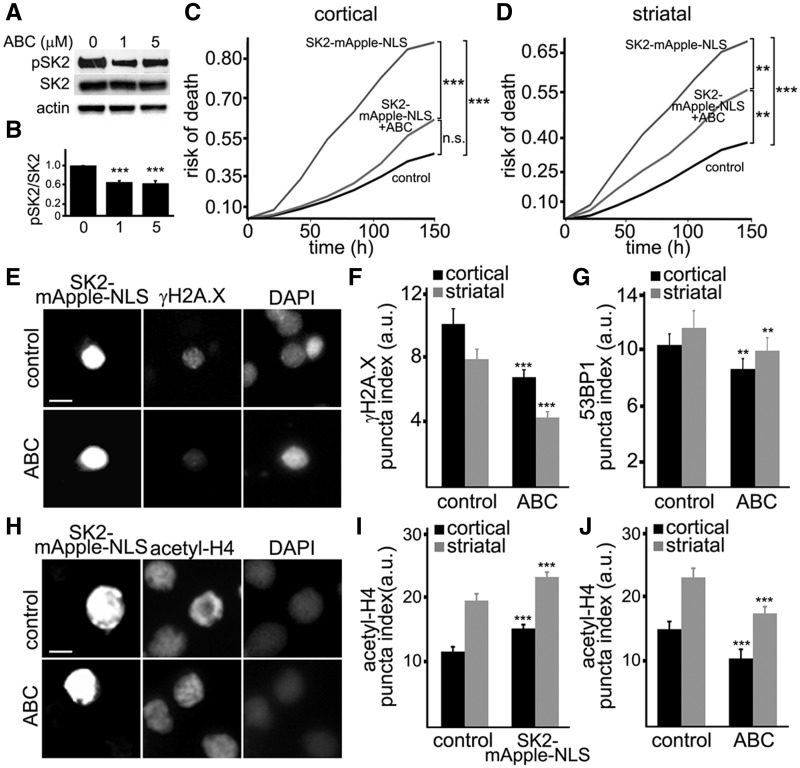
SK2 has basal phosphorylation activity towards sphingosine that is enhanced by phosphorylation of SK2 by ERK1/2 (31). The ABC294640 drug specifically inhibits SK2 in non-neuronal cells (32–36). To test if this inhibitor works in cultured neurons, we treated neurons with different doses of ABC294640 and analyzed SK2 phosphorylation, which indicates the catalytic activity of the enzyme (31). Expectedly, ABC294640 reduced the levels of phospho-SK2, indicating that SK2 is partially inhibited (Fig. 5A and B).
Figure 5.
An inhibitor of SK2, ABC294640, mitigates neurotoxicity induced by ectopically expressed SK2. (A) Primary cortical neurons were treated with a vehicle or with different concentrations of ABC294640 (ABC, 1 and 5 µM). Neuronal lysates were then analyzed by western blotting with antibodies against phosphorylated SK2 (pSK2) and pan-SK2 (SK2). Actin was used as a loading control. (B) Quantification of pSK2 levels normalized to SK2 from (A). Results were pooled from three independent experiments. (C) Primary cortical neurons transfected with GFP and mApple (control) or with GFP and SK2-mApple-NLS constructs. Two cohorts of neurons that express SK2-mApple-NLS and GFP were treated with a vehicle (SK2-mApple-NLS) or 1 μM ABC294640 (SK2-mApple-NLS + ABC). Risk of death was calculated from Kaplan–Meier curves. The ABC294640 drug reduces the risk of death in SK2-mApple-NLS-expressing neurons. ***P < 0.001 (Log-Rank test). n.s., not significant (P = 0.06). (D) Primary striatal neurons transfected with GFP and mApple (control) or with GFP and SK2-mApple-NLS constructs. Neurons that express SK2-mApple-NLS and GFP were treated with a vehicle (SK2-mApple-NLS) or 1 μM ABC294640 (SK2-mApple-NLS + ABC). Risk of death was calculated from Kaplan–Meier curves. The ABC294640 drug reduces the risk of death in SK2-mApple-NLS-expressing striatal neurons. ***P < 0.001 (log-rank test), **P = 0.0058 (control vs SK2-mApple-NLS + ABC), and **P = 0.0061 (SK2-mApple-NLS vs SK2-mApple-NLS + ABC). One hundred fifty neurons were analyzed. Results were pooled from three independent experiments. (E) Cortical neurons were transfected with SK2-mApple-NLS, and treated with a vehicle (control) or 5 μM ABC294640 (ABC) for 24 h. Cells were fixed, stained with an antibody against γH2A.X and with the nuclear Hoechst dye (DAPI), and imaged. Note that SK2 inhibitor reduces the γH2A.X puncta index in SK2-mApple-NLS-expressing neurons compared to vehicle-treated transfected cells. Scale bar is 5 μm. (F) Quantification of the γH2A.X puncta index in cortical and striatal neurons transfected with SK2-mApple-NLS and treated with a vehicle (control) or 5 μM ABC294640 (ABC) for 24 h. ***P < 0.0001 (t-test). A.u., Arbitrary units. Three hundred neurons were analyzed from three independent experiments. (G) Quantification of the 53BP1 puncta index in cortical and striatal neurons transfected with SK2-mApple-NLS, treated with a vehicle (control) or 5 μM ABC294640 (ABC) for 24 h. **P = 0.0013 and **P = 0.0027 (t-test) for cortical and striatal neurons, respectively. A.u., Arbitrary units. Two hundred neurons were analyzed from three independent experiments. (H) Cortical neurons were transfected with SK2-mApple-NLS, and treated with a vehicle (control) or 5 μM ABC294640 (ABC) for 24 h. Cells were fixed, stained with an antibody against histone H4 (acetyl K5 + K8 + K12 + K16, acetyl-H4) and with the nuclear Hoechst dye (DAPI), and imaged. Scale bar is 5 μm. (I) Quantification of the acetyl-H4 puncta index in cortical and striatal neurons transfected with mApple (control) or with SK2-mApple-NLS construct. ***P = 0.0002 and ***P < 0.0001 (t-test) for cortical and striatal neurons, respectively. A.u., Arbitrary units. Two hundred neurons were analyzed from three independent experiments. (J) Quantification of the acetyl-H4 puncta index in cortical and striatal neurons transfected with SK2-mApple-NLS and treated with a vehicle (control) or with 5 μM ABC294640 (ABC). ***P = 0.003 and ***P = 0.0038 (t-test) for cortical and striatal neurons, respectively. A.u., Arbitrary units. Two hundred neurons were analyzed from three independent experiments.
Next, to examine if ABC294640 reduces neurotoxicity of ectopically expressed SK2-mApple-NLS, cortical and striatal neurons were transfected with GFP + SK2-mApple-NLS and treated with a vehicle or ABC294640. Two neuronal cohorts were tracked for 6 days with an automated microscope, and the cumulative risk of neuronal death was analyzed. Treatment with the ABC294640 drug significantly reduced neurodegeneration associated with SK2 overexpression in both cortical and striatal neurons (Fig. 5C and D).
