Abstract
Cryptophthalmos (CO, MIM: 123570) is rare congenital anomalies of eyelid formation, which can occur alone or in combination with multiple congenital anomalies as part of Fraser syndrome (FS) or Manitoba Oculotrichoanal syndrome. Causal mutations have been identified for these syndromes but not in the isolated cases. Here, we described two patients from two unrelated Chinese families: one with unilateral isolated CO, while the other with unilateral CO and renal agenesis. A novel homozygous mutation (c.6499C>T: p.Arg2167Trp) and compound heterozygote mutations (c.15delG; c.6499C>T: p.Arg2167Trp) in FREM2 (NM_172862) were identified for the two patients, respectively. The deletion mutation c.15delG resulted in a frameshift and triggered the nonsense-mediated mRNA decay. For the shared missense mutation, p.Arg2167Trp altered a conserved residue and was predicted to affect protein structure by in silico analysis. Functional analysis revealed that Arg2167Trp mutant decreased its interaction with FRAS1 related extracellular matrix 1 (FREM1) and impaired the function of the FRAS1–FRAS1 related extracellular matrix 1 (FREM2)–FREM1 ternary complex required for normal embryogenesis. Furthermore, considering that mutation (c.5914C>T: p.Glu1972Lys) in FREM2 causes FS, a severe systemic disorder, we also compared these two different missense mutations. Our results showed that p.Arg2167Trp had a weaker effect in interrupting interactions between FREM2 and FREM1 than FS-associated missense mutation p.Glu1972Lys. Overall, our data demonstrate that the homozygous mutation p.Arg2167Trp in FREM2 causes isolated CO, which will facilitate our better understanding of the molecular mechanisms underlying the disease.
Introduction
Cryptophthalmos (CO), first described by Zehender in 1872 (1), is a congenital malformation characterized by various degrees of abnormalities of eyelid development. It can occur bilateral or unilateral (2–4). CO can be classified into complete (typical), incomplete (atypical) and abortive (congenital symblepharon) forms (5). CO is usually seen with multiple congenital malformation as a part of the so-called ‘syndromic CO’, including Fraser syndrome (FS, MIM: 219000) (3) and Manitoba oculotrichoanal (MOTA, MIM: 248450) syndrome (6,7), while isolated CO in the absence of other anomalies has infrequently been reported (3,8,9). FS is an autosomal recessive congenital disorder characterized by CO, syndactyly, ambiguous genitalia, ear malformations and renal defects (3,8). In human, mutations in Fraser extracellular matrix complex subunit 1 (FRAS1), FREM2 or glutamate receptor interacting protein 1 (GRIP1) result in classic FS phenotype (10–12). Mutations in FREM1 lead to a less severe phenotype as seen in MOTA syndrome, which is typically with upper eyelid colobomas and anophthalmia/micro-phthalmia, triangular growths of hair extending from scalp to eyebrow, a bifid or broad nasal tip, and gastrointestinal anomalies (6,7). FRAS1, FREM1 and FREM2 genes encode members of extracellular matrix proteins characterized by 12 consecutive CSPG repeats and several Calx-β domains (13). These proteins are secreted and form a ternary complex at basement membrane (BM), acting as a functional extracellular unit that ensures epithelial–mesenchymal interactions during organogenesis (13–16). GRIP1 encodes a multi-PDZ domain-containing protein, an FRAS1- and FREM2-interacting adaptor protein, involved in the basolateral trafficking and export of FRAS1 and FREM2 in epithelial cells (17,18). Defects of any of these components result in a destabilization of the whole complex, causing dermal–epidermal detachment (13).
Thus far, approximately half of the patients with syndromic CO have been shown to have mutations in FRAS1, FREM1, FREM2 or GRIP1 (12,19,20), while the molecular defect underlying the other half, particularly for isolated cases, remains unknown. In this study, we reported two patients: one with unilateral isolated CO, while the other with unilateral cryptophthalmos with renal agenesis (CORA). A novel homozygous mutation (c.6499C>T: p.Arg2167Trp) and compound heterozygote mutations (c.15delG; c.6499C>T: p.Arg2167Trp) in FREM2 were identified, respectively. Both of mutations were proved pathogenic via in silico and in vitro techniques. Furthermore, we presented a molecular mechanism to explain the clinical difference between syndromic and isolated CO patients who bear different mutations of FREM2.
Results
Clinical features
This research received approval from the Ethics Committee of ZhongShan Ophthalmic Center, Sun Yat-sen University (2017KYPJ046), and adhered to the tenets of the Declaration of Helsinki. Clinical data and biological specimens were obtained from the families with written informed consent. Two patients were recruited: one with isolated CO and the other with CO and unilateral renal agenesis (CORA).
