Abstract
In humans, fetal erythropoiesis takes place in the liver whereas adult erythropoiesis occurs in the bone marrow. Fetal and adult erythroid cells are not only produced at different sites, but are also distinguished by their respective transcriptional program. In particular, whereas fetal erythroid cells express γ-globin chains to produce fetal hemoglobin (HbF), adult cells express β-globin chains to generate adult hemoglobin. Understanding the transcriptional regulation of the fetal-to-adult hemoglobin switch is clinically important as re-activation of HbF production in adult erythroid cells would represent a promising therapy for the hemoglobin disorders sickle cell disease and β-thalassemia. We used RNA-sequencing to measure global gene and microRNA (miRNA) expression in human erythroblasts derived ex vivo from fetal liver (n = 12 donors) and bone marrow (n = 12 donors) hematopoietic stem/progenitor cells. We identified 7829 transcripts and 402 miRNA that were differentially expressed (false discovery rate <5%). The miRNA expression patterns were replicated in an independent collection of human erythroblasts using a different technology. By combining gene and miRNA expression data, we developed transcriptional networks which show substantial differences between fetal and adult human erythroblasts. Our analyses highlighted the miRNAs at the imprinted 14q32 locus in fetal erythroblasts and the let-7 miRNA family in adult erythroblasts as key regulators of stage-specific erythroid transcriptional programs. Altogether, our results provide a comprehensive resource to prioritize genes that may modify clinical severity in red blood cell (RBC) disorders, or genes that might be implicated in erythropoiesis by genome-wide association studies of RBC traits.
Introduction
The human fetal-to-adult hemoglobin (HbA) switch is a major developmental event occurring after birth. During this transition, HbA progressively replaces fetal hemoglobin (HbF) as the main hemoglobin produced by erythroblasts (1,2). HbF accounts generally for <1% of the total hemoglobin in healthy adults. The β-chain of hemoglobin tetramers are mainly composed of γ- and β-globin in fetal and adult RBCs, respectively (1,3). Sickle cell disease (SCD) and β-thalassemia are genetic disorders resulting from mutations at the β-globin gene (HBB) locus. These mutations normally do not affect HBG1/HBG2, which encode the γ-globin chains. The most important modifier of severity for SCD and β-thalassemia is HbF: patients that naturally continue to produce high HbF levels have a longer life expectancy and suffer from fewer complications (4). Re-activation of HbF is a highly promising therapy for the treatment of these blood disorders (1,2).
Loci associated with HbF levels have been identified through candidate-gene DNA sequencing, linkage scans, and genome-wide association studies (GWASs). The association of this trait with variants at the HBB, BCL11A and HBSIL-MYB loci has been well replicated (5–9). More recently, whole-genome sequencing in the Sardinian population has revealed an association with a rare intronic genetic variant in the NFIX gene, although this variant has not been replicated (10). Genetic studies can help prioritize attractive therapeutic targets for HbF re-activation, but further functional characterization is needed to understand the role of these variants and genes on the regulation of HbF. For instance, variants act on BCL11A, a transcriptional repressor of HbF, by disrupting an erythroid developmental stage-specific enhancer of the gene (11). At the HBS1L-MYB locus, HbF-associated variants act by modulating expression of MYB, resulting in erythroid differentiation defects (12).
The fetal-to-adult erythropoietic developmental transition is marked by several transcriptomic changes. The adult erythroid transcriptional program is characterized by increased expression of BCL11A, CA1 and GCNT2 (13–15). Studies have also implicated microRNAs (miRNAs) in the regulation of HbF production. Specifically, several members of the let-7 miRNA family are up-regulated in adult erythroblasts (16–19). Overexpression of LIN28A and LIN28B, which degrades let-7 miRNAs, leads to increased HbF levels (16,20,21). miR-96 is up-regulated in adult reticulocytes and its overexpression inhibits γ-globin gene expression (17). Expression of miR-486 increases during erythropoiesis, and the mature miRNA generated from its 3p arm precursor directly targets BCL11A in erythroid cells (22). Elevated HbF levels in partial trisomy 13 cases have been linked to additional copies of miR-15a and miR-16–1, which themselves modulate MYB expression levels (23). Finally, elevated expression of miR-26b, miR-151–3p, miR-148a and miR-494 has been observed in SCD patients treated with the HbF-inducing agent hydroxyurea (24,25). In particular, overexpression of miR-26b in K562 cells increased γ-globin gene expression (26).
Given the paramount role that transcriptional regulation plays in the fetal-to-HbA switch in human erythrocytes and its direct implication for the development of therapies for the β-hemoglobinopathies, it is important to comprehensively describe and compare the transcriptome—including coding and non-coding RNAs—of fetal and adult human erythroblasts. Previous attempts were limited by the technology used (e.g. microarrays), the number of samples tested, or the absence of fetal erythroblasts in the final study design (17,18,27–32). Here, we performed RNA- and small RNA-sequencing (RNA-seq) of erythroblasts from 12 fetal and 12 adult samples from human donors. We validated the differential expression of 72 miRNAs by quantitative polymerase chain reaction (qPCR) in independent samples. The transcriptome of fetal and adult erythroblast captured known developmental stage-specific transcriptional events, and displayed substantial differences in terms of mRNA and miRNA expression. We confirmed that let-7 miRNA family members were predominantly expressed in adult erythroblasts. Conversely, the chromosome 14q32 miRNA cluster was substantially up-regulated in fetal erythroblasts, suggesting a novel role for this imprinted region in the regulation of the fetal erythroid transcriptional program.
