Abstract
An infant presented with fatal infantile lactic acidosis and cardiomyopathy, and was found to have profoundly decreased activity of respiratory chain complex I in muscle, heart and liver. Exome sequencing revealed compound heterozygous mutations in NDUFB10, which encodes an accessory subunit located within the PD part of complex I. One mutation resulted in a premature stop codon and absent protein, while the second mutation replaced the highly conserved cysteine 107 with a serine residue. Protein expression of NDUFB10 was decreased in muscle and heart, and less so in the liver and fibroblasts, resulting in the perturbed assembly of the holoenzyme at the 830 kDa stage. NDUFB10 was identified together with three other complex I subunits as a substrate of the intermembrane space oxidoreductase CHCHD4 (also known as Mia40). We found that during its mitochondrial import and maturation NDUFB10 transiently interacts with CHCHD4 and acquires disulfide bonds. The mutation of cysteine residue 107 in NDUFB10 impaired oxidation and efficient mitochondrial accumulation of the protein and resulted in degradation of non-imported precursors. Our findings indicate that mutations in NDUFB10 are a novel cause of complex I deficiency associated with a late stage assembly defect and emphasize the role of intermembrane space proteins for the efficient assembly of complex I.
Introduction
Mitochondrial diseases are disorders caused by deficient mitochondrial bioenergetics due to genetic compromise of the activities of the respiratory chain complexes. The respiratory chain is composed of five complexes, and deficiencies can affect either a single complex or a combination of several complexes when the genetic cause affects overall mitochondrial biogenesis. Deficiencies in complex I (nicotinamide adenine dinucleotide (NADH):ubiquinone oxidoreductase, EC 1.6.5.3) represent the most common cause of an isolated primary complex deficiency. Isolated complex I deficiency (OMIM 252010) is a clinically heterogeneous disorder, identified in approximately 30% of patients with mitochondrial disease (1,2). Common clinical presentations include optic atrophy (OMIM 53500) and mitochondrial encephalopathy with lactic acidosis and stroke-like episodes (OMIM 540000) particularly for patients with mitochondrial DNA encoded genetic defects. Other common clinical syndromes, particularly with nuclear encoded genetic defects, include Leigh disease (OMIM 256000), leukoencephalopathy, neonatal cardiomyopathy, and fatal infantile lactic acidosis with multisystem disease (2–7).
Mitochondrial complex I is large and complex with a combined mass of 1 MDa. The holoenzyme is comprised of 44 unique subunits of which 7 subunits are encoded by the mitochondrial genome, and 37 subunits are encoded by the nuclear genome (8). It forms an L-shaped complex with a hydrophilic core that protrudes into the mitochondrial matrix and a hydrophobic core that resides within the mitochondrial membrane (9). It is functionally divided into three modules (9,10). The N-module is present in the matrix-protruding arm and is responsible for the oxidation of NADH into NAD+ with the transfer of electrons over the flavin and iron-sulfur centers to the final N2-iron-sulfur center. The Q module, located at the interface of the membrane and matrix-arm, transfers these electrons further into the ubiquinone reduction site, which includes the ND1 subunit (11,12). The membrane embedded P module contains the remaining mitochondrial DNA encoded subunits and links NADH oxidation to proton translocation from the matrix into the intermembrane space (IMS) via a conformational mechanism (7,9,10,13). This P-module is subdivided into two domains, referred to as the proximal PP domain which contains ND2, ND3, ND6 and ND4L, and the distal domain PD, which contains ND4 and ND5, making up the proton transporter pathways (9,10,12,14). The catalytic core of complex I, which in prokaryotes is able to fulfill the complete function of complex I, comprises the mtDNA encoded subunits and seven nuclear encoded subunits (10). The remaining 30 subunits are referred to as accessory subunits whose presumed role in the activity, assembly, stabilization or in protection from reactive oxygen species is poorly understood (7,15,16).
The biogenesis of complex I has been unraveled by studies of genetic deficiencies (16,17), although some controversies remain. The assembly occurs in a stepwise process with complex I subassemblies residing in the mitochondrial matrix and inner membrane (18). First a subcomplex of NDUFS2, NDUFS3, and NDUFS8 join via NDUFS7 with ND1 present in the membrane to form a 400 kDa subcomplex of the Q module. This subcomplex then acquires additional subunits of the PP module including ND2, ND3, ND4L, ND6 and NDUFB6 to form a 460 kDa subcomplex. Next ancillary subunits are added such as NDUFB8, NDUFA9, and NDUFA10, followed by the addition of a subcomplex of the N-module consisting of NDUFV1, NDUFV2, NDUFV3, NDUFS1, NDUFS4, NDUFS6, and NDUFA12 to form an 830 kDa subcomplex. Subunits of the PD module such as ND4 and ND5 are added either before the 830 kDa subcomplex (19) or after the 830 kDa subcomplex (16), to a final assembled complex at 980 kDa (19).
The extensive number of genetic defects responsible for complex I-related disease lends itself to next-generation sequencing. Exome sequencing in individuals with complex I deficiency has provided an increased rate of genetic diagnosis and led to the identification of disease-associated genes (20–22). Genetic defects in each of the mtDNA encoded subunits and in 26 nuclear encoded subunits of complex I (14 core subunits and 18 accessory subunits) have been associated with mitochondrial disease (5,8,23,24). Mutations in nine chaperones of complex I assembly factors have been associated with human disease (5,8). Genotype-phenotype relations have been difficult to make. Mutations in genes associated with human disease have been identified in nearly all genes of subunits of both the N and the Q module, but in contrast, few mutations in genes in the accessory subunits of the P module have been associated with human disease: only 4 of 9 subunits associated with the PP module and 3 of 12 accessory subunits of the PD module, namely NDUFB3, NDUFB9 and NDUFB11 (8,20,21,24–26). Little is known about the function of the accessory subunits of the PD module.
In this study, we describe a newborn who had a prenatal diagnosis of cardiomyopathy and presented with fatal infantile lactic acidosis. Following a metabolic autopsy, isolated reduced complex I activity was identified in muscle, heart and liver tissue. Using trio-based exome sequencing, we identified compound heterozygous mutations in NDUFB10, which encodes an accessory subunit located within the hydrophobic arm of complex I. NDUFB10 is one of four complex I subunits that have been proposed as substrates of the intermembrane space (IMS) oxidoreductase CHCHD4 (also Mia40) (27–29). We confirm this and demonstrate that the identified cysteine mutation in NDUFB10 prevents its efficient CHCHD4-dependent mitochondrial import and oxidative protein folding. Evidence is presented that implicates this accessory subunit as required for complex I assembly.
Case Report
This female patient was the second child born of non-consanguineous parents of Irish and German descent. The first child, a full brother, had prenatal hydrops, died on the first day of life from a cardiac defect, and reportedly had Noonanoid features. This second child was born at 36 weeks gestation by planned Cesarean section following a pregnancy complicated by fetal cardiomyopathy, non-immune hydrops, lung hypoplasia, and symmetric intrauterine growth retardation. Fetal echocardiograms showed moderate cardiac enlargement and biventricular dysfunction with depressed output. At birth, she had excessive nuchal skin, but otherwise had a normal exam including normal tone. Due to hypoxia she was transferred to the neonatal intensive care unit, where bilateral pneumothoraxes and pulmonary hypertension were noted. She was intubated and bilateral chest tubes were placed which initially improved the patient’s respiratory status. Her postnatal echocardiogram revealed severe pulmonary hypertension, low-normal systolic function of the left ventricle, and hypertrophy of both the left and right myocardium. She developed a rapidly progressive metabolic acidosis, refractory hypotension, and a notable decline in cardiac systolic function by repeat echocardiography. She had rapidly worsening lactic acidosis, unresponsive to treatment with biotin, thiamin, and riboflavin, starting at three hours with a lactic acidosis of 6.89 mM (range 0.5–2.0 mM) and gradually increasing to a peak lactate of 29.19 mM, with a simultaneous lactate-to-pyruvate ratio of 33. Plasma amino acid analysis showed elevated alanine 1403 µM (range 131–710 µM) and proline 1034 µM (range 110–417 µM). Urine organic acid analysis showed increased lactate and 3-hydroxybutyric, 2-hydroxybutyric, and glyoxalic species. An acylcarnitine profile identified elevations of long-chain acylcarnitines and long-chain 3-hydroxyacylcarnitines in a nonspecific pattern. Due to lack of therapeutic response, persistent acidosis, and overall poor prognosis, life sustaining measures were withdrawn and the patient died at 27 hours of life. Following identification of decreased complex I activity, clinical sequencing of mitochondrial DNA and of a panel of complex I deficiency related genes was performed. Sequencing of mtDNA in skeletal muscle noted a rare homoplasmic variant in MTND5, m.13762T > G, p.S476A, which had been previously reported as a common polymorphism in the Finnish population (30,31), and maternal blood testing noted that the subject’s asymptomatic mother was also homoplasmic for this variant. Next-generation sequencing of nuclear genes associated with complex I deficiency was negative for the following genes: C20ORF7, FOXRED1, NDUFA1, NDUFA10, NDUFA11, NDUFA13, NDUFA2, NDUFA7, NDUFA8, NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, NDUFB6, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS5, NDUFS6, NDUFS7, NDUFS8, NDUFV1, NDUFV3, NUBPL.