Ectopic SK2 expression induces DNA damage (Fig. 4). Could ABC294640 reduce the formation of DNA DSBs associated with the expression of the SK2 construct? To test this, cortical and striatal neurons were transfected with SK2-mApple-NLS construct and treated with a vehicle or ABC294640. Neurons were fixed and analyzed for γH2A.X and 53BP1. Expectedly, ABC294640 reduced the formation of DNA DSBs associated with SK2 expression (Fig. 5E–G).
Next, we tested if ABC294640 decreased acetylation levels of histones that were likely enhanced due to SK2 overexpression. Cortical and striatal neurons were transfected with mApple or with SK2-mApple-NLS and then treated with a vehicle or ABC294640. At 24 h after transfection, neurons were fixed and stained with an antibody against histone H4 (acetyl K5 + K8 + K12 + K16). HDAC1/2 deacetylate these histone H4 lysines (37). As expected, ABC294640 decreased histone acetylation that was enhanced due to overexpression of SK2-mApple-NLS (Fig. 5H–J). Therefore, we conclude that ABC294640 inhibits SK2 in cultured primary neurons.
SK2 is upregulated in brain samples from BACHD mice
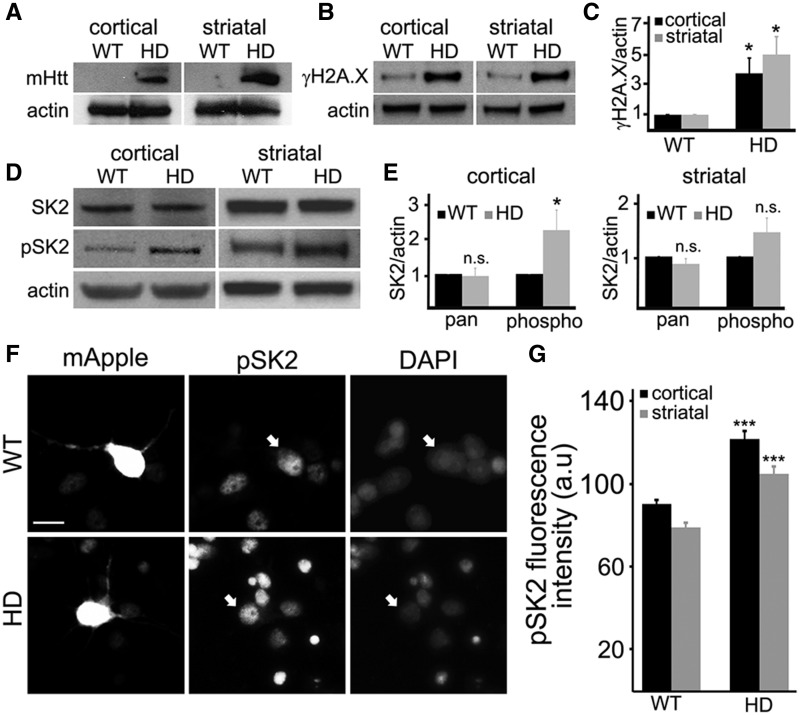
We next wondered if SK2 is altered in an HD mouse model. BACHD mice contain a bacterial artificial chromosome with a 170-kb human mHtt (97 glutamines) genomic locus and flanking sequences. The BACHD model recapitulates many molecular and cellular and behavior features observed in HD patients (38). The symptomatic aged BACHD (8–10 months old) mice exhibit an increase in DNA damage in the brain (39). Cortex and striatum brain areas were dissected from those mice. mHtt and DSB levels and the phosphorylation status of SK2 were assessed with antibodies against mHtt, γH2A.X, phospho-SK2 and pan-SK2. As expected, we found that the BACHD mice expressed mHtt (Fig. 6A) and exhibited more γH2A.X than WT mice (Fig. 6B and C). Although the levels of pan-SK2 were not altered, the levels of phospho-SK2 were elevated in BACHD mice cortices (Fig. 6D and E). The changes in levels of phospho-SK2 in the striata did not reach statistical significance, but there was a clear trend towards an increase in the phosphorylation status of SK2 (Fig. 6D and E). Since overactive SK2 relocalizes to the cytoplasm in some types of cancer cells (40), we confirmed that SK2 is still nuclear in BACHD neurons. Indeed, hyperphosphorylated SK2 is localized to the nucleus in BACHD neurons (Fig. 6F and G). Our data suggest that SK2 has a negative role in the pathogenesis of HD.
Figure 6.
SK2 is hyperphosphorylated in an HD mouse model. (A) The cortex and striatum were obtained from wild-type (WT) and BACHD (HD) mouse brains, homogenized and processed by immunoblotting with antibodies against mHtt. Actin was used as a loading control. (B) Cortices and striata were dissected from wild-type (WT) and BACHD (HD) mouse brains, homogenized, and processed by immunoblotting with antibodies against phosphorylated histone H2A.X, γH2A.X. Actin was used as a loading control. (C) Levels of γH2A.X were normalized to actin in WT and BACHD brain samples. *P = 0.018 and *P = 0.046 (t-test) for cortical and striatal neurons, respectively. Results were pooled from three WT and three BACHD mice. (D) The cortex and striatum were obtained from wild-type (WT) and BACHD (HD) mouse brains, homogenized and processed by immunoblotting with antibodies against pan-SK2 (SK2) and phosphorylated SK2 (pSK2). Actin was used as a loading control. (E) Levels of pan SK2 (pan) and phosphorylated SK2 (phospho) were normalized to actin in WT and BACHD brain samples. *P = 0.046 (t-test). n.s., not significant (P = 0.15). Results were pooled from three WT and three BACHD mice. (F) Cortical wild-type (WT) and BACHD (HD) neurons were transfected with mApple, fixed, and immunostained with an antibody against phosphorylated SK2 (pSK2), and with the nuclear Hoechst dye (DAPI). Scale bar is 10 μm. White arrows depict transfected neurons. (G) Quantification of the nuclear phosphorylated SK2 fluorescence intensity from (F). ***P < 0.001 (t-test). A.u., arbitrary units. Two hundred neurons were analyzed from two independent experiments.
