Abstract
Tuberculosis is a distinctive disease in which the causative agent, Mycobacterium tuberculosis, can persist in humans for decades by avoiding clearance from host immunity. During infection, M. tuberculosis maintains viability by extracting and utilizing essential nutrients from the host, and this is a prerequisite for all of the pathogenic activities that are deployed by the bacterium. In particular, M. tuberculosis preferentially acquires and metabolizes host-derived lipids (fatty acids and cholesterol), and the bacterium utilizes these substrates to cause and maintain disease. In this review, we discuss our current understanding of lipid utilization by M. tuberculosis, and we describe how these pathways promote pathogenesis to fuel metabolic processes in the bacillus. Finally, we highlight weaknesses in these pathways that potentially can be targeted for drug discovery.
Keywords: cholesterol, fatty acid, lipid, LucA, Mce1, Mce4
Mycobacterium tuberculosis uses cholesterol and fatty acids together to cause disease, we are just now beginning to understand how this process occurs.
INTRODUCTION
In 2015, Mycobacterium tuberculosis caused 1.8 million deaths and initiated ∼10.4 million new cases of tuberculosis (TB; World Health Organization 2016). TB is an insidious disease and ∼90% of infected individuals develop an asymptomatic or latent form of TB in which immunity contains, but does not eliminate, the bacteria from the host. These asymptomatic M. tuberculosis infections persist for the entire lifespan of the infected individual; meanwhile M. tuberculosis maintains a keen ability to cause debilitating or fatal disease. Due to factors that are not completely understood, roughly ∼10% of these infected individuals will develop active TB disease immediately following initial infection or sometime later in life (Pai et al.2016). Without antibiotic intervention, active TB can be deadly with fatality rates of 53–86 with a weighted mean of 70% in HIV-negative individuals (Tiemersma et al.2011) and this phase of the disease facilitates the aerosol transmission of the bacteria between humans. Thus, the bacterium's ability to persist for long periods of time ensures that M. tuberculosis will maintain a pandemic grip on the human population for many years to come.
Humans are the only known reservoir for M. tuberculosis, and it is likely that thousands of years of co-evolution in this host have uniquely shaped the bacterium's physiology and pathogenicity (Wirth et al.2008; Comas et al.2013). Many studies have determined that M. tuberculosis has a unique ability to assimilate and to utilize host lipids (fatty acids and cholesterol), and this is a defining characteristic of this pathogen (Cole et al.1998). Host-derived lipids are important carbon sources for M. tuberculosis, which are catabolized to fuel central metabolic pathways to facilitate the bacterium's persistence (Russell et al.2009). However, lipids are more than simple carbon sources and recent reports have revealed that lipid utilization by M. tuberculosis is more complex than was thought previously. Mycobacterium tuberculosis imports and utilizes fatty acids and cholesterol to convert both these lipids into bacterial end products that mediate bacterial pathogenesis. These bacterial lipid end products regulate bacterial replication, drug tolerance and virulence.
In this review, we focus on our understanding of the lipid assimilation and utilization pathways in M. tuberculosis with a special emphasis on how these pathways contribute to M. tuberculosis pathogenesis. Further, we highlight potential targets in these pathways that may be perturbed with drugs to enhance current and future TB antibiotic treatment(s).
CHOLESTEROL UTILIZATION BY M. TUBERCULOSIS
Over the past 10 years, numerous labs have demonstrated that cholesterol is required for the optimal growth and persistence of M. tuberculosis in various animal models of infection (Chang et al.2007; Van der Geize et al.2007; Yam et al.2009; Hu et al.2010; Nesbitt et al.2010; VanderVen et al.2015) and that cholesterol accumulates in human granulomas (Kim et al.2010). The complete degradation of cholesterol by bacteria or any organism is unusual, and only a relatively few gram-negative and Actinomyces species are known to do this (Yam et al.2011; Wipperman, Sampson and Thomas 2014). The M. tuberculosis genome contains a cluster of ∼80 genes (Van der Geize et al.2007) that encode proteins dedicated to the complex processes of cholesterol import, degradation and regulation (Capyk et al.2009, 2011; Yam et al.2009; Dresen et al.2010; Driscoll et al.2010; Lack et al.2010; Nesbitt et al.2010; Ouellet et al.2010; Griffin et al.2011, 2012; Thomas et al.2011; Casabon et al.2013a,b; Casabon et al.2014; Wipperman et al.2013; Frank, Madrona and Ortiz de Montellano 2014; Yang et al.2014, 2015; Lu, Schmitz and Sampson 2015; Ho et al.2016; Crowe et al.2017).
CHOLESTEROL IMPORT
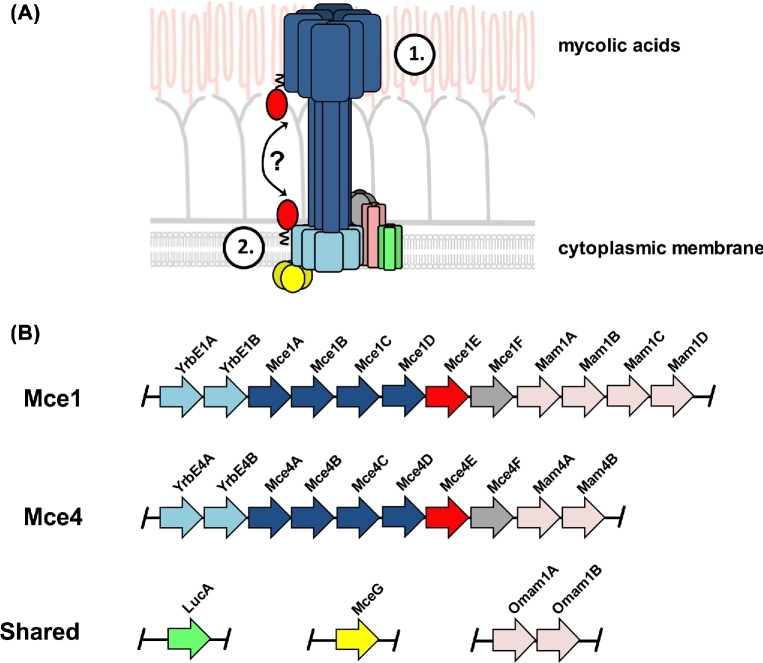
The mycobacterial cell envelope represents a significant physical barrier for the import of any molecule, and this structure is comprised of a hydrophobic cytoplasmic membrane bilayer, an aqueous pseudo-periplasmic space and an unusually thick layer of hydrophobic mycolic acids (Brennan 2003). To import cholesterol across this problematic atypical diderm cell envelope, M. tuberculosis relies on the multiprotein complex termed Mce4. Mutant studies with M. tuberculosis have confirmed that the Mce4 complex is required for cholesterol import (Pandey and Sassetti 2008; Nazarova et al.2017) and for cholesterol utilization when M. tuberculosis is cultured on cholesterol as the sole carbon source (Pandey and Sassetti 2008; Griffin et al.2011; Perkowski et al.2016; Nazarova et al.2017). The Mce4 complex is one of four closely related, ATP-binding cassette (ABC) transporter-like complexes that are encoded by the mce1–4 operons in the M. tuberculosis genome (Casali and Riley 2007). The mce4 operon spans the genes rv3492c-rv3501c and encodes 10 putative ‘core’ proteins that make up the Mce4 complex (Fig. 1). This core complex is comprised of two putative, integral membrane permease subunits (Rv3501/YrbE4 and Rv3502/YrbE4B), which are thought to translocate cholesterol across the cytoplasmic membrane (Casali and Riley 2007). Additionally, the Mce4 complex is comprised of six putative cell wall proteins (Rv3499/Mce4A, Rv3498/Mce4B, Rv3497/Mce4C, Rv3496/Mce4D, Rv3495/Mce4E, Rv3494/Mce4F), all of which conserve distinct Mce domains that probably facilitate cholesterol transport across the mycolic acid layer and/or the pseudoperiplasmic space (Casali and Riley 2007). In addition, the mce4 operon encodes two accessory proteins (Rv3493/Mam4A, Rv3492/Mam4B), which are required for cholesterol import (Casali and Riley 2007). These accessory proteins likely play a regulatory role to control stability or assembly of the Mce4 complex (Nazarova et al.2017). Although a structural and mechanistic understanding of how the Mce4 complex functions is lacking, the collective data are consistent with the idea that Mce4 mediates cholesterol import. We propose a hypothetical model for the function of proteins in the Mce4 complex, which we describe in more detail below.
Figure 1.
Hypothetical model for Mce-mediated lipid transport in M. tuberculosis and organization of the mce1 and mce4 operons. (A) Stage 1 depicts a process where Mce proteins bind and transport the lipid substrates across the exterior portion of the mycobacterial cell wall and pseudoperiplasmic space. Stage 2 illustrates the final translocation of lipid substrates across the cytoplasmic membrane by a putative permease complex. (B) The substrate-specific or ‘core proteins’ of the Mce1 and Mce4 complexes are encoded within the mce1 and mce4 operons. The putative subunits shared by the Mce1 and Mce4 complexes (LucA, MceG and OmamAB) are encoded by genes outside of the mce1 and mce4 operons.