Results
Pathology findings
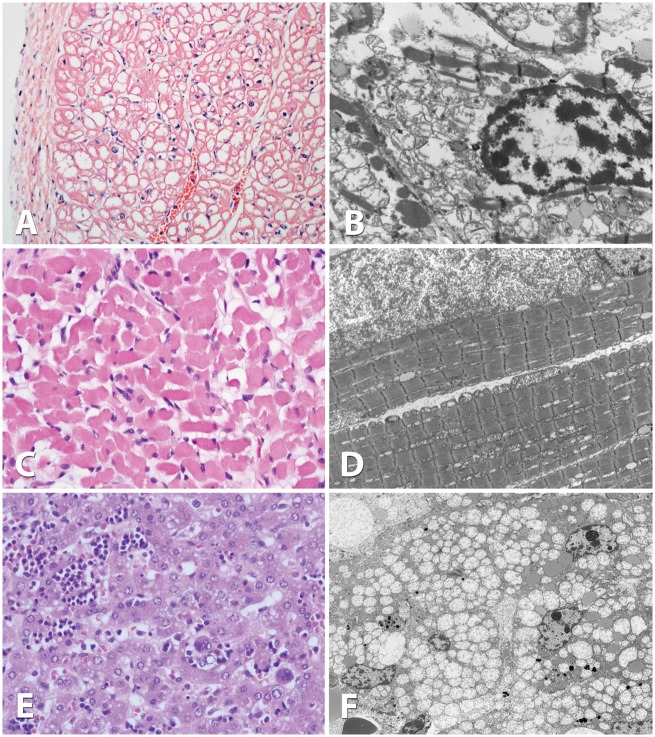
Light microscopic examination of cardiac muscle revealed decreased contractile elements and mildly increased lipids, and liver showed moderate microvesicular steatosis, whereas skeletal muscle appeared normal (Fig. 1A,C,E). Electron microscopy of muscle did not show major alterations, but cardiac muscle revealed extensive proliferation of mitochondria displacing the contractile elements and mild increase in lipid, and liver showed extensive proliferation of mitochondria, often enlarged and occupying a significant portion of the cytoplasm of the hepatocytes in addition to some microvesicular steatosis (Fig. 1B,D,F). In all three tissues, the mitochondria seemed rarefied with decreased cristae, appeared most likely to be due to the pathology, although a postmortem artifact could not be excluded.
Figure 1.
Pathology of skeletal muscle, heart muscle, and liver tissue. Light microscopy examination using eosin-hematoxylin staining of cardiac muscle at 20x magnification (A), skeletal muscle at 40x (C) and liver at 40x (E). Electron microscopic evaluation of cardiac muscle at 2000x magnification (B), skeletal muscle at 2000x (D), and liver at 1500x (F). Whereas light microscopy did not reveal substantive changes, electron microscopy showed extensive proliferation of mitochondria displacing cytosolic elements in heart, muscle and liver, and mitochondria appear rarefied with poor cristae in all three tissues.
Analysis of the respiratory chain enzyme activities revealed isolated complex I deficiency
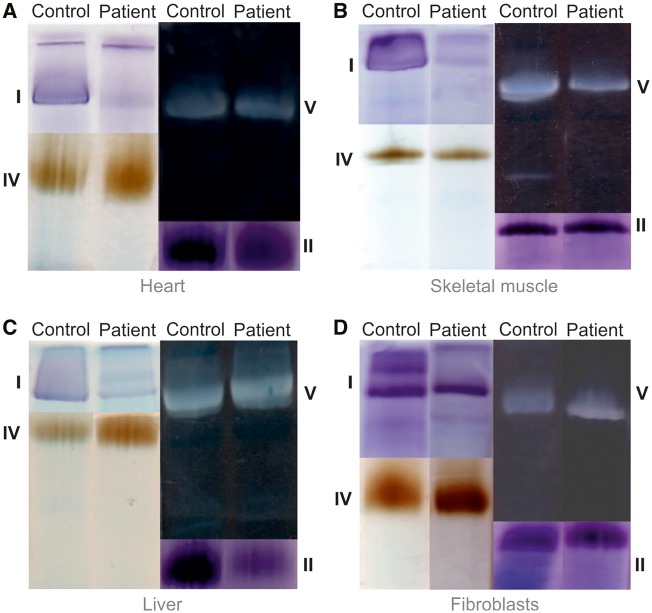
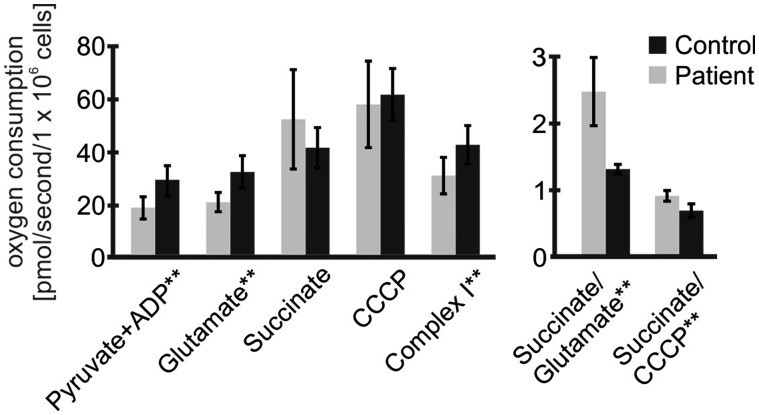
Respiratory chain enzyme analysis revealed an isolated complex I deficiency in skeletal muscle and heart and complex I and complex II deficiencies in the liver, the latter possibly due to postmortem artifact (Table 1). In contrast, enzyme activities in fibroblasts were normal. Likewise, blue native polyacrylamide gel electrophoresis (BN-PAGE) with in-gel activity staining showed near absent complex I activity in muscle, heart and liver, whereas the assembly and activities of complexes II, IV, and V appeared normal in comparison to the intra-assay control (Fig. 2). In fibroblasts enzyme activities and complex assembly appeared normal with this assay. However, in permeabilized fibroblasts high-resolution respirometry revealed a deficiency in complex I derived respiratory capacity (Fig. 3, Supplementary Material, Table S2). The oxygen consumption rate in stage 3 under ADP stimulation with the addition of complex I substrate pyruvate was significantly decreased in the patient and remained decreased with added glutamate. The succinate/glutamate ratio (Q junction) was significantly increased indicating greater than the normal flux through complex II compared to complex I. Rotenone-inhibited isolated maximal complex I respiratory capacity was significantly reduced. These results are consistent with isolated complex I deficiency in fibroblasts identified using high-resolution respirometry, which proved a sensitive indicator.
Table 1.
Respiratory chain enzyme activities in multiple tissues
| Tissue/complex | Activity� (controls) | SD | Ratio/CS� (controls) | SD | Ratio/complex II� (controls) | SD |
|---|---|---|---|---|---|---|
| MUSCLE | ||||||
| Complex I | 5�.0� (23�.6–74�.8) | −6�.0 | 37� (98–271) | −6�.5 | 67� (285–767) | −7�.2 |
| Complex II | 74�.2� (49�.0–133�.4) | −0�.4 | 550� (251–573) | 1�.4 | NA | NA |
| Complex III | 16�.1� (5�.7–31�.4) | 0�.2 | 119� (19–172) | 0�.7 | 216� (45–369) | 0�.3 |
| Complex II–III | 101�.6� (34�.2–107�.6) | 1�.4 | 753� (172–472) | 0�.3 | 1370� (549–1226) | 2�.1 |
| Complex IV | 1�.5� (1�.1–3�.8) | −1�.1 | 11� (4–23) | −1�.0 | 20� (11–68) | −1�.1 |
| Citrate synthase | 134�.9� (159�.8–353�.3) | −2�.3 | NA | NA | NA | NA |
| HEART | ||||||
| Complex I | 5�.9� (146�.9–366�.9) | −11�.6 | 5� (144–376) | −12�.5 | 14� (506–1064) | −13�.7 |
| Complex II | 431�.7� (176�.4–412�.5) | 1�.4 | 387� (235–356) | 2�.3 | NA | NA |
| Complex III | 208�.7� (16�.5–260�.4) | 1�.3 | 187� (12–210) | 1�.1 | 483� (45–680) | 0�.9 |
| Complex II–III | 467�.3(179�.2–546�.0) | 1�.2 | 418� (189–433) | 1�.3 | 1082� (680–1468) | 0�.4 |
| Complex IV | 17�.0� (12�.0–32�.0) | −0�.4 | 15� (11–32) | −0�.5 | 39� (36–100) | −1�.0 |
| Citrate synthase | 1117� (497–1473) | 1�.6 | NA | NA | NA | NA |
| LIVER | ||||||
| Complex I | 7�.3� (14�.4–56�.0) | −3�.0 | 76� (162–730) | −3�.1 | 60� (68–252) | −2�.6 |
| Complex II | 122�.2� (174�.7–309�.8) | −2�.4 | 1264� (2304–3311) | −4�.3 | NA | NA |
| Complex III | 26�.4� (13�.8–27�.6) | 0�.9 | 273� (128–315) | 0�.6 | 216� (50–118) | 1�.9 |
| Complex II–III | 146�.7� (10�.8–107�.3) | 1�.5 | 1519� (138–1062) | 1�.5 | 1202� (62–383) | 2�.7 |
| Complex IV | 4�.5� (0�.5–3�.2) | 1�.9 | 47� (6–35) | 1�.8 | 37� (3–13) | 2�.3 |
| Citrate synthase | 96�.5� (59�.5–109�.3) | 0�.6 | NA | NA | NA | NA |
| FIBROBLAST | ||||||
| Complex I | 79�.4� (48�.9–120�.1) | 0�.0 | 133� (151–387) | −1�.3 | 181� (255–539) | −1�.9 |
| Complex II | 438�.2� (144�.4–349�.2) | 2�.1 | 736� (471–919) | 0�.8 | NA | NA |
| Complex III | 9�.0� (7�.92–30�.38) | −1�.0 | 15� (21–79) | −1�.5 | 20� (37–118) | −1�.6 |
| Complex II–III | 188�.3� (102�.1–210�.6) | 0�.9 | 316� (265–571) | −0�.6 | 430� (437–812) | −1�.5 |
| Complex IV | 10�.8� (2�.13–6�.84) | 2�.3 | 18� (6–25) | 1�.9 | 25� (12–37) | 1�.8 |
| Citrate synthase | 595�.5� (245�.4–540�.4) | 1�.6 | NA | NA | NA | NA |
Legend: Activities with range of control values are provided�. The patient value is listed as the Z-score expressed in SD of the log transformed normal value population, with bolded values outside 2 SD�. CS is citrate synthase�.
Figure 2.