Inhibitor of SK2 is protective in neuron models of HD
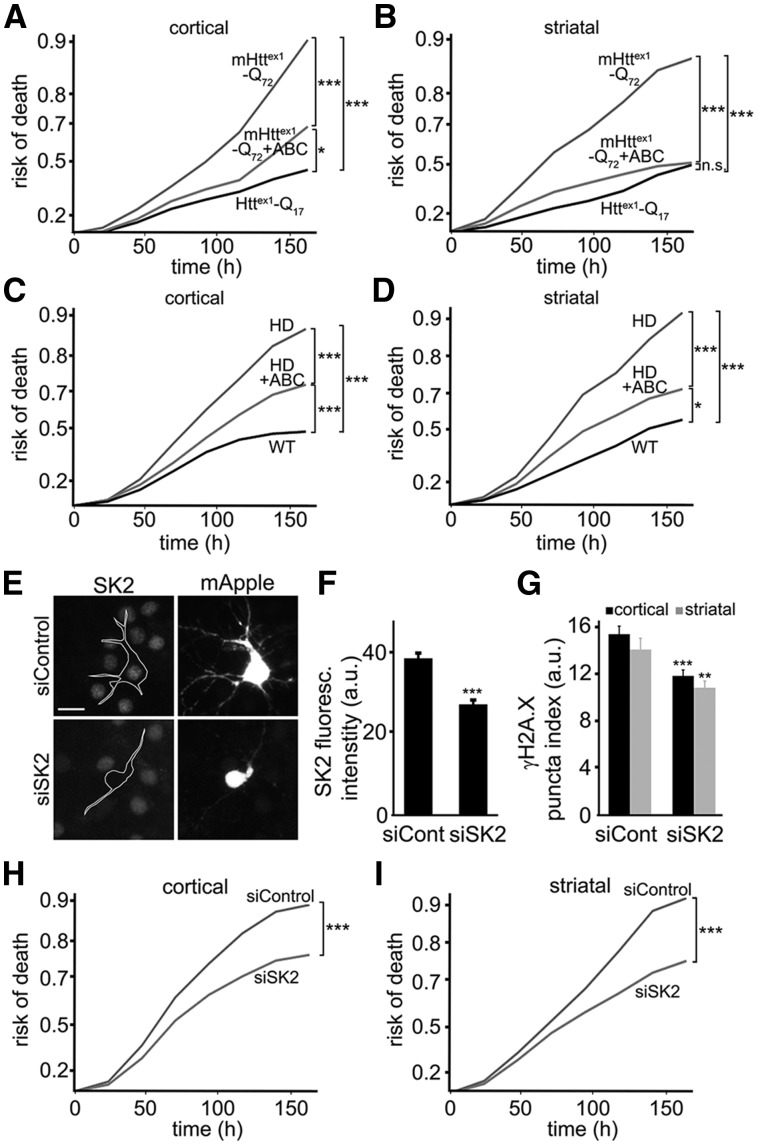
ABC294640 reduces SK2 kinase activity and mitigates DNA damage and neurotoxicity induced by SK2 nuclear expression (Fig. 5). We hypothesized that ABC294640 might be protective in primary neuron models of HD. To test our hypothesis, we used a neuron model of HD based on the expression of an N-terminal fragment of mHttex1 fused to GFP (17–21,41,42). A similar fragment is generated in HD by aberrant splicing of mHtt mRNA and by proteolytic cleavage of mHtt protein. Expression of mHttex1 produces HD-like features in mice (43–45). Cortical and striatal neurons were transfected with mApple (a morphology and viability marker) and mHttex1-Q72-GFP. Neurons were then treated with a vehicle or ABC294640, and imaged longitudinally. The risk of death plots revealed that mHttex1-Q72-GFP-expressing neurons that were treated with ABC294640 survived better than control neurons (Fig. 7A and B).
Figure 7.
ABC294640 mitigates mHtt-associated neurodegeneration in two primary neuron HD models. (A) Primary rat cortical neurons transfected with Httex1-Q17-GFP + mApple (a viability and morphology marker), or with mHttex1-Q72-GFP + mApple. Neurons transfected with mHttex1-Q72-GFP + mApple were treated with a vehicle or 1 μM ABC294640 (ABC), and tracked longitudinally. Exponential risk of death was calculated from Kaplan–Meier curves (JMP software). Risk of death curves demonstrate that ABC is neuroprotective and reduces the risk of death in mHttex1-Q72-GFP-expressing neurons. ***P < 0.001, and *P = 0.04 (log-rank test). One hundred fifty neurons were analyzed. Results were pooled from at least three independent experiments. (B) Primary striatal neurons transfected Httex1-Q17-GFP + mApple or with mHttex1-Q72-GFP + mApple. Neurons transfected with mHttex1-Q72-GFP + mApple were treated with a vehicle or 1 μM ABC294640 (ABC). Exponential risk of death was calculated from Kaplan–Meier curves. Risk of death curves show that ABC294640 is also neuroprotective for striatal neurons that express mHttex1. ***P < 0.001 (log-rank test). n.s., not significant (P = 0.1). One hundred fifty neurons were analyzed. Results were pooled from at least three independent experiments. (C) Primary cortical neurons were cultured from wild-type (WT) BACHD (HD) mouse pups, transfected with GFP (a viability and morphology marker), and treated with a vehicle or 1 μM ABC294640 (ABC). Exponential risk of death was calculated from Kaplan–Meier curves. Risk of death curves show that ABC reduces the risk of death in HD cortical neurons. ***P < 0.001, *P = 0.038 (log-rank test). Fifty neurons were analyzed from 3 WT mice and 3 HD mice. Results were pooled from two independent experiments. (D) Primary striatal neurons were cultured from wild-type (WT) and BACHD (HD) mouse pups, transfected with GFP (a viability and morphology marker), and treated with a vehicle or 1 μM ABC294640 (ABC). Exponential risk of death was calculated from Kaplan–Meier curves. Risk of death curves reveal that ABC is neuroprotective for HD striatal neurons. ***P < 0.001 (log-rank test). Fifty neurons were analyzed from 3 WT mice and 3 HD mice. Results were pooled from two independent experiments. (E) Cortical BACHD neurons were transfected with mApple (a viability and morphology marker) and scrambled siRNA (siControl) or with mApple and SK2-targeted siRNA (siSK2). Neurons were then fixed and immunostained with an antibody against SK2 and with the nuclear Hoechst dye. Scale bar is 10 μm. (F) Quantification of SK2 fluorescence intensity in neurons transfected with mApple and scrambled siRNA (siCont) or with mApple and SK2-targeted siRNA (siSK2). ***P < 0.001 (t-test). A.u., arbitrary units. Two hundred neurons were analyzed from two independent experiments. (G) Cortical and striatal neurons were transfected with mApple, mHttex1-Q46 and scrambled siRNA (siCont) or with mApple, mHttex1-Q46 and siRNA against SK2 mRNA (siSK2). Neurons were fixed 72 h after transfection and immunostained against γH2A.X. Neurons were imaged and analyzed. SK2 downregulation reduces γH2A.X puncta index, compared to controls. **P = 0.0036 and ***<0.0001 (t-test). A.u., arbitrary units. One hundred fifty neurons per condition were analyzed from two independent experiments. (H) Cortical BACHD neurons were transfected with mApple and scrambled siRNA (siControl) or with mApple and siRNA against SK2 mRNA (siSK2), and then tracked longitudinally. Risk of death was calculated from Kaplan–Meier curves. Risk of death curves demonstrates that reducing SK2 expression is neuroprotective and reduces the risk of death. ***P < 0.001 (log-rank test). (I) Striatal BACHD neurons were transfected with mApple and scrambled siRNA (siControl) or with mApple and siRNA against SK2 mRNA (siSK2), and then tracked longitudinally. Risk of death was calculated from Kaplan–Meier curves. Risk of death curves demonstrates that reducing SK2 expression is neuroprotective and reduces the risk of death in striatal cultures. ***P < 0.001 (log-rank test). Results were pooled from two independent experiments.