Results
Fetal and adult erythroblasts have different transcriptomic profiles
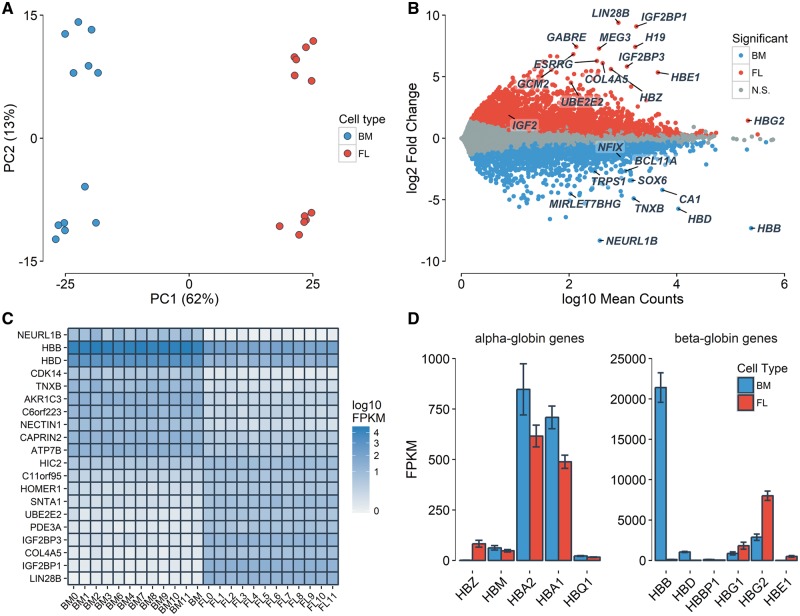
We first measured global gene expression levels in erythroblasts from 12 fetal liver and 12 adult bone marrow human donors. We had previously described genome-wide DNA methylation patterns in these samples (33). Principal component (PC) analysis of gene expression profiles recapitulated the erythroblast developmental stage (fetal versus adult, first PC = 62% of the variance explained) and the sex of the donors (second PC = 13% of the variance explained) (Fig. 1A). In total, 3687 and 4142 transcripts were significantly [false discovery rate (FDR) < 0.05] up-regulated in adult and fetal erythroblasts, respectively (Fig. 1B, Supplementary Material, Table S1). Of these, 1129 adult and 2000 fetal up-regulated genes had a fold change (FC) ≥ 2. As expected, adult β-globin (HBB) and fetal γ-globin (HBG1/HBG2) were highly expressed, and both were DE in the expected direction (Fig. 1B–D, Supplementary Material, Table S1). Consistent with previous observations (33), we noted the unexpected expression of some of the globin genes in these ex vivo differentiated erythroblasts, such as the expression of the embryonic globin genes (HBZ and HBE) or the low-level expression of adult HBB in fetal erythroblasts (Fig. 1D, Supplementary Material, Table S1).
Figure 1.
Differential gene expression between fetal and adult human erythroblast. Fetal and adult erythroblasts were differentiated ex vivo from CD34+ HSPCs collected from the fetal liver (FL) and adult bone marrow (BM). (A) PC analysis of the 500 most variable genes. The first PC captures the developmental stage, whereas the second PC is explained by the sex of the donors (genes expressed on the X-chromosome). (B) DE genes that are up-regulated in fetal (red) and adult (blue) erythroblasts (FDR <5%). The y-axis represents the log2 of fetal-to-adult expression FC. The x-axis represents the mean normalized gene counts calculated by DESeq2. N.S., not significant. (C) Expression of the top 20 most significantly DE genes in order of FC. (D) Mean FPKM of genes at the α- and β-globin loci. Error bars represent the S.E.M.
Genes marking the fetal-to-adult erythroid transition such as CA1 and GCNT2, as well as genes implicated in γ-globin gene silencing such as BCL11A and SOX6, were up-regulated in adult erythroblasts (Fig. 1B and C, Supplementary Material, Table S1). LIN28B was strongly up-regulated in fetal erythroblasts (log2 FC = 9.4, P = 2 × 10−254). LIN28B has been shown to regulate the expression and translation of IGF2, which is highly expressed in fetal hepatocytes supporting hematopoietic stem/progenitor cells (HSPCs) (16,34,35). IGF2 (log2 FC = 1.9, P = 5 × 10−5), as well as its binding partners IGF2BP1 (log2 FC = 9.1, P < 1 × 10−300) and IGF2BP3 (log2 FC = 5.8, P < 3 × 10−267) were also strongly up-regulated in fetal erythroblasts (Fig. 1B and C, Supplementary Material, Table S1). Overexpression of IGF2BP1 in adult RBC progenitors increased HbF levels (36). As expected given its role in erythroid cell proliferation, MYB was not differentially expressed (DE) (log2 FC = 0.08, P = 0.54). An intronic variant of NFIX was recently associated with HbF levels in the Sardinian population (10). This gene was up-regulated in adult erythroblasts (log2 FC = −1.5, P = 5 × 10−31). We had observed that CpGs near this gene show differential DNA methylation between fetal and adult erythroblasts, and its binding motif was enriched in differentially methylated regions (33). We noted that two long non-coding RNA, MEG3 and H19, were also strongly up-regulated in fetal erythroblasts (P < 5 × 10−68) (Fig. 1B, Supplementary Material, Table S1). We detected the strong differential expression of the ubiquitin ligase NEURL1B (log2 FC = −8.3, P = 5 × 10−112) and the ubiquitin conjugating enzyme UBE2E2 (log2 FC = 4.5, P = 1 × 10−154), respectively up-regulated in adult and fetal erythroblasts (Fig. 1B and C). This supports an emerging role for protein degradation in erythropoiesis (37).
By considering genes that are DE, we identified 75 and 29 biological pathways that were enriched (FDR < 5%) in fetal and adult erythroblasts, respectively (Supplementary Material, Table S2). Most of these pathways were not highlighted in a previous transcriptomic analysis of terminal erythroid differentiation (28), suggesting that the transcriptomic differences observed here are more likely due to the developmental stage of the cells, as opposed to differences in their kinetics of differentiation. DE genes that were up-regulated in fetal erythroblasts were enriched in pathways such as Rap1 signaling (P = 6 × 10−15), cAMP signaling (P = 4 × 10−14), and calcium signaling (P = 5 × 10−14) (Supplementary Material, Table S2). The cAMP pathway has been implicated in sustaining high levels of HbF (38). In adult erythroblasts, up-regulated genes highlighted the adipocytokine signaling (P = 2 × 10−13), Fanconi anemia (P = 5 × 10−10) and butyrate (P = 8 × 10−6) and fatty acid metabolism (P = 8 × 10−5) pathways (Supplementary Material, Table S2). Mutations in genes that lead to Fanconi anemia are associated with increased HbF levels (39,40). Moreover, short-chain fatty acids, like butyrate, induce HbF synthesis (41), and butyrate has been used in a phase 2 clinical trial to re-activate HbF production in SCD patients (42).