In-gel activity staining reveals isolated complex I deficiency. Heart (A), skeletal muscle (B), liver (C), and fibroblasts (D) of patient and of a control sample were homogenized, and solubilized respiratory chain enzyme complexes from a mitochondrial membrane pellet were separated on a non-denaturing blue native polyacrylamide gel and the activity assessed with in gel activity stains. In comparison to control samples, the patient had decreased complex I activity in muscle, heart, and liver, whereas activity appeared normal in fibroblasts. The activities of complexes II, IV and V were normal.
Figure 3.
High-resolution respirometry in fibroblasts reveals mild complex I defects. Results of high-resolution respirometry in fibroblasts from the patient (n = 5) compared to 20 control fibroblast samples. There are decreased parameters relating to complex I activity: when stimulated with ADP the rates with the substrates pyruvate and glutamate are decreased, as is the rotenone-inhibited component of the uncoupled rate, which is complex I specific. In contrast, the rate is excessively increased when a complex II substrate succinate is added as reflected in the succinate/glutamate ratio. * p < 0.05, ** p < 0.01 by Student t-testing.
Exome sequencing identified compound heterozygous mutations in NDUFB10
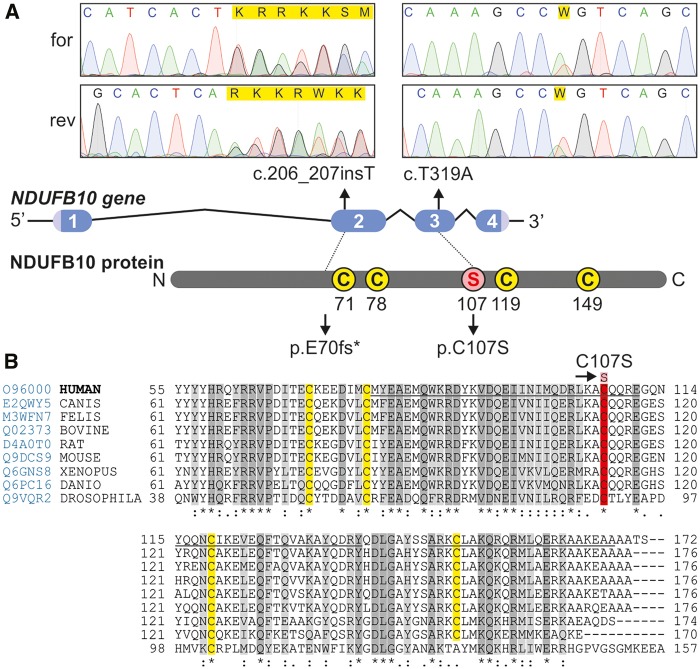
After clinical molecular testing failed to identify a genetic cause, exome sequencing was performed in the proband and unaffected parents, and analyzed under various inheritance models. Using a recessive inheritance model, we identified compound heterozygous, non-synonymous sequence variants in NDUFB10 (NADH-ubiquinone oxidoreductase 1 beta subcomplex 10), which were verified by Sanger sequencing (Fig. 4). The patient has a paternally inherited nonsense mutation c.206_207insT (p.E70*) and a maternally inherited missense mutation (c.319T > C, p.C107S). The nonsense mutation is predicted to result in a premature stop codon and absent protein, and to be pathogenic. Indeed, RT-PCR of fibroblast NDUFB10 mRNA showed only the presence of the c.319C allele (data not shown). The missense mutation replaces a highly conserved cysteine residue with a serine residue (SIFT score = 0, GERP score =5.39, conserved in all 60 species reviewed in Polyphen2) (Fig. 4). This mutation is not present in the Exome Aggregation Consortium database or 1000 genomes, and was predicted to be pathogenic by bioinformatic software (Polyphen2 = 1.0, Mutation-Taster = 0.9, Grantham Score =112). NDUFB10 (NM_004539.1) contains 4 exons and encodes for a 20.7 kDa protein (NP_004539.1). The variants were deposited in the Leiden Open Variation Database accessible at http://databases.lovd.nl/shared/genes/NDUFB10. This protein is 172 amino acids long and is located in the inner mitochondrial membrane within the hydrophobic arm (Iβ) of complex I (17). The NDUFB10 protein does not appear to have a mitochondrial import signal according to the bioinformatics programs MitoProt, Target-P, Predotar and Wolf-PSORT, but it does contain two atypical dicysteine motifs CX6C (Cys61-Cys78) and CX11C (Cys107-Cys119). Mutations within this subunit have not been previously reported and the role of this protein in complex I remains unknown. We therefore examined the impact of these mutations on NDUFB10 import, folding and homeostasis and on complex I assembly.
Figure 4.
Mutations in NDUFB10 and sequence alignment over multiple species. The position of the mutations in NDUFB10 is shown on a diagram of the gene, and respective Sanger sequencing results from the forward (for) and reverse (rev) sequencing are shown. The homology of NDUFB10 over multiple species (Homo sapiens, Canis lupus, Felis catus, Bos taurus, Rattus norvegicus, Mus musculus, Xenopus laevis, Danio rerio, Drosophila melanogaster) was built using Clustal Omega available online at uniprotkb, accessed at www.uniprot.org/align on 7/16/2016. Prediction of secondary structure helical motifs (underlined regions) was done according to Jpred4 prediction software accessed at www.compbio.dundee.ac.uk/jpred4 on 7/16/2016. Highly conserved amino acids are shown in dark grey and moderately conserved amino acids in light grey, whereas in yellow are shown the cysteines of the presumptive CXnC structures. The mutated amino acid C107S is shown in red.
NDUFB10 is a substrate of the IMS machinery for oxidation-dependent protein import into the mitochondrial intermembrane space
The conserved cysteine motifs of NDUFB10 and the absence of an N-terminal mitochondrial import signal implied import of NDUFB10 into the IMS by the oxidative folding machinery of the IMS (32–34). This machinery consists of two enzymes: CHCHD4 and ALR. CHCHD4 is an IMS-localized oxidoreductase that serves as import receptor and chaperone for small soluble proteins with conserved cysteine motifs. To function properly CHCHD4 has to be constantly reoxidized by the sulfhydryl oxidase ALR (augmenter of liver regeneration; in yeast Erv1) which eventually transfers electrons via cytochrome c into complex IV of the respiratory chain.
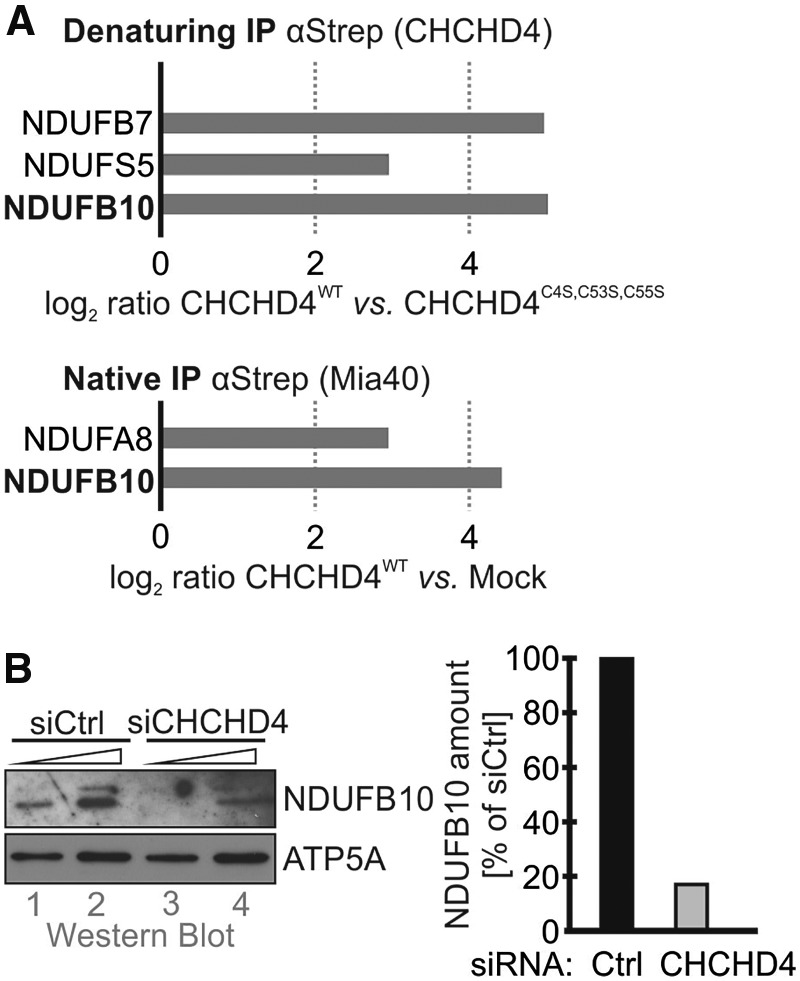
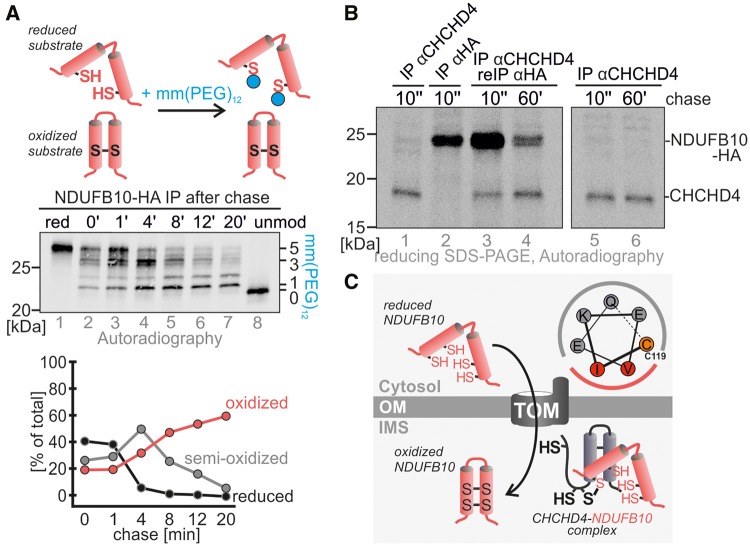
To facilitate the import, CHCHD4 interacts with cysteine residues and hydrophobic patches in its substrates, and promotes thereby directed diffusion of the protein over the outer membrane and prevents backsliding to the cytosol. During its action, CHCHD4 introduces disulfide bonds into its substrates. Earlier in silico studies and the recently determined interactome of CHCHD4 showed four subunits of complex I as probable CHCHD4 interaction partners: NDUFS5, NDUFA8, NDUFB7, and NDUFB10, all accessory subunits (Fig. 5A) (27–29). In line with a role of CHCHD4 in NDUFB10 import, we found a decrease in NDUFB10 levels upon siRNA-mediated knockdown of CHCHD4 (Fig. 5B). Such a correlation between CHCHD4 and levels of its substrates has been found for most of its substrates (35–38). We next investigated directly the maturation of NDUFB10. We tested whether it becomes oxidized during import and whether it interacts with CHCHD4 during this time. To follow oxidation of newly synthesized NDUFB10 we employed a pulse-chase-based oxidation assay (Fig. 6A). Directly after synthesis, the NDUFB10 protein was almost fully reduced (Fig. 6A, compare lanes 1 and 2). With kinetics comparable to other CHCHD4 substrates NDUFB10 then became oxidized within 20 minutes (27,39). In a second pulse-chase experiment we analyzed the disulfide-dependent interaction between CHCHD4 and NDUFB10 (Fig. 6B). This demonstrated that a large portion of both proteins interacted at a time when NDUFB10 acquired its disulfides, and that this interaction was to a large extent lost after NDUFB10 was fully oxidized (Fig. 6B, lanes 3 and 4). Taken together, these data show that NDUFB10 is imported and folded by CHCHD4, a process that relies on the cysteines in NDUFB10 (Fig. 6C). Interestingly, the predicted CHCHD4 interaction site in NDUFB10 (which is a cysteine residue in a helix flanked on the same face by hydrophobic residues) appears to localize to cysteine residue 119 (Fig. 6C). Thus, initial mixed disulfide formation with CHCHD4 likely does not involve C107.