Notably, the mce4 genes are required for optimal growth and persistence of M. tuberculosis in vivo. Early genetic screens predicted that the mce4 genes were required for M. tuberculosis's survival when passaged in mice during 2–4 weeks of infection (Sassetti and Rubin 2003). This observation was confirmed subsequently using an intravenous, competitive infection assay with a mutant lacking the putative Mce4 permease subunit (Rv3501/YrbE4A) (Pandey and Sassetti 2008). In this competition assay, the Mce4 mutant replicated slower in lung tissues relative to wild type beginning 4 weeks post-infection, and this growth defect worsened progressively through 14 weeks post-infection. Additionally, a M. tuberculosis mutant that lacks the entire mce4 operon grows more slowly in a murine, low-dose, aerosol infection model, and this growth defect is most apparent during the persistent phase of infection (Senaratne et al.2008). Thus, Mce4 is required for optimal bacterial growth and persistence in murine lung tissue regardless of the route of infection. The slow growth phenotype of the Mce4 mutants in vivo correlates with the onset of adaptive immunity and the generation of antigen-specific, interferon gamma (IFN-γ)-secreting CD4 + T cells. Consistent with this, M. tuberculosis Mce4 mutants replicate more poorly in IFN-γ activated macrophages relative to wild-type bacteria (Pandey and Sassetti 2008). Given that M. tuberculosis resides within macrophages and the bacteria appear to preferentially rely on cholesterol in his host cell type (VanderVen et al.2015) blocking cholesterol utilization in M. tuberculosis could target intracellular bacteria and augment current therapies.
DEGRADATION OF CHOLESTEROL
The degradation of cholesterol by M. tuberculosis can be categorized into three main processes: (i) β-oxidation of the cholesterol side chain, (ii) oxygen-dependent cleavage of the A and B rings and (iii) oxidative degradation of the C and D rings. Our understanding of cholesterol degradation by M. tuberculosis is not complete, but the current model indicates that the cholesterol degradation principally yields acetyl-CoA, propionyl-CoA, succinyl-CoA and pyruvate that feed intobacterial central metabolism.
β-OXIDATION OF THE SIDE CHAIN
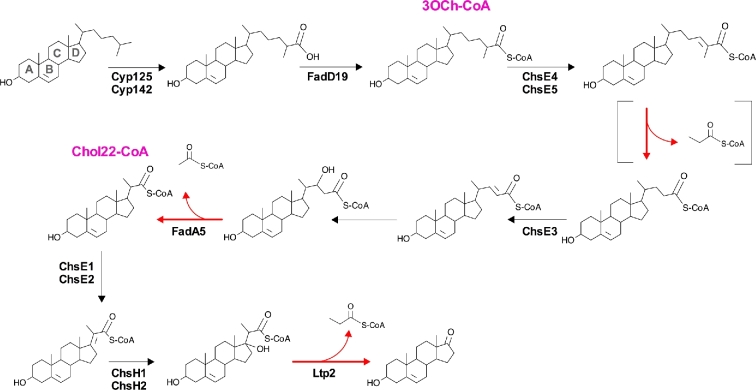
Degradation of the cholesterol side chain results in the release of two propionyl-CoA units and one acetyl-CoA unit (Fig. 2), and this process begins with hydroxylation of the terminal C26 or C27 carbon of cholesterol by the monoxygenases, Rv3545/Cyp125 or Rv3518/Cyp142 (Capyk et al.2009; Driscoll et al.2010; Ouellet et al.2010). After side chain hydroxylation, the acyl-CoA synthase, Rv3515/FadD19, ligates a coenzyme-A (CoA) onto the side chain (Casabon et al.2014). This step is necessary to initiate the first cycle of β-oxidation and the subsequent release of a propionyl-CoA unit. Completion of the first cycle of β-oxidation requires the multimeric acyl-CoA dehydrogenase complex that is comprised of Rv3504/ChsE4 and Rv3505/ChsE5, but the additional enzymes that are involved in this cycle have not been identified (Wipperman et al.2013; Yang et al.2015). The dehydrogenase Rv3572/ChsE3 (Yang et al.2015) and the thiolase Rv3546/FadA5 (Nesbitt et al.2010) participate in the second cycle of side chain β-oxidation to release a single acetyl-CoA unit. The final cycle of side chain β-oxidation releases a propionyl-CoA unit in three discrete stages. Dehydrogenation of the cholest-22-CoA (Chol22-CoA) is catalyzed by the multimeric dehydrogenase complex (Rv3543/ChsE1 and R3544/ChsE2) (Thomas and Sampson 2013). Next, the hydratase complex (Rv3541/ChsH1 and Rv3542/ChsH2) hydroxylates the C17 carbon in the D ring of the cholesterol (Yang et al.2014). The last carbon–carbon bond between the side chain and the D ring is cleaved by the aldolase Rv3540/Ltp2 to release the final propionyl-CoA unit from the side chain (Gilbert, Hood and Seah 2017).
Figure 2.
Cholesterol side chain degradation in M. tuberculosis. Confirmed enzymes are denoted with the enzyme name(s) next to the reaction arrows. Reactions that are indicated with red arrows indicate the release of either acetyl-CoA or propionyl-CoA. Brackets indicate stages in the pathway requiring multiple enzymatic reactions. Pink text denotes chemical name abbreviations of the catabolic intermediates.
DEGRADATION OF THE A AND B RINGS
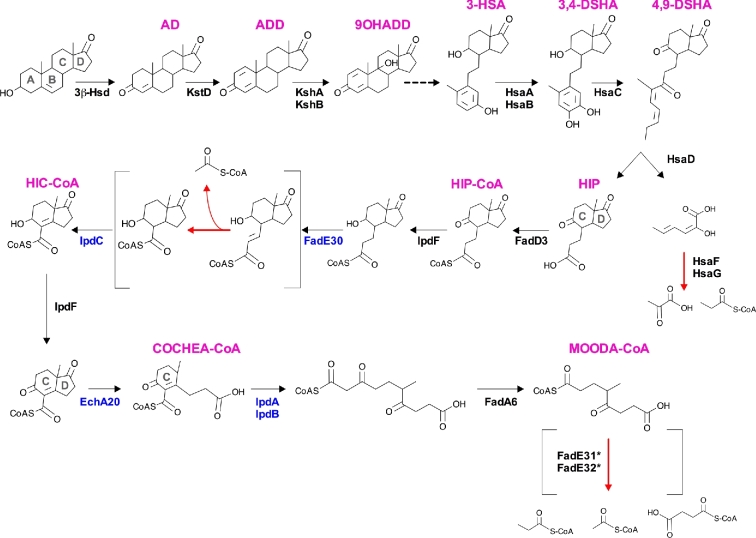
Degradation of the cholesterol A and B rings results in the release of one propionyl-CoA unit and one pyruvate unit (Fig. 3) The enzymes responsible for degrading the A and B rings of cholesterol have been characterized over the past 10 years in a series of elegant studies. To simplify the names of the cholesterol degradation intermediates, this description assumes that the side chain has been degraded. It should be noted that degradation of the side chain and rings can occur simultaneously (Thomas et al.2011). Catabolic degradation of the cholesterol A and B rings begins with a dehydrogenation reaction, which is catalyzed by Rv1106/3β-HSD to form 4-androstendione (AD) (Yang et al.2007). Next, desaturation of the A ring by Rv3537/KstD produces 1,4-androstendione (ADD) (Knol et al.2008). Subsequent hydroxylation by the monoxygenase/reductase complex Rv3526/KshA and Rv3571/KshB at the C9 position of the sterol forms 9-hydroxy-androsta-1,4-diene-3,17-dione (9OHADD) (Capyk et al.2009, 2011). The precise order of the preceding desaturation and hydroxylation reactions is unclear. Irrespective of the reaction order, once formed, 9OHADD undergoes non-enzymatic cleavage in the B ring spontaneously, and the A ring is aromatized. Together, these reactions form 3-hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3-HSA). Rv3570/HsaA and Rv3567/HsaB comprise an oxygenase/reductase complex that hydroxylates 3-HSA to form 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4-DSHA) (Dresen et al.2010). Next, the A ring is cleaved through the activity of the dioxygenase, Rv3568/HsaC; this introduces two oxygen atoms into the molecule and opens the dihydroxylated A ring to generate 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid (4,9-DSHA) (Yam et al.2009). 4,9-DSHA is cleaved further by the carbon–carbon hydrolase Rv3569/HsaD, which produces (3aα-H-4α(3’-propanate)-7aβ-methylhexahydro-1,5-indanedione (HIP) and 2-hydroxy-hexa-2,4-dienoic acid (HHD) (Lack et al.2010). Finally, HHD is degraded by the aldolase/dehydrogenase complex Rv3534/HsaF and Rv3535/HsaG, which produces pyruvate and propionyl-CoA (Carere et al.2013).
Figure 3.
Degradation of the cholesterol rings in M. tuberculosis. Confirmed enzymes are denoted with the enzyme name(s) next to the reaction arrow. Pink text denotes the abbreviation of the catabolic intermediate chemical name. Black text indicates enzymes from M. tuberculosis that catalyze the specific reactions, and blue text indicates confirmed homologous enzymes from related Actinomyces spp. that catalyze the specific reactions. Asterisks indicate a tentative assignment, given that MOODA-CoA accumulates in a cholesterol-grown mutant of M. smegmatis that lacks FadE32 (Crowe et al.2017). The dashed arrow indicates a spontaneous, non-enzymatic, ring cleavage event in 9OHADD. Reactions indicated with red arrows indicate the release of acetyl-CoA, propionyl-CoA, pyruvate or succinyl-CoA. Brackets highlight stages in the pathway that require multiple enzymatic reactions.
