Abstract
A diagnosis of brucellosis can be difficult because routine culture and serological methods exhibit variable sensitivity and specificity. We present the use of a metagenomic next- generation sequencing assay to diagnose a case of neurobrucellosis from cerebrospinal fluid, resulting in the institution of appropriate antibiotic treatment and a favorable clinical outcome.
Keywords: diagnostic microbiology, encephalitis, meningitis, metagenomic next-generation sequencing, neurobrucellosis
Diagnosis of brucellosis remains a clinical challenge because of its protean manifestations and imperfect diagnostic methods [1]. At present, the standard laboratory diagnosis of brucellosis is based on isolation of the bacteria from clinical specimens and/or serological detection of Brucella antibodies. We describe a case of neurobrucellosis in which the diagnosis was made using unbiased metagenomic next-generation sequencing (mNGS) of cerebrospinal fluid (CSF).
Case Report
An 11-year-old female patient from Mexico presented in February 2015 to a local hospital in Mexico with headache, back pain, nausea, and emesis. She had been well until 4 weeks before admission when headaches and nausea developed, followed by abnormal jerking movements of her fingers, hands, arms, and legs. She was admitted to the hospital and a lumbar puncture was performed; microbiological testing was positive for Epstein-Barr virus and human herpesvirus 7 from CSF (Table 1). Testing for other pathogens, including Brucella by immunoglobulin (Ig)M antibody, was negative. She was treated with parenteral acyclovir, followed by oral ganciclovir to complete a 14-day course. Two weeks after hospital discharge, she developed back pain and worsening headache. She returned to the hospital where a physician suggested a repeat lumbar puncture. However, her mother refused and brought her to Los Angeles for further evaluation.
Table 1.
Patient’s Microbiological Workup
| Variable | Mexico | Los Angeles (Children’s Hospital Los Angeles) | ||
|---|---|---|---|---|
| First Admission (February 26, 2015) | Hospital Day 8 (March 6, 2015) | Second Admission (March 9, 2016) | ||
| Cerebrospinal fluid | ||||
| Color | Colorless | Colorless | Colorless | Colorless |
| Turbidity | Slight | Clear | Hazy | Clear |
| Red cell count (per mm3) | - | 0 | 1 | 0 |
| White cell count (per mm3)a | 183 | 137 | 176 | 0 |
| Differential count (%) | ||||
| Neutrophils | 0 | 5 | 53 | 0 |
| Lymphocytes | 0 | 91 | 38 | 0 |
| Monocytes | 100 | 4 | 9 | 0 |
| Protein (mg/dL)b | 270 | 200 | 131 | 23 |
| Glucose (mg/dL)c | 25 | 25 | 25 | 45 |
| Blood glucose (mg/dL)d | 84 | 86 | - | 87 |
| Microbiological tests | ||||
| Staininge | Rare Gram-positive cocci | No organisms | No organisms | No organisms |
| Culturef | No growth | No growth | No growth | No growth |
| Cytomegalovirus DNA (PCR) | Negative | - | - | - |
| Enterovirus DNA (PCR) | Negative | Negative | - | - |
| Epstein-Barr virus DNA (PCR) | Detected | Negative | Detected | - |
| Herpes simplex virus (PCR) | Negative | Negative | - | - |
| Human herpesvirus 6 (PCR) | Negative | - | Negative | - |
| Human herpesvirus 7 (PCR)g | Detected | - | Detected | - |
| Mycobacterial DNA (MTB-PCR)h | - | Negative | Negative | - |
| VDRL | Nonreactive | - | - | - |
| mNGS testing (UCSF Clinical Microbiology Laboratory) | - | - | Brucella detected | - |
| Blood | ||||
| Brucella ELISA IgM | Negative | - | - | - |
| Brucella EIA IgMi | - | Negative | - | Negative |
| Brucella EIA IgGi [<0.80, negative; 0.8–1.09, equivocal; ≥1.10, positive] |
- | Positive (4) | - | Positive (4.68) |
| Brucella agglutininsj [≥1:80, positive] |
- | - | - | Positive (1:80) |
| Coccidioidal antibodyk | - | Negative | - | - |
| Cryptococcal antigen | - | Negative | - | - |
| HIV antibody and p24 antigen | - | Negative | - | - |
| Interferon gamma release assay (QuantiFERON-TB Gold) | - | Negative | - | Negative |
| RPR | Negative | Negative | - | - |
| Urine | ||||
| Histoplasma antigen | - | Negative | - | - |
Abbreviations: CSF, cerebrospinal fluid; DNA, deoxyribonucleic acid; EIA, enzyme assay; ELISA, enzyme-linked immunosorbent assay; HIV, human immunodeficiency virus; Ig, immunoglobulin; mNGS, metagenomic next-generation sequencing; MTB, Mycobacterium tuberculosis complex; PCR, polymerase chain reaction; RPR, rapid plasma reagin; UCSF, University of California, San Francisco; VDRL, Venereal Disease Research Laboratory.