To confirm a neuroprotective effect of SK2 in a more physiologic HD model, we cultured cortical and striatal cultures from the wild-type and BACHD newborn pups. Neurons were transfected with mApple to visualize morphology, treated with a vehicle or ABC294640, and tracked over time. Survival data revealed that BACHD neurons treated with ABC294640 survived better than vehicle-treated BACHD neurons (Fig. 7C and D).
To ensure that downregulating SK2 with a genetic tool would also result in neuroprotection, we used the RNA interference method. BACHD cortical and striatal neurons were transfected with mApple and scrambled siRNA or with mApple and siRNA against SK2 mRNA. To determine if SK2 was indeed downregulated, transfected neurons were fixed and stained for SK2 (Fig. 7E and F). Expression of SK2 was reduced 30–35% in neurons transfected with siRNA against SK2 mRNA than in neurons transfected with scrambled siRNA (Fig. 7F).
mHtt induces DNA DSBs (46–50). To test if SK2 downregulation reduces mHtt-induced DNA damage, cortical and striatal neurons were transfected with mHttex1-Q46, mApple and scrambled siRNA or with mHttex1-Q46, mApple and siRNA against SK2 mRNA. Neurons were fixed and analyzed for γH2A.X. SK2 downregulation reduced the γH2A.X puncta index (Fig. 7G). Next, we cultured BACHD cortical and striatal neurons, transfected them with mApple and scrambled siRNA or with mApple and siRNA against SK2 mRNA, and followed them with our automated microscope. SK2 downregulation reduced neurotoxicity in BACHD cultures (Fig. 7H and I). Overall, our findings indicate that inhibiting SK2 has therapeutic benefit in HD.
Discussion
In this study, we sought to investigate the roles of SK2, a kinase that generates S1P in the nucleus, in neuronal physiology and in the pathogenesis of HD. Overexpression of SK2 is neurotoxic to cultured cortical and striatal neurons in a dose-dependent manner and leads to the formation of DNA DSBs. A small molecule inhibitor of SK2, ABC294640, mitigates DNA damage and neurotoxicity caused by SK2 overexpression. DNA damage is enhanced and SK2 is hyperphosphorylated in brain samples from the BACHD mouse model. These findings indicate that overactive SK2 may be a pathogenic mechanism in HD. Remarkably, the SK2 inhibitor is neuroprotective in two neuron models of HD, which are based on the expression of the exon-1 mHtt fragment and full-length mHtt. Our data suggest that the SK2 pathway may be a target for therapy development in HD.
Different types of phospholipids, such as phosphoinositides and sphingolipids, are abundant within the interior of the nucleus, but their functions are not clear (51). Interestingly, incubation of purified non-neuronal nuclei with phospholipids alters chromatin structure. Phospholipids may target many proteins in the neuronal nucleus that directly control transcription, splicing, and DNA damage responses. For example, increasing levels of nuclear S1P by overexpressing SK2 inhibits HDAC1/2. SK2 forms a complex with HDAC1/2, and S1P directly binds to HDAC1/2 and inhibits their deacetylase activity (10,12). Since HDACs have been vigorously investigated as a therapeutic target in HD, one may suggest that SK2 could be neuroprotective in HD. Our results show that the SK2 pathway in neurons may be more complex and SK2 may have partners other than HDAC1/2.
Various transcription factors interact with lipids. An example of a large protein/lipid complex that regulates transcription is a transcription complex that consists of a transcription factor SF-1, sphingosine bound to it, ceramidase, Sin3A, and HDACs. In non-neuronal cells, once ceramidase is activated by PKA, it metabolizes sphingosine, leading to the release of HDACs from the complex and the activation of transcription (52,53). Lipids, lipid-modifying enzymes and zinc-finger proteins also interact (54–56). Some zinc-finger proteins, such as the components of the chromatin remodeling and transcription machinery, DNA-binding SAP30L and SAP30, bind phospholipids directly (54). Given that all these nuclear lipid pathways are not studied in neurons, our findings open a new research avenue. Our future studies will investigate what partners are specifically targeted by SK2/S1P in neurons.
HD is caused by a single mutation in the Htt protein, but neurodegenerative processes associated with the expression of mHtt are highly complex and intertwined (1). Htt, being a scaffold protein, interacts with hundreds of proteins, and the mutation either enhances those interactions or disrupts the Htt-protein complexes, altering signaling cascades and inducing cytotoxicity. However, throughout evolution, cells have developed numerous mechanisms to cope with stressors, including misfolded proteins. For example, ERK1/2 is hyperphosphorylated in HD cells and pharmacological activation of ERK1/2 is cytoprotective, suggesting that the ERK1/2 cascade may be a cellular coping mechanism (57). We recently found that SK1 regulates protective autophagy in neurons (8,9). Here, we showed that SK2 promotes neurotoxicity. Remarkably, ERK1/2 regulates both SK1 and SK2 (31,58), at least in non-neuronal cells, suggesting that coping and pathogenic mechanisms can unfold in parallel. Future studies will investigate how these two arms of the ERK1/2 cascade function in mice and assess their temporal regulation during disease progression.
Our study is the first to investigate if SK2 is involved in HD. Previously, upregulated SK2 was found to be pathogenic in Alzheimer’s disease and in the hypoxic brain (59,60). In general, modulating S1P levels has shown benefits in animal models of cancer, sepsis, inflammation and other conditions (11). Intriguingly, increasing the activity of SK2 has been proposed for treating anxiety disorders (12). These studies underscore how targeting a fundamental biologic signaling pathway can yield new therapeutic avenues with applications to many challenging diseases.
Our data unexpectedly showed that striatal neurons are more sensitive to the expression of SK2 than cortical neurons (Fig. 3C and D). Notably, the levels of SK2 predict stronger neurotoxicity in striatal neurons than in cortical neurons (Fig. 3E and F). In HD, extensive human pathology studies have been performed, and the literature shows that medium spiny neurons of the striatum are among the first to die and are more severely affected than cortical neurons (61–65). Importantly, the levels of mHtt are stronger predictors of death in striatal neurons than in cortical neurons (17). Our experiments mirror studies of the pathology in HD patients and the regional differences in cell loss and cell susceptibility to mHtt in the cortex and striatum.
Is there a connection between the SK2 pathway and the protein Htt? Htt forms intranuclear protein complexes that consist of transcription repressors, such as HDACs, that are disrupted by the polyQ expansions (66). We hypothesize that SK2 is a part of a large protein complex that also contains Htt, and that the polyQ expansion disperses this complex. As a result, SK2 and mHtt may be the components of the same pathogenic signaling cascade, which first affects striatal neurons. This question is being pursued in our laboratory.
There has been a considerable interest in DNA damage in neurons over the last decade. DNA damage is observed in some extent in post-mitotic neurons under physiological conditions (67). However, abnormally increased numbers of DSBs are associated with neuronal dyshomeostasis such as synaptic dysfunction and aging (67,68). Models of Alzheimer’s disease and HD exhibit more DNA damage (67,69). Numerous pathways regulate DNA damage and repair in neurons (70–73). Interestingly, in addition to regulating transcription and chromatin structure, HDACs regulate DNA damage response. In some models, knockdown or pharmacological inhibition of HDAC1 results in the formation of DNA DSBs and cell death (4). HDAC1 and 2 accumulate at the sites of DNA damage, promoting DNA repair, at least in non-neuronal cells; therefore, over-inhibiting HDACs may result in enhanced DNA damage and neurodegeneration (74). Our results suggest that under some pathological circumstances, SK2 may play a direct role in DNA damage in neurons as an inhibitor of HDACs. Therefore, SK2 might be a promising target for therapeutic development in HD, in which DNA damage is present (69).