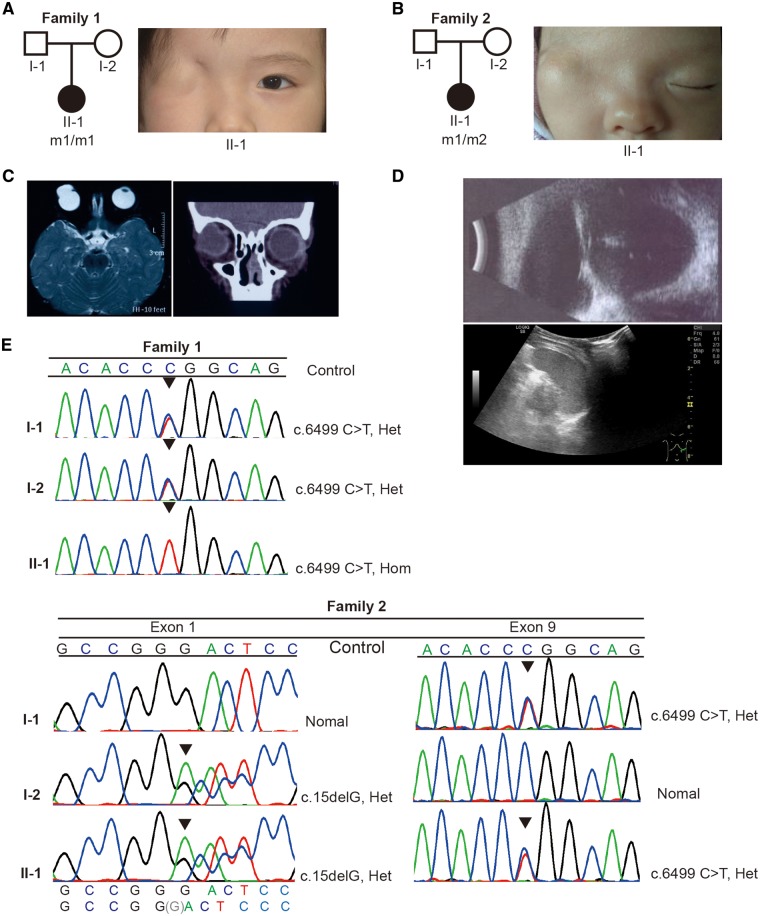
The Patient 1, a 3-year-old female, is the first child of healthy non-consanguineous parents (Fig. 1A, left panel) from Guangxi province of China. The child was delivered at term after an uneventful pregnancy and noted to have typical facial features of complete CO, where the right eye was completely covered by intact skin with the absence of eyelid, eyelashes or eyebrows (Fig. 1A, right panel). However, the contralateral eye was normal. Furthermore, magnetic resonance imaging (MRI) and ocular CT scan demonstrated a unilateral gourd-shaped cyst without a clear internal structure, markedly enlarged cornea and anterior chamber depth (Fig. 1C). There were no umbilical hernia, mental retardation, genito-urinary defects or other congenital defects of fingers, ears, nose or larynx (Supplementary Material, Fig. S1A). This patient underwent an exploratory surgery at 4 years old. A gourd-shaped cyst was separated, but no iris tissue, anatomical eyelid structure or conjunctival sac structure was identified.
Figure 1.
Pedigree, clinical features and identification of mutations of patients. (A, B) Pedigrees and facial appearance of the patients. m1: c.6499C>T: p.Arg2167Trp, m2: c.15delG. Squares: males; circles: females. Filled symbols represent affected individuals. (C) MRI (left panel) and ocular CT scanning (right panel) of the Patient 1. (D) CT scanning Ocular (top panel) and of the Patient 2 showing the gourd-shaped cyst with the absence of lens of the right eye (top panel) and left-sided renal agenesis (bottom panel). (E) Sequence chromatograms from Sanger sequencing analysis of FREM2 gene showing the homozygous mutation c.6499T>C in the Patient 1, and the heterozygous state in her mother and father (top panel); the Patient 2 is compound heterozygote for the maternal deletion c.15delG and the paternal c.6499T>C variants. The mutations are indicated by triangle.
The Patient 2, a 1-month-old female, is the first child of a healthy unrelated couple, from Guangdong province of China (Fig. 1B, left panel). Although a routine antenatal scan showed left renal agenesis, the parents decided to continue the pregnancy. The proband was delivered by cesarean sections at 39 weeks, with right-sided complete CO (Fig. 1B, right panel), whereas her left eye was normal. At the last visit at the age of 11 months, renal ultrasound confirmed left-sided renal agenesis (Fig. 1D, bottom panel). There was no syndactyly, mental retardation, omphalocele and other congenital defects of ears, nose, larynx or anus (Supplementary Material, Fig. S1B). Her phenotype is milder and less recognizable owing to fewer malformations compared with the distinct syndromic entities of FS or MOTA syndrome.
Identification of pathogenic mutations
We applied trio whole-exome sequencing (21) to the Patient 1 as well as her unaffected parents. Genetic variants were prioritized based on sequencing quality, inheritance mode, mutant allele frequency and damage potential to gene product. Among the 16 candidates that met our analytical criteria (Supplementary Material, Table S1), we found a homozygous missense variant (c.6499C>T: p.Arg2167Trp) in FREM2; the parents were heterozygotes (Fig. 1E, top panel). The variant is absent from the following available annotation databases: LOVD (22), NCBI dbSNP (23) and 1000 Genomes (24), with the exception of Genome Aggregation Database (gnomAD) (25), in which the variant is present in heterozygous state at a frequency of 8/17 202 in East Asians. This alteration was predicted to be deleterious by different predicted software, including SIFT (26), Polyphen2 (27), MutationTaster (28) and GERP++ (29) (Supplementary Material, Table S1). In addition, this variant has not been found in the in-house database of around 8000 whole exome sequencing (WES) unrelated healthy control individual of Chinese (provided by MyGenostics, Inc., China), suggesting it is not a common local variant.