aReference range (0–10/mm3).
bReference range (20–40 mg/dL).
cReference range (40–80 mg/dL).
dReference range (60–115 mg/dL).
eGram staining for bacteria, acid-fast staining for mycobacteria, and wet preparation with Calcofluor staining for fungi.
fBacterial, mycobacterial, and fungal.
gTest was performed by Viracor-IBT Laboratories.
hTest was performed by Focus Diagnostics.
iTest was performed by Quest Diagnostics.
jTest was performed by Mayo Medical Laboratories after mNGS testing revealing Brucella spp DNA in the CSF specimen.
kTest was performed by University of California Davis Coccidioidomycosis Serology Laboratory.
The patient’s history was significant for an episode of Salmonella infection 3 months before admission, during which she developed fever and headache, but no diarrhea. Her family lived in the city of Gómez Palacio, Durango in the state of Coahuila, Mexico, and they owned several pet dogs and a turtle. There was no history of exposure to farm animals. Family history was notable for active pulmonary tuberculosis (TB) in her father and sister 20 and 15 years before admission, respectively.
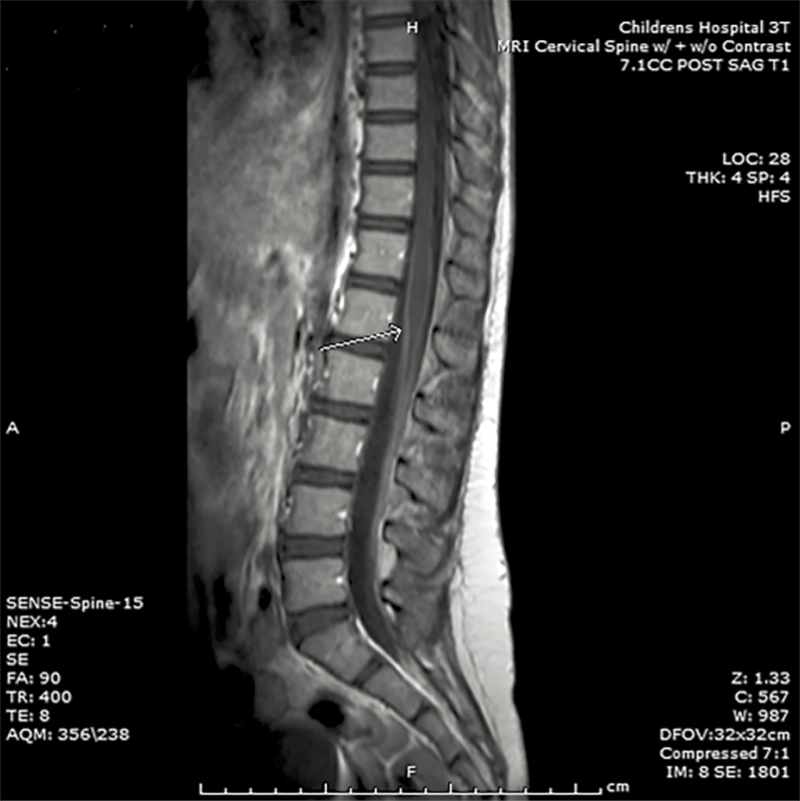
Upon admission to Children’s Hospital Los Angeles in late February 2015, the patient was moderately distressed due to back pain but alert and oriented to person, place, and time. Her vital signs were normal without fever. Physical examination was remarkable for multifocal myoclonus throughout her body. There was hyperreflexia and decreased proprioceptive sensation of bilateral lower limbs. The rest of the physical examination was unremarkable. A complete blood count was notable for a white blood cell count of 4.82 × 109/L (44% neutrophils, 44% lymphocytes, 10% monocytes). Hemoglobin, platelet counts, and comprehensive metabolic panel were normal, as was magnetic resonance imaging (MRI) of the head. An MRI of the spine demonstrated leptomeningeal enhancement along the conus medullaris involving the cauda equina without underlying signal abnormality of the spinal cord (Figure 1). A repeat lumbar puncture was performed, with additional microbiological testing unrevealing (Table 1). Cytologic examination of the CSF revealed no malignant cells. Despite a negative tuberculin skin test and QuantiFERON-TB test, she was started on 4-drug therapy with isoniazid (INH), rifampin, pyrazinamide, and ethambutol given a high concern for TB disease based on the patient’s suggestive CSF profile and risk factors for TB.