In summary, we demonstrated that nuclear SK2 is a part of pathogenic mechanisms in HD. Our data support previous findings that mHtt acts in the nucleus to induce neuronal death (75). Future studies in mice will determine if targeting SK2 alleviates the disease phenotypes associated with the expression of mHtt.
Materials and Methods
Chemicals and plasmids
Antibodies against SK1 were from Novus Biologicals (#NBP1-22974; 1:100). Antibodies against SK2 (#sc-366378; 1:100), mouse antibodies against MAP2c (#sc-74421, 1:100), and rabbit antibodies against MAP2c (#sc-20172, 1:100) were from Santa Cruz Biotechnology. Anti-rabbit Alexa Fluor 488-labeled, anti-mouse Alexa Fluor 546-labeled, anti-rabbit Alexa 546-labeled, and anti-mouse Alexa 488-labeled secondary antibodies were from Life Technologies. Antibodies against γH2A.X (mouse monoclonal antibody, clone JBW301; 1:100-1:1000), mouse IgG(H + L) conjugated with HRP (#AP308P, 1:3000), and rabbit IgG(H + L) conjugated with HRP (#AP307P, 1:3000) were from EMD Millipore. Antibodies against phospho-SK2(Thr614) (rabbit polyclonal antibody) was from Avivasysbio (#OAAF00574; 1:1000). Antibodies against 53BP1 (#ab172580, 1:200) and antibodies against histone H4 (acetyl K5 + K8 + K12 + K16) (#ab177790, 1:1000) were from Abcam. An antibody against mHtt (mouse monoclonal antibody) was from Sigma (clone 3B5H10; 1:1000). Rabbit antibodies against phospho-SK1(Ser225) were custom made by Yenzym Antibodies, LLC. Antibody against β-actin (mouse monoclonal antibody) was from Cell Signalling (#12262, 1:3000). Hoechst dye was from Santa Cruz Biotechnology (#sc-394039). (1s,3r,5R,7S)-3-(4-chlorophenyl)-N-(pyridin-4-ylmethyl)adamantane-1-carboxamide (ABC294640) was from Active Biochem (#A-1269). Etoposide was from Selleckchem (#S1225). pGW1-Httex1-Q17-GFP and pGW1-Httex1-Q72-GFP were described (18,19). pGW1-SK2-GFP and pGW1-SK2-mApple-NLS were cloned from pCMV6-XL4-SPHK2 (#SC113181, OriGene). pGW1-mHttex1-Q46 was derived from pGW1-mHttex1-Q46-GFP plasmid (17). Non-targeting siRNA (#D-001810) and siRNA against SK2 (#L-041258) were from Dharmacon.
Animals
Pregnant Long-Evans rats were purchased from Charles River Laboratories. Noncarrier female mice and male mice hemizygous for Tg(HTT*97Q)IXwy were purchased from the Jackson Laboratory. Rats and mice were maintained in accordance with guidelines and regulation of the University of Texas, Houston (protocol numbers #AWC-13-121 and #AWC-13-122). All experimental protocols were approved by the University of Texas, Houston. The methods were carried out in accordance with the approved guidelines.
Neuronal cultures and transfection
Cortices and striata from rat embryos (E17–19) or from WT and BACHD mouse new-born pups (P0) were dissected, dissociated, and plated on 24-well tissue-culture plates (7×105/well) coated with poly-D-lysine (BD Biosciences) as described (17–19,41). Cultures were made from individual pups delivered by a pair of WT female and BACHD male mice. The tails were cut and sent for genotyping to identify WT and BACHD cultures (Transnetyx).
Neurons were grown in a modified neuronal growth medium made from Neurobasal Medium (Life Technologies), B-27 supplement (Life Technologies), GlutaMAX (Life Technologies), and penicillin-streptomycin (Life Technologies). Some cultures were transfected after 4 days with Lipofectamine 2000 (Life Technologies) and with a total of 2–3 μg of plasmid DNA per well in 24-well plates, as described (17–19,41,76).
In some experiments, neurons were electroporated with the Neon Transfection System from Thermo Fisher Scientific (1000 V pulse, pulse width: 30, pulse #2). We routinely had up to 60% transfection efficiency.
Fluorescence microscopy and image analysis
Imaging of fixed neurons was performed with the EVOS microscopy system (Life Technologies). Briefly, the plate was placed on the EVOS microscope stage, which automatically positions the 20x objective to the center of the first well and collects fluorescence images with the RFP filter (mApple; MAP2c), the GFP filter (GFP; MAP2c; γH2A.X; 53BP1; acetylated H4; SK2 and phosphorylated SK2), and the DAPI filter (Hoechst), thereafter moving the stage to each adjacent field in the well. These steps are repeated until all required wells are imaged. Images were analyzed with the EVOS software and ImageJ software.
γH2A.X, 53BP1, and H4 Ac-K fluorescence was analyzed by the puncta index. The puncta index is estimated by measuring the standard deviation of fluorescence intensity in a region corresponding to the neuronal nucleus (8,30,77). Diffuse localization corresponds to a low puncta index, whereas punctate localization indicates a high puncta index.
Longitudinal fluorescent microscopy and survival analysis
Cortical and striatal cultures were transfected with plasmid DNA or with plasmid DNA and siRNA. Some cultures were treated with a vehicle or with 1 μM ABC294640. Cultures were then imaged every 24 h for a week. The plate was placed on the microscope stage, which automatically moves the 20x objective to the center of the first well and collects fluorescence images with the red filter (mApple) only or with the red filter (mApple, SK2-mApple-NLS) and the green filter (GFP, mHtt-GFP), thereafter moving the stage to each adjacent field in the well. These steps are repeated until all required wells are imaged. For tracking the same neurons over time, an image of the fiduciary field with neurons on the plate was collected at the first time-point and used as a reference image. Each time the same plate was imaged thereafter, the fiduciary image was aligned with the reference image. Neurons that died during the imaging interval were assigned a survival time. These event times were used to obtain the exponential cumulative survival graphs and analyzed for statistical significance by Log-Rank test with JMP software (SAS Institute) as described (8,17,18). Fifty to three hundred neurons were analyzed from three independent experiments per condition. Curves were generated in JMP. Experiments were repeated at least 3 times.
Immunocytochemistry
Neurons were fixed with 4% formaldehyde for 15 min at room temperature, permeabilized in PBS containing 0.1% Triton X-100, and blocked for 1 h in PBS containing 10% serum from the host species of a secondary antibody. Cells were incubated with a primary antibody diluted in blocking buffer at 4°C overnight. Cells were then washed with blocking buffer, and incubated with a secondary antibody in blocking buffer for 1 h at room temperature. Nuclei were stained with Hoechst dye in PBS. Cells were washed again three times for 5 min with PBS and analyzed.