The 14q32 miRNA cluster is up-regulated in fetal erythroblasts
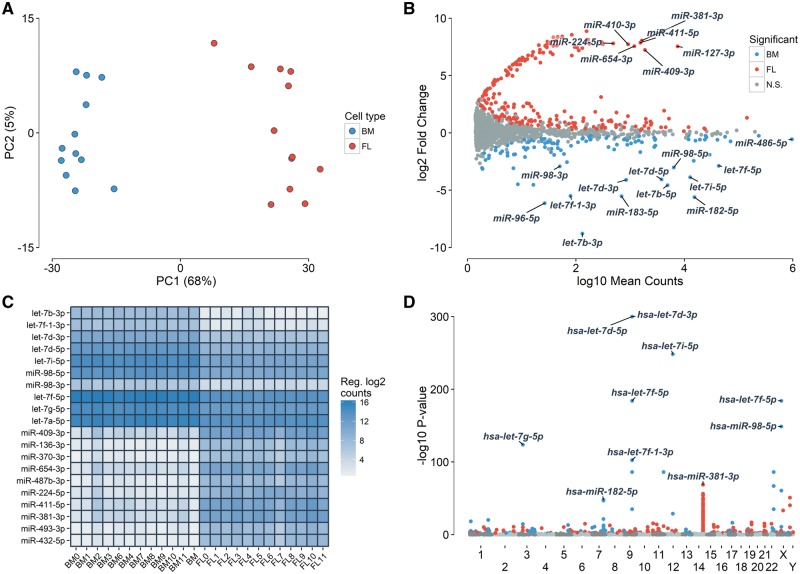
We next carried out the first comprehensive survey of miRNAs expressed in human erythroblasts by sequencing small RNAs in all 24 erythroblast samples. We measured expression of 1, 481 known miRNAs and identified 214 novel miRNAs based on in silico predictions from the miRDeep2 software (Supplementary Material, Tables S3 and S4) (43). PC analysis of miRNA expression captured cell type (first PC = 68% of the variance explained) (Fig. 2A). In total, 139 and 263 miRNAs were up-regulated in adult and fetal erythroblasts, respectively (Fig. 2B, Supplementary Material, Table S3). Among the DE miRNAs, a few had previously been implicated in erythropoiesis: miR96–5p, miR-15a and miR-16–1 were up-regulated in adult erythroblasts, whereas miR-26b-3p and mir-494–3p were significantly up-regulated in fetal erythroblasts (Supplementary Material, Table S3). miR-486–5p was the most expressed miRNA in both fetal and adult erythroblasts, contributing ∼34% of all reads mapping to mature miRNAs (Fig. 2B). This miRNA is regulated by the erythroid master regulator GATA1 and its expression increases during erythropoiesis (44). Members of the let-7 miRNA family, which are expressed during the adult-stage erythroid program, represented the most up-regulated family of miRNAs in adult erythroblasts (Fig. 2B–D, Supplementary Material, Table S3) (16,18). The let-7 miRNAs contributed 6.8 and 1.5% of all reads mapping to mature miRNAs in adult and fetal erythroblasts, respectively. When mapping each mature miRNA to the genomic position of their precursor sequences, we found that the large miRNA cluster in the chromosome 14q32 imprinted region was strongly up-regulated in fetal erythroblasts (Fig. 2D, Supplementary Material, Fig. S1). The most significant miRNA of this locus was miR-381–3p (log2 FC = 8.1, P = 1.5 × 10−69). These 14q32 miRNAs contributed 1.4% of the total reads mapping to mature sequences in fetal erythroblasts, whereas they contributed to only 0.005% of reads in adult-stage cells. Although we identified a sub-group of 14 miRNAs that share a highly similar motif (ACAACAGG) (Supplementary Material, Fig. S2), most of the 77 up-regulated 14q32 miRNAs do not share a seed sequence (first 2–7 nucleotides) and therefore likely target multiple genes.
Figure 2.
Many miRNAs are DE between fetal and adult human erythroblast. Fetal and adult erythroblasts were differentiated ex vivo from CD34+ HSPCs collected from the fetal liver (FL) and adult bone marrow (BM). (A) PC analysis of the 500 most variable miRNAs. The first PC captures the developmental stage of the erythroblasts. (B) DE miRNAs that are up-regulated in fetal (red) and adult (blue) erythroblasts (FDR < 5%). The y-axis represents the log2 of fetal-to-adult expression FC. The x-axis represents the mean normalized gene counts calculated by DESeq2. Most of the outstanding miRNA up-regulated in fetal erythroblasts are from the 14q32 locus. N.S., not significant. (C) Expression of the top 10 most up-regulated miRNAs in fetal erythroblasts, and of the 10 most up-regulated miRNAs in adult erythroblasts. Expression is reported in counts normalized using DESeq2’s regularized log2 transformation. (D) Manhattan plot of DE miRNAs P-values (y-axis) based on the genomic location of their precursor sequences (x-axis).
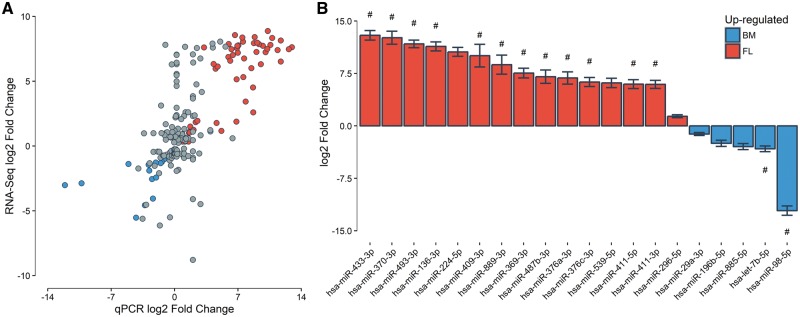
We took advantage of an independent qPCR dataset of miRNAs measured in three fetal and three adult erythroblast samples to validate our miRNA RNA-seq results (see ‘Materials and Methods’ section). Of 202 miRNAs that were DE in our RNA-seq analysis and tested in the smaller qPCR experiment, 72 showed a consistent and nominally significant differential expression pattern (one tailed P < 0.05), and 160 miRNAs were up-regulated in the expected cell type (binomial P = 2 × 10−17). There was a significant correlation between the miRNA expression FCs of both assays (fetal versus adult, Pearson’s correlation r = 0.73, P = 1.1 × 10−34, Fig. 3A). miRNAs consistently up-regulated in adult erythroblasts in both datasets were over-represented by the let-7 miRNA family (7/14, one-tailed Fisher’s exact test P = 0.01), whereas miRNA up-regulated in fetal erythroblasts were predominantly from the chromosome 14q32 imprinted locus (36/58, one-tailed Fisher’s exact test P = 0.02) (Fig. 3B, Supplementary Material, Table S5). Altogether, the qPCR experiment in a limited number of human erythroblast represents a strong and independent validation of our miRNA RNA-seq results.