Figure 1.
Contrast-enhanced T1-weighted magnetic resonance imaging of the lumbar spine from the case patient. Smooth and linear leptomeningeal enhancement is observed along the margins of the conus medullaris (arrow) and involves the cauda equina without underlying signal abnormality of the spinal cord or vertebral bodies.
On day 8 of hospitalization, CSF testing by polymerase chain reaction (PCR) for Mycobacterium tuberculosis complex was negative. Cerebrospinal fluid testing after a repeat lumber puncture was again unrevealing (Table 1). Given the lack of response to empiric TB therapy, and the region of Mexico in which she lived, there was a concern for drug-resistant TB [2]. Ethambutol was thus changed to ethionamide, and levofloxacin was added to the regimen. She improved substantially after her antibiotic regimen was changed and she was discharged home on 5 anti-TB medications. At follow up 1 week and 1 month after discharge, her headache had resolved, but she continued to have fatigue, mild back pain, and intermittent episodes of shaking of her extremities. When the result of the mycobacterial culture was finalized as negative at 6 weeks, she returned to Mexico to continue her anti-TB therapy with INH and rifampin alone.
METHODS
Sample Collection
After discharge, the patient’s CSF sample was submitted for metagenomic sequencing under a research protocol for comprehensive identification of potential pathogens approved by the institutional review boards of Children’s Hospital Los Angeles and University of California, San Francisco. Informed consent for publication was obtained from the patient’s mother.
Metagenomic Library Construction
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) metagenomic libraries were constructed from the patient’s CSF sample as previously described [3, 4]. After bead-beating using Lysis matrix B (MP Biomedicals, Santa Ana, CA) at 6 m/s for 30 seconds, total nucleic acid was extracted using the QIAGEN EZ1 viral kit (QIAGEN, Valencia, CA). Half of the nucleic acid from CSF was treated with Turbo DNase (Ambion, Waltham, MA), followed by reverse transcription of the RNA to cDNA using random hexamers and NGS library preparation using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA). The remaining half was treated with the NEBNext Microbiome DNA Enrichment Kit to enrich for microbial DNA (New England Biosciences, Ipswich, MA), followed by Nextera XT library preparation. Dual-indexed, barcoded NGS libraries were quantitated on the BioAnalyzer (Agilent, Santa Clara, CA) and run on the Illumina HiSeq as 1 × 140 base pair (bp) single- end sequencing.
The patient’s CSF sample was processed and sequenced as part of a recently developed standardized operating procedure (SOP) for clinical mNGS testing from patient samples in the University of California, San Francisco Clinical Microbiology Laboratory, a Clinical Laboratory Improvement Amendments (CLIA)-licensed laboratory (Naccache et al, manuscript in preparation). For each sequencing run, the SOP includes running 2 external controls: (1) a negative “no-template” control (NTC) sample consisting of elution buffer, (2) and a positive control (PC) sample consisting of a quantified mixture of 7 representative pathogens (cytomegalovirus, human immunodeficiency virus, Streptococcus agalactiae, Klebsiella pneumoniae, Cryptococcus neoformans, Aspergillus niger, and Toxoplasma gondii). Each is spiked into negative CSF matrix at a concentration 1–2 log above the estimated limits of detection for that microorganism by probit analysis.