DEGRADATION OF C AND D RINGS
The final stages of cholesterol degradation release at least one acetyl-CoA unit, and probably a unit of propionyl-CoA, pyruvate or succinyl-CoA. Degradation of the sterol C and D rings commences when the acyl-CoA synthase, Rv3561/FadD3, ligates a CoA molecule onto degradation intermediate HIP, which forms Hip-CoA (Casabon et al. 2013b) (Fig. 3). Recent studies on Hip-CoA degradation in M. tuberculosis have made it possible to propose a model of C ring and D ring degradation (Crowe et al.2017). Briefly, a cholesterol-grown mutant of M. tuberculosis that lacked Rv3551/IpdA and Rv3552/IpdB accumulated (R)-2-(2-carboxyethyl)-3-methyl-6-oxycyclohex-1-ene-1-carboxy-CoA (COCHEA-CoA). Accumulation of COCHEA-CoA indicates that the D ring is opened prior to the opening of the C ring (Fig. 3). The same study also established that recombinant enzymes from M. tuberculosis (Rv3559/IpdF and Rv3556/FadA6) and homologous enzymes from related Actinomyces spp. (IpdC, EchA20, IpdA and IpdB) transform 3aα-H-4α(3΄-carboxyal-CoA)-5-hydroxy-7aβ-methylhexahydro-1-indanedione (HIC-CoA) into COCHEA-CoA and 4-methyl-5-oxo-octanedioic acid (MOODA-CoA) (Crowe et al.2017). Furthermore, a cholesterol-grown mutant of M. smegmatis lacking a FadE32 homolog accumulates MOODA-CoA, supporting the C and D ring degradation model in M. tuberculosis (Fig. 3).
Although the end products of MOODA-CoA degradation are unknown currently, it is likely that acetyl-CoA, propionyl-CoA and/or succinyl-CoA are released from this metabolic intermediate. Moreover, this model of cholesterol degradation is consistent with metabolomic analyses which demonstrated that propionyl-CoA pools increase when M. tuberculosis is grown on cholesterol (Jain et al.2007; Yang et al.2009; Griffin et al.2012). Interestingly, cholesterol-grown M. tuberculosis also accumulates high levels of methyl-succinate (Griffin et al.2012). It is tempting to speculate that methyl-succinate could be released from MOODA-CoA, but this remains to be determined.
TRANSCRIPTIONAL REGULATION OF CHOLESTEROL DEGRADATION
The TetR-like transcriptional repressors Rv3574/KstR1 and Rv3557/KstR2 regulate cholesterol degradation in M. tuberculosis. These repressors are de-repressed in a ‘feed forward’ manner after binding with specific intermediates of cholesterol degradation. KstR1 is de-repressed by the catabolic intermediate 3-hydroxy-cholest-5-ene-26-oyl-CoA (3OCh-CoA) (Fig. 2), which promotes transcription of the genes encoding enzymes required for degrading the side chain and the A and B rings of cholesterol (Ho et al.2016). KstR1 also binds putative promoter sequences throughout the M. tuberculosis genome (Galagan et al.2013) and controls expression of numerous genes outside the main cholesterol catabolic gene cluster (Kendall et al.2007; Wipperman, Sampson and Thomas 2014). This suggests that cholesterol utilization is integrated into a larger metabolic network, but how these genes/proteins participate in cholesterol utilization is unknown currently. Similarly, the KstR2 repressor is de-repressed by HIP-CoA (Fig. 3), which specifically activates transcription of genes in the KstR2 regulon that encode enzymes required for C and D ring degradation (Casabon et al. 2013).
CHOLESTEROL FUELS CENTRAL METABOLIC PATHWAYS DURING INFECTION
Propionyl-CoA is a major metabolite that is released during cholesterol degradation, and this three-carbon intermediate lies at a critical axis in M. tuberculosis metabolism (Savvi et al.2008). Mycobacterium tuberculosis can generate propionyl-CoA through β-oxidation of odd-chain fatty acids, degradation of branched-chain amino acids (leucine, isoleucine and valine) and cholesterol degradation (Savvi et al.2008; Venugopal et al.2011). The major lipids in mammalian cells are typically even chain and thus not likely a source of propionyl-CoA for M. tuberculosis during infection. Although M. tuberculosis can import and metabolize host-derived amino acids (Gouzy, Poquet and Neyrolles 2014), it is unknown how branched chain amino acids contribute to propionyl-CoA pools during infection. Recent studies indicate that cholesterol is the main source of propionyl-CoA for M. tuberculosis during infection. For example, the inhibitor 3-nitropropionate (3-NP) induces a propionyl-CoA-dependent toxicity in M. tuberculosis by inhibiting bacterial isocitrate lyase enzymes (Munoz-Elias and McKinney 2005; Eoh and Rhee 2013). Treatment with 3-NP reduces bacterial fitness during infection in macrophages (Griffin et al.2012; Eoh and Rhee 2013), and this 3-NP induced toxicity can be partially reversed by deleting the Mce4 cholesterol transporter in M. tuberculosis. Additionally, we developed a reporter construct for M. tuberculosis that expresses GFP in response to increases in propionyl-CoA pools. Using this tool we found that mutants of M. tuberculosis lacking Mce4 and Rv3723/LucA poorly express GFP during infection in macrophages relative to wild-type bacteria (Nazarova et al.2017).
The three metabolic fates of propionyl-CoA are as follows: (i) anaplerotic assimilation by the methylcitrate cycle (MCC) (Muñoz-Elías et al.2006; Eoh and Rhee 2014), (ii) anaplerotic assimilation through the methylmalonyl pathway (MMP) (Savvi et al.2008) and (iii) biosynthetic incorporation into methyl-branched, polyketide lipids (Fig. 4). The MCC condenses propionyl-CoA with oxaloacetate to ultimately generate succinate and pyruvate for central metabolism. In sequence, the methylcitrate synthase Rv1131/PrpC, methylcitrate dehydratase Rv1130/PrpD, aconitase Rv1475/Acn and isocitrate lyase Rv0467/Icl1 or Rv1915–16/Icl2 enzymes all make up the MCC (Munoz-Elias and McKinney 2005; Muñoz-Elías et al.2006; Upton and McKinney 2007). The enzymes Icl1 and/or Icl2 catalyze the last reaction of the MCC in M. tuberculosis and these enzymes are bifunctional and necessary in the glyoxylate shunt for sustain bacterial growth on even chain fatty acids (McKinney et al.2000; Munoz-Elias and McKinney 2005). Consequently, Icl1 and Icl2 are involved in two anaplerotic pathways that assimilate both propionyl-CoA and acetyl-CoA. When a M. tuberculosis Icl1/2 double mutant was provided acetate or propionate as sole carbon sources the bacteria were unable to assimilate neither carbon source, which depleted pools of central metabolic intermediates, disrupted membrane potential and perturbed pH homeostasis (Eoh and Rhee 2014). Based on this, it is likely that the severe attenuation of the Icl1/2 double mutant in animal models is multifactorial and could be partly due to this mutant's inability to assimilate propionyl-CoA into central metabolism (Munoz-Elias and McKinney 2005).
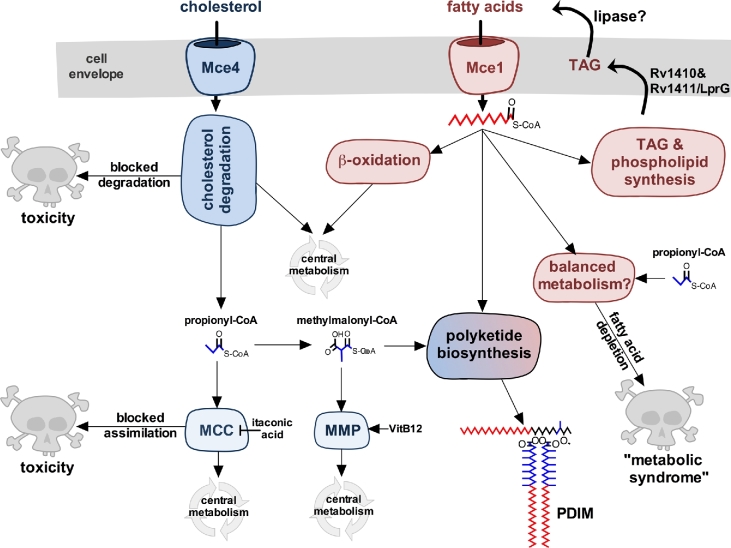
Figure 4.
Cholesterol and fatty acid utilization in M. tuberculosis. Cholesterol and fatty acid degradation fuels central metabolism and supplies pathways required for pathogenesis. Cholesterol-derived propionyl-CoA can be assimilated by the MCC or converted into methylmalonyl-CoA. Methylmalonyl-CoA can be assimilated by the MMP or used for polyketide lipid synthesis. Inactivating cholesterol degradation enzymes or the MCC can induce metabolic intoxication that inhibits bacterial growth. Salvaged fatty acids can also be used in biosynthetic reactions to generate membrane lipids, TAG and polyketide lipids. Cholesterol-derived propionyl-CoA induces a ‘metabolic syndrome’ that is associated with an inability to export TAG and/or when fatty acid pools are depleted. Vitamin B12 is indicated as VitB12.