Figure 3.
Validation of DE miRNAs by qPCR. (A) Correlation between miRNA expression FCs obtained by RNA-seq (Nadult = 12, Nfetal = 12) and qPCR (Nadult=3, Nfetal = 3). Only DE miRNA in the RNA-seq experiment are included. Red and blue dots represent DE miRNAs with one-tailed P < 0.05 in the qPCR experiment that are up-regulated in fetal and adult erythroblasts, respectively. (B) qPCR log2 FC for the 20 most DE miRNAs in the qPCR experiment (all one-tailed P < 5 × 10−3). Each bar represents the mean log2 FC for adult (blue) and fetal (red) erythroblasts. Error bars represent the S.E.M. Bars marked with # correspond to miRNAs from the let-7 family (blue bars) or the 14q32 locus (red bars).
Integration of miRNA and mRNA expression data
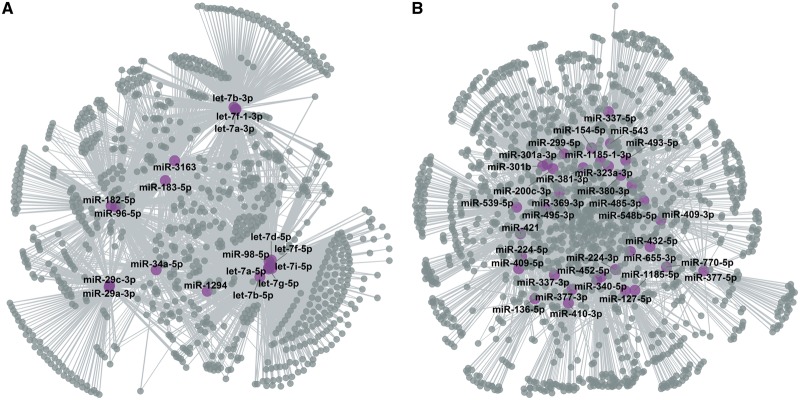
miRNA can modulate gene expression by inhibiting translation and promoting the degradation of their target genes. Accordingly, it should be possible to identify pairs of miRNA-gene in which both are DE, but in the opposite direction. In other words, we looked for miRNAs that are up-regulated in adult or fetal erythroblasts, and anti-correlated with their target genes in the same cell type. We linked miRNAs to their target genes using an aggregated list of validated and predicted miRNA–target interactions (see ‘Materials and Methods’ section). Using the information in Supplementary Material, Table S6, simple queries can identify which DE miRNAs target which genes, or which DE genes are targeted by which miRNAs. We found 18 and 34 adult and fetal up-regulated miRNAs that were enriched for down-regulated targets (hypergeometric test FDR < 0.05; Fig. 4, Supplementary Material, Table S7). Of the 18 adult-stage miRNAs, ten were from the let-7 miRNA family—an enrichment since only 15 of the 106 adult-stage miRNAs included in this analysis were from the let-7 miRNA family (Fisher’s exact test P = 0.0003). This highlights the importance that this family of miRNA plays in the adult erythroid program. Another adult-stage enriched miRNA, miR-96–5p, is an inhibitor of γ-globin gene expression (17). In fetal erythroblasts, 25 of the 34 up-regulated miRNAs belong to the 14q32 miRNA cluster (Fisher’s exact test P = 6 × 10−5). Moreover, seven of these fetal-stage miRNAs were predicted to target BCL11A, including miR-381–3p—the most significantly up-regulated miRNA in fetal erythroblasts (Fig. 2B–D).
Figure 4.
miRNA target networks. Using validated and predicted connections between miRNAs and their target genes, we identified genes that are down-regulated when their targeting miRNA is up-regulated. Gray and purple circles correspond to DE target genes and miRNAs, respectively. (A) The network in adult erythroblasts includes 18 DE miRNAs. Most adult-stage miRNAs target the same genes, consistent with an over-representation of members of the let-7 family. (B) Network of 34 up-regulated fetal miRNAs enriched for down-regulated targets.
In the previous section, we identified DE miRNAs that target more DE genes than expected by the null hypothesis. To complement that analysis, we also explored whether some DE genes were enriched targets of DE miRNAs. We found 27 adult- and seven fetal-stage down-regulated genes that were targeted more often by DE miRNAs than what was expected by chance (FDR < 0.05, Table 1). The higher number of adult-stage down-regulated genes was again due to the over-representation of let-7 miRNAs, which often target the same genes. Indeed, 26 of these 27 adult-stage down-regulated genes were targeted by at least one let-7 miRNA family member (Supplementary Material, Table S7). The most enriched target gene that was down-regulated in fetal erythroblasts was TRPS1 (P < 1 × 10−4), a transcriptional repressor that specifically binds GATA sequences (45). BCL11A was a potential target of 17 fetal up-regulated miRNAs, but was only nominally enriched (P = 0.01, q-value = 0.18).
Table 1.