Bioinformatic Analysis
Metagenomic NGS data were analyzed for pathogens using a modified version of the “sequence-based ultra-rapid pathogen identification” (SURPI) computational pipeline, which identifies pathogen sequences on the basis of nucleotide (nt) alignments to National Center for Biotechnology Information nt reference database (March 2015 build) [5]. A clinical version of the SURPI pipeline, named SURPI+, which uses taxonomic classification for more accurate read assignments and to establish normalized metrics and thresholds for clinical results reporting, was used for automated interpretation. In brief, for identification of viral sequences, mNGS data from both DNA and RNA sample libraries were used, whereas only mNGS data from DNA libraries were used for identification of sequences from bacteria, fungi, and parasites. An edit distance cutoff of 16, indicating the number of single bp insertions, deletions, or mismatches allowed between the read and the reference sequence, was used for virus detection, whereas a more stringent edit distance cutoff of 1 was used for bacterial, fungal, and parasitic detection [3, 4]. After alignment, a rapid taxonomic classification algorithm based on the lowest common ancestor algorithm was used to assign viral, bacterial, and nonchordate eukaryotic (fungal or parasitic) NGS reads to the species, genus, or family level, as previously described [3, 4].
Results Reporting
As part of clinical validation of mNGS testing for pathogen detection, threshold cutoffs were established for automated reporting of positive results (Naccache et al, manuscript in preparation). In brief, for reporting of bacteria, fungi, and parasites, the cutoff is defined as a reads per million (RPM) ratio of ≥10, where the RPM ratio is defined as the RPMsample/RPMNTC for any given taxon (species, genus, or family). If the taxon is not present in the NTC, then the RPMNTC is 1. For reporting of viruses, criteria include coverage of at least 3 noncontiguous/non-overlapping gene regions of size >140 bp (the designated read length for the Illumina HiSeq run). Viruses with nonvertebrate hosts that are found in the NTC or that constitute normal body flora (eg, anelloviruses) are not reported.
Brucella Polymerase Chain Reaction Confirmation
Confirmation of Brucella detection was performed by Brucella genus-specific PCR using the following primer set targeting the IS711 gene [6]: BrucellaGenus_F-2 GCCTTGGATCTGAGCCGTT; BrucellaGenus_IS711_R GGCCTACCGCTGCGAAT. The reaction was carried out using the QIAGEN One-Step RT-PCR Kit in a 25 µL total reaction volume by addition of 4 µL Q solution, 4 µL 5 × buffer, 1 µL dNTP, 1 µL enzyme, 1 µL of each primer at 12 pmol, 2 µL of template extracted DNA, and water. Cycling conditions were as follows: 40 cycles of 94°C, 30 s/53°C, 30 s/72°C, 30. The sequence of the PCR amplicon by Sanger sequencing corresponded to Brucella spp.
RESULTS
RNA and DNA mNGS libraries from 600 μL of the patient’s CSF sample were constructed. From a total of 23,638,587 raw reads in the DNA library, there were 277 (0.0012%) reads corresponding to the Brucella genus, corresponding to an RPM ratio of 15.6, with all species-specific reads aligning to Brucella melitensis (Supplementary Tables 1–6). It is notable that no reads aligning to Brucella among 9,161,626 reads in the corresponding RNA library from CSF were detected, and Brucella reads were absent in both negative NTC and PC samples. Other bacterial reads present in the DNA library were also present in the NTC sample processed in parallel on the same run, and thus they did not meet the pre-established threshold for reporting using an RPM ratio of ≥10 and were attributed to laboratory reagent contamination. Likewise, no viruses, fungi, or parasites met criteria for reporting. In contrast, all 7 microorganisms in the PC were detected at levels above the pre-established reporting threshold. Importantly, no Brucella isolates or positive clinical samples from suspected or confirmed cases had been present in the clinical laboratory before the mNGS testing, nor had Brucella sequences ever been detected in the NTC.