Inactivating Icl1 activity drives the accumulation of propionyl-CoA derived intermediates of the MCC that are also capable of intoxicating M. tuberculosis and inhibiting bacterial growth in rich media containing cholesterol or propionate (Lee et al.2013; VanderVen et al.2015). This metabolic toxicity is not a starvation response because the growth inhibition occurs in the presence of other nutrients (glucose, glycerol and fatty acids), which together are capable of sustaining anaplerotic metabolism in M. tuberculosis. In an Icl1-deficient strain of M. tuberculosis, this cholesterol-dependent toxicity can be reversed by introducing secondary mutations in Rv1131/PrpC which indicates that the observed toxicity is associated with an inactive MCC (Nazarova et al.2017). Furthermore, chemical inhibition of Rv1131/PrpC also reverses the cholesterol-dependent metabolic toxicity in an Icl1-deficient strain of M. tuberculosis (VanderVen et al.2015). Similarly, chemical or genetic inactivation of enzymes in the cholesterol breakdown pathway also reverses the metabolic intoxication in an Icl1-deficient strain of M. tuberculosis when the bacteria are grown in cholesterol-containing media (VanderVen et al.2015; Nazarova et al.2017).
Propionyl-CoA can also be converted to methylmalonyl-CoA, which then feeds into central metabolism through the MMP (Fig. 4). The MMP assimilates methylmalonyl-CoA through the activity of the methylmalonyl-CoA mutase complex that is comprised of Rv1492/MutA and Rv1493/MutB. The MutAB complex requires the cofactor vitamin B12 for enzymatic activity (Savvi et al.2008). In the presence of vitamin B12, propionyl-CoA can assimilated into anaplerotic metabolism. Importantly, the addition of vitamin B12 can reverse the cholesterol-dependent toxicity in a M. tuberculosis strain lacking Icl1 (Lee et al.2013). Vitamin B12 also partially rescues growth of the Icl1 mutant during infection in macrophages following treatment with 3-NP (Griffin et al.2012). Therefore, if cholesterol is a major source of propionyl-CoA for the bacterium during infection, there are also unfavorable consequences associated with cholesterol metabolism when M. tuberculosis is unable to efficiently assimilate propionyl-CoA into anaplerotic metabolism.
CHOLESTEROL-DERIVED, PROPIONYL-COA FUELS VIRULENCE LIPID BIOSYNTHESIS
The propionyl-CoA that is converted to methylmalonyl-CoA can also be used for the synthesis of methyl-branched, polyketide virulence lipids in M. tuberculosis (Rainwater and Kolattukudy 1983; Jackson, Stadthagen and Gicquel 2007; Quadri 2014) (Fig. 4). Polyketide synthases incorporate methylmalonyl-CoA to generate the methyl-branched moieties of phthiocerol-dimycocerosate (PDIM), polyacylated trehalose and sulfolipid (SL) (Quadri 2014). It is well known that M. tuberculosis grown in axenic media without cholesterol still synthesizes methyl-branched polyketide lipids (Layre et al.2011; Sartain et al.2011). In axenic media lacking cholesterol, branched-chain amino acids probably serve as the source of propionyl-CoA (Venugopal et al.2011). However, when M. tuberculosis is cultured in media that contains propionate or cholesterol, the absolute amount of PDIM and SL increases dramatically, reflecting increases in the bacterial propionyl-CoA pools (Jain et al.2007; Yang et al.2009; Griffin et al.2012). Additionally, the methyl-branched acyl chains of PDIM and SL are longer when the bacteria are supplied with propionate or cholesterol (Jain et al.2007; Yang et al.2009; Griffin et al.2012). These ‘high mass forms’ of PDIM and SL have additional methylmalonyl-CoA units incorporated into their acyl chains, resulting in more methyl-branched subunits in the lipids. Given the role of PDIM and SL in pathogenesis, these observations indicate that an expansion of propionyl-CoA pools in M. tuberculosis modulates important virulence factors of the bacterium (Lee et al.2013). Importantly, these ‘high mass forms’ of PDIM and SL are also produced by M. tuberculosis in vivo (Jain et al.2007), which further supports the idea that cholesterol is a dominant nutrient for M. tuberculosis which feeds propionyl-CoA pools during infection.
It has long been known that M. tuberculosis lipids in general, and PDIM in particular, play important roles in pathogenesis (Camacho et al.1999; Cox et al.1999), so the coupling of cholesterol degradation to the biosynthesis of methyl-branched lipids such as PDIM may have important consequences in pathogenesis. PDIM has been implicated in macrophage invasion and recruitment (Astarie-Dequeker et al.2009; Cambier et al.2014), resistance to immune-mediated stress (Rousseau et al.2004; Kirksey et al.2011; Day et al.2014), masking cell wall antigens (Cambier et al.2014) and facilitating bacterial escape from macrophage phagosomes (Barczak et al.2017; Quigley et al.2017). All of these activities could be influenced by fluctuating abundances or structural alterations of PDIM or other polyketide virulence lipids in the bacterial cell wall.
DEGRADATION AND METABOLISM OF FATTY ACIDS
In addition to cholesterol, fatty acids are also a highly abundant lipid found in human granulomas (Kim et al.2010). Seminal work by Segal and Bloch published in 1956 demonstrated that fatty acids, but not carbohydrates, specifically stimulated ex vivo respiration rates of M. tuberculosis harvested from murine lung tissue (Segal and Bloch 1956). Although M. tuberculosis's ability to assimilate and to metabolize fatty acids has long been appreciated, genome sequencing revealed that the bacterium has an expanded set of putative fatty acid β-oxidation genes (Cole et al.1998). Initially, it was thought that M. tuberculosis contained roughly ∼250 enzymes dedicated to fatty acid β-oxidation (Cole et al.1998). Subsequent work demonstrated that many of these putative lipid oxidation enzymes are involved in lipid biosynthesis (Trivedi et al.2004; Krithika et al.2006), function as multimeric complexes or are not involved in fatty acid β-oxidation at all (Casabon et al. 2013; Thomas and Sampson 2013; Yang et al.2014, 2015). Nonetheless, M. tuberculosis still possesses a remarkable number of proteins that are involved in the breakdown of lipids, but the precise functions/substrates for most of these putative enzymes remain uncharacterized.
Although cholesterol appears only to be degraded by M. tuberculosis to generate metabolic intermediates, fatty acids can either be degraded to fuel metabolism or used for biosynthesis directly (Fig. 4). During infection, M. tuberculosis likely scavenges fatty acids but can also synthesize C16–22 fatty acids through the fatty acid synthase-I (FAS-I) enzyme (Takayama, Wang and Besra 2005). However, given the energetic cost associated with de novo fatty acid synthesis, the bacteria likely β-oxidizes scavenged fatty acids or incorporates these lipids directly into biosynthetic pathways (Fig. 4). Fatty acids in the form of acyl-AMP intermediates are donated to polyketide synthases which iteratively incorporate malonyl-CoA and/or methylmalonyl-CoA to synthesize polyketide lipids (Quadri 2014). Full-length mycolic acids are synthesized by elongating fatty acids through the specialized fatty acid synthase-II complex (FAS-II) to a length of C22–62 (Marrakchi, Laneelle and Daffe 2014). The FAS-II complex elongates either salvaged or de novo synthesized fatty acids to generate the full-length acyl-chains of mycolic acids (Odriozola, Ramos and Bloch 1977; Portevin et al.2004). Fatty acids can also be assimilated directly into membrane phospholipids or converted into TAG. Phospholipids are required to maintain integrity of cytoplasmic membranes, and TAG functions as a carbon reserve that can be catabolized when nutrients are limiting (Daniel et al.2004, 2011). While these pathways for utilizing fatty acids in M. tuberculosis are relatively well known, recent evidence indicates that utilization of fatty acids and cholesterol is coordinated, and the co-utilization of these substrates influences pathogenesis, summarized in more detail below.
FATTY ACID IMPORT
The mechanism responsible for fatty acid import across the M. tuberculosis cell envelope has remained enigmatic until recently. Using a series of unbiased approaches, we discovered that the Mce1 complex imports fatty acids into M. tuberculosis (Nazarova et al.2017). Similar to the mce4 operon, the mce1 operon encodes two putative permease subunits (Rv0167/YrbE1A and Rv0168/YrbE1B), six Mce proteins (Rv0169/Mce1A, Rv0170/Mce1B, Rv0171/Mce1C, Rv0172/Mce1D, Rv0173/Mce1E and Rv0174/Mce1F) and four accessory subunits (Rv0175/Mam1A, Rv0176/Mam1B, Rv0177/Mam1C and Rv0178/Mam1D) (Fig. 1). In our studies, we found that in M. tuberculosis lacking Rv3723/LucA, the Mce1A, Mce1D and Mce1E proteins are degraded, and the Rv3723/LucA mutant is unable to import fatty acids during infection and in axenic media. Based on this observation, we next confirmed that M. tuberculosis mutants lacking genes in the mce1 operon are also unable to import fatty acids in axenic media and during infection in macrophages. Because of the similarities between the genes in the mce1 and mce4 operons, it was previously hypothesized that Mce1 imports fatty acids (Forrellad et al.2014), and our data strongly support this idea. Added support that Mce1 functions as a fatty acid transporter is inferred from studies in M. leprae. The minimal genome of M. leprae conserves a single mce operon, which is most closely related to the mce1 operon from M. tuberculosis (Wiker et al.1999). Importantly, M. leprae is capable of importing and metabolizing fatty acids ex vivo, suggesting that Mce1 also imports fatty acids in related mycobacterial pathogens (Franzblau 1988).