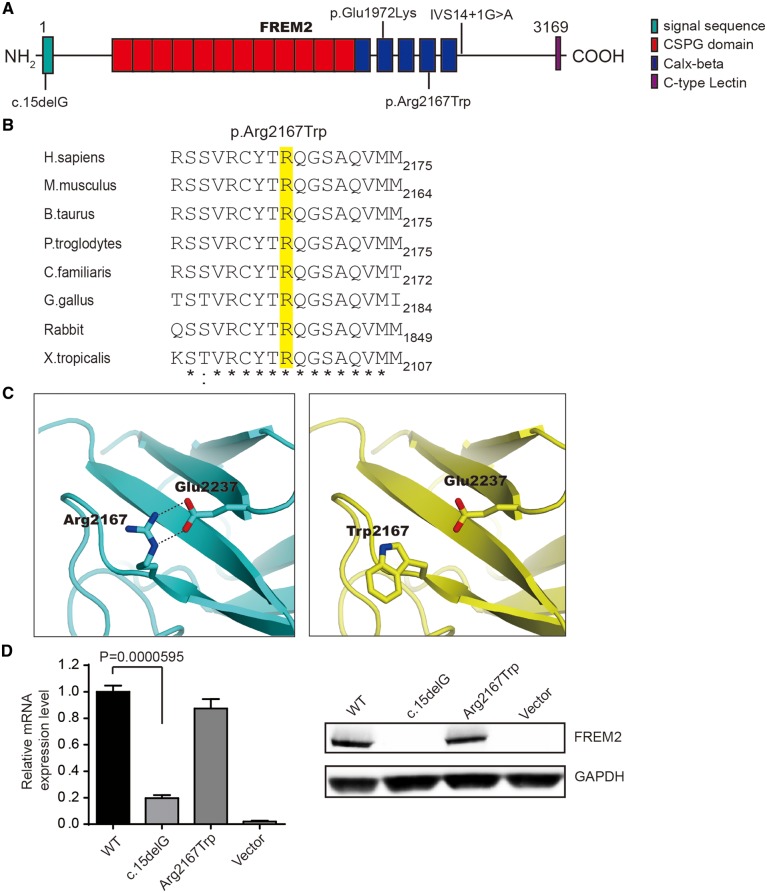
The variant p.Arg2167Trp occurs in the fourth of five consecutive Calx-β domains (Fig. 2A) and substitutes a highly conserved positively charged arginine to a non-polar tryptophan at position 2167 (Arg2167Trp) (Fig. 2B). To predict the effects of the p.Arg2167Trp variant on the three-dimensional structure of the Calx-β domain in FREM2, we performed a computational modeling analysis. The results showed that the Arg2167-Glu2237 salt bridge was disrupted by the substitution (Fig. 2C), suggesting that this missense mutation is likely to disrupt the folding of FREM2.
Figure 2.
Characterization of identified novel mutations in FREM2. (A) A schematic of the FREM2 protein and mutations in FREM2 (reported, upside; identified, downside). The functional domains are indicated by colored boxes. (B) Evolutionary conservation of the Arg2167 residue (highlighted in yellow) in FREM2 across species. (C) The expression of mutants was assessed by q-PCR (left panel) and WB (right panel). Results of q-PCR were presented as means±SEM from six independent experiments. (D) Three-dimensional structure of the Calx-beta domain with the missense mutation site Arg2167 in FREM2. A salt bridge between Arg2167 and Glu2237 was disrupted by the mutation p.Arg2167Trp.
For the Family 2, we directly sequenced all coding exons of FRAS1, FREM2, FREM1 and GRIP1 genes, including exon–intron boundaries. A compound heterozygote (c.15delG; c.6499C>T: p.Arg2167Trp) for the maternal deletion c.15delG and the paternal p.Arg2167Trp in FREM2 was identified for the Patient 2 (Fig. 1E, bottom panel). The novel deletion c.15delG is located in the signal domain (Fig. 2A) and predicted to lead to the formation of an aberrant transcript with a frameshift that starts at codon Gly5 followed by a premature STOP codon at 36 positions downstream (p.Trp6Leufs*36). Similarly, this deletion is also absent from the publicly available databases (LOVD, NCBI dbSNP, 1000 Genomes, gnomAD and the in-house database of 8000 WES samples). The missense mutation p.Arg2167Trp inherited from her father is the same as the Patient 1.
Depletion of endogenous FREM2 in HEK293T cells and cloning strategy
Since FREM2 is expressed and performs function in early embryonic stages and no longer detectable after birth (15,16), the in vitro mutagenesis model which consisted of HEK293T cells expressing wild-type (WT) or mutated FREM2 variants is necessary to study the function of mutant FREM2. In the HEK293T cells, a low basal expression level of FREM2 was detected. To minimize impact of endogenous FREM2, we depleted it by small interfering RNA (siRNA), resulting in an 80% inhibition of mRNA expression (Supplementary Material, Fig. S2A and B). It showed that depletion of endogenous FREM2 had no effect on cell morphology, cell proliferation and cell adhesion of HEK293T cells (Supplementary Material, Fig. S2C–E). Accordingly, we constructed siRNA-resistant plasmid pcDNA–FREM2 (WT) and siRNA-resistant pcDNA–FREM2 (mut) and transfected each of them into HEK293T cells, of which the endogenous FREM2 was depleted by siRNA.
c.15delG mutation causes a functional null allele
To examine whether the mutations affect the protein’s stability, we tested the expression levels of FREM2 in HEK293T cells transfected with siRNA and siRNA-resistant pcDNA–FREM2 or pcDNA–FREM2 (mut). For FREM2–Arg2167Trp, there were no significant changes in FREM2 mRNA or protein level compared with WT control (Fig. 2D). Whereas for FREM2-c.15delG, the mRNA level was severely reduced, and no full-length proteins were detected (Fig. 2D), suggesting that the deletion was most likely to trigger nonsense-mediated decay (NMD). Furthermore, the mutant FREM2-c.15delG would produce a polypeptide chain containing 40 amino acid residues and would therefore lead to a complete loss of function. These results suggest that the deletion c.15delG results in a functional null allele.