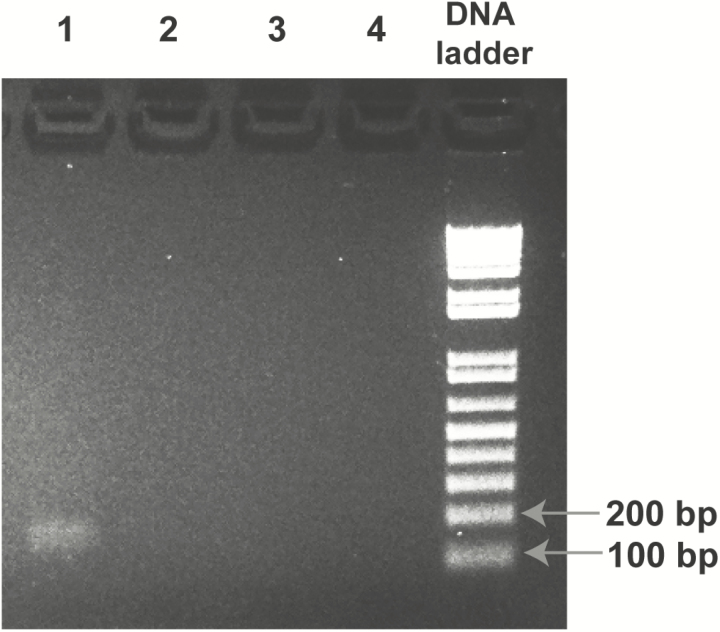
After detection of Brucella reads in her CSF sample, the patient was contacted and instructed to seek further medical evaluation. She returned to the hospital in Los Angeles for a second admission in March 2016. Although she had completed INH and rifampin therapy 1 week prior, she reported persistent back pain, nausea, and fatigue. Repeat MRI of the brain and spine was remarkable, and CSF was normal (Table 1). A confirmatory serum Brucella agglutinin titer was positive at 1:80. Because of persistent symptoms, mNGS and PCR testing showing Brucella, and positive confirmatory serology, she was diagnosed with chronic neurobrucellosis and started on targeted therapy with doxycycline and rifampin. Two weeks after starting therapy, she reported that her symptoms had fully resolved. It is notable that Brucella spp was not isolated from either CSF or blood culture, nor was Brucella DNA detected in CSF submitted for clinical 16S rRNA gene sequencing. Follow-up PCR testing for Brucella in the research laboratory was positive (Figure 2), with Sanger sequencing of the amplicon confirming the presence of Brucella spp.
Figure 2.
Polymerase chain reaction (PCR) confirmation of Brucella in the case patient’s cerebrospinal fluid (CSF). Shown is a globally contrast- enhanced image of the 2% agarose gel from Brucella PCR targeting a 177-base pair region of the IS711 gene. The lanes correspond to (1) patient CSF sample extract, (2) no-template control (NTC), (3) positive control (PC) mixture of 7 organisms, (4) water, and (“DNA ladder”) 1 kb Plus DNA ladder (Invitrogen). The PCR amplicon from the patient’s CSF extract was sequenced and identified as Brucella spp. Abbreviation: DNA, deoxyribonucleic acid.
DISCUSSION
In this study, we highlight the clinical impact of mNGS for diagnosis of infections such as neurobrucellosis that are challenging to accurately diagnose and treat. Although initially covered for TB meningitis using antibiotics that partially treat neurobrucellosis (rifampin ± levofloxacin), the patient continued to be symptomatic and was not placed on targeted therapy for Brucella until comprehensive metagenomic sequencing revealed the presence of bacterial DNA in her CSF and neurobrucellosis was confirmed by Brucella agglutinin testing.
We do not know precisely how the patient was exposed to Brucella and became infected. Mexico remains a country where brucellosis is endemic [7]. Although exposure to farm animals is a known risk factor, brucellosis is re-emerging as a foodborne disease transmitted to humans through the consumption of fresh cheeses made from unpasteurized milk. The patient and her family did admit to consuming fresh cheeses while in Mexico, and we believe that this is the most likely route by which our patient acquired her illness.
Neurobrucellosis is a known complication of systemic Brucella infection. It remains a difficult diagnosis to make and can mimic other fastidious infections such as TB [1]. Manifestations of neurobrucellosis are widely variable and include meningoencephalitis, cerebrovascular disease, peripheral and cranial neuropathies, or myelitis [8]. Neuroimaging studies of neurobrucellosis can also vary greatly among individuals [9]. A previous study from Turkey reported a high prevalence of neurobrucellosis at 37% among 128 patients with brucellosis [10]. Although culture is the gold standard for diagnosis, Brucella species are relatively fastidious and slow- growing; cultures fail to recover the organism 30%–90% of the time, and they also present a risk of laboratory-acquired infection [11]. Serology is more sensitive for detection, but it can lead to false-positive results and may not distinguish between active and prior infection. Molecular methods based on detection of nucleic acid such as PCR [12] and now potentially metagenomic NGS [13] can offer increased sensitivity and specificity over conventional diagnostic testing.