HOW DOES MCE1 CONTRIBUTE TO M. TUBERCULOSIS PATHOGENESIS?
Proteins of the Mce1 complex have long been regarded as virulence factors in M. tuberculosis, despite conflicting evidence regarding their specific functions. Early studies concluded that Mce1 proteins are virulence factors, which mediate bacterial entry into mammalian cells. Surface expression of the M. tuberculosis Mce1A protein in a non-pathogenic strain of Escherichia coli endows the bacterium with an ability to invade mammalian epithelial cells; consequently, proteins related to Mce1A were named mammalian cell entry or Mce proteins (Arruda et al.1993). The domain of Mce1A that interacts with mammalian cells was mapped to the Mce domain of Mce1A (amino acids 106–177), and this is the minimal region responsible for the E. coli invasion phenotype (Casali et al.2002). In M. tuberculosis, Mce1A is secreted and found on the outer surface of the bacterial cell envelope (Chitale et al.2001). Since it is now understood that proteins conserving Mce domains bind lipids (Krachler, Ham and Orth 2011; Ekiert et al.2017), it is plausable that surface Mce1A binds lipids for import into M. tuberculosis. Expressing Mce1A on the surface of E. coli may promote close interactions between the bacteria and mammalian cell membrane lipids to facilitate host cell invasion by non-pathogenic E. coli. Given the multitude of cell entry pathways that are exploited by M. tuberculosis (Russell 2011), we hypothesize that the principle function of Mce1 is to import fatty acids to promote M. tuberculosis pathogenesis.
Analysis of transposon mutants passaged within mice found that mutations throughout the mce1 operon confer in vivo growth defects, primarily early in infection (Sassetti and Rubin 2003). Supporting this, both targeted and unbiased studies also reported that Mce1 mutants displayed fitness defects in mice and macrophages, in addition to causing less severe lung pathology in animals (Gioffre et al.2005; Stewart et al.2005; Joshi et al.2006; McCann et al.2011). In contrast, M. tuberculosis Mce1 mutants can display hyper-virulent phenotypes in mice. Additionally, macrophages infected with Mce1 mutants produced less TNF-α and nitric oxide, and these infected macrophages were unable to control intracellular growth of an Mce1 mutant relative to wild-type M. tuberculosis (Shimono et al.2003). The reported anti-inflammatory phenotype of the Mce1 mutant in macrophage infection models has been attributed to free mycolic acids (Sequeira, Senaratne and Riley 2014) that accumulate in the cell envelope of a Mce1 mutant (Cantrell et al.2013; Forrellad et al.2014; Queiroz et al.2015). The apparent contradictions of the Mce1 mutant phenotypes in vivo may be explained by differences in the genetic background of mice, routes of infection, location/type of genetic mutations or the duration of the experiments.
Other data have been reported that are consistent with Mce1 functioning as a M. tuberculosis fatty acid importer. For example, cell wall lipids and membrane phospholipid levels are reduced in an Mce1 mutant, and the mutant overexpresses the FAS-I enzyme (Queiroz et al.2015). This indicates that fatty acid pools are depleted in the Mce1 mutant and the bacteria compensate for the fatty acid depletion by increasing de novo fatty acid synthesis. Free mycolic acids also accumulate in the cell envelope of Mce1 mutants; therefore, it was proposed that Mce1 functions to recycle mycolic acids (Cantrell et al.2013; Forrellad et al.2014). Importing mycolic acids through Mce1 seems unlikely given that a Mce1 mutant had no growth defect when the bacteria were supplied exogenous mycolic acids as a sole carbon source in axenic culture (Dunphy et al.2010); however, the substrate specificity of Mce1 has yet to be defined. Together, these results are consistent with the idea that Mce1 functions as a fatty acid importer and fatty acids are required for M. tuberculosis to establish and maintain optimal infections in animals.
EXPANDING MODEL FOR MCE1 AND MCE4 TRANSPORTERS
The Mce1 and Mce4 transporters are analogous to multisubunit, ABC transporters that are comprised of putative substrate binding proteins (SBP), permease proteins and an ATPase subunit that provides energy for the transport process (Wilkens 2015). Gram-negative bacteria shuttle lipids across their cell envelope, and these processes also rely on proteins that conserve Mce domains (Malinverni and Silhavy 2009; Thong et al.2016; Isom et al.2017; Nakayama and Zhang-Akiyama 2017). Mce proteins make up the MlaD superfamily (cl27420) and participate in trafficking lipids across double membrane structures (Malinverni and Silhavy 2009; Sutterlin et al.2016; Thong et al.2016; Ekiert et al.2017; Isom et al.2017; Nakayama and Zhang-Akiyama 2017). The Mce proteins in gram-negative bacteria form multimeric structures and are predicted to bind and facilitate lipid transport (Ekiert et al.2017). In M. tuberculosis, the Mce or SBP proteins of the Mce1 and Mce4 complexes are Mce1A-F and Mce4A-F (Fig. 1). Mce1A-E and Mce4A-E conserve canonical, secretory signal sequences, and they are abundant in M. tuberculosis cell envelope protein fractions (Feltcher et al.2015). In addition, Mce1A has been shown to localize on the cell surface of the bacteria (Chitale et al.2001). Mce1F and Mce4F proteins conserve a putative N-terminal transmembrane domain and are probably inserted within the cytoplasmic membrane or cell wall.
We hypothesize that these Mce proteins bind and shuttle lipid substrates across the mycolic acid layer and the pseudoperiplasmic space of the M. tuberculosis cell envelope. The large number of putative Mce proteins in the Mce1/4 complexes (six each) suggests that these proteins form a pore or channel and allow substrate shuttling from the bacterial surface to deliver substrates to the permease subunits within the cytoplasmic membrane. The putative permease subunits (YrbE1A/B and YrbE4A/B) are probably embedded in the cytoplasmic membrane and complete lipid translocation across the cytoplasmic membrane. Finally, accessory proteins (Mam1A-D and Mam4A-B) encoded within the mce1 and mce4 operons are required for lipid import, but the function of these proteins is unclear. Our recent work demonstrated that the accessory protein Rv3492/Mam4B is required for cholesterol import by the Mce4 complex, but the Rv3492/Mam4B protein appears to be involved in regulating the activity or assembly of the Mce4 transporter (Nazarova et al.2017).
It has been proposed that the Mce systems in E. coli use two distinct mechanisms for transporting lipid substrates across the aqueous periplasm. One system relies on a soluble protein that likely binds and shuttle lipids across the periplasm; the other system employs a closed channel that spans the periplasmic space, and lipids are likely transported internally within the protein complex (Ekiert et al.2017). It will be exciting to learn how the Mce1 and Mce4 complexes function mechanically and structurally. Given the similarities between the putative subunits in Mce1 and Mce4 complexes, these transporters probably use very similar mechanisms to import substrates, and the differences between these complexes likely define the substrate specificities of these transporters. Finally, assembly and regulation of the Mce1 and Mce4 transporters in the bacterial cell envelope are poorly understood processes.
Our hypothetical model proposes that proteins encoded in mce1 and mce4 operons comprise the ‘core proteins’ of Mce1 and Mce4 transporters, respectively. However, it is now clear that unlinked genes that are located outside the mce1 and mce4 operons also encode proteins that are required for Mce1- and Mce4-mediated nutrient transport (Fig. 1). The putative ATPase subunit Rv0655/MceG is predicted to hydrolyze ATP and provide energy for substrate import, and MceG is required for cholesterol import (Mohn et al.2008; Pandey and Sassetti 2008). It has been hypothesized that Rv0655/MceG may also function as the ‘common’ ATPase required for the Mce1, Mce2 and Mce3 complexes (Joshi et al.2006). We have recently confirmed that Rv0655/MceG is involved in fatty acid uptake in M. tuberculosis (unpublished). It is currently unknown if Rv0655/MceG participates in Mce2- and Mce3-mediated lipid import, and the enzymatic activity of Rv0655/MceG has not yet been confirmed.
Recently, it was reported that the orphaned Mce accessory protein Rv0199/OmamA plays a role in cholesterol utilization and that Rv0199/OmamA stabilizes the Mce1 complex (Perkowski et al.2016). Another protein common to both transporters is Rv3723/LucA, which interacts with accessory subunits (Mam1C, Mam4B and OmamA) of the Mce1 and Mce4 transport complexes (Nazarova et al.2017). Importantly, Rv3723/LucA is the first known M. tuberculosis protein that is required for both cholesterol and fatty acid import (Nazarova et al.2017). Based on this observation, our hypothetical model for Mce1 and Mce4 transporters in M. tuberculosis also includes proteins that are shared by both complexes. Based on these findings, it is likely that additional unknown proteins mediate or coordinate nutrient import using Mce1 and Mce4. Our current hypothesis is that a network of proteins including the ‘core proteins’ of the Mce1 and Mce4 transporters coordinates fatty acid and cholesterol import. Characterizing the network of lipid import proteins will be important to understand M. tuberculosis pathogenesis and persistence fully.