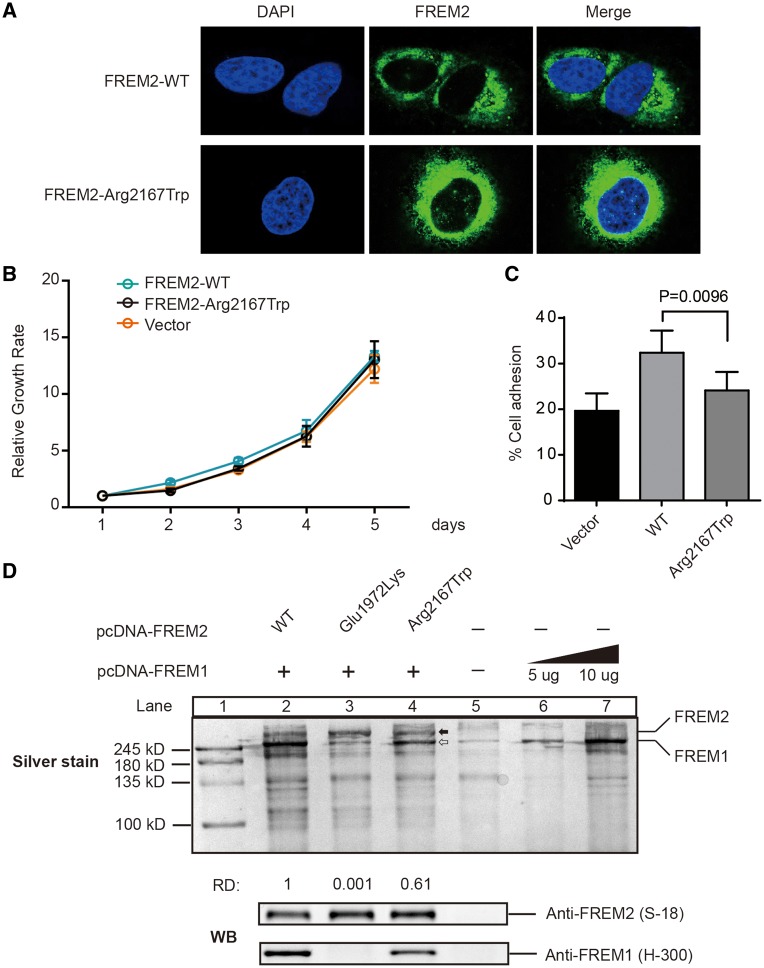
However, as shown in Figure 3A and B, the mutation p.Arg2167Trp had no effect on the subcellular localization of FREM2 or cell proliferation.
Figure 3.
Function of FREM2 mutation p.Arg2167Trp. (A) Cellular localization of FREM2–WT and FREM2–Arg2167Trp mutant in HEK293T treated with the siRNA and reintroduced it by transfecting siRNA-resistant plasmids: pFREM2–WT or pFREM2–Arg2167Trp. Cells were stained with anti-FREM2 antibody (S-12, Santa Cruz). Nuclei was stained by DAPI (Blue). (B) Growth curve analysis of HEK293T cells transfected with either FREM2–WT or FREM2–Arg2167Trp. Control was done in cells transfected with pcDNA3.1. Error bars represent SEM of six independent experiments. (C) Fibronectin adhesion assay of HEK293T treated with WT or mutant FREM2 as described in Materials and Methods. Error bars represent SEM of six independent experiments. (D) Silver stained and immunoblotted SDS-PAGE gel. Lane 1 is ladder; Lanes 2–5 are immunoprecipitated FREM2 using monoclonal antibody anti-FREM2 (F-1, Santa Cruz); Lanes 6 and 7 immunoprecipitated FREM1 using anti-FREM1 (H-300), solid arrow indicates FREM1 and open arrow indicates FREM2, which is confirmed by immunoblotting with anti-FREM1 (H-300, Santa Cruz) and FREM2 (S-18, Santa Cruz) antibodies (bottom panel). Quantification of the band's density was performed using ImageJ. The capacity of binding of FREM1 was calculate using formulas: . The capacity of binding of FREM1 in Lane 2 was set as 1, and the number under Lanes 3 and 4 indicates the relative capacity of the mutant FREM2 as compared with WT (Lane 2). RD, relative capacity of coimmunoprecipitated FREM1.
p.Arg2167Trp mutation affects the function of FRAS1/FREM complex
Since FREM2, FRAS1 and FREM1 proteins have been experimentally shown to secrete into plasma membrane and form a mutually stabilized FRAS1/FREM proteins complex governing epidermal–dermal interactions during morphogenetic processes (13,15), the function of the ternary complex containing mutant Arg2167Trp was examined. We co-cultured 293F cells transfected with pcDNA–FREM1 and cells transfected with pcDNA–FRAS1 and pcDNA–FREM2 or mutant FREM2–Arg2167Trp, and then performed cell adhesion experiments using supernatants of 293F cells. As shown in Figure 3C, the Arg2167Trp mutant significantly reduced cell adhesion compared with WT, but slightly increased the adhesion in comparison to the blank vector, suggesting that mutation p.Arg2167Trp does impair the function of FRAS1/FREM complex.
p.Arg2167Trp mutation impairs FREM2–FREM1 interaction
In the ternary complex, FREM2 is an intermediate link that could interact with FRAS1 and FREM1 (13,15). Therefore, we further performed co-immunoprecipitation (Co-IP) experiments to investigate whether mutation p.Arg2167Trp impaired its interaction with the other two proteins. We transiently expressed FREM2–Arg2167Trp mutant with pcDNA–FREM1 in 293F cells and then immunoprecipitated FREM2 from the conditioned medium to assess their capacity of binding FREM1. FREM1 was coprecipitated with FREM2 and resolved by SDS-PAGE, then visualized by silver staining and immunoblotted. Meanwhile, we constructed a recombinant FREM2-Glu1972Lys mutant that has been shown to be associated with FS (10). Interestingly, compared with WT (Lane 2), mutant Arg2167Trp (Lane 4) exhibited a lower binding capacity with FREM1, while the Glu1972Lys mutant almost abolished the interaction with FREM1 (Lane 3) (Fig. 3D). These findings strongly indicate that both of the missense mutations affect the function of FREM2 and the FS-associated mutation p.Glu1972Lys leads to complete loss of the function of FREM2, whereas p.Arg2167Trp causes a partial loss of the function. It may help explain why the Patient 1 with homozygous mutation p.Arg2167Trp presents relatively weaker phenotype (isolated CO) than FS patients with p.Glu1972Lys. Accordingly, the clinical phenotypes observed in the Patient 2, severer than isolated CO but milder than FS, are likely to result from the additive effects of a severer mutation c.15delG and a mild missense mutation p.Arg2167Trp.