Preparation of nuclear extracts
To harvest nuclei, neurons were swelled and burst in 10 mM HEPES (pH 7.8), 10 mM KCl, 0.1 mM EDTA, 1 mM Na3VO4, 1 mM DTT, 0.2 mM PMSF, protease inhibitors (#10810400, Roche) and phosphatase inhibitors (#P5726, Sigma). Neurons were incubated on ice for 15 min, and Triton X-100 was added (0.75%). A plate was vortexed for 10 sec, and the supernatant was carefully collected. Nuclei were collected by centrifugation at 3000 rpm, for 3 min at 4°C. Pellets were resuspended in 20 mM HEPES (pH 7.8), 0.4 M NaCl, 1 mM EDTA, 1 mM Na3VO4, 1 mM DTT, and protease and phosphatase inhibitors.
Immunoblotting
BACHD and WT mice (8–10 months of age) were deeply anesthetized with sodium pentobarbital (100 mg/kg) and transcardially perfused with PBS containing a cocktail with phosphatase and protease inhibitors. Brains were removed and cryoprotected.
Brain tissue or neuronal cultures were lysed in RIPA buffer (150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris/HCl (pH 8.0), containing phosphatase and protease inhibitors cocktail) at 4 °C. Lysates were vortexed and centrifuged at 14000 g for 10 min at 4 °C. Supernatants were collected, and protein concentrations were determined using the Bicinchoninic Acid Protein Assay Kit (Thermo Scientific), according to the manufacturer’s protocol. Samples were analyzed by SDS/PAGE (10–16% gels), and proteins were transferred on to PVDF membranes by the iBlot2 system (Life Technologies). Membranes were blocked with 5% (w/v) non-fat dried skimmed milk and incubated overnight with the corresponding primary antibodies. Then membranes were washed with TBS (Tris-buffered saline; 10 mM Tris/HCl and 150 mM NaCl (pH 7.4)) and probed with secondary antibodies for 1 h. Signals were visualized using Pierce ECL Western Blotting Substrate (Thermo Scientific) on Medical X-Ray Film (Kodak).
Metabolic profiling by liquid chromatography and mass spectrometry
S1P was measured using Agilent 1290 HPLC coupled with the 6495 triple quadrupole (QQQ) mass spectrometer. A HILIC column (SeQuant ZIC-HILIC, 100 × 2.1 mm, 3.5 μm, 100A) was used to separate the compound. The mobile phase A was 50 mM ammonium formate in water with 0.2% formic acid, and the mobile phase B was 5% mobile phase A and 95% acetonitrile with 0.2% formic acid. The initial gradient B was set at 100%, then decreased to 50% at 1.9 min and kept constant until 4 min. After that, the mobile phase B increased to 100% for equilibrium at 4.1 min and stayed until 6 min. The flow rate was 0.5 ml per min, and the injection volume was 20 μl. The MRM transition for S1P is 378.2–79.2 at the negative mode. C17-S1P was used as an internal standard.
The nitrogen drying gas was set with a flow rate of 14 l/min at 275 °C. The pressure of the nitrogen nebulizing gas was set at 20 psi. The sheath gas temperature was 300 °C, and the sheath gas flow was 12 l/min. The capillary voltage was set at 3000 volts. The Nozzle voltage was 1500 volts. The optimized high pressure RF was set as 150 volts, and the low pressure RF was set as 60 volts.
Acknowledgements
We thank Dr. Jing Zhao for help with mice perfusions. We thank Dr. Hengameh Zahed for advice on nucleofection with the Neon system. Raquel Cornell, Sharon Gordon, Summer Hensley, Diana Parker, and Martha Belmares provided administrative assistance.
Conflict of Interest statement. None declared.
Funding
The University of Texas, McGovern Medical School at Houston, the Department of Neurobiology and Anatomy, CPRIT Core Facility Support Award RP120092 "Proteomic and Metabolomic Core Facility, NCI/2P30CA125123-09 Shared Resources Metabolomics core and funds from Dan L. Duncan Cancer Center (DLDCC), Baylor College of Medicine, Alkek Center for Molecular Discovery, Metabolomic Core Facility, Hereditary Disease Foundation.
References
- 1. Finkbeiner S. (2011) Huntington's Disease. Cold Spring Harb. Perspect. Biol., 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burli R.W., Luckhurst C.A., Aziz O., Matthews K.L., Yates D., Lyons K.A., Beconi M., McAllister G., Breccia P., Stott A.J., et al. (2013) Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease. J. Med. Chem., 56, 9934–9954. [DOI] [PubMed] [Google Scholar]
- 3. Thomas E.A., Coppola G., Desplats P.A., Tang B., Soragni E., Burnett R., Gao F., Fitzgerald K.M., Borok J.F., Herman D., et al. (2008) The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc. Natl. Acad. Sci. U S A, 105, 15564–15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim D., Frank C.L., Dobbin M.M., Tsunemoto R.K., Tu W., Peng P.L., Guan J.S., Lee B.H., Moy L.Y., Giusti P., et al. (2008) Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron, 60, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bardai F.H., Price V., Zaayman M., Wang L., D'Mello S.R. (2012) Histone deacetylase-1 (HDAC1) is a molecular switch between neuronal survival and death. J. Biol. Chem., 287, 35444–35453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spiegel S., Milstien S. (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell. Biol., 4, 397–407. [DOI] [PubMed] [Google Scholar]
- 7. Choi J.W., Chun J. (2013) Lysophospholipids and their receptors in the central nervous system. Biochim. Biophys. Acta, 1831, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moruno Manchon J.F., Uzor N.E., Dabaghian Y., Furr-Stimming E.E., Finkbeiner S., Tsvetkov A.S. (2015) Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci. Rep., 5, 15213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moruno Manchon J.F., Uzor N.E., Finkbeiner S., Tsvetkov A.S. (2016) SPHK1/sphingosine kinase 1-mediated autophagy differs between neurons and SH-SY5Y neuroblastoma cells. Autophagy, 12, 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hait N.C., Allegood J., Maceyka M., Strub G.M., Harikumar K.B., Singh S.K., Luo C., Marmorstein R., Kordula T., Milstien S., et al. (2009) Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science, 325, 1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maceyka M., Harikumar K.B., Milstien S., Spiegel S. (2012) Sphingosine-1-phosphate signaling and its role in disease. Trends Cell. Biol., 22, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hait N.C., Wise L.E., Allegood J.C., O'Brien M., Avni D., Reeves T.M., Knapp P.E., Lu J., Luo C., Miles M.F., et al. (2014) Active, phosphorylated fingolimod inhibits histone deacetylases and facilitates fear extinction memory. Nat. Neurosci., 17, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Igarashi N., Okada T., Hayashi S., Fujita T., Jahangeer S., Nakamura S. (2003) Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J. Biol. Chem., 278, 46832–46839. [DOI] [PubMed] [Google Scholar]