DE genes that are significantly targeted by DE miRNAs in human erythroblasts
| Gene symbol | Annotation | Number of miRNAs targeting this gene (number of DE miRNAs) | P-value | q-value |
|---|---|---|---|---|
| Genes down-regulated in fetal erythroblasts | ||||
| TRPS1 | Transcriptional repressor GATA binding 1 | 137 (32) | <1 × 10−4 | <0.05 |
| HGF | Hepatocyte growth factor | 28 (10) | 0.0001 | 0.0097 |
| JAZF1 | JAZF zinc finger 1 | 108 (23) | 0.0005 | 0.026 |
| ERRFI1 | ERBB receptor feedback inhibitor 1 | 36 (11) | 0.0006 | 0.029 |
| SLC46A3 | Solute carrier family 46 member 3 | 33 (10) | 0.0006 | 0.029 |
| RIMKLB | Ribosomal modification protein rimK like family member B | 93 (20) | 0.0009 | 0.039 |
| THRB | Thyroid hormone receptor beta | 63 (15) | 0.0011 | 0.046 |
| Genes down-regulated in adult erythroblasts | ||||
| GYG2 | Glycogenin 2 | 14 (7) | <1 × 10−4 | <0.05 |
| PLCB4 | Phospholipase C beta 4 | 47 (16) | <1 × 10−4 | <0.05 |
| DAGLA | Diacylglycerol lipase alpha | 39 (15) | <1 × 10−4 | <0.05 |
| DZIP1 | DAZ interacting zinc finger protein 1 | 25 (11) | <1 × 10−4 | <0.05 |
| GPAT3 | Glycerol-3-phosphate acyltransferase 3 | 13 (8) | <1 × 10−4 | <0.05 |
| PTPRO | Protein tyrosine phosphatase | 39 (15) | <1 × 10−4 | <0.05 |
| SLAMF6 | SLAM family member 6 | 13 (8) | <1 × 10−4 | <0.05 |
| LIPH | Lipase H | 13 (8) | <1 × 10−4 | <0.05 |
| COL4A6 | Collagen type IV alpha 6 chain | 26 (12) | <1 × 10−4 | <0.05 |
| GRID2IP | Grid2 interacting protein | 13 (7) | <1 × 10−4 | <0.05 |
| STARD13 | StAR related lipid transfer domain containing 13 | 70 (20) | 0.0001 | 0.0097 |
| CCR7 | C-C motif chemokine receptor 7 | 16 (8) | 0.0002 | 0.015 |
| STX3 | Syntaxin 3 | 23 (9) | 0.0002 | 0.015 |
| NAT8L | N-acetyltransferase 8 like | 34 (12) | 0.0002 | 0.015 |
| ADRB3 | Adrenoceptor beta 3 | 17 (8) | 0.0002 | 0.015 |
| PHACTR2 | Phosphatase and actin regulator | 87 (20) | 0.0003 | 0.021 |
| PLD3 | Phospholipase D family member 3 | 15 (7) | 0.0004 | 0.023 |
| C1orf21 | Chromosome 1 open reading frame 21 | 87 (21) | 0.0004 | 0.023 |
| HIF3A | Hypoxia inducible factor 3 alpha subunit | 29 (10) | 0.0004 | 0.023 |
| FBN1 | Fibrillin 1 | 36 (11) | 0.0004 | 0.023 |
| MYCN | v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog | 88 (20) | 0.0005 | 0.026 |
| ELOVL4 | ELOVL fatty acid elongase 4 | 47 (13) | 0.0007 | 0.031 |
| NRARP | NOTCH-regulated ankyrin repeat protein | 32 (10) | 0.0007 | 0.031 |
| HES1 | Hes family bHLH transcription factor 1 | 23 (8) | 0.0011 | 0.045 |
| COL1A1 | Collagen type I alpha 1 chain | 39 (11) | 0.0012 | 0.046 |
| ASIC1 | Acid sensing ion channel subunit 1 | 30 (9) | 0.0012 | 0.046 |
| ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif 6 | 71 (16) | 0.0013 | 0.048 |
Discussion
In summary, gene expression patterns of fetal and adult human erythroblasts largely capture cell type specificity. Using comprehensive RNA-seq, we found >7800 transcripts that are DE between fetal and adult erythroblast. In comparison with previous transcriptomic studies (17,18,27–32), we identified a larger number of DE genes between fetal and adult human erythroblasts because of our larger sample size, allowing us to detect more accurately small difference in gene expression levels. DE genes include known genes associated with erythroid developmental stages, and are enriched in pathways that have been implicated in HbF production. We also found 139 and 263 miRNAs that are up-regulated in adult and fetal erythroblasts, respectively. For the vast majority of these miRNAs, our study is the first to report their differential expression in human erythroblasts. Finally, we used computational approaches to integrate mRNA and miRNA sequencing to create co-expression networks in erythroid cells.
To our knowledge, we are the first to report up-regulation in human fetal erythroblasts of miRNAs (Fig. 2B) and long non-coding RNAs (MEG8) that map to the imprinted region on chromosome 14q32. Uniparental disomy (UPD), that is the aberrant expression of imprinted genes at the 14q32 locus, is responsible for the Temple [UDP14(maternal), MIM no. 616222] and Kagami-Ogata [UDP14(paternal), MIM no. 608149] syndromes. Both of these syndromes are characterized by impaired intrauterine development, growth issues following birth, and reduced survival (46,47). Hematopoietic defects have not been reported in UPD14 patients, and it is unclear what might be the roles that small and long non-coding RNAs encoded at the 14q32 locus have in fetal erythropoiesis. Epigenetic dysregulation at 14q32 has been linked to tumorigenesis, including lung cancer and osteosarcoma (48,49). Interestingly, loss of heterozygosity analyses of this locus in acute lymphoblastic leukemia has revealed that a reduction of the expression of the miRNAs located at 14q32 is associated with increased expression of BCL11A (50). If this observation translates from lymphocytes to erythroid cells, we can hypothesize that one important role for the miRNAs at 14q32 in fetal erythroblasts is to reduce or block BCL11A expression and allow HbF production. Thus, modulating the 14q32-controlled transcriptional dynamics in erythroblasts might represent an interesting strategy to re-activate HbF production.
To understand how gene expression is developmentally regulated at 14q32, we retrieved Hi-C and epigenomic annotations for this locus from fetal and adult erythroblasts (Supplementary Material, Fig. S3) (32,51). Hi-C data can be used to partition the genome of erythroid cells between A (open chromatin) and B (repressive chromatin) compartments. Chromatin compartments at 14q32 seem to switch in the expected A-to-B direction between fetal and adult erythroblasts (see PC1 and D.I. in Supplementary Material, Fig. S3). When we focus on histone tail marks at the locus, we find that there is a group of erythroid enhancers near the cluster of DE 14q32 miRNAs and genes, and that these enhancers seem more active (as evidenced by increased H3K4me1/me2, H3K9ac, and H3K27ac histone tail marks) in fetal erythroblasts (Supplementary Material, Fig. S3). In addition, we note stronger Hi-C contacts between these erythroid enhancers and the 14q32 miRNA cluster in fetal erythroblasts. Genome editing experiments of these erythroid enhancers may yield interesting insights into how miRNAs and genes at 14q32 impact fetal erythropoiesis.