Discussion
Over the past two decades, a total of 41 mutations have been identified in approximately half of the patients suffering from FS or MOTA syndrome (Supplementary Material, Tables S2 and S3). In both of FS and MOTA syndrome, ocular abnormalities of the CO spectrum occur. Complete CO is more common in FS, while MOTA syndrome comes with milder eyelid abnormalities, including incomplete CO with an ill-defined upper eyelid completely fused with an abnormal globe and a keratinized cornea (7,30). Of the total 41 mutations, 29 were identified in FS families and 12 mutations in MOTA syndrome families (Supplementary Material, Table S2). In FS, 79% (23/29) of the mutations were in FRAS1 gene reported in 19 (19/28, 68%) families, 7% (2/29) of the mutations were in FREM2 gene reported in four (4/28, 14%) families and 14% (4/29) of mutations in GRIP1 gene reported in five (5/28, 18%) families. For FRAS1 gene, 20 mutations are truncating and three are missense mutations. For FREM2 gene, two are truncating and two are missense mutations. For GRIP1 gene, all of mutations are truncating. In MOTA syndrome, three causing missense mutations and nine causing truncating mutations in FREM1 gene have been described (Supplementary Material, Table S2).
Notably, all 41 mutations identified just account for syndromic CO, but not for isolated CO. In this study, we reported a patient with isolated CO and identified a novel homozygous mutation p.Arg2167Trp in FREM2 gene using trio-based WES strategy. This mutation p.Arg2167Trp affects a key amino acid in the conserved Calx-β domain and alters the structure of FREM2. We also show experimentally that this substitution leads to a partial loss of function of FREM2, impairing FREM2–FREM1 interaction, which in turn may affect FRAS1/FREM complex formation.
To the best of our knowledge, only two homozygous mutations (c.5914G>A: p.Glu1972Lys) and IVS14+1G>A in FREM2 have been reported in FS to date (10,19,31) (Supplementary Material, Table S2). Previous evidence for FREM2-Glu1972Lys being responsible for human FS was only based on sequence and structure alignments but without experimental confirmation (10). It could not be determined whether FREM2-Glu1972Lys leads to a complete or a partial loss of the function of FREM2. In this study, we introduced Glu1972Lys into WT FREM2 and performed CO-IP experiments. Results reveal that p.Glu1972Lys transition leads to a complete loss of function. The other mutation IVS14+1G>A, reported on two sibling fetuses with FS, generates a premature termination codon and triggers NMD (31). Consequently, the present report provides additional evidence that FS may indeed be caused by complete lack or loss of function of the FREM2 protein.
Curiously, FS-associated mutation Glu1972Lys and isolated CO-associated mutation Arg2167Trp are both located in the Calx-β cadherin motif of FREM2. These findings raise an important question on how missense mutations in the same gene can be associated with distinct clinical features. Based on our experimental results, it is likely that the novel mutation Arg2167Trp exhibits a less severe impact on FREM2 protein versus mutation Glu1972Lys. Thus, the mutation Glu1972Lys results in more severe phenotypes than the mutation Arg2167Trp which only affects the eyelid development.
In the Patient 2, compound heterozygous mutations (c.15delG; c.6499: p.Arg2167Trp) were identified: one allele was affected by mutation p.Arg2167Trp, same as the Patient 1, while the other allele was affected by the severe truncating mutation c.15delG. Consistent with the genotype, the patient has a milder clinical phenotype than p.Glu1972Lys homozygous patients (10), but not as mild as the Patient 1 with p.Arg2167Trp homozygote. This case also indirectly proves that Arg2167Trp affects the function of FREM2, otherwise the Patient 2 would present the same phenotype as her unaffected mother with heterozygous mutation c.15delG.
Biallelic loss of function mutations in FREM1 have been not only identified in patients with MOTA syndrome but also associated with the phenotype of bifid nose with or without anorectal and renal anomalies (BNAR, OMIM: 608980) syndrome, a rare disorder without ocular abnormalities (Supplementary Material, Table S2). To explore whether the FREM1-related conditions owing to the breakdown of FRAS–FREM complex, we selected two missense mutations of FREM1 and constructed pcDNA–FREM1 (mut) (Supplementary Material, Fig. S3A) followed by Co-IP experiments to assess the interaction with FREM2. One of missense mutations (c.3971T>G: p.Leu1324Arg) occurred in two unrelated MOTA families (F-ID#36 and #39 in Supplementary Material, Table S2), and the other (c.1945C>T: p.Arg649Trp) was identified in a BNAR family (F-ID#45). As shown in Supplementary Material, Figure S3B, both of the FREM1 mutants (Lanes 3 and 4) almost failed to be coprecipitated with WT FREM2 and showed more deleterious effects on FREM1–FREM2 interaction compared with the identified missense mutant in our study (Lane 2), supporting failure of complex formation contributes to the FREM1-related conditions. This is consistent with the previous study that molecular cause of MOTA and BANR is closely tied to the pathogenesis of FS (32). Although FRAS1, FREM1 and FREM2 are components of the ternary complex, they have been associated with different clinical phenotype when deficient (Supplementary Material, Tables S2 and S3). In murine research, loss of Fras1 or Frem2 from the BM has been shown to abolish diminish or the BM deposition of the remaining two proteins (13,32), while marginal levels of Fras1 and Frem2 remain detectable in the skin BM of Frem1 deficient embryos (13). This could provide an explanation for the quantitatively milder phenotypes associated with FREM1 deficiency compared with FS resulting from FRAS1 and FREM2 deficiency in human (32,33).