- 14. Sheng R., Zhang T.T., Felice V.D., Qin T., Qin Z.H., Smith C.D., Sapp E., Difiglia M., Waeber C. (2014) Preconditioning stimuli induce autophagy via sphingosine kinase 2 in mouse cortical neurons. J. Biol. Chem., 289, 20845–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagen N., Van Veldhoven P.P., Proia R.L., Park H., Merrill A.H. Jr., van Echten-Deckert G. (2009) Subcellular origin of sphingosine 1-phosphate is essential for its toxic effect in lyase-deficient neurons. J. Biol. Chem., 284, 11346–11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barmada S.J., Serio A., Arjun A., Bilican B., Daub A., Ando D.M., Tsvetkov A., Pleiss M., Li X., Peisach D., et al. (2014) Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol., 10, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsvetkov A.S., Arrasate M., Barmada S., Ando D.M., Sharma P., Shaby B.A., Finkbeiner S. (2013) Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat. Chem. Biol., 9, 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsvetkov A.S., Ando D.M., Finkbeiner S. (2013) Longitudinal imaging and analysis of neurons expressing polyglutamine-expanded proteins. Methods Mol. Biol., 1017, 1–20. [DOI] [PubMed] [Google Scholar]
- 19. Tsvetkov A.S., Miller J., Arrasate M., Wong J.S., Pleiss M.A., Finkbeiner S. (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. U S A, 107, 16982–16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arrasate M., Finkbeiner S. (2005) Automated microscope system for determining factors that predict neuronal fate. Proc. Natl. Acad. Sci. U S A, 102, 3840–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature, 431, 805–810. [DOI] [PubMed] [Google Scholar]
- 22. Hagen N., Hans M., Hartmann D., Swandulla D., van Echten-Deckert G. (2011) Sphingosine-1-phosphate links glycosphingolipid metabolism to neurodegeneration via a calpain-mediated mechanism. Cell Death Differ., 18, 1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colombo E., Di Dario M., Capitolo E., Chaabane L., Newcombe J., Martino G., Farina C. (2014) Fingolimod may support neuroprotection via blockade of astrocyte nitric oxide. Ann. Neurol., 76, 325–337. [DOI] [PubMed] [Google Scholar]
- 24. Miller J., Arrasate M., Shaby B.A., Mitra S., Masliah E., Finkbeiner S. (2010) Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. J. Neurosci., 30, 10541–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai T.H., Ewald B., Zecevic A., Liu C., Sulda M., Papaioannou D., Garzon R., Blachly J.S., Plunkett W., Sampath D. (2016) HDAC Inhibition Induces MicroRNA-182, which Targets RAD51 and Impairs HR Repair to Sensitize Cells to Sapacitabine in Acute Myelogenous Leukemia. Clin. Cancer. Res., 22, 3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makita N., Ninomiya I., Tsukada T., Okamoto K., Harada S., Nakanuma S., Sakai S., Makino I., Kinoshita J., Hayashi H., et al. (2015) Inhibitory effects of valproic acid in DNA double-strand break repair after irradiation in esophageal squamous carcinoma cells. Oncol. Rep., 34, 1185–1192. [DOI] [PubMed] [Google Scholar]
- 27. Min A., Im S.A., Kim D.K., Song S.H., Kim H.J., Lee K.H., Kim T.Y., Han S.W., Oh D.Y., Kim T.Y., et al. (2015) Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res., 17, 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vashishta A., Hetman M. (2014) Inhibitors of histone deacetylases enhance neurotoxicity of DNA damage. Neuromolecular Med., 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walles S.A., Zhou R., Liliemark E. (1996) DNA damage induced by etoposide; a comparison of two different methods for determination of strand breaks in DNA. Cancer Lett., 105, 153–159. [DOI] [PubMed] [Google Scholar]
- 30. Moruno Manchon J.F., Dabaghian Y., Uzor N.E., Kesler S.R., Wefel J.S., Tsvetkov A.S. (2016) Levetiracetam mitigates doxorubicin-induced DNA and synaptic damage in neurons. Sci. Rep., 6, 25705.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hait N.C., Bellamy A., Milstien S., Kordula T., Spiegel S. (2007) Sphingosine kinase type 2 activation by ERK-mediated phosphorylation. J. Biol. Chem., 282, 12058–12065. [DOI] [PubMed] [Google Scholar]
- 32. White M.D., Chan L., Antoon J.W., Beckman B.S. (2013) Targeting ovarian cancer and chemoresistance through selective inhibition of sphingosine kinase-2 with ABC294640. Anticancer Res., 33, 3573–3579. [PubMed] [Google Scholar]
- 33. Gao P., Peterson Y.K., Smith R.A., Smith C.D. (2012) Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS One, 7, e44543.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chumanevich A.A., Poudyal D., Cui X., Davis T., Wood P.A., Smith C.D., Hofseth L.J. (2010) Suppression of colitis-driven colon cancer in mice by a novel small molecule inhibitor of sphingosine kinase. Carcinogenesis, 31, 1787–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. French K.J., Schrecengost R.S., Lee B.D., Zhuang Y., Smith S.N., Eberly J.L., Yun J.K., Smith C.D. (2003) Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res., 63, 5962–5969. [PubMed] [Google Scholar]
- 36. French K.J., Zhuang Y., Maines L.W., Gao P., Wang W., Beljanski V., Upson J.J., Green C.L., Keller S.N., Smith C.D. (2010) Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase-2. J. Pharmacol. Exp. Ther., 333, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fritah S., Col E., Boyault C., Govin J., Sadoul K., Chiocca S., Christians E., Khochbin S., Jolly C., Vourc'h C. (2009) Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol. Biol. Cell, 20, 4976–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gray M., Shirasaki D.I., Cepeda C., Andre V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E., et al. (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci., 28, 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu X.H., Mattis V.B., Wang N., Al-Ramahi I., van den Berg N., Fratantoni S.A., Waldvogel H., Greiner E., Osmand A., Elzein K., et al. (2014) Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington's disease. Sci. Transl. Med., 6, 268ra178.. [DOI] [PubMed] [Google Scholar]
- 40. Sankala H.M., Hait N.C., Paugh S.W., Shida D., Lepine S., Elmore L.W., Dent P., Milstien S., Spiegel S. (2007) Involvement of sphingosine kinase 2 in p53-independent induction of p21 by the chemotherapeutic drug doxorubicin. Cancer Res., 67, 10466–10474. [DOI] [PubMed] [Google Scholar]
- 41. Mitra S., Tsvetkov A.S., Finkbeiner S. (2009) Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in huntington disease. J. Biol. Chem., 284, 4398–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mitra S., Tsvetkov A.S., Finkbeiner S. (2009) Protein turnover and inclusion body formation. Autophagy, 5, 1037–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wellington C.L., Hayden M.R. (2000) Caspases and neurodegeneration: on the cutting edge of new therapeutic approaches. Clin. Genet., 57, 1–10. [DOI] [PubMed] [Google Scholar]
- 44. Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., Paganetti P., Muchowski P.J., Wilson S., Bates G.P. (2010) Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum. Mol. Genet., 19, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell, 87, 493–506. [DOI] [PubMed] [Google Scholar]
- 46. Illuzzi J., Yerkes S., Parekh-Olmedo H., Kmiec E.B. (2009) DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J. Neurosci. Res., 87, 733–747. [DOI] [PubMed] [Google Scholar]