CONNECTIONS BETWEEN CHOLESTEROL AND FATTY ACID UTILIZATION
The first indication that cholesterol and fatty acid utilization is coordinated was gleaned from in vivo genetic screens that used M. tuberculosis mutants lacking Mce1 and Mce4 (Joshi et al.2006). This work used attenuation phenotypes in mice as a readout to predict proteins that potentially function in the same pathways as Mce1, Mce4, or both Mce1 and Mce4 together (Fig. 5). Rv3723/LucA was among the proteins identified as being involved in the same pathway as Mce4 (Joshi et al.2006). We independently discovered that Rv3723/LucA is required for Mce4-mediated cholesterol import. Surprisingly Rv3723/LucA is also required for Mce1-mediated fatty acid import (Nazarova et al.2017). Rv0655/MceG was also identified as being in the same pathways as Mce4 and Mce1, and this observation has been subsequently confirmed (unpublished) (Joshi et al.2006; Pandey and Sassetti 2008). These observations are consistent with our hypothesis that a network of proteins facilitates and coordinates cholesterol and fatty acid import in M. tuberculosis. Additionally, several proteins are involved in the downstream utilization of fatty acids and cholesterol and function in the same pathways as Mce1 and Mce4. Characterization of these common proteins involved in the utilization of fatty acids and cholesterol downstream of Mce1 and Mce4 have demonstrated a ‘metabolic codependency’ of these two substrates in M. tuberculosis metabolism.
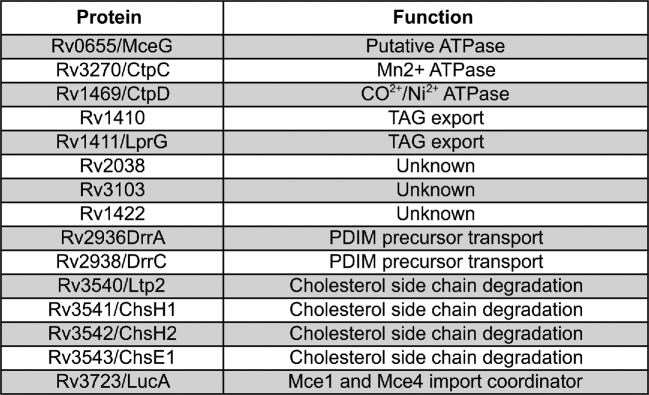
Figure 5.
Proteins involved in pathways linked to Mce1 and Mce4. Proteins identified by genetic epistasis mapping from in vivo attenuation phenotypes in mice.
FATTY ACIDS BUFFER THE METABOLIC COSTS OF CHOLESTEROL UTILIZATION
Rv1410 and Rv1411/LprG are two proteins that function in the same pathways as both Mce1 and Mce4 and are involved in the downstream utilization of lipids in M. tuberculosis. Specifically, these two proteins facilitate the export of TAG from the bacterial cytosol (Martinot et al.2016). These proteins are required for full virulence of M. tuberculosis (Gaur et al.2014; Martinot et al.2016) and, notably, these proteins have been linked to mycobacterial growth on cholesterol (Ramon-Garcia et al.2015). Importantly, a cholesterol-grown M. tuberculosis mutant lacking Rv1410 and Rv1411/LprG experiences a cholesterol-derived, propionyl-CoA-dependent growth defect that can be reversed by supplying the bacteria exogenous free fatty acids, but not vitamin B12 (Martinot et al.2016). Chemically blocking the release of free fatty acids from TAG exacerbated the growth defect in an Rv1410 and Rv1411/LprG-deficient mutant when the bacteria were grown in cholesterol media. Together, these data demonstrate that in the absence of Rv1410 and Rv1411/LprG M. tuberculosis can become intoxicated by cholesterol, and expanding the pool size of free fatty acids alleviates the intoxication or ‘metabolic syndrome’. It remains to be determined precisely how Mce1 is involved in the ‘metabolic syndrome’, but it is conceivable that Mce1 is required to recycle fatty acids released from TAG stored in the cell envelope (Fig. 4).
Cholesterol degradation in M. tuberculosis increases the flux of methylmalonyl-CoA (derived from propionyl-CoA) into polyketide lipid biosynthesis (Jain et al.2007; Yang et al.2009; Griffin et al.2012). Increased biosynthesis of polyketide lipids requires sufficient levels of free fatty acids to serve as fatty acid-AMP primers for polyketide synthase enzymes (Quadri 2014) (Fig. 4). Synchronizing fatty acid and cholesterol import by Mce1 and Mce4 may help to ensure that a balanced supply of biosynthetic precursors is maintained to efficiently synthesize polyketide lipids. Biosynthesis of polyketide lipids also serves as a sink to prevent accumulation of toxic metabolic intermediates generated by the MCC (Lee et al.2013). Metabolic detoxification requires the conversion of propionyl-CoA into methylmalonyl-CoA and excess fatty acid-AMP primers to synthesize polyketide lipids. For instance, growth inhibition of an M. tuberculosis mutant lacking Icl1 is reversed by supplying excess fatty acids during infection, and these fatty acids are preferentially incorporated into PDIM (Lee et al.2013). This suggests that the amount of available free fatty acids impacts the flux of methylmalonyl-CoA (derived from propionyl-CoA) into polyketide lipid biosynthesis. During infection, fatty acids could also help to counter growth inhibition due to itaconic acid (Michelucci et al.2013), which is produced by IFN-γ-activated macrophages cells and inhibits Icl1/2 in M. tuberculosis (Eoh and Rhee 2014) (Fig. 4).
Therefore, cholesterol metabolism is associated with numerous detrimental burdens on bacterial metabolism, but these can be buffered by increasing the pools of available fatty acids. Given that M. tuberculosis probably encounters fatty acids and cholesterol simultaneously in vivo (Kim et al.2010), perhaps the metabolic pathways in the bacterium evolved to operate most efficiently when cholesterol and fatty acids are co-metabolized. Finally, coordinating the import of these lipid substrates through Mce1 and Mce4 may ensure that metabolism in the bacterium remains balanced.
THERAPEUTIC POTENTIAL OF LIPID UTILIZATION PATHWAYS IN M. TUBERCULOSIS
TB antibiotic therapy will always require multidrug treatments to prevent drug resistance and to treat the different bacterial subpopulations that differentiate in vivo (Evangelopoulos, da Fonseca and Waddell 2015). Thus, new drugs capable of targeting bacterial subpopulations that are not effectively eliminated by current antibiotics could enhance current TB drug regimens and shorten therapy, prevent drug resistance and reduce relapse. Our recent drug discovery efforts to identify compounds that inhibit M. tuberculosis replication in macrophages have found a large number of compounds that inhibit processes related to cholesterol utilization in the bacterium (VanderVen et al.2015). We predict that compounds capable of blocking cholesterol utilization in M. tuberculosis could specifically inhibit the growth of bacterial subpopulations within macrophages. Chemical perturbation of cholesterol utilization in M. tuberculosis can induce carbon starvation, metabolic intoxication and unbalanced central metabolism. Consequently, there are a large number of potential vulnerabilities in the cholesterol utilization pathways of M. tuberculosis that can be exploited for drug discovery.
TARGETING CHOLESTEROL AND FATTY ACID UTILIZATION PATHWAYS
Mycobacterium tuberculosis can simultaneously metabolize cholesterol from ‘both ends’ to release metabolic intermediates from both the rings and side chain of the molecule (Thomas et al.2011). This suggests that not all cholesterol degradation enzymes are equally good targets for drug development if the goal is to prevent the release of metabolic intermediates from the sterol. We have found that compounds that inhibit the cholesterol degradation enzymes (HsaAB) can block intracellular replication of the bacteria (VanderVen et al.2015), indicating that these enzymes are a weakness in the pathway. Alternatively, inactivating the ability of M. tuberculosis to import cholesterol efficiently may induce a type of carbon starvation in the bacterium. For example, inhibiting the Mce4 cholesterol transporter may starve the bacteria by restricting bacterial access to cholesterol, particularly at specific stages of infection such as during persistence (Pandey and Sassetti 2008).
Inhibiting key enzymes in the cholesterol degradation pathways may also promote the accumulation of toxic cholesterol-derived degradation intermediates. For example, M. tuberculosis mutants lacking Cyp125, HsaC and IpdAB are intoxicated by cholesterol-derived degradation intermediates and exhibit growth defects even in the presence of additional carbon sources (Chang et al.2009; Yam et al.2009; Ouellet et al.2010; Crowe et al.2017). Thus, inhibiting proteins in cholesterol metabolism is compelling since inhibiting key steps in the cholesterol degradation pathway of M. tuberculosis could potentially transform a preferred nutrient of the bacterium into a growth-restricting toxin. Mycobacterium tuberculosis mutants lacking the TAG transporter suffer from a cholesterol-dependent ‘metabolic syndrome’ which is associated with unbalanced metabolism (Martinot et al.2016) (Fig 5). This cholesterol-dependent metabolic intoxication could also be exploited with inhibitors of Rv1410/Rv1411/LprG.
The pathways for fatty acid degradation in M. tuberculosis are heavily redundant, which is a significant hurdle for drug development. However, chemical inhibition of Mce1 may limit the availability of free fatty acids and perturb M. tuberculosis metabolism. The encouraging realization that proteins such as Rv3723/LucA are required for the function of both Mce1 and Mce4 indicates that it may be possible to develop compounds that disable both transporters simultaneously.
ROLE OF CAMP IN CHOLESTEROL UTILIZATION
We recently identified a novel class of compounds that stimulated 3΄,5΄-cyclic adenosine monophosphate (cAMP) production in M. tuberculosis by activating the bacterial adenylyl cyclase Rv1625c/Cya (VanderVen et al.2015). Activating cAMP production with these compounds blocks cholesterol utilization in M. tuberculosis and inhibits bacterial replication in macrophages. Although the mechanism explaining how cAMP blocks cholesterol utilization in M. tuberculosis is still incomplete, our analyses indicate that cAMP blocks early stages in the cholesterol degradation pathway, and that this metabolic blockade is responsible for the inhibition of bacterial replication in macrophages (VanderVen et al.2015).