In conclusion, this is the first time to identify a homozygous mutation p.Arg2167Trp in FREM2 in a patient with the completed isolated CO. In addition, this study expands the field of genotype–phenotype correlations and the mutational spectrum of CO and increases our understanding of molecular mechanisms of the disease.
Materials and Methods
Exome sequencing and data analysis
Genomic DNA was extracted from peripheral blood (QIAamp DNA Blood Mini Kit, Qiagen) and subsequently sequenced on an Illumina HiSeq2000 platform with 100 bp paired-end reads at a minimum coverage of 100×. Raw reads were mapped to the NCBI Build 37 reference genome using Burrows–Wheeler Aligner software (Version 0.7.15) (34). Sequences with a mapping Phred quality score under 20 were excluded from future analysis. Local realignment around insertions and deletions was performed via GATK (Version 3.6-0-g89b7209) (35). Samtools (Version 1.3.1) (36) was used to call variations relative to the human genome reference and variants observed with fewer than ten reads or a quality score <30 were omitted. Exclusion of frequently observed variants (minor allele frequency >0.05) was achieved by comparison with publicly available and proprietary databases of variants. Genes were found harboring single nucleotide polymorphisms (SNPs) or insertions and deletions (indels), potentially rendering the genes to encode non-functional proteins, which were predicted pathogenic by SIFT, Polyphen2, MutationTaster and GERP++, through wANNOVAR online (37). Screening strategies: variants would be retained if the frequencies in 1000 Genomes Project <0.01 and they should be predicted to be more ‘D’: either at least 3 ‘D’ or 2 ‘D’ and GERP score above 3. Variants were prioritized as de novo or recessive inheritance (including compound heterozygotes).
We have submitted the new c.15delG and (c.6499C>T: p.Arg2167Trp) mutations to the Leiden Open Variation Database (LOVD 3.0) as genomic variant #00111704 (databases.lovd.nl/shared/individuals/00111704).
Computational modeling analysis
Computational modeling analysis was performed to predict the effects of the p.Arg2167Trp variant on three-dimensional structure of the Calx-β domain in FREM2. The structural models of human FREM2 were generated based on the crystal structure of CBD12 from drosophila CALX1.2 (Protein Data Bank code 3RB7) using the SWISS-MODEL server (38). Structural illustrations were prepared using the PyMOL Molecular Graphic Systems (Version0.99, Schrödinger LLC).
Sanger sequencing
Validation of the identified mutation c.6499C>T in FREM2 was performed by PCR amplification via specific primer pairs followed by Sanger sequencing. The entire coding sequence including exon–intron boundaries of FREM2 were analyzed by direct sequencing in the second family. Oligonucleotide primer sequences and PCR conditions are available upon request.
Mutagenesis and plasmid construction
Human WT FRAS1, FREM1 and FREM2 coding sequences (CDS) were amplified by PCR from HEK293T total RNA (primer sequences are available on request). PCR products were subcloned into the mammalian expression vector pcDNATM3.1 (+) using In-fusion HD cloning kit (Clontech, Japan). All constructs were verified by Sanger sequencing. siRNA-resistant forms of FREM2 were created by subcloning an fragment of FREM2 700–1475 containing silent mutations 1072DEQLIV1077 into BspEI and BmgBI sites of the WT pcDNA–FREM2. The resultant siRNA-resistant pcDNA–FREM2 was used as a template for mutagenesis by PCR with mutagenic primers (sequences are available upon request) to introduce the mutations (subsequently by DNA sequencing). Similarly, we constructed pcDNA–FREM1 (Leu1324Arg) and pcDNA–FREM1 (Arg649Trp).
Cell culture and transfection
HEK293T cells were seeded at a density of 2.5e5 cells/ml and cultured in Dulbecco's modified Eagle’s Medium (DMEM, Gibco) containing l-glutamine, 1% penicillin–streptomycin (Invitrogen) and 10% fetal bovine serum (FBS, Gibico) at 37°C and at 5% CO2 atmosphere. Cell were transfected 2 h after plating with siRNA targeting FREM2 (5ʹ-GGUGAACAGUUGAUAGUAA-3ʹ) or negative control (5ʹ-CUGCCUGCGUGAGAUUCUC-3ʹ) at 20 nM concentration using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. After 48 h, cells were reseeded and transfected again with siRNA and DNA using Lipofectamine-3000; 48 h later, cells were assayed.
Cell proliferation assay
For the cell proliferation, HEK293T cells were transfected with siRNA and siRNA resistant pcDNA–FREM2 or pcDNA–FREM2 (Arg2167Trp) as described above, and they were cultured for 48 h. Cells were then seeded at 1e4 cells/well in 96-well plates (Corning) coated with collagen IV and incubated the plate at 37°C for 20 min. Wash off any non-adherent cells for four times. Add 10 μl Thiazolyl Blue Tetrazolium Blue (MTT) (Sigma, 5.0 mg/ml in PBS) to the cells at a final concentration of 0.5 mg/ml. After incubation for 3 h at 37°C, the metabolized MTT was solubilized with 100 μl of DMSO. The absorbance at 570 nm was determined on a microplate reader (Synergy H1, Biotek), and percentages of untreated controls were reported.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde and permeabilized with PBS 0.1% Triton X-100 for 5 minutes for 4 times and blocked with 10% FBS and 1% BSA in PBS for 1 hour, then stained with primary monoclonal anti-FREM2 antibody (1:200, F-1, Santa Cruz) in blocking buffer for 1 hour. Alexa488 conjugated anti-rabbit IgG secondary antibodies (Invitrogen) were applied in blocking buffer for 1 hour. Coverslips were mounted in VECTASHIED antifade mounting medium with DAPI (H-1200, VECTOR). Slides were imaged using a 60× objective lens on an OLYMPUS fluorescent microscope.