Of particular relevance for this case, 2 chronic granulomatous infections, TB and brucellosis, have not only overlapping clinical or radiographic features but also histologic characteristics in common [14]. As such, misdiagnosis of TB in patients with brucellosis has been reported in the literature [14]. This matter is further complicated by the fact that neither a negative CSF mycobacterial culture nor an M tuberculosis PCR-based assay excludes the diagnosis of TB meningitis if the clinical suspicion is high [15]. In addition, false-positive Brucella seroreactivity, enzyme-linked immunosorbent assay, and agglutination titer in patients with active TB have also been reported [16]. In this patient’s case, the negative Brucella IgM but positive IgG was incorrectly attributed to false-positive Brucella seroreactivity in the setting of TB and not to active Brucella infection. It is possible that the patient may not have mounted a detectable IgM antibody response, or that Brucella IgM levels had waned by the time of hospital admission. Confirmatory agglutinin testing may have been helpful in making the diagnosis of Brucella earlier.
This patient had temporary clinical improvement after initiation of anti-TB medications. Rifampin and levofloxacin are 2 anti-TB agents that are also active against Brucella spp [1]. However, rifampin, although active against Brucella, should always be used in combination with other agents (eg, doxycycline, trimethoprim-sulfamethoxazole, or quinolones) since monotherapy, as was inadvertently administered to this patient initially, has been associated with high relapse rates [1]. In hindsight, the patient’s prolonged and indolent course was more likely to be associated with Brucella, because more rapid clinical deterioration would have been expected for patients with active TB who are inadequately treated [17]. Knowledge of these pitfalls is essential for clinicians to reduce diagnostic errors.
The extraneous reads in the NTC and PC samples reflect (1) presumed bacterial environmental or laboratory contamination (eg, Ralstonia picketti [18]), (2) reagent contamination (eg, murine leukemia virus sequences in nucleic acid extraction columns [19]), (3) skin or other body flora (eg, papillomavirus), (4) misidentifications resulting from misannotations and/or misassemblies in the reference databases used for alignment (eg, reads corresponding to Gongylonema pulchrum, a parasitic nematode, correspond in actuality to bovine reads that likely represent contamination from fetal bovine serum, a common reagent in nearly all clinical microbiological laboratories), and (5) incomplete taxonomic classification such as lack of sufficient genomic representation in the reference database (eg, Streptococcus suis and Streptococcus salivarius reads in the PC, which contains only S agalactiae). The presence of multiple sources of contamination or inadvertent errors in interpretation underscore the importance of (1) establishing rigorous normalization threshold metrics used for making positive calls, (eg, “RPM ratio ≥ 10”), (2) pre-established validated cutoffs for distinguishing between coinfections from different microbial species and incomplete classification (eg, we rank bacterial species by RPM ratio and require that a minority species have at least 10% of the number of reads corresponding to the predominant species to call a coinfection), and (3) manual review and signoff of results by experienced laboratory directors, if the results of the mNGS assay are to be used clinically.
CONCLUSIONS
Metagenomic next-generation sequencing is an emerging approach in diagnostic microbiology with the ability to detect all microorganisms—viruses, bacteria, fungi, and parasites—in a single assay [3, 5, 13, 20, 21]. In this study, mNGS was used to provide an accurate diagnosis of neurobrucellosis and to guide the institution of targeted therapy, leading to complete resolution of the patient’s illness. This test has now been validated in the CLIA-certified University of California, San Francisco clinical microbiology laboratory as of June of 2016, and efforts to establish clinical utility for diagnosis of meningitis and encephalitis are ongoing as part of a precision medicine initiative [22].
Supplementary Data
Supplementary materials are available at Journal of The Pediatric Infectious Diseases Society online.
Supplementary Material
Notes
Acknowledgments. None.
Financial support. This study was supported in part by National Institutes of Health grants R01-HL105704 and R21-AI120977 (to C. Y. C.), an award from Abbott Laboratories, Inc. (to C. Y. C.), and funding from the California Institute to Advance Precision Medicine (to C. Y. C.).