- 47. Enokido Y., Tamura T., Ito H., Arumughan A., Komuro A., Shiwaku H., Sone M., Foulle R., Sawada H., Ishiguro H., et al. (2010) Mutant huntingtin impairs Ku70-mediated DNA repair. J. Cell. Biol., 189, 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Illuzzi J.L., Vickers C.A., Kmiec E.B. (2011) Modifications of p53 and the DNA damage response in cells expressing mutant form of the protein huntingtin. J. Mol. Neurosci., 45, 256–268. [DOI] [PubMed] [Google Scholar]
- 49. Jeon G.S., Kim K.Y., Hwang Y.J., Jung M.K., An S., Ouchi M., Ouchi T., Kowall N., Lee J., Ryu H. (2012) Deregulation of BRCA1 leads to impaired spatiotemporal dynamics of gamma-H2AX and DNA damage responses in Huntington's disease. Mol. Neurobiol., 45, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura T., Sone M., Iwatsubo T., Tagawa K., Wanker E.E., Okazawa H. (2011) Ku70 alleviates neurodegeneration in Drosophila models of Huntington's disease. PloS One, 6, e27408.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiume R., Keune W.J., Faenza I., Bultsma Y., Ramazzotti G., Jones D.R., Martelli A.M., Somner L., Follo M.Y., Divecha N., et al. (2012) Nuclear phosphoinositides: location, regulation and function. Subcell. Biochem., 59, 335–361. [DOI] [PubMed] [Google Scholar]
- 52. Lucki N.C., Sewer M.B. (2012) Nuclear sphingolipid metabolism. Annu. Rev. Physiol., 74, 131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Urs A.N., Dammer E., Sewer M.B. (2006) Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology, 147, 5249–5258. [DOI] [PubMed] [Google Scholar]
- 54. Viiri K.M., Janis J., Siggers T., Heinonen T.Y., Valjakka J., Bulyk M.L., Maki M., Lohi O. (2009) DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell. Biol., 29, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matthews J.M., Sunde M. (2002) Zinc fingers–folds for many occasions. IUBMB Life, 54, 351–355. [DOI] [PubMed] [Google Scholar]
- 56. Stenmark H., Aasland R., Driscoll P.C. (2002) The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett., 513, 77–84. [DOI] [PubMed] [Google Scholar]
- 57. Apostol B.L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E.S., Olson J.M., Kazantsev A., Marsh J.L., et al. (2006) Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum. Mol. Genet., 15, 273–285. [DOI] [PubMed] [Google Scholar]
- 58. Pitson S.M., Moretti P.A., Zebol J.R., Lynn H.E., Xia P., Vadas M.A., Wattenberg B.W. (2003) Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. embo J., 22, 5491–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takasugi N., Sasaki T., Suzuki K., Osawa S., Isshiki H., Hori Y., Shimada N., Higo T., Yokoshima S., Fukuyama T., et al. (2011) BACE1 activity is modulated by cell-associated sphingosine-1-phosphate. J. Neurosci., 31, 6850–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wacker B.K., Park T.S., Gidday J.M. (2009) Hypoxic preconditioning-induced cerebral ischemic tolerance: role of microvascular sphingosine kinase 2. Stroke, 40, 3342–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Penney J.B. Jr, Vonsattel J.P., MacDonald M.E., Gusella J.F., Myers R.H. (1997) CAG repeat number governs the development rate of pathology in Huntington's disease. Ann. Neurol., 41, 689–692. [DOI] [PubMed] [Google Scholar]
- 62. Sapp E., Ge P., Aizawa H., Bird E., Penney J., Young A.B., Vonsattel J.P., DiFiglia M. (1995) Evidence for a preferential loss of enkephalin immunoreactivity in the external globus pallidus in low grade Huntington's disease using high resolution image analysis. Neuroscience, 64, 397–404. [DOI] [PubMed] [Google Scholar]
- 63. Myers R.H., Vonsattel J.P., Paskevich P.A., Kiely D.K., Stevens T.J., Cupples L.A., Richardson E.P. Jr., Bird E.D. (1991) Decreased neuronal and increased oligodendroglial densities in Huntington's disease caudate nucleus. J. Neuropathol. Exp. Neurol., 50, 729–742. [DOI] [PubMed] [Google Scholar]
- 64. Myers R.H., Vonsattel J.P., Stevens T.J., Cupples L.A., Richardson E.P., Martin J.B., Bird E.D. (1988) Clinical and neuropathologic assessment of severity in Huntington’s disease. Neurology, 38, 341–347. [DOI] [PubMed] [Google Scholar]
- 65. Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Jr. (1985) Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol., 44, 559–577. [DOI] [PubMed] [Google Scholar]
- 66. Bardai F.H., Verma P., Smith C., Rawat V., Wang L., D'Mello S.R. (2013) Disassociation of histone deacetylase-3 from normal huntingtin underlies mutant huntingtin neurotoxicity. J. Neurosci., 33, 11833–11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suberbielle E., Sanchez P.E., Kravitz A.V., Wang X., Ho K., Eilertson K., Devidze N., Kreitzer A.C., Mucke L. (2013) Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat. Neurosci., 16, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suberbielle E., Djukic B., Evans M., Kim D.H., Taneja P., Wang X., Finucane M., Knox J., Ho K., Devidze N., et al. (2015) DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat. Commun., 6, 8897.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shiwaku H., Okazawa H. (2015) Impaired DNA damage repair as a common feature of neurodegenerative diseases and psychiatric disorders. Curr. Mol. Med., 15, 119–128. [DOI] [PubMed] [Google Scholar]
- 70. Madabhushi R., Gao F., Pfenning A.R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T.X., Yamakawa H., Pao P.C., et al. (2015) Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell, 161, 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patrick G.N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L.H. (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature, 402, 615–622. [DOI] [PubMed] [Google Scholar]
- 72. Qiu H., Lee S., Shang Y., Wang W.Y., Au K.F., Kamiya S., Barmada S.J., Finkbeiner S., Lui H., Carlton C.E., et al. (2014) ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J. Clin. Invest., 124, 981–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang W.Y., Pan L., Su S.C., Quinn E.J., Sasaki M., Jimenez J.C., Mackenzie I.R., Huang E.J., Tsai L.H. (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci., 16, 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P. (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol., 17, 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saudou F., Finkbeiner S., Devys D., Greenberg M.E. (1998) Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell, 95, 55–66. [DOI] [PubMed] [Google Scholar]
- 76. Moruno-Manchon J.F., Uzor N.E., Kesler S.R., Wefel J.S., Townley D.M., Nagaraja A.S., Pradeep S., Mangala L.S., Sood A.K., Tsvetkov A.S. (2016) TFEB ameliorates the impairment of the autophagy-lysosome pathway in neurons induced by doxorubicin. Aging (Albany NY), 8, 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bradley J., Carter S.R., Rao V.R., Wang J., Finkbeiner S. (2006) Splice variants of the NR1 subunit differentially induce NMDA receptor-dependent gene expression. J. Neurosci., 26, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]