Quantitative real-time PCR analysis
Total RNA was extracted using TRIzol (Ambion™) according to the manufacturer’s recommendations, and RNA was reverse transcribed using the PrimerscriptTM RT reagent kit with gDNA Eraser (TAKARA). Real-time qPCR for FREM2 was performed using the IQ-SYBR master mixture (BioRad) (sequences are available upon request). All data were normalized to GAPDH, and quantitative measures were obtained using the ΔΔCT method.
Cell adhesion assay
293F were cultured in FreeStyle 293 Expression Medium (Invitrogen) transfected by pcDNA–FREM1 and changed with fresh medium 8 h later and followed by co-culture with HEK293F cells transfected with pcDNA-FRAS1 and pcDNA–FREM2 or pcDNA–FREM2 (Arg2167Trp) for 3 days. The conditioned medium was collected by centrifugation at 1000g for 15 min, and at 20 000g for 15 min to remove cell debris. Transfer the supernatant into an Amicon Ultra-15 centrifugal filter unit at 5000g for 30 min and concentrate down to 500 µl. The 96-well plates were coated with collagen IV at 100 µg/ml in 0.1% acetic acid, overnight at 4°C. Dried coated dishes were sterilized by exposure to UV light in a sterile culture hood. Take 20 μl concentrated protein solution into coated well and seed HEK293T cells at 5e4 cells/well. Twenty minutes later, wash two times with PBS to remove non-adherent cells. Remaining cells were measured by MTT. Cell adhesion was calculated as percentage with plates that not washed (set to 100% for each sample).
Western blotting
Cells were co-transfected by siRNA and siRNA resistant pcDNA–FREM2 or pcDNA–FREM2 (mutant) as described above, and then lysed in lysis buffer [20 mM Tris (Sigma), pH 8.0; 150 mM NaCl (Sigma); 1% NP-40; 10 μg/ml leupeptin (Sigma); 10 μg/ml aprotinin (Sigma); 1 mM Na3VO4 (Sigma); and 1 mM NaF (Sigma)]. An equal amount of protein from each sample was separated on a 7.5% SDS-PAGE gel (Bio-Rad) and then transferred to a PVDF membrane (0.45 μm, Millipore). The primary antibodies included rabbit anti-FREM2 (1:300, S-18, Santa Cruz), rabbit anti-FREM1 (1:300, H-300, Santa Cruz) and rabbit anti-GAPDH (1:1000, CST). After three washes, the PVDF membranes were blotted with the HRP-linked anti-rabbit IgG (1:10 000, CST), followed by the detection with ECL (Millipore) according to the manufacturer’s recommendations.
Co-immunoprecipitation assays
To evaluated the effects of deleterious variants on FREM2–FREM1 protein interactions, we co-transfected HEK293F cells with pcDNA–FREM1 and pcDNA–FREM2 or pcDNA–FREM2 (mutant) plasmids and changed with fresh medium 8 h later. After 48 h, supernatant was collected and incubated with monoclonal anti-FREM2 (F-1, Santa Cruz) antibody overnight at 4°C followed by binding with protein A/G magnetic beads for 1 hour at room temperature, then the beads were washed five times with Tris-buffered saline. To reveal the band position of FREM1, we transfected HEK293F cells with 5 μg or 10 μg pcDNA–FREM1 and immunoprecipitated using anti-FREM1 antibody (197-120, Sigma). Bead-bound proteins were resolved by 7.5% SDS-PAGE and examined by silver staining and subsequent immunoblotting with anti-FREM1 (1:300, H-300, Santa Cruz) and FREM2 (1:300, S-18, Santa Cruz) antibodies.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We would like to thank the patients and the families for their participation and full cooperation with this study. We would like to thank Dr Yuming Wang and Dr Yao He for inputs to the manuscript, and Dr Hao Hu (Guangzhou Women and Children's Medical Center) and Dr Zibing Jin (Wenzhou Medical University) for the content discussions.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Natural Science Foundation of China (31471232) and Major Program of Science and Technology of Guangzhou (201607020001).
References
- 1. Zehender W. (1872) Eine Missgeburt mit hautueberwachsenen Augen oder Kryptophthalmus. Klin. Monatsbl. Augenheilkd, 10, 225–249. [Google Scholar]
- 2. Egier D., Orton R., Allen L., Siu V.M. (2005) Bilateral complete isolated cryptophthalmos: a case report. Ophthalmic. Genet., 26, 185–189. [DOI] [PubMed] [Google Scholar]
- 3. Slavotinek A., Tifft C. (2002) Fraser syndrome and cryptophthalmos: review of the diagnostic criteria and evidence for phenotypic modules in complex malformation syndromes. J. Med. Genet., 39, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabra M., Gulati S., Ghosh M., Menon P. (2000) Fraser-cryptophthalmos syndrome. Indian J. Pediat., 67, 775–778. [DOI] [PubMed] [Google Scholar]
- 5. Francois J. (1969) Syndrome malformatif avec cryptophtalmie. Acta Genet. Med. Gemellol. (Roma), 18, 18–50. [DOI] [PubMed] [Google Scholar]
- 6. Li C., Slavotinek A., Chudley A.E. (2008) Manitoba Oculotrichoanal Syndrome. University of Washington Press, Washington. [Google Scholar]
- 7. Chacon-Camacho O.F., Zenker M., Schanze D., Ledesma-Gil J., Zenteno J.C. (2017) Novel FREM1 mutations in a patient with MOTA syndrome: clinical findings, mutation update and review of FREM1-related disorders literature. Eur. J. Med. Genet., 60, 190–194. [DOI] [PubMed] [Google Scholar]
- 8. Thomas I., Frias J., Felix V., De Leon L.S., Hernandez R., Jones M., Reynolds J.F. (1986) Isolated and syndromic cryptophthalmos. Am. J. Med. Genet. A, 25, 85–98. [DOI] [PubMed] [Google Scholar]