Mycobacterium tuberculosis is capable of producing at least 10 active adenylyl cyclases, including Rv1625/Cya, all of which convert ATP into cAMP (Knapp and McDonough 2014). These adenylyl cyclases are diverse structurally, and each protein contains a unique-sensing domain. The putative-sensing domains of these proteins are thought to respond to environmental stimuli or ligands which control adenylyl cyclase activity (Agarwal and Bishai 2009; Bai, Knapp and McDonough 2011). The natural ligand or stimuli that activate Rv1625/Cya is unknown, but our model suggests that Rv1625/Cya can be activated by a small molecule that binds at one of two active sites in the enzyme complex (Johnson et al.2017). Because elevated levels of cAMP can impact central metabolism (Xu, Hegde and Blanchard 2011; Lee et al.2012; Knapp et al.2015; VanderVen et al.2015), transcription (Kahramanoglou et al.2014), pathogenicity (Agarwal et al.2009), dormancy (Shleeva et al.2013) and stress responses (Choudhary, Bishai and Agarwal 2014) in M. tuberculosis, activating production of cAMP could be a novel approach to target bacterial subpopulations that reside within macrophages.
CONCLUDING REMARKS
Recent insights into the lipid utilization pathways of M. tuberculosis have introduced some new lines of questioning. Given that Mce1 and Mce4 import fatty acids and cholesterol, respectively, it is likely that Mce2 and Mce3 also import hydrophobic lipid substrates that are yet unknown. The finding that the fatty acid and cholesterol importers rely on common proteins (Rv3723/LucA and Rv0655/MceG) suggests that additional bottlenecks of these two pathways exist, which could be targeted with drugs to disable multiple processes simultaneously. Mycobacterium tuberculosis causes a heterogeneous disease that involves various organs, cell types, immune responses and tissue pathology (Cadena, Fortune and Flynn 2017). It will be exciting to determine how M. tuberculosis utilizes lipid nutrients in different environments, cell type and time points during infection, ultimately to understand how the immune response constrains or modulates bacterial lipid metabolism and the available nutrients. This is an exciting time to continue investigating fundamental aspects of the physiology and pathogenesis of M. tuberculosis. With new insights and tools, we are optimistic that novel approaches and findings from current M. tuberculosis research will apply the brakes to slow or to stop the well-greased wheels of M. tuberculosis pathogenesis.
FUNDING
This work was supported by the NIH grants (AI119122 and AI130018) to BCV.
Conflict of interest. None declared.
REFERENCE
- Agarwal N, Bishai WR. cAMP signaling in Mycobacterium tuberculosis. Indian J Exp Biol 2009;47:155–68. [PubMed] [Google Scholar]
- Agarwal N, Lamichhane G, Gupta R et al. . Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature 2009;460:98–102. [DOI] [PubMed] [Google Scholar]
- Arruda S, Bomfim G, Knights R et al. . Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 1993;261:1454–7. [DOI] [PubMed] [Google Scholar]
- Astarie-Dequeker C, Le Guyader L, Malaga W et al. . Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog 2009;5:e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Knapp GS, McDonough KA. Cyclic AMP signalling in mycobacteria: redirecting the conversation with a common currency. Cell Microbiol 2011;13:349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczak AK, Avraham R, Singh S et al. . Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 2017;13:e1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 2003;83:91–7. [DOI] [PubMed] [Google Scholar]
- Cadena AM, Fortune SM, Flynn JL. Heterogeneity in tuberculosis. Nat Rev Immunol 2017;17:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho LR, Ensergueix D, Perez E et al. . Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 1999;34:257–67. [DOI] [PubMed] [Google Scholar]
- Cambier CJ, Takaki KK, Larson RP et al. . Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 2014;505:218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell SA, Leavell MD, Marjanovic O et al. . Free mycolic acid accumulation in the cell wall of the mce1 operon mutant strain of Mycobacterium tuberculosis. J Microbiol 2013;51:619–26. [DOI] [PubMed] [Google Scholar]
- Capyk JK, Casabon I, Gruninger R et al. . Activity of 3-ketosteroid 9alpha-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J Biol Chem 2011;286:40717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capyk JK, D’Angelo I, Strynadka NC et al. . Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem 2009;284:9937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capyk JK, Kalscheuer R, Stewart GR et al. . Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. J Biol Chem 2009;284:35534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carere J, McKenna SE, Kimber MS et al. . Characterization of an aldolase-dehydrogenase complex from the cholesterol degradation pathway of Mycobacterium tuberculosis. Biochemistry 2013;52:3502–11. [DOI] [PubMed] [Google Scholar]
- Casabon I, Crowe AM, Liu J et al. . FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol Microbiol 2013a;87:269–83. [DOI] [PubMed] [Google Scholar]
- Casabon I, Swain K, Crowe AM et al. . Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J Bacteriol 2014;196:579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabon I, Zhu SH, Otani H et al. . Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol Microbiol 2013b;89:1201–12. [DOI] [PubMed] [Google Scholar]
- Casali N, Konieczny M, Schmidt MA et al. . Invasion activity of a Mycobacterium tuberculosis peptide presented by the Escherichia coli AIDA autotransporter. Infect Immun 2002;70:6846–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 2007;8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Harik NS, Liao RP et al. . Identification of mycobacterial genes that alter growth and pathology in macrophages and in mice. J Infect Dis 2007;196:788–95. [DOI] [PubMed] [Google Scholar]
- Chang JC, Miner MD, Pandey AK et al. . igr Genes and Mycobacterium tuberculosis cholesterol metabolism. J Bacteriol 2009;191:5232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S, Ehrt S, Kawamura I et al. . Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell Microbiol 2001;3:247–54. [DOI] [PubMed] [Google Scholar]
- Choudhary E, Bishai W, Agarwal N. Expression of a subset of heat stress induced genes of Mycobacterium tuberculosis is regulated by 3',5'-cyclic AMP. PLoS One 2014;9:e89759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J et al. . Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537–44. [DOI] [PubMed] [Google Scholar]
- Comas I, Coscolla M, Luo T et al. . Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 2013;45:1176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M et al. . Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 1999;402:79–83. [DOI] [PubMed] [Google Scholar]
- Crowe AM, Casabon I, Brown KL et al. . Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other Bacteria. mBio 2017;8:pii:e00321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Deb C, Dubey VS et al. . Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 2004;186:5017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Maamar H, Deb C et al. . Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 2011;7:e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TA, Mittler JE, Nixon MR et al. . Mycobacterium tuberculosis strains lacking surface lipid phthiocerol dimycocerosate are susceptible to killing by an early innate host response. Infect Immun 2014;82:5214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresen C, Lin LY, D’Angelo I et al. . A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J Biol Chem 2010;285:22264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll MD, McLean KJ, Levy C et al. . Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J Biol Chem 2010;285:38270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy KY, Senaratne RH, Masuzawa M et al. . Attenuation of Mycobacterium tuberculosis functionally disrupted in a fatty acyl-coenzyme A synthetase gene fadD5. J Infect Dis 2010;201:1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Isom GL et al. . Architectures of lipid transport systems for the bacterial outer membrane. Cell 2017;169:273–85.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. P Natl Acad Sci USA 2013;110:6554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H, Rhee KY. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. P Natl Acad Sci USA 2014;111:4976–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelopoulos D, da Fonseca JD, Waddell SJ. Understanding anti-tuberculosis drug efficacy: rethinking bacterial populations and how we model them. Int J Infect Dis 2015;32:76–80. [DOI] [PubMed] [Google Scholar]
- Feltcher ME, Gunawardena HP, Zulauf KE et al. . Label-free quantitative proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Mol Cell Proteomics 2015;14:1501–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrellad MA, McNeil M, Santangelo Mde L et al. . Role of the Mce1 transporter in the lipid homeostasis of Mycobacterium tuberculosis. Tuberculosis 2014;94:170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DJ, Madrona Y, Ortiz de Montellano PR. Cholesterol ester oxidation by mycobacterial cytochrome P450. J Biol Chem 2014;289:30417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzblau SG. Oxidation of palmitic acid by Mycobacterium leprae in an axenic medium. J Clin Microbiol 1988;26:18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Minch K, Peterson M et al. . The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 2013;499:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur RL, Ren K, Blumenthal A et al. . LprG-mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 2014;10:e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S, Hood L, Seah SYK. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis. J Bacteriol 2017;200:pii:e00512-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioffre A, Infante E, Aguilar D et al. . Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect 2005;7:325–34. [DOI] [PubMed] [Google Scholar]
- Gouzy A, Poquet Y, Neyrolles O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat Rev Microbiol 2014;11:729–37. [DOI] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, Dejesus MA et al. . High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 2011;7:e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Pandey AK, Gilmore SA et al. . Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 2012;19:218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho NA, Dawes SS, Crowe AM et al. . The structure of the transcriptional repressor KstR in complex with CoA thioester cholesterol metabolites sheds light on the regulation of cholesterol catabolism in Mycobacterium tuberculosis. J Biol Chem 2016;291:7256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, van der Geize R, Besra GS et al. . 3-Ketosteroid 9alpha-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol Microbiol 2010;75:107–21. [DOI] [PubMed] [Google Scholar]
- Isom GL, Davies NJ, Chong ZS et al. . MCE domain proteins: conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci Rep 2017;7:8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis 2007;87:78–86. [DOI] [PubMed] [Google Scholar]