GWAS have identified hundreds of variants associated with RBC indices, including HbF (52–54). Most of these variants fall within non-coding regions, and therefore likely modulate phenotypic variation by regulating gene expression specifically in RBCs (55). Our comprehensive transcriptomic analyses of fetal and adult human erythroblasts provide a critical resource to link together variants associated with erythrocytes indices and gene expression, facilitating the identification of causal genes at these GWAS loci. Further, our comparison of transcriptomes from fetal and adult human erythroblasts highlight several genes and biological pathways, including DE miRNAs at the 14q32 locus, that could be functionally characterized for a potential role in the fetal-to-HbA switch. Our results could impact our strategies to develop new therapies in order to re-activate HbF production in SCD and β-thalassemia patients.
Materials and Methods
Differential gene expression analysis
We differentiated erythroblasts from HSPCs as previously described in (15,33). We purchased human fetal (fetal liver, N = 12) and adult (bone marrow, n = 12) CD34+ HSPCs from DV Biologics and Lonza, respectively. We also described the RNA extraction and RNA-seq protocols elsewhere (33,55). Briefly, we performed RNA-seq with an Illumina HiSeq2000 sequencer using a stranded cDNA library and a paired-ends 100-bp protocol. We mapped reads to the genome (hg19) using Tophat2 (v.2.0.9, with options –library-type fr-firststrand –microexon-search –coverage-search) (56). Per gene read counts were obtained using htseq-count (v. 0.6.0, with options -f bam -r name -s reverse -t exon -i gene_id) on Ensembl gene sets (release 75). We tested differential expression of genes using the R package DESeq2 (v.1.12.4) (57). For visualization, we calculated Fragments Per Kilobase per Million mapped reads (FPKM) using DESeq2’s fpkm function and Ensembl General Transfer Format file release 75 (hg19). Data are available from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (accession number: GSE90878).
Differential miRNA expression analysis
We performed RNA-seq of small RNAs on an Illumina HiSeq2000 sequencer using a cDNA library with a single-end 50-bp protocol (TruSeq small RNA library kit). We removed adapter sequences using the fastx-clipper tool (v.0.0.14). We removed reads that mapped to ribosomal genes using bowtie (v.1.0.0, with options -n 0 -S –best –chunkmbs 200). We mapped reads to both mature and precursor miRNA sequences from miRBASE release 21 using mirDeep2 (v.0.0.5) (43). We considered potentially novel miRNA with a mirDeep2 score >3, which had an estimated 88 ± 2% true positive probability. We tested differential expression of miRNAs using the R package DESeq2 (v.1.12.4) (57). We looked for motif enrichment in the first nine nucleotides of miRNAs using MEME (MEME suite 4.12.0) (58). Data are available from the NCBI GEO repository (accession number: GSE90878).
Enrichment analyses
To identify miRNA that were enriched for DE target genes, we downloaded for each miRNA all validated targets and the top 35% predicted targets using multiMiR (v.2.1.1) (59). We only considered miRNA–target pairs that were either functionally validated in at least one resource or that were predicted in at least three databases out of the 11 databases that we queried (miRecords, miRTarBase, TarBase, DIANA-microT-CDS, ElMMo, MicroCosm, miRanda, miRDB, PicTar, PITA and TargetScan). For each miRNA, we counted how many target genes were both DE between fetal and adult erythroblasts and had their expression levels inversely correlated with the miRNA expression levels (Pearson’s correlation tests, FDR < 0.05). We calculated statistical enrichments using hypergeometric tests.
In addition, we evaluated if DE genes were overrepresented in miRNA targets by assessing how many times each gene was targeted by a DE miRNA. Since the number of miRNA targeting a specific gene was often small, we permuted miRNA DE labels 10 000 times to assess significance. We counted the number of times a gene was targeted by more DE miRNAs in permutations than what was observed, and derived an empirical P-value by calculating the ratio of this value over the total number of permutations. We restricted this analysis to genes targeted by at least 10 miRNAs with a FC >2. We used the Generally Applicable Gene-set Enrichment (v.2.22.0) method to calculate the enrichment of Kyoto Encyclopedia of Genes and Genomes (57,60,61).
miRNA qPCR validation
We differentiated erythroblasts from HSPCs from the fetal liver (n = 3) or the bone marrow (n = 3) as previously described in (15). We measured expression using a TaqMan Low Density Array covering 768 different miRNAs (ThermoFisher Scientific). We derived expression levels using the 2−ΔΔCt method (62). We tested for differential expression using a one-sided t-test on the ΔΔCt values for 202 miRNAs that were found to be DE in the RNA-seq dataset. miRNAs with differential expression one-tailed P < 0.05 in the consistent direction were considered as replicated.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
S.L. held fellowships from the Canadian Institute of Health Research (CIHR) and the ‘Fondation Pierre Lavoie’. D.E.B. was supported by NIDDK (K08DK093705), NHLBI (P01HL032262, DP2OD022716), and Burroughs Wellcome Fund. GL was supported by grants from the CIHR [123382], the Doris Duke Charitable Foundation, the National Sciences and Engineering Research Council of Canada [RGPIN-2016–04597], the Montreal Heart Institute Foundation and the Canada Research Chair Program.