Figure 5.
NDUFB10 is a substrate of CHCHD4. (A) NDUFB10 was detected in quantitative proteomics approaches as disulfide-linked interaction partner of CHCHD4. HEK293 cell lines stably expressing either CHCHD4-Strep wild type or the inactive C4S,C53S,C55S mutant were subjected to native and denaturing immunoprecipitation against the Strep tag. Subsequently, CHCHD4 interaction partners were identified by quantitative proteomics (data from (29)). (B) siRNA-mediated depletion of CHCHD4 results in decreased levels of NDUFB10. HeLa cells were transfected with siRNA directed against CHCHD4. 72 h after transfection cells were lysed and analyzed by immunoblot against NDUFB10. Depletion of CHCHD4 results in loss of NDUFB10.
Figure 6.
CHCHD4 oxidizes NDUFB10 during maturation. (A) NDUFB10 forms two disulfide bonds during its maturation. Proteins were radioactively labelled, and followed for different chase times. After the chase proteins were modified with the alkylating agent mmPEG12 to distinguish reduced (i.e. modifiable) from oxidized protein. Then, NDUFB10 was isolated by immunoprecipitation and the precipitate analyzed by SDS-PAGE and autoradiography. (B) CHCHD4 interacts with NDUFB10 during NDUFB10 maturation in pulse chase analysis. A denaturing immunoprecipitation against CHCHD4 was followed by a re-immunoprecipitation against NDUFB10, thus isolating CHCHD4-NDUFB10 complexes. Interaction between both proteins takes place very early during NDUFB10 maturation and is decreasing with time. (C) Model for maturation of NDUFB10 by CHCHD4. The potential CHCHD4 recognition motif in NDUFB10 allows for the initial interaction of NDUFB10 and CHCHD4. This motif is localized around cysteine 119. A mixed disulfide intermediate between both proteins is resolved once the disulfides in NDUFB10 are formed.
Mutation of cysteine 107 of NDUFB10 results in impaired mitochondrial import of the protein
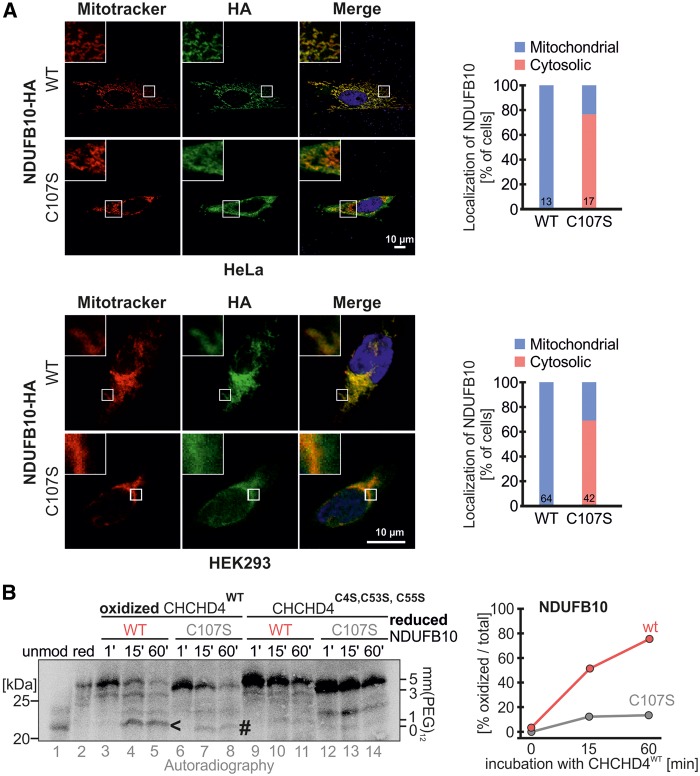
To understand the role of the C107S patient mutation, we next tested where NDUFB10C107S would localize compared to wild type protein (Fig. 7A). We found that NDUFB10C107S localized largely to the cytosol, while the wild type protein was completely localized to mitochondria. To assess the reason for this mislocalization we tested whether NDUFB10C107S still constituted a CHCHD4 substrate using an in vitro oxidation assay. We mixed purified CHCHD4 variants with radioactive NDUFB10 variants and followed the oxidation of NDUFB10 by an mmPEG12 gel shift assay (Fig. 7B). Wild type NDUFB10 became efficiently oxidized by wild type CHCHD4 but not the redox-inactive version of the oxidoreductase during the time of the assay. Conversely, the C107S mutant was only slowly oxidized and acquired a presumably non-native disulfide bond that included the non-conserved cysteine 149. Taken together, this shows that NDUFB10C107S is not imported into mitochondria anymore because it does not constitute a CHCHD4 substrate. Instead, NDUFB10C107S accumulates in the cytosol and is there likely degraded by the proteasome.
Figure 7.
NDUFB10C107S is not efficiently oxidized and accumulates in the cytosol. (A) NDUFB10C107S accumulates in the cytosol. HeLa cells were transfected with plasmids coding either for NDUFB10-HA or NDUFB10C107S-HA. 24 h after transfection cells were fixed and analyzed by immunofluorescence. HEK293 cells stably expressing the respective NDUFB10 variants were induced with doxycycline for 24 h, then fixed and analyzed by immunofluorescence. The colocalization of NDUFB10-HA variants with Mitotracker was quantified and the fraction of cells exhibiting cytosolic or mitochondrial localization was plotted for HeLa and HEK293 cells. Bars contain the number of cells assessed for quantification. (B) Oxidation kinetics of NDUFB10 variants with CHCHD4 variants show that C107S is oxidized slower and presumably to a misoxidized product. Purified oxidized CHCHD4 wild type or a redox-inactive variants (C4S, C53S, C55S) were incubated with radioactive lysate of reduced NDUFB10 variants. At the indicated times the reaction was stopped by addition of mmPEG12-containing buffer. Samples were analyzed by SDS-PAGE and autoradiography. The arrowhead indicates the correctly oxidized NDUFB10, while the hashtag indicates the presumable mis-oxidized NDUFB10C107S.
NDUFB10 protein expression is reduced in the affected tissues and is required for the assembly of complex I
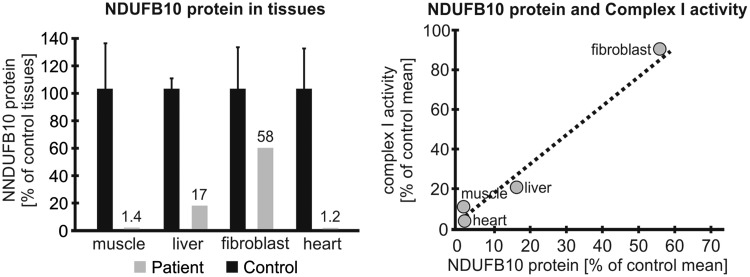
Next, we assessed how this impaired import process affected the NDUFB10 protein levels in the tissues of the patient. When normalized to citrate synthase protein amount, NDUFB10 protein expression on Western blot was strongly reduced in muscle (1.4% of control n = 6), heart (1.2% of control n = 6), reduced to a lesser extent in the liver (17% of control n = 5), but only mildly affected in fibroblasts (58% of control n = 5) (Fig. 8, Supplementary Material, Fig. S1). Across tissues, there was a direct relation between the amount of NDUFB10 protein and the residual complex I activity (Fig. 8). This residual protein was due to the C107S mutant as the paternal allele was absent in fibroblast mRNA.
Figure 8.
NDUFB10 amounts are decreased in affected patient tissues. The decrease in the amount of NDUFB10 protein assessed by Western blotting differs between tissues (left). NDUFB10 levels directly relate to the residual complex I activity (right).