- 9. Kulkami M., Sureshkumar C., Venkataraman V. (1995) Syndromic and isolated cryptophthalmos. Indian Pediatr., 32, 1112–1115. [PubMed] [Google Scholar]
- 10. Jadeja S., Smyth I., Pitera J.E., Taylor M.S., van Haelst M., Bentley E., McGregor L., Hopkins J., Chalepakis G., Philip N.. et al. (2005) Identification of a new gene mutated in Fraser syndrome and mouse myelencephalic blebs. Nat. Genet., 37, 520–525. [DOI] [PubMed] [Google Scholar]
- 11. Slavotinek A., Li C., Sherr E., Chudley A. (2006) Mutation analysis of the FRAS1 gene demonstrates new mutations in a propositus with Fraser syndrome. Am. J. Med. Genet. A, 140A, 1909–1914. [DOI] [PubMed] [Google Scholar]
- 12. Vogel M.J., van Zon P., Brueton L., Gijzen M., van Tuil M.C., Cox P., Schanze D., Kariminejad A., Ghaderi-Sohi S., Blair E.. et al. (2012) Mutations in GRIP1 cause Fraser syndrome. J. Med. Genet., 49, 303–306. [DOI] [PubMed] [Google Scholar]
- 13. Kiyozumi D., Sugimoto N., Sekiguchi K. (2006) Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects. Proc. Natl. Acad. Sci. USA, 103, 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Timmer J.R., Mak T.W., Manova K., Anderson K.V., Niswander L. (2005) Tissue morphogenesis and vascular stability require the Frem2 protein, product of the mouse myelencephalic blebs gene. Proc. Natl. Acad. Sci. USA, 102, 11746–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pavlakis E., Chiotaki R., Chalepakis G. (2011) The role of Fras1/Frem proteins in the structure and function of basement membrane. Int. J. Biochem. Cell. Biol., 43, 487–495. [DOI] [PubMed] [Google Scholar]
- 16. Chiotaki R., Petrou P., Giakoumaki E., Pavlakis E., Sitaru C., Chalepakis G. (2007) Spatiotemporal distribution of Fras1/Frem proteins during mouse embryonic development. Gene. Expr. Patterns, 7, 381–388. [DOI] [PubMed] [Google Scholar]
- 17. Long J., Wei Z., Feng W., Yu C., Zhao Y., Zhang M. (2008) Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1. J. Mol. Biol., 375, 1457–1468. [DOI] [PubMed] [Google Scholar]
- 18. Takamiya K., Kostourou V., Adams S., Jadeja S., Chalepakis G., Scambler P.J., Huganir R.L., Adams R.H. (2004) A direct functional link between the multi-PDZ domain protein GRIP1 and the Fraser syndrome protein Fras1. Nat. Genet., 36, 172–177. [DOI] [PubMed] [Google Scholar]
- 19. van Haelst M.M., Maiburg M., Baujat G., Jadeja S., Monti E., Bland E., Pearce K., Fraser Syndrome Collaboration G., Hennekam R.C., Scambler P.J. (2008) Molecular study of 33 families with Fraser syndrome new data and mutation review. Am. J. Med. Genet. A, 146A, 2252–2257. [DOI] [PubMed] [Google Scholar]
- 20. Schanze D., Kayserili H., Satkin B.N., Altunoglu U., Zenker M. (2014) Fraser syndrome due to mutations in GRIP1–clinical phenotype in two families and expansion of the mutation spectrum. Am. J. Med. Genet. A, 164, 837–840. [DOI] [PubMed] [Google Scholar]
- 21. Crowe S., Westbrook A., Bourke M., Lyons B., Russell J. (2004) Impossible laryngeal intubation in an infant with Fraser syndrome. Paediatr. Anaesth., 14, 276–278. [DOI] [PubMed] [Google Scholar]
- 22. Fokkema I.F., Taschner P.E., Schaafsma G.C., Celli J., Laros J.F., den Dunnen J.T. (2011) LOVD v. 2.0: the next generation in gene variant databases. Hum. Mutat., 32, 557–563. [DOI] [PubMed] [Google Scholar]
- 23. Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids. Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E.. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60, 706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 27. Adzhubei I., Jordan D.M., Sunyaev S.R. (2013) Predicting functional effect of human missense mutations using PolyPhen‐2. Curr. Protoc. Hum. Genet., Chapter 7, Unit 7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods, 11, 361–362. [DOI] [PubMed] [Google Scholar]
- 29. Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol., 6, e1001025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mitter D., Schanze D., Sterker I., Müller D., Till H., Zenker M. (2012) MOTA syndrome: molecular Genetic confirmation of the diagnosis in a newborn with previously unreported clinical features. Mol. Syndromol., 3, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shafeghati Y., Kniepert A., Vakili G., Zenker M. (2008) Fraser syndrome due to homozygosity for a splice site mutation of FREM2. Am. J. Med. Genet. A, 146A, 529–531. [DOI] [PubMed] [Google Scholar]
- 32. Slavotinek A.M., Baranzini S.E., Schanze D., Labelle-Dumais C., Short K.M., Chao R., Yahyavi M., Bijlsma E.K., Chu C., Musone S.. et al. (2011) Manitoba-oculo-tricho-anal (MOTA) syndrome is caused by mutations in FREM1. Am. J. Med. Genet., 48, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petrou P., Makrygiannis A.K., Chalepakis G. (2008) The Fras1/Frem family of extracellular matrix proteins: structure, function, and association with Fraser syndrome and the mouse bleb phenotype. Connect. Tissue Res., 49, 277–282. [DOI] [PubMed] [Google Scholar]
- 34. Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang H., Wang K. (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc., 10, 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res., 42, W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.