Supplementary Material
References
- 1. Bauer D.E., Kamran S.C., Orkin S.H. (2012) Reawakening fetal hemoglobin: prospects for new therapies for the beta-globin disorders. Blood, 120, 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lettre G., Bauer D.E. (2016) Fetal haemoglobin in sickle-cell disease: from genetic epidemiology to new therapeutic strategies. Lancet, 387, 2554–2564. [DOI] [PubMed] [Google Scholar]
- 3. Stamatoyannopoulos G. (2005) Control of globin gene expression during development and erythroid differentiation. Exp. Hematol., 33, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Platt O.S., Brambilla D.J., Rosse W.F., Milner P.F., Castro O., Steinberg M.H., Klug P.P. (1994) Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med., 330, 1639–1644. [DOI] [PubMed] [Google Scholar]
- 5. Menzel S., Garner C., Gut I., Matsuda F., Yamaguchi M., Heath S., Foglio M., Zelenika D., Boland A., Rooks H.. et al. (2007) A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet., 39, 1197–1199. [DOI] [PubMed] [Google Scholar]
- 6. Lettre G., Sankaran V.G., Bezerra M.A., Araujo A.S., Uda M., Sanna S., Cao A., Schlessinger D., Costa F.F., Hirschhorn J.N.. et al. (2008) DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. U. S. A., 105, 11869–11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thein S.L., Menzel S., Peng X., Best S., Jiang J., Close J., Silver N., Gerovasilli A., Ping C., Yamaguchi M.. et al. (2007) Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc. Natl. Acad. Sci.U. S. A., 104, 11346–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uda M., Galanello R., Sanna S., Lettre G., Sankaran V.G., Chen W., Usala G., Busonero F., Maschio A., Albai G.. et al. (2008) Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. U. S. A., 105, 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galarneau G., Palmer C.D., Sankaran V.G., Orkin S.H., Hirschhorn J.N., Lettre G. (2010) Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet., 42, 1049–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Danjou F., Zoledziewska M., Sidore C., Steri M., Busonero F., Maschio A., Mulas A., Perseu L., Barella S., Porcu E.. et al. (2015) Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat. Genet., 47, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L.. et al. (2013) An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science, 342, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Canver M.C., Lessard S., Pinello L., Wu Y., Ilboudo Y., Stern E.N., Needleman A.J., Galacteros F., Brugnara C., Kutlar A.. et al. (2017) Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat. Genet., 49, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brady H.J., Edwards M., Linch D.C., Knott L., Barlow J.H., Butterworth P.H. (1990) Expression of the human carbonic anhydrase I gene is activated late in fetal erythroid development and regulated by stage-specific trans-acting factors. Br. J. Haematol., 76, 135–142. [DOI] [PubMed] [Google Scholar]
- 14. Inaba N., Hiruma T., Togayachi A., Iwasaki H., Wang X.H., Furukawa Y., Sumi R., Kudo T., Fujimura K., Iwai T.. et al. (2003) A novel I-branching beta-1, 6-N-acetylglucosaminyltransferase involved in human blood group I antigen expression. Blood, 101, 2870–2876. [DOI] [PubMed] [Google Scholar]
- 15. Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science, 322, 1839–1842. [DOI] [PubMed] [Google Scholar]
- 16. Lee Y.T., de Vasconcellos J.F., Yuan J., Byrnes C., Noh S.J., Meier E.R., Kim K.S., Rabel A., Kaushal M., Muljo S.A.. et al. (2013) LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood, 122, 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Azzouzi I., Moest H., Winkler J., Fauchere J.C., Gerber A.P., Wollscheid B., Stoffel M., Schmugge M., Speer O. (2011) MicroRNA-96 directly inhibits gamma-globin expression in human erythropoiesis. PLoS One, 6, e22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noh S.J., Miller S.H., Lee Y.T., Goh S.H., Marincola F.M., Stroncek D.F., Reed C., Wang E., Miller J.L. (2009) Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. J. Transl. Med., 7, 98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Vasconcellos J.F., Byrnes C., Lee Y.T., Allwardt J.M., Kaushal M., Rabel A., Miller J.L. (2017) Tough decoy targeting of predominant let-7 miRNA species in adult human hematopoietic cells. J. Transl. Med., 15, 169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piskounova E., Viswanathan S.R., Janas M., LaPierre R.J., Daley G.Q., Sliz P., Gregory R.I. (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem., 283, 21310–21314. [DOI] [PubMed] [Google Scholar]
- 21. Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. (2008) Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell, 32, 276–284. [DOI] [PubMed] [Google Scholar]
- 22. Lulli V., Romania P., Morsilli O., Cianciulli P., Gabbianelli M., Testa U., Giuliani A., Marziali G. (2013) MicroRNA-486-3p regulates gamma-globin expression in human erythroid cells by directly modulating BCL11A. PloS One, 8, e60436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sankaran V.G., Menne T.F., Scepanovic D., Vergilio J.A., Ji P., Kim J., Thiru P., Orkin S.H., Lander E.S., Lodish H.F. (2011) MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc. Natl. Acad. Sci. U. S. A., 108, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saki N., Abroun S., Soleimani M., Kavianpour M., Shahjahani M., Mohammadi-Asl J., Hajizamani S. (2016) MicroRNA expression in beta-thalassemia and sickle cell disease: a role in the induction of fetal hemoglobin. Cell J., 17, 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker A.L., Steward S., Howard T.A., Mortier N., Smeltzer M., Wang Y.D., Ware R.E. (2011) Epigenetic and molecular profiles of erythroid cells after hydroxyurea treatment in sickle cell anemia. Blood, 118, 5664–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alijani S., Alizadeh S., Kazemi A., Khatib Z.K., Soleimani M., Rezvani M., Minayi N., Karami F., Tayebi B. (2014) Evaluation of the Effect of miR-26b Up-Regulation on HbF Expression in Erythroleukemic K-562 Cell Line. Avicenna J. Med. Biotechnol., 6, 53–56. [PMC free article] [PubMed] [Google Scholar]
- 27. Goh S.H., Josleyn M., Lee Y.T., Danner R.L., Gherman R.B., Cam M.C., Miller J.L. (2007) The human reticulocyte transcriptome. Physiol. Genomics, 30, 172–178. [DOI] [PubMed] [Google Scholar]