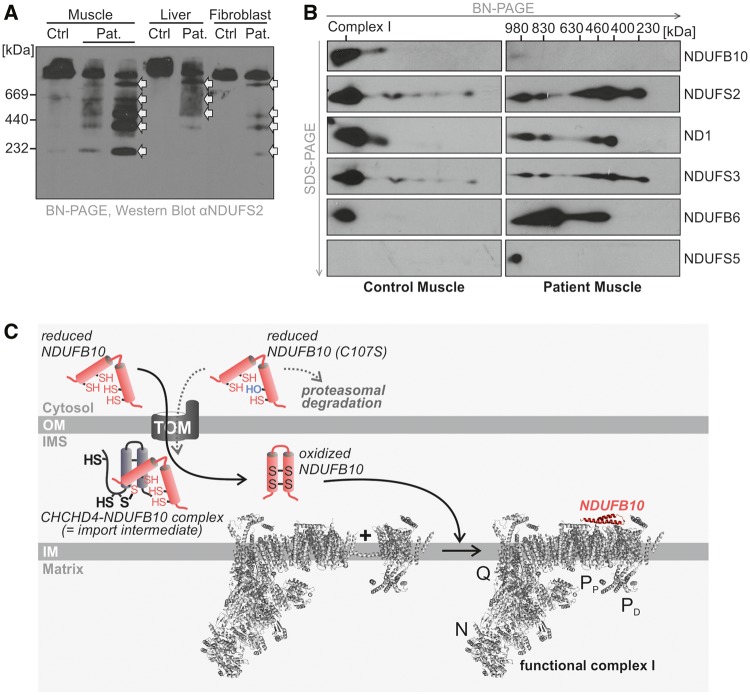
Probing the assembly of complex I with an antibody against NDUFS2, an early component, on non-denaturing BN-PAGE, five complex I intermediates (230 kDa, 400 kDa, 460 kDa, 630 kDa, and 830 kDa) were observed in the patient’s muscle and liver and some amount of apparently completely assembled holocomplex I was also present (Fig. 9A). This method could not discern by size if an individual subunit such as NDUFB10 would be absent from the holocomplex. Trace amounts of these intermediates were also observed in fibroblasts. These intermediates were not present in 10 control samples of each tissue (data not shown). On 2D gel analysis, the first 230 kDa subunit showed the presence of NDUFS3, the 400 kDa intermediate showed the presence of ND1, and the 830 kDa intermediate showed presence of NDUFS5 as expected according to the current understanding of these intermediates (Fig. 9B) (19). NDUFB10 protein appeared mostly in the final 980 kDa assembled complex in control samples of all tissues examined (muscle, liver, fibroblasts) with small amount present in the 830 kDa subcomplex (Fig. 9B and Supplementary Material, Fig. S1). In the patient, the location was similar albeit in a more or less reduced amount. This analysis provides evidence that NDUFB10 may be involved in the late stage of complex I assembly. To further validate this finding, HepG2 cells were transduced with lentiviruses corresponding to four different shRNAs that targeted NDUFB10 mRNA expression. The shRNA1 construct reduced expression of NDUFB10 protein to 69% compared to untransfected HepG2 cells. In these mildly deficient cells, complex I subcomplexes were present, similar to what was observed in the patient’s fibroblasts, and that were not detected in controls of empty vector and scrambler shRNAs (Supplementary Material, Fig. S2). These data fit to a model in which NDUFB10 serves as a clamp to hold different membrane parts of complex I together (Fig. 9C).
Figure 9.
Assembly of complex I is impaired in cells harboring NDUFB10C107S. (A) The assembly of complex I is evaluated by separation on a native gel followed by Western blotting and probing with an antibody against NDUFS2, a subunit which is present in the early stage of assembly. The patient (Pat.) muscle has subassembly complexes present at 230 kDa, 400 kDa, 460 kDa, 630 kDa, and 830 kDa before the fully assembled complex (white arrows). These are less evident in liver and fibroblasts, but still present in the patient, and not in controls. (B) Two-dimensional gel analysis with blue native PAGE in the first dimension followed by SDS-PAGE in the second dimension of muscle tissue of the patient. After Western blotting, the membrane was probed with antibodies against subunits known to be present in various subcomplexes. Subcomplexes at 230 kDa, 400 kDa, 460 kDa, 830 kDa and the mature complex at 980 kDa are visible in the patient, with only traces visible in control sample. NDUFS2 and NDUFS3 are identified from the first subassembly, whereas ND1 is present from the 400 kDa subocmplex, NDUFB6 from the 460 kDa and NDUFS5 from the fully assembled complex only. NDUFB10 is present from the 830 kDa subcomplex on. (C) Model for the role of NDUFB10 in the assembly of complex I of the respiratory chain. Structure of complex I modified from (47) (pdb 4UQ8).
Discussion
Complex I deficiency is a phenotypically and genetically heterogeneous disorder. The role of the catalytic core of complex I in oxidizing NADH, reducing ubiquinone, and transporting protons across the mitochondrial membrane has been well described. The role of the accessory subunits within complex I is not that well understood, and it has been assumed that some play a role in the assembly and stability of complex I. Here we describe a patient with a profound complex I deficiency who was found to be compound heterozygous for mutations in an accessory subunit of complex I, NDUFB10. The clinical presentation in this case is compatible with neonatal cardiomyopathy and fatal infantile lactic acidosis, a presentation well reported before in isolated complex I deficiency (4). This is the first report of mutations in NDUFB10, encoding an accessory subunit of the PD part. The paternal mutation creates a frameshift and stop mutation likely triggering nonsense-mediated mRNA decay and was absent in the fibroblast mRNA. The maternal missense mutation affects a highly conserved cysteine resulting in reduced protein levels, but to a variable degree across tissues. Across different tissues, the decrease in NDUFB10 protein correlates directly with the decrease in complex I activity. Even in fibroblasts, with a high amount of residual protein, a mild functional deficiency of complex I related oxygen consumption could be demonstrated with high-resolution respirometry. Of note, the mild decrease in complex II only observed in the liver is likely due to the fact that complex II is the most labile enzyme complex postmortem (40).
Mutations in accessory subunits of mitochondrial complex I can affect complex I in four ways: by disrupting the assembly or destabilizing complex I resulting in assembly intermediates, by affecting the rate of catalysis, by accelerating superoxide production, or by affecting the coupling of the redox reaction to proton transport (9). Knockdown of most accessory subunits, including NDUFB10, in HEK293T cells resulted in decrease in fully assembled complex I (41). NDUFB10 is added early in evolution to the ancestral eukaryotic core and has homologs in all eukaryotic species with an intact complex I (42). Disruption of its homolog nuo12.3 in Neurospora crassa resulted in incomplete assembly of the membrane arm, as evidenced by elution in an earlier sucrose fraction of the NADH:ferricyanide oxidoreducing part constituting the peripheral arm, whereas the membrane arm protein components were present but not assembled (43). Chlamydomonas rheinhardtii mutants with a disrupted homologous NUOB10 gene (mutants amc5 and amc7) have isolated deficiency of complex I activity, and incomplete assembly of complex I to a 700 kDa complex with absence of the fully assembled 950 kDa enzyme (44). Thus in these two species, absence of the homolog of NDUFB10 disrupted the assembly of the membrane component of complex I, and similarly in our patient multiple assembly intermediates accumulated up to the large 830 kDa subassembly form. In current models of complex I assembly, compatible with our 2-D gel analysis, elements of the PD part of complex I are assembled either at or beyond the 830 kDa subassembly stage, which is compatible with the findings presented here, where the majority of NDUFB10 is found in the completely assembled complex and a minority in the 830 kDa subcomplex. Interestingly, even in the mildly affected tissue of fibroblasts, some subcomplexes accumulated, a finding never observed in controls, illustrating that this represents a very sensitive finding. Thus, our findings indicate that NDUFB10 is important for the assembly of complex I at the late stage where the PD domain is being assembled, and that genetic disruption of this gene results in incomplete assembly. It should be noted that defects in the assembly of complex I are often associated with increased oxidative stress from the formation of superoxide when incubated with NADH (11). NDUFB10 has several posttranslational modifications which our study does not address such as removal of initiation methionine, Src-mediated phosphorylation on Ser145 associated with an effect on complex I activity, and possible ubiquination sites at Lys91, Lys121, and Lys131 (7,45).
NDUFB10 had been identified as a subunit located in the PD domain of the Iβ part in the peripheral membrane arm (14), and has been assigned to the ND4 module (41). In contrast to most PD accessory subunits, it does not contain a transmembrane domain (46–48). In the recently resolved molecular structure of bovine complex I, the bovine homolog PDSW of human NDUFB10 appears to be located parallel to the membrane on the IMS face spanning ND4 and ND5 (46–48). This IMS-facing location is similar to bovine B18, PGIV and 15 kDa (human NDUFB7, NDUFA8, NDUFS5, respectively) the remaining three complex I proteins that are imported, oxidized and folded by CHCHD4 (Supplementary Material, Fig. S3). While NDUFA8, NDUFB7 and NDUFS5 contain regular twin-CXnC motifs and CHCHD4 recognition sequences, NDUFB10 harbors an atypical CX6C/CX11C motif. NDUFB10 and PDSW also deviate from the common structure of twin CXnC proteins: two antiparallel helices that are fixed by two disulfide bonds, which are formed between the cysteines of the different CXnC motifs and not within the motif (data not shown and (47)). Instead, it appears that in NDUFB10 and PDSW the disulfides are formed between consecutive cysteines, one disulfide in the CX6C motif and one in the CX11C motif, respectively. Importantly, atypical CXnC configurations have been described before as targets of the pathway for oxidative import into the IMS (27,34) and we demonstrate here the involvement of CHCHD4 in NDUFB10 maturation. In addition, NDUFB10 also contains a CHCHD4-recognition motif, an α-helix with a hydrophobic face also containing a cysteine residue (Fig. 6C). In summary, we propose from our data that all four complex I subunits that are CHCHD4 substrates line the IMS face of complex I to serve as clamps holding together and protecting individual units of complex I. We think that this provides the mechanistic explanation for the involvement of the CHCHD4-dependent protein import pathway in complex I assembly that has been experimentally confirmed (27,28,34,49).