- Jain M, Petzold CJ, Schelle MW et al. . Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. P Natl Acad Sci USA 2007;104:5133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Bai G, DeMott CM et al. . Chemical activation of adenylyl cyclase Rv1625c inhibits growth of Mycobacterium tuberculosis on cholesterol and modulates intramacrophage signaling. Mol Microbiol 2017;105:294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SM, Pandey AK, Capite N et al. . Characterization of mycobacterial virulence genes through genetic interaction mapping. P Natl Acad Sci USA 2006;103:11760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Cortes T, Matange N et al. . Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res 2014;42:8320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall SL, Withers M, Soffair CN et al. . A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol 2007;65:684–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Wainwright HC, Locketz M et al. . Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med 2010;2:258–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirksey MA, Tischler AD, Siméone R et al. . Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect Immun 2011;79:2829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp GS, Lyubetskaya A, Peterson MW et al. . Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res 2015;43:5377–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp GS, McDonough KA. Cyclic AMP signaling in mycobacteria. Microbiol Spectr 2014;2:MGM2-001102013. [DOI] [PubMed] [Google Scholar]
- Knol J, Bodewits K, Hessels GI et al. . 3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J 2008;410:339–46. [DOI] [PubMed] [Google Scholar]
- Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. P Natl Acad Sci USA 2011;108:11614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithika R, Marathe U, Saxena P et al. . A genetic locus required for iron acquisition in Mycobacterium tuberculosis. P Natl Acad Sci USA 2006;103:2069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack NA, Yam KC, Lowe ED et al. . Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J Biol Chem 2010;285:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre E, Sweet L, Hong S et al. . A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol 2011;18:1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lang PT, Fortune SM et al. . Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat Struct Mol Biol 2012;19:811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, VanderVen BC, Fahey RJ et al. . Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 2013;288:6788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Schmitz W, Sampson NS. alpha-Methyl Acyl CoA racemase provides Mycobacterium tuberculosis catabolic access to cholesterol esters. Biochemistry 2015;54:5669–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann JR, McDonough JA, Sullivan JT et al. . Genome-wide identification of Mycobacterium tuberculosis exported proteins with roles in intracellular growth. J Bacteriol 2011;193:854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ et al. . Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 2000;406:735–8. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. P Natl Acad Sci USA 2009;106:8009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H, Laneelle MA, Daffe M. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 2014;21:67–85. [DOI] [PubMed] [Google Scholar]
- Martinot AJ, Farrow M, Bai L et al. . Mycobacterial metabolic syndrome: LprG and Rv1410 regulate triacylglyceride levels, growth rate and virulence in Mycobacterium tuberculosis. PLoS Pathog 2016;12:e1005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A, Cordes T, Ghelfi J et al. . Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. P Natl Acad Sci USA 2013;110:7820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn WW, van der Geize R, Stewart GR et al. . The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem 2008;283:35368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 2005;11:638–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, Upton AM, Cherian J et al. . Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol 2006;60:1109–22. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Zhang-Akiyama QM. pqiABC and yebST, putative mce operons of escherichia coli, encode transport pathways and contribute to membrane integrity. J Bacteriol 2017;199:pii:e00606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarova EV, Montague CR, La T et al. . Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 2017;6:pii:e26969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt NM, Yang X, Fontán P et al. . A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect Immun 2010;78:275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odriozola JM, Ramos JA, Bloch K. Fatty acid synthetase activity in Mycobacterium smegmatis characterization of the acyl carrier protein-dependent elongating system. BBA- Lipid Lipid Met 1977;488:207–17. [DOI] [PubMed] [Google Scholar]
- Ouellet H, Guan S, Johnston JB et al. . Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol Microbiol 2010;77:730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M, Behr MA, Dowdy D et al. . Tuberculosis. Nat Rev Dis Primers 2016;2:16076. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. P Natl Acad Sci USA 2008;105:4376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkowski EF, Miller BK, McCann JR et al. . An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol Microbiol 2016;100:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portevin D, De Sousa-D’Auria C, Houssin C et al. . A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. P Natl Acad Sci USA 2004;101:314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri LE. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit Rev Biochem Moll 2014;49:179–211. [DOI] [PubMed] [Google Scholar]
- Queiroz A, Medina-Cleghorn D, Marjanovic O et al. . Comparative metabolic profiling of mce1 operon mutant vs wild-type Mycobacterium tuberculosis strains. Pathog Dis 2015;73:ftv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley J, Hughitt VK, Velikovsky CA et al. . The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. PLoS Pathog 2017;8:pii:e00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater DL, Kolattukudy PE. Synthesis of mycocerosic acids from methylmalonyl coenzyme A by cell-free extracts of Mycobacterium tuberculosis var. bovis BCG. J Biol Chem 1983;258:2979–85. [PubMed] [Google Scholar]
- Ramon-Garcia S, Stewart GR, Hui ZK et al. . The mycobacterial P55 efflux pump is required for optimal growth on cholesterol. Virulence 2015;6:444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Winter N, Pivert E et al. . Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell Microbiol 2004;6:277–87. [DOI] [PubMed] [Google Scholar]
- Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev 2011;240:252–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG, Cardona PJ, Kim MJ et al. . Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol 2009;10:943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain MJ, Dick DL, Rithner CD et al. . Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J Lipid Res 2011;52:861–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. P Natl Acad Sci USA 2003;100:12989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvi S, Warner DF, Kana BD et al. . Functional characterization of a vitamin B12-Dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J Bacteriol 2008;190:3886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal W, Bloch H. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J Bacteriol 1956;72:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senaratne RH, Sidders B, Sequeira P et al. . Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol 2008;57:164–70. [DOI] [PubMed] [Google Scholar]
- Sequeira PC, Senaratne RH, Riley LW. Inhibition of toll-like receptor 2 (TLR-2)-mediated response in human alveolar epithelial cells by mycolic acids and Mycobacterium tuberculosis mce1 operon mutant. Pathog Dis 2014;70:132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono N, Morici L, Casali N et al. . Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. P Natl Acad Sci USA 2003;100:15918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleeva M, Goncharenko A, Kudykina Y et al. . Cyclic AMP-dependent resuscitation of dormant Mycobacteria by exogenous free fatty acids. PLoS One 2013;8:e82914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GR, Patel J, Robertson BD et al. . Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog 2005;1:269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin HA, Shi H, May KL et al. . Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. P Natl Acad Sci USA 2016;113:E1565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev 2005;18:81–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ST, Sampson NS. Mycobacterium tuberculosis utilizes a unique heterotetrameric structure for dehydrogenation of the cholesterol side chain. Biochemistry 2013;52:2895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ST, VanderVen BC, Sherman DR et al. . Pathway Profiling in Mycobacterium tuberculosis: elucidation of a cholesterol-derived catabolite and the enzymes that catalyze its metabolism. J Biol Chem 2011;286:43668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong S, Ercan B, Torta F et al. . Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife 2016;5:pii:e19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemersma EW, van der Werf MJ, Borgdorff MW et al. . Natural history of tuberculosis: duration and fatality of untreated pulmonary tuberculosis in HIV negative patients: a systematic review. PLoS One 2011;6:e17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Sridharan V et al. . Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 2004428:441–5. [DOI] [PubMed] [Google Scholar]
- Upton AM, McKinney JD. Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 2007;153:3973–82. [DOI] [PubMed] [Google Scholar]
- Van der Geize R, Yam K, Heuser T et al. . A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. P Natl Acad Sci USA 2007;104:1947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVen BC, Fahey RJ, Lee W et al. . Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS Pathog 2015;11:e1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A, Bryk R, Shi S et al. . Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe 2011;9:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiker HG, Spierings E, Kolkman MA et al. . The mammalian cell entry operon 1 (mce1) of Mycobacterium leprae and Mycobacterium tuberculosis. Microb Pathog 1999;27:173–7. [DOI] [PubMed] [Google Scholar]
- Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep 2015;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol 2014;49:269–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipperman MF, Yang M, Thomas ST et al. . Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an alpha2beta2 heterotetrameric acyl coenzyme A dehydrogenase family. J Bacteriol 2013;195:4331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Hildebrand F, Allix-Béguec C et al. . Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 2008;4:e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Global Tuberculosis Report. WHO, 2017. http://www.who.int/tb/publications/global_report/en/. [Google Scholar]
- Xu H, Hegde SS, Blanchard JS. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 2011;50:5883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam KC, D’Angelo I, Kalscheuer R et al. . Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog 2009;5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam KC, Okamoto S, Roberts JN et al. . Adventures in Rhodococcus - from steroids to explosives. Can J Microbiol 2011;57:155–68. [DOI] [PubMed] [Google Scholar]
- Yang M, Guja KE, Thomas ST et al. . A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem Biol 2014;9:2632–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lu R, Guja KE et al. . Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha2beta2 Acyl-CoA dehydrogenase initiates beta-oxidation of 3-oxo-cholest-4-en-26-oyl CoA. ACS Infect Dis 2015;1:110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Dubnau E, Smith I et al. . Rv1106c from Mycobacterium tuberculosis is a 3beta-hydroxysteroid dehydrogenase. Biochemistry 2007;46:9058–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Nesbitt NM, Dubnau E et al. . Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 2009;48:3819–21. [DOI] [PMC free article] [PubMed] [Google Scholar]