Potential conflicts of interest. S. N. N., S. M., and C. Y. C. are coinventors of sequence-based ultra-rapid pathogen identification (SURPI)+ and associated algorithms related to pathogen detection, interpretation, and visualization from metagenomic next-generation sequencing data, and a patent pending for SURPI+ has been filed by the University of California, San Francisco (UCSF). C. Y. C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center and receives research support from Abbott Laboratories.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med 2005; 352:2325–36. [DOI] [PubMed] [Google Scholar]
- 2. Zazueta-Beltran J, León-Sicairos C, Canizalez-Roman A. Drug resistant Mycobacterium tuberculosis in Mexico. J Infect Dev Ctries 2009; 3:162–8. [DOI] [PubMed] [Google Scholar]
- 3. Greninger AL, Messacar K, Dunnebacke T, et al. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med 2015; 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greninger AL, Naccache SN, Messacar K, et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis 2015; 15:671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naccache SN, Federman S, Veeraraghavan N, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 2014; 24:1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hinić V, Brodard I, Thomann A, et al. Novel identification and differentiation of Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B. neotomae suitable for both conventional and real-time PCR systems. J Microbiol Methods 2008; 75:375–8. [DOI] [PubMed] [Google Scholar]
- 7. Morales-Garcia MR, Lopez-Mendez J, Pless R, et al. Brucellosis outbreak in a rural endemic region of Mexico - a comprehensive investigation. Vet Ital 2015; 51:185–90. [DOI] [PubMed] [Google Scholar]
- 8. Gul HC, Erdem H, Bek S. Overview of neurobrucellosis: a pooled analysis of 187 cases. Int J Infect Dis 2009; 13:e339–43. [DOI] [PubMed] [Google Scholar]
- 9. Al-Sous MW, Bohlega S, Al-Kawi MZ, et al. Neurobrucellosis: clinical and neuroimaging correlation. AJNR Am J Neuroradiol 2004; 25:395–401. [PMC free article] [PubMed] [Google Scholar]
- 10. Guven T, Ugurlu K, Ergonul O, et al. Neurobrucellosis: clinical and diagnostic features. Clin Infect Dis 2013; 56:1407–12. [DOI] [PubMed] [Google Scholar]
- 11. Moyer NP, Murray PR, Baron EJ, et al. Brucella. In: Manual of Clinical Microbiology. Washington, D.C: ASM Press, 1995: pp 549–55. [Google Scholar]
- 12. Yu WL, Nielsen K. Review of detection of Brucella spp. by polymerase chain reaction. Croat Med J 2010; 51:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiu CY, Miller S. Next-generation sequencing. In: Persing DH, Tenover FC, Hayden RTet al. eds. Molecular Microbiology, Diagnostic Principles and Practice, 3rd Edition Washington, D.C: ASM Press, 2016: pp 68–79. [Google Scholar]
- 14. Dasari S, Naha K, Prabhu M. Brucellosis and tuberculosis: clinical overlap and pitfalls. Asian Pac J Trop Med 2013; 6:823–5. [DOI] [PubMed] [Google Scholar]
- 15. Marx GE, Chan ED. Tuberculous meningitis: diagnosis and treatment overview. Tuberc Res Treat 2011; 2011:798764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varshochi M, Majidi J, Amini M, et al. False positive seroreactivity to brucellosis in tuberculosis patients: a prevalence study. Int J Gen Med 2011; 4:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verdon R, Chevret S, Laissy JP, Wolff M. Tuberculous meningitis in adults: review of 48 cases. Clin Infect Dis 1996; 22:982–8. [DOI] [PubMed] [Google Scholar]
- 18. Laurence M, Hatzis C, Brash DE. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 2014; 9:e97876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erlwein O, Robinson MJ, Dustan S, et al. DNA extraction columns contaminated with murine sequences. PLoS One 2011; 6:e23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naccache SN, Peggs KS, Mattes FM, et al. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 2015; 60:919–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014; 370:2408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. University of California San Francisco. Precision Diagnosis of Acute Infectious Diseases. Available at: http://www.ciapm.org/project/precision-diagnosis-acute- infectious-diseases. Accessed 2 May 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.