The loss of the first cysteine of the second CXnC motif (NDUFB10C107S) interferes with both the targeting and the stability of NDUFB10 resulting in a decrease in NDUFB10 levels. Interestingly, the NDUFB10C107S loss and the resulting complex I deficiency exhibit a large tissue heterogeneity. While the protein is absent in muscle, its levels are half that observed in control fibroblasts, compatible with the absence of the product of the paternal allele, similar to carrier status. In line with this, complex I activity is preserved. This indicates that in fibroblasts NDUFB10C107S can become imported, correctly folded and integrated into complex I. From our in silico analyses, we concluded that the CHCHD4 interaction motif of NDUFB10 is around cysteine residue 119. Thus, the initial CHCHD4-NDUFB10 interaction likely still takes place in the NDUFB10C107S mutant. However, subsequent steps in oxidation and folding are apparently differentially affected by losing the conserved cysteine 107 in different tissues. It is thus tempting to speculate that the tissue differences in the IMS quality control, the redox conditions of the IMS as well as the activity of CHCHD4 are responsible for these variations. Redox conditions in the IMS are mainly controlled by the glutathione redox potential and limiting glutaredoxin amounts (32,50). In Drosophila melanogaster and in mouse differences in the glutathione redox potential of the cytosol (that likely translate to differences in the IMS) of different tissues have been reported (51,52). In addition, it has also been shown that the steady-state levels of CHCHD4 vary across different tissues (37); however, it remains unclear whether also the redox state of the redox-active cysteine motif in CHCHD4 differs between tissues.
Of the 12 accessory subunits in the PD part of complex I, mutations have thus far only been described in three other genes: NDUFB3, NDUFB9 and NDUFB11 (8,20,21,24–26). Patients with NDUFB3 (OMIM 603839) and NDUFB9 (OMIM 601445) mutations presented with fatal infantile lactic acidosis and had deficient complex I activity (20,21). In NDUFB9, in addition to near absent NDUFB9 protein, the levels of other subunits NDUFS1, NDUFS3, NDUFA9 and NDUFB8 were also decreased, indicating lack of assembly or of stability of complex I. Mutations in the X-linked gene NDUFB11 (OMIM 300403) are reported most commonly in females with skewed X-inactivation, who present with various combinations of linear skin defects, eye abnormalities, histicytoid cardiomyopathy, sideroblastic anaemia, and lactic acidosis (24–26). Most patients had no biochemical abnormalities as presumably affected cells were lost (26), but one patient had decreased NDUFB11 protein levels with decreased holocomplex I (25). Decreasing the NDUFB11 amount by siRNA in HeLa cells reduced complex I activity, and also reduced levels of NDUFB8 in the PD region, but preserved normal amounts of NDUFS3, a subunit of the N-module. Two-dimension gel analyses showed multiple subassembly complexes similar to that observed in our case with NDUFB10 deficiency (26). Thus, defects in accessory subunits of the PD unit of complex I result in decreased complex I enzyme activity, decreased holocomplex I assembly, and decreased amounts of multiple subunits, regardless of whether the mutation affects a transmembrane helix such as NDUFB3 and NDUFB11, a structural role such as the LYR4 motif for NDUFB9, or a membrane parallel structure for NDUFB10 (48).
In summary, we describe a patient with a new genetic cause of severe complex I deficiency: mutations in NDUFB10, encoding a subunit of the PD part of complex I. Clinically, our patient presented with cardiomyopathy and lethal infantile lactic acidosis. Molecularly, this deficiency resulted in a late assembly defect at the level of the 830 kDa subcomplex. Tissue specific differences may be related to the impact of the mutation of a conserved cysteine, which may affect disulfide bond formation. Further, this illustrates that the oxidative pathway for IMS protein import is essential for the import of accessory subunits of complex I, which are critical for its assembly and stability, and mutations interfering with this import pathway can result in severe human disease.
Materials and Methods
The parents and subject posthumously, were enrolled into an IRB-approved research protocol (COMIRB 07-0386).
Pathology and tissue procurement
With consent, a metabolic autopsy was performed within 45 minutes of death and heart, liver, and skeletal muscle were obtained, snap frozen, and stored at −80 °C. For histology, hematoxylin and eosin stained sections were prepared from both ventricles of the heart, liver, and quadriceps muscle. For electron microscopic examination, tissues were stored in glutaraldehyde, fixed with osmium tetroxide, embedded in epoxy resin, and ultrathin sections (80 nm) post-stained with uranyl acetate and lead citrate. Images were acquired using a Hitachi H-7650 transmission electron microscope at 60 kV accelerating voltage (Hitachi High-Technologies Corp., Tokyo, Japan).
Analysis of respiratory chain enzyme activities and functions
Respiratory chain enzyme activities for complexes I, II, II + III, III, IV, and citrate synthase were assayed in post-600 g homogenates spectrophotometrically on a Cary 300 spectrophotometer at 30 °C in muscle, liver, heart, and skin fibroblasts, and activities calculated as initial rates for complexes I, II, II + III and citrate synthase, and as first order rate constants for complexes III and IV as previously described (53). Activities were normalized for sample total protein content, and also expressed as ratios over activity of citrate synthase and of complex II. Normal ranges were determined from 25 muscle control samples, 16 liver control samples, 10 heart control samples, and 33 primary fibroblast control cell lines. In control samples, the natural log of the activities and of the ratios was normally distributed and the patient results were expressed as Z-scores from this control distribution. Isolated mitochondrial membrane fractions of muscle, heart, liver, and fibroblast samples were analyzed by blue native PAGE with in-gel activity staining as described (53,54).
High resolution respirometry was employed to measure oxygen consumption in permeabilized fibroblast cells using the Oroboros Oxygraph 2K system following a SUIT protocol as previously described and modified as follows (55). Fibroblasts were cultured in α-MEM medium supplemented with 10% FBS, non-essential amino acids, and antibiotic antimycotic solution (reagents from Hyclone) at 37° C and 5% CO2 until they reached 70–80% confluency. Cells were trypsinized (0.25%; Hyclone), resuspended in mitochondrial respiration buffer MIR05 at 5 × 105 cells/ml, 2 ml were placed into each O2K chamber, and oxygen consumption (pmol O2/second/1 × 106 cells) measured following a SUIT protocol in sequential order: routine, malate, pyruvate, digitonin, ADP, glutamate, succinate, CCCP (uncoupler), rotenone, antimycin A, tetramethyl-p-phenylenediamine (TMPD) + ascorbate, and azide. Calculated derived parameters from these data included: acceptor control ratio (ADP-pyruvate), [glutamate – ADP]/glutamate (contribution of glutamate beyond pyruvate), succinate/glutamate (Q junction), uncoupled control ratio (succinate/CCCP), complex I ratio (uncoupled – rotenone), and complex IV ratio ([TMPD + ascorbate] - azide). Patient fibroblasts were analyzed 5 times and compared to normal ranges established from 20 control human fibroblasts.
DNA extraction and exome sequencing
Exome sequencing using a trio-based analysis was performed on the patient and both parents. High-quality, unfragmented genomic DNA was extracted from whole blood using the Puregene Blood Kit (Qiagen). The genomic DNA was sheared, size selected (∼400–600 bp), ligated to sequencing adapters, and PCR amplified following standard library preparation, and enriched for exonic sequences using SureSelect Human All Exon 51 Mb Kit (Agilent Technologies, Santa Clara, California, USA). The exome enriched samples were sequenced to 100 bp paired-end on a HiSeq2000 (Illumina, San Diego, California, USA). The reads were aligned to the human genome assembly GRCh37 using GSNAP (Genomic Short-read Nucleotide Alignment Program, version 2012-07-20) and variants identified by GATK (Genome Analysis Toolkit, v2.1-8-g5efb575). Only non-synonymous coding variants, coding indels and variants affecting splice sites were retained for further analysis. Common variants present in dbSNP and 1000 Genomes data were filtered out. Parental exome sequencing data was used to analyze variants following various inheritance models including dominant (de novo mutations) and recessive (compound heterozygous, homozygous, and X-linked hemizygous mutations) models. Variants were confirmed by Sanger sequencing using primers as listed (Supplementary Material, Table S1). To evaluate the expressed allele, RNA was extracted from patient fibroblasts, reverse transcribed, and the NDUFB10 cDNA PCR amplified and Sanger sequenced.
Western blot analysis
Western blot analysis of the NDUFB10 protein was performed on post-600g mitochondrial homogenates isolated from the patients' muscle, heart, liver and cultured skin fibroblasts in triplicate and compared to 5 to 6 normal controls from each tissue type. Ten µg of protein were separated on 5–20% gradient polyacrylamide gels containing sodium dodecyl sulfate (SDS-PAGE), and after blotting probed with primary antibodies obtained from Abcam (Cambridge, MA, USA), and detected using the appropriate HRP-conjugated secondary antibody followed by enhanced chemiluminescence. Quantification of bands was done using Bio-Rad Quantity One 1-D Analysis software. The amount of NDUFB10 protein was normalized to both loading controls: COXIV an inner mitochondrial membrane protein, and citrate synthase a mitochondrial matrix protein. The assembly of complex I was evaluated by Western blotting following non-denaturing BN-PAGE of isolated mitochondrial membrane fractions of muscle, liver and fibroblasts probed with the antibody for NDUFS2, a component of the earliest complex I assembly intermediate (17). To determine the effects of reduced or absent expression of NDUFB10 on the assembly of complex I, two dimensional gel analysis was performed as described (56). The intact enzyme complexes were separated by BN-PAGE using a non-denaturing gradient 5–13% polyacrylamide gel from isolated membrane fractions of patient and control muscle and the entire gel lane was cut and soaked in 1% SDS (w/v) and 1% β-mercaptoethanol (v/v), and the gel piece rotated 90°, and separated on a 10% polyacrylamide SDS gel to resolve the individual subunits. Different intermediates of complex I assembly were detected with a series of antibodies against NDUFS2, NDUFS3, NDUFS4, NDUFB6, NDUFS5, NDUFA2, and MTND1. The assessment of the location of NDUFB10 in the intermediates of complex I assembly was similarly performed. NDUFB10 knockdown experiments were performed using shRNAs cloned into a pLKO.1-puro vector (Functional Genomics, University of Colorado, Aurora, CO) which contained the puromycin selection cassette and the RSV-LTR promoter system. HepG2 cells were transduced with viral supernatant corresponding to the shRNAs and controls (empty vector and scrambled vector), selected with puromycin, and the cell line with the strongest reduction in NDUFB10 by Western blot analysis and RT-qPCR was expanded, and assembly of complex I evaluated as above.