- 28. An X., Schulz V.P., Li J., Wu K., Liu J., Xue F., Hu J., Mohandas N., Gallagher P.G. (2014) Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood, 123, 3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doss J.F., Corcoran D.L., Jima D.D., Telen M.J., Dave S.S., Chi J.T. (2015) A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics, 16, 952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi L., Lin Y.H., Sierant M.C., Zhu F., Cui S., Guan Y., Sartor M.A., Tanabe O., Lim K.C., Engel J.D. (2014) Developmental transcriptome analysis of human erythropoiesis. Hum. Mol. Genet., 23, 4528–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X., Zhang Y., Ni M., Cao H., Signer R.A.J., Li D., Li M., Gu Z., Hu Z., Dickerson K.E.. et al. (2017) Regulation of mitochondrial biogenesis in erythropoiesis by mTORC1-mediated protein translation. Nat. Cell. Biol., 19, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang P., Keller C.A., Giardine B., Grevet J.D., Davies J.O.J., Hughes J.R., Kurita R., Nakamura Y., Hardison R.C., Blobel G.A. (2017) Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Dev., 31, 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lessard S., Beaudoin M., Benkirane K., Lettre G. (2015) Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med., 7, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A. (2007) Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev., 21, 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chou S., Lodish H.F. (2010) Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A., 107, 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tumburu L., Byrnes C., Lee Y.T., de Vasconcellos J.F., Rabel A., Miller J.L. (2015) IGF2BP1 reverses hemoglobin switching in adult erythroblasts. Blood, 126, 639–639. [Google Scholar]
- 37. Nguyen A.T., Prado M.A., Schmidt P.J., Sendamarai A.K., Wilson-Grady J.T., Min M., Campagna D.R., Tian G., Shi Y., Dederer V.. et al. (2017) UBE2O remodels the proteome during terminal erythroid differentiation. Science, 357, eaan0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ikuta T., Kuroyanagi Y., Odo N., Liu S. (2013) A common signaling pathway is activated in erythroid cells expressing high levels of fetal hemoglobin: a potential role for cAMP-elevating agents in beta-globin disorders. J. Blood Med., 4, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krauss J.S., Hahn D.A., Jonah M.H., Trinh M. (1985) Estimation of highly increased concentrations of fetal hemoglobin in Fanconi’s anemia. Clin. Chem., 31, 1737–1738. [PubMed] [Google Scholar]
- 40. Alter B.P., Rosenberg P.S., Day T., Menzel S., Giri N., Savage S.A., Thein S.L. (2013) Genetic regulation of fetal haemoglobin in inherited bone marrow failure syndromes. Br. J. Haematol., 162, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhatia H., Hallock J.L., Dutta A., Karkashon S., Sterner L.S., Miyazaki T., Dean A., Little J.A. (2009) Short-chain fatty acid-mediated effects on erythropoiesis in primary definitive erythroid cells. Blood, 113, 6440–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reid M.E., El Beshlawy A., Inati A., Kutlar A., Abboud M.R., Haynes J. Jr., Ward R., Sharon B., Taher A.T., Smith W.. et al. (2014) A double-blind, placebo-controlled phase II study of the efficacy and safety of 2, 2-dimethylbutyrate (HQK-1001), an oral fetal globin inducer, in sickle cell disease. Am. J. Hematol., 89, 709–713. [DOI] [PubMed] [Google Scholar]
- 43. Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res., 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shaham L., Vendramini E., Ge Y., Goren Y., Birger Y., Tijssen M.R., McNulty M., Geron I., Schwartzman O., Goldberg L.. et al. (2015) MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood, 125, 1292–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malik T.H., Shoichet S.A., Latham P., Kroll T.G., Peters L.L., Shivdasani R.A. (2001) Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J., 20, 1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howard M., Charalambous M. (2015) Molecular basis of imprinting disorders affecting chromosome 14: lessons from murine models. Reproduction, 149, R237–R249. [DOI] [PubMed] [Google Scholar]
- 47. Rosenfeld J.A., Fox J.E., Descartes M., Brewer F., Stroud T., Gorski J.L., Upton S.J., Moeschler J.B., Monteleone B., Neill N.J.. et al. (2015) Clinical features associated with copy number variations of the 14q32 imprinted gene cluster. Am. J. Med. Genet. A, 167, 345–353. [DOI] [PubMed] [Google Scholar]
- 48. Hill K.E., Kelly A.D., Kuijjer M.L., Barry W., Rattani A., Garbutt C.C., Kissick H., Janeway K., Perez-Atayde A., Goldsmith J.. et al. (2017) An imprinted non-coding genomic cluster at 14q32 defines clinically relevant molecular subtypes in osteosarcoma across multiple independent datasets. J. Hematol. Oncol., 10, 107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Enfield K.S., Martinez V.D., Marshall E.A., Stewart G.L., Kung S.H., Enterina J.R., Lam W.L. (2016) Deregulation of small non-coding RNAs at the DLK1-DIO3 imprinted locus predicts lung cancer patient outcome. Oncotarget, 7, 80957–80966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Agueli C., Cammarata G., Salemi D., Dagnino L., Nicoletti R., La Rosa M., Messana F., Marfia A., Bica M.G., Coniglio M.L.. et al. (2010) 14q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a. Am. J. Hematol., 85, 575–578. [DOI] [PubMed] [Google Scholar]
- 51. Xu J., Shao Z., Glass K., Bauer D.E., Pinello L., Van Handel B., Hou S., Stamatoyannopoulos J.A., Mikkola H.K., Yuan G.C.. et al. (2012) Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev. Cell, 23, 796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A.. et al. (2016) The allelic landscape of human blood cell trait variation and links to common complex disease. Cell, 167, 1415–1429 e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mousas A., Ntritsos G., Chen M.-H., Song C., Huffman J.E., Tzoulaki I., Elliott P., Psaty B.M., Auer P.L., Johnson A.D.. et al. (2017) Rare coding variants pinpoint genes that control human hematological traits. PLoS Genet., 13, e1006925.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chami N., Chen M.H., Slater A.J., Eicher J.D., Evangelou E., Tajuddin S.M., Love-Gregory L., Kacprowski T., Schick U.M., Nomura A.. et al. (2016) Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am. J. Hum. Genet., 99, 8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lessard S., Gatof E.S., Beaudoin M., Schupp P.G., Sher F., Ali A., Prehar S., Kurita R., Nakamura Y., Baena E.. et al. (2017) An erythroid-specific ATP2B4 enhancer mediates red blood cell hydration and malaria susceptibility. J. Clin. Invest., 127, 3065–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15, 550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bailey T.L., Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 59. Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., Mahaffey S., Rossi S., Calin G.A., Bemis L.. et al. (2014) The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res., 42, e133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics, 10, 161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.