Analysis of oxidative protein folding
Experiments were performed as described (27). In short, HEK293 cells were treated with 1 µg/ml doxycycline to induce protein expression two hours prior to the experiment. The cells were incubated with Cys- and Met-free medium (Sigma-Aldrich, St. Louis, MO) for 15 min at 37 °C. Newly synthesized proteins were pulse labelled for 5 min at 37 °C with Cys- and Met-free medium containing [35S]-Met at a concentration of 200 μCi/ml (Perkin Elmer, Waltham, MA). Pulse labelling was stopped by removing the medium and adding chase medium containing 20 mM Met. The chase was performed for variable times at 37 °C, when it was stopped by adding ice-cold 8% trichloroacetic acid (TCA). TCA precipitation was performed by centrifugation at 13,000g for 15 min and washing with 5% ice-cold TCA. Protein precipitates were dissolved in modification buffer (0.2 M Tris, pH 7.5, 6 M urea, 10 mM EDTA, 2% SDS). Samples were modified with a final mmPEG12 concentration of 15 mM for 1 h at room temperature. Reduced controls were treated with a 2 mM Tris(2-carboxyethyl)phosphine (TCEP; final concentration) for 5 min at 96 °C before mmPEG12 modification; oxidized controls were untreated. After modification samples were filled to 250 μl using lysis buffer A (30 mM Tris-Cl, pH 8, 100 mM NaCl, 5 mM EDTA, 2% SDS) and incubated for 5 min at 96 °C. Then 750 μl of lysis buffer B (30 mM Tris-Cl, pH 8, 100 mM NaCl, 5 mM EDTA, 2.5% Triton X-100) was added and the mixture incubated at 4 °C for 1 h. Samples were cleared by centrifugation at 25,000 × g for 1 h. The supernatant was subjected to immunoprecipitations (IP) with antibodies conjugated to protein A–Sepharose beads at 4 °C overnight under gentle shaking. The samples were washed three times using lysis buffer C (30 mM Tris-Cl, pH 8, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100) and once using lysis buffer D (30 mM Tris-Cl, pH 8, 100 mM NaCl, 5 mM EDTA). Proteins were eluted by adding Laemmli buffer (2% SDS, 60 mM Tris, pH 6.8, 10% glycerol, 0.0025% bromophenolblue) to the dried beads and subsequent boiling for 5 min at 95 °C. Samples were analyzed by SDS-PAGE and autoradiography.
Analysis of NDUFB10-CHCHD4 interaction
HEK293 cells were starved with Cys- and Met-free medium (Sigma) for 15 min at 37 °C. Newly synthesized proteins were pulse labelled for 3 hours at 37 °C with Cys- and Met-free medium containing [35S]-Met at a radionuclide concentration of 200 μCi/ml. Pulse labelling was stopped by removing the medium and adding chase medium containing 20 mM cold Met. The chase was performed for variable times at 37 °C. For denaturing IPs to visualize disulfide-linked interaction partners, the chase was stopped by the addition of ice cold PBS containing 20 mM NEM to preserve disulfide-linked interactions. Cells were harvested and resuspended in 250 µl denaturing lysis buffer (30 mM Tris-HCl pH 8.1, 150 mM NaCl, 1 mM EDTA, 1.6% SDS) and treated for 20 min at 96 °C. Then, 750 µl Triton X-100 was added to the lysate and kept on ice for 1 h. The lysates were cleared by centrifugation at 25,000g for 1 h, and the supernatant was used for IPs with antibodies conjugated to protein A beads at 4 °C overnight. The samples were washed three times using lysis buffer containing Triton X-100 and once without any detergent. Proteins were eluted by adding 8% SDS and the volume was adjusted to 250 µl with denaturing lysis buffer and total samples were taken. Then, 750 µl Triton X-100 was added to the lysate and kept on ice for 1 h. The lysate was used for another round of IP with antibodies conjugated to protein A beads at 4 °C overnight. The samples were washed three times using lysis buffer containing Triton X-100 and once without any detergent. Proteins were eluted by the addition of Laemmli buffer to the dried beads and subsequent boiling for 5 min at 96 °C. Samples were analyzed by SDS-PAGE and autoradiography.
Immunofluorescence experiments
HEK293 and HeLa cells were cultured on poly-L-lysine-coated cover slips for 24 h. Cells were incubated with Mitotracker red (Thermo Fisher) for 1 h. Fixation was performed with 4% paraformaldehyde for 15 min. Then cell membranes were permeabilized with blocking buffer [20 mM HEPES pH 7.4, 3% BSA, 0.3% Triton X-100] for 1 hour. Cells were washed and incubated with primary (anti-HA) and secondary antibodies (anti-mouse, ALEXA488) for 1 h, respectively. Cells were stained for 15 min with 2 ng/ml DAPI in PBS. Then, cells were washed, cover slips transferred to microscope slides (cover medium: 30% glycerol, 12% polyvinyl alcohol, 60 mM TRIS, 2.5% 1,4-diazabicyclo-2,2,2-octan) and analyzed by immunofluorescence microscopy. Colocalization analysis of the HA signal and Mitotracker signal was performed using CellProfiler with correlation pipeline set to 15% threshold. As a cytosolic control, a cytosolic GFP was used. The colocalization of NDUFB10C107S-HA was normalized to the mean value of cytosolic control and mean value of NDUFB10WT-HA (mitochondrial). Cells expressing NDUFB10C107S-HA with colocalization scores below 50% of the normalized mean were counted as cytosolic, and the scores above 50% were counted as mitochondrial. Representative pictures were processed for brightness and contrast using ImageJ.
Protein purification and in vitro reconstitution assay
Experiments were performed as described (57). In short, coding sequences of human CHCHD4 (full length) and its redox-inactive cysteine mutant were cloned into pGEX-6p-1 (GE Healthcare) expression vector containing N-terminal cleavable GST tag. The proteins were overproduced in E. coli Rosetta™(DE3) cells. After lysis of cells using a French pressure cell in buffer containing 20 mM Tris-HCl, 200 mM NaCl, 0.2 mM PMSF, pH 7.4 and clearing centrifugation, GST-fused CHCHD4 protein was obtained in soluble form. The purification was performed using Glutathione Sepharose 4B beads, and GST tag was cleaved off from CHCHD4 using previously immobilized PreScission Protease.
Radiolabelled NDUFB10 variants were synthesized in vitro with the TNT Quick Coupled Transcription/Translation System (Promega; Mannheim, Germany) in a hypoxic chamber (Sigma). After mixing the radiolabelled NDUFB10 and CHCHD4 variants, the reactions were stopped at indicated times by addition of sample buffer containing 20 mM mmPEG12 and incubated for 1 h at 25 °C. Samples were analyzed by non-reducing SDS-PAGE and autoradiography.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by funding from Miracles for Mito, the Children’s Hospital Colorado Foundation and University of Colorado Foundation to JVH. This study was supported by National Institutes of Health/National Center for Advancing Translational Sciences Colorado CTSA [Grant Number UL1 TR001082]. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views. The work was also supported by grants from the German research foundation (DFG) RI2150/1-2 and SFB1218 TP B02 to JR.
Supplementary Material
References
- 1. Rodenburg R.J. (2011) Biochemical diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis., 34, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirby D.M., Crawford M., Cleary M.A., Dahl H.H., Dennett X., Thorburn D.R. (1999) Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology, 52, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 3. Koene S., Rodenburg R.J., van der Knaap M.S., Willemsen M.A., Sperl W., Laugel V., Ostergaard E., Tarnopolsky M., Martin M.A., Nesbitt V., et al. (2012) Natural disease course and genotype-phenotype correlations in Complex I deficiency caused by nuclear gene defects: what we learned from 130 cases. J. Inherit. Metab. Dis., 35, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bugiani M., Invernizzi F., Alberio S., Briem E., Lamantea E., Carrara F., Moroni I., Farina L., Spada M., Donati M.A., et al. (2004) Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta, 1659, 136–147. [DOI] [PubMed] [Google Scholar]
- 5. Fassone E., Rahman S. (2012) Complex I deficiency: clinical features, biochemistry and molecular genetics. J. Med. Genet., 49, 578–590. [DOI] [PubMed] [Google Scholar]
- 6. Distelmaier F., Koopman W.J., van den Heuvel L.P., Rodenburg R.J., Mayatepek E., Willems P.H., Smeitink J.A. (2009) Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain, 132, 833–842. [DOI] [PubMed] [Google Scholar]
- 7. Janssen R.J., Nijtmans L.G., van den Heuvel L.P., Smeitink J.A. (2006) Mitochondrial complex I: structure, function and pathology. J. Inherit. Metab. Dis., 29, 499–515. [DOI] [PubMed] [Google Scholar]
- 8. Rodenburg R.J. (2016) Mitochondrial complex I-linked disease. Biochim. Biophys. Acta, 1857, 938–945. [DOI] [PubMed] [Google Scholar]
- 9. Hirst J. (2013) Mitochondrial complex I. Annu. Rev. Biochem., 82, 551–575. [DOI] [PubMed] [Google Scholar]
- 10. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem., 75, 69–92. [DOI] [PubMed] [Google Scholar]
- 11. Leman G., Gueguen N., Desquiret-Dumas V., Kane M.S., Wettervald C., Chupin S., Chevrollier A., Lebre A.S., Bonnefont J.P., Barth M., et al. (2015) Assembly defects induce oxidative stress in inherited mitochondrial complex I deficiency. Int. J. Biochem. Cell Biol., 65, 91–103. [DOI] [PubMed] [Google Scholar]
- 12. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science, 329, 448–451. [DOI] [PubMed] [Google Scholar]
- 13. Ohnishi T., Salerno J.C. (2005) Conformation-driven and semiquinone-gated proton-pump mechanism in the NADH-ubiquinone oxidoreductase (complex I). FEBS Lett., 579, 4555–4561. [DOI] [PubMed] [Google Scholar]
- 14. Drose S., Krack S., Sokolova L., Zwicker K., Barth H.D., Morgner N., Heide H., Steger M., Nubel E., Zickermann V., et al. (2011) Functional dissection of the proton pumping modules of mitochondrial complex I. PLoS Biol., 9, e1001128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheffler I.E. (2015) Mitochondrial disease associated with complex I (NADH-CoQ oxidoreductase) deficiency. J. Inherit. Metab. Dis., 38, 405–415. [DOI] [PubMed] [Google Scholar]
- 16. Lazarou M., Thorburn D.R., Ryan M.T., McKenzie M. (2009) Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta, 1793, 78–88. [DOI] [PubMed] [Google Scholar]
- 17. Vogel R.O., Smeitink J.A., Nijtmans L.G. (2007) Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim. Biophys. Acta, 1767, 1215–1227. [DOI] [PubMed] [Google Scholar]
- 18. Dieteren C.E., Willems P.H., Vogel R.O., Swarts H.G., Fransen J., Roepman R., Crienen G., Smeitink J.A., Nijtmans L.G., Koopman W.J. (2008) Subunits of mitochondrial complex I exist as part of matrix- and membrane-associated subcomplexes in living cells. J. Biol. Chem., 283, 34753–34761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. (2012) Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta, 1817, 851–862. [DOI] [PubMed] [Google Scholar]
- 20. Haack T.B., Haberberger B., Frisch E.M., Wieland T., Iuso A., Gorza M., Strecker V., Graf E., Mayr J.A., Herberg U., et al. (2012) Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J. Med. Genet., 49, 277–283. [DOI] [PubMed] [Google Scholar]
- 21. Calvo S.E., Compton A.G., Hershman S.G., Lim S.C., Lieber D.S., Tucker E.J., Laskowski A., Garone C., Liu S., Jaffe D.B., et al. (2012) Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med., 4, 118ra110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y., Hirata T., Yatsuka Y., Yamashita-Sugahara Y., Nakachi Y., et al. (2016) A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet., 12, e1005679.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angebault C., Charif M., Guegen N., Piro-Megy C., Mousson de Camaret B., Procaccio V., Guichet P.O., Hebrard M., Manes G., Leboucq N., et al. (2015) Mutation in NDUFA13/GRIM19 leads to early onset hypotonia, dyskinesia and sensorial deficiencies, and mitochondrial complex I instability. Hum. Mol. Genet., 24, 3948–3955. [DOI] [PubMed] [Google Scholar]
- 24. Shehata B.M., Cundiff C.A., Lee K., Sabharwal A., Lalwani M.K., Davis A.K., Agrawal V., Sivasubbu S., Iannucci G.J., Gibson G. (2015) Exome sequencing of patients with histiocytoid cardiomyopathy reveals a de novo NDUFB11 mutation that plays a role in the pathogenesis of histiocytoid cardiomyopathy. Am. J. Med. Genet. A, 167, A, 2114–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torraco A., Bianchi M., Verrigni D., Gelmetti V., Riley L., Niceta M., Martinelli D., Montanari A., Guo Y., Rizza T., et al. (2016) A novel mutation in NDUFB11 unveils a new clinical phenotype associated with lactic acidosis and sideroblastic anemia. Clin. Genet., doi: 10.1111/cge.12790. [DOI] [PubMed] [Google Scholar]
- 26. van Rahden V.A., Fernandez-Vizarra E., Alawi M., Brand K., Fellmann F., Horn D., Zeviani M., Kutsche K. (2015) Mutations in NDUFB11, encoding a complex I component of the mitochondrial respiratory chain, cause microphthalmia with linear skin defects syndrome. Am. J. Hum. Genet., 96, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fischer M., Horn S., Belkacemi A., Kojer K., Petrungaro C., Habich M., Ali M., Kuttner V., Bien M., Kauff F., et al. (2013) Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell, 24, 2160–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cavallaro G. (2010) Genome-wide analysis of eukaryotic twin CX9C proteins. Mol. Biosyst., 6, 2459–2470. [DOI] [PubMed] [Google Scholar]
- 29. Petrungaro C., Zimmermann K.M., Kuttner V., Fischer M., Dengjel J., Bogeski I., Riemer J. (2015) The Ca(2+)-Dependent Release of the Mia40-Induced MICU1-MICU2 Dimer from MCU Regulates Mitochondrial Ca(2+) Uptake. Cell Metab., 22, 721–733. [DOI] [PubMed] [Google Scholar]
- 30. Lehtonen M.S., Moilanen J.S., Majamaa K. (2003) Increased variation in mtDNA in patients with familial sensorineural hearing impairment. Hum. Genet., 113, 220–227. [DOI] [PubMed] [Google Scholar]
- 31. Soini H.K., Moilanen J.S., Finnila S., Majamaa K. (2012) Mitochondrial DNA sequence variation in Finnish patients with matrilineal diabetes mellitus. BMC Res. Notes, 5, 350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riemer J., Bulleid N., Herrmann J.M. (2009) Disulfide formation in the ER and mitochondria: two solutions to a common process. Science, 324, 1284–1287. [DOI] [PubMed] [Google Scholar]
- 33. Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell, 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Modjtahedi N., Tokatlidis K., Dessen P., Kroemer G. (2016) Mitochondrial Proteins Containing Coiled-Coil-Helix-Coiled-Coil-Helix (CHCH) Domains in Health and Disease. Trends Biochem. Sci., 41, 245–260. [DOI] [PubMed] [Google Scholar]
- 35. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L.K., Schulze-Specking A., Truscott K.N., Guiard B., Meisinger C., Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. Embo J., 23, 3735–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naoe M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. (2004) Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem., 279, 47815–47821. [DOI] [PubMed] [Google Scholar]
- 37. Hofmann S., Rothbauer U., Muhlenbein N., Baiker K., Hell K., Bauer M.F. (2005) Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J. Mol. Biol., 353, 517–528. [DOI] [PubMed] [Google Scholar]
- 38. Weckbecker D., Longen S., Riemer J., Herrmann J.M. (2012) Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. Embo J., 31, 4348–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki Y., Ali M., Fischer M., Riemer J. (2013) Human copper chaperone for superoxide dismutase 1 mediates its own oxidation-dependent import into mitochondria. Nat. Commun., 4, 2430.. [DOI] [PubMed] [Google Scholar]
- 40. Thorburn D.R., Chow C.W., Kirby D.M. (2004) Respiratory chain enzyme analysis in muscle and liver. Mitochondrion, 4, 363–375. [DOI] [PubMed] [Google Scholar]
- 41. Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., et al. (2016) Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature, 538, 123–126. [DOI] [PubMed] [Google Scholar]
- 42. Gabaldon T., Rainey D., Huynen M.A. (2005) Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I). J. Mol. Biol., 348, 857–870. [DOI] [PubMed] [Google Scholar]
- 43. Duarte M., Sousa R., Videira A. (1995) Inactivation of genes encoding subunits of the peripheral and membrane arms of neurospora mitochondrial complex I and effects on enzyme assembly. Genetics, 139, 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barbieri M.R., Larosa V., Nouet C., Subrahmanian N., Remacle C., Hamel P.P. (2011) A forward genetic screen identifies mutants deficient for mitochondrial complex I assembly in Chlamydomonas reinhardtii. Genetics, 188, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hebert-Chatelain E., Jose C., Gutierrez Cortes N., Dupuy J.W., Rocher C., Dachary-Prigent J., Letellier T. (2012) Preservation of NADH ubiquinone-oxidoreductase activity by Src kinase-mediated phosphorylation of NDUFB10. Biochim. Biophys. Acta, 1817, 718–725. [DOI] [PubMed] [Google Scholar]
- 46. Vinothkumar K.R., Zhu J., Hirst J. (2014) Architecture of mammalian respiratory complex I. Nature, 515, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu J., Vinothkumar K.R., Hirst J. (2016) Structure of mammalian respiratory complex I. Nature, 536, 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu J., King M.S., Yu M., Klipcan L., Leslie A.G., Hirst J. (2015) Structure of subcomplex Ibeta of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc. Natl. Acad. Sci. U. S. A, 112, 12087–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meyer K., Buettner S., Ghezzi D., Zeviani M., Bano D., Nicotera P. (2015) Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death Dis., 6, e1814.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kojer K., Peleh V., Calabrese G., Herrmann J.M., Riemer J. (2015) Kinetic control by limiting glutaredoxin amounts enables thiol oxidation in the reducing mitochondrial intermembrane space. Mol. Biol. Cell, 26, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujikawa Y., Roma L.P., Sobotta M.C., Rose A.J., Diaz M.B., Locatelli G., Breckwoldt M.O., Misgeld T., Kerschensteiner M., Herzig S., et al. (2016) Mouse redox histology using genetically encoded probes. Sci. Signal, 9, rs1. [DOI] [PubMed] [Google Scholar]
- 52. Albrecht S.C., Barata A.G., Grosshans J., Teleman A.A., Dick T.P. (2011) In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab., 14, 819–829. [DOI] [PubMed] [Google Scholar]
- 53. Chatfield K.C., Coughlin C.R. 2nd, Friederich M.W., Gallagher R.C., Hesselberth J.R., Lovell M.A., Ofman R., Swanson M.A., Thomas J.A., Wanders R.J., et al. (2015) Mitochondrial energy failure in HSD10 disease is due to defective mtDNA transcript processing. Mitochondrion, 21, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coughlin C.R. 2nd, Scharer G.H., Friederich M.W., Yu H.C., Geiger E.A., Creadon-Swindell G., Collins A.E., Vanlander A.V., Coster R.V., Powell C.A., et al. (2015) Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J. Med. Genet., 52, 532–540. [DOI] [PubMed] [Google Scholar]
- 55. Ye F., Hoppel C.L. (2013) Measuring oxidative phosphorylation in human skin fibroblasts. Anal. Biochem., 437, 52–58. [DOI] [PubMed] [Google Scholar]
- 56. Sunderhaus S., Eubel H., Braun H.P. (2007) Two-dimensional blue native/blue native polyacrylamide gel electrophoresis for the characterization of mitochondrial protein complexes and supercomplexes. Methods Mol. Biol., 372, 315–324. [DOI] [PubMed] [Google Scholar]
- 57. Bien M., Longen S., Wagener N., Chwalla I., Herrmann J.M., Riemer J. (2010) Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell, 37, 516–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.