Abstract
Biofilms are formed by communities of microorganisms living in a self-produced extracellular polymeric matrix attached to a surface. When living in a biofilm microorganisms change phenotype and thus are less susceptible to antibiotic treatment and biofilm infections can become severe. The aim of this study was to determine if the presence of multikingdom microorganisms alters the virulence of a biofilm infection in a host organism. The coexistence of Candida albicans and Staphylococcus epidermidis in biofilm was examined in the nematode model Caenorhabditis elegans. It was evaluated if the hyphal form of C. albicans and extracellular polymeric substances (EPS) formed by S. epidermidis increases biofilm virulence. Survival assays were performed, where C. elegans nematodes were exposed to S. epidermidis and C. albicans. Single inoculation assays showed a decreased survival rate after 2 days following exposure, while dual inoculation assays showed that a clinical S. epidermidis strain together with C. albicans significantly increased the virulence and decreased nematode survival. EPS seem to interfere with the bacterial attachment to hyphae, since the EPS overproducing S. epidermidis strain was most virulent. The clinical S. epidermidis paired with C. albicans led to a severe infection in the nematodes resulting in reduced survival.
Keywords: biofilm, infection, virulence, Staphylococcus epidermidis, Candida Albicans, Caenorhabditis elegans
The study illustrates the effect of EPS in polymicrobial biofilm infections by using a simple but efficient invertebrate model.
INTRODUCTION
Most microorganisms exist in biofilms that can be defined as communities of microorganisms set in a self-produced extracellular polymeric matrix that is attached to a surface (Fey and Olson 2010; Grant and Hung 2013). For many patients with medical implants or indwelling medical devices, biofilms pose a risk of implant-associated infections (O’Gara and Humphreys 2001). Bacteria from the patient's skin microflora, healthcare personnel or contaminated equipment can colonize medical devices, such as central venous catheters, prosthetic joints and urinary catheters during implantation (Donlan 2001; Mekni et al. 2012). It is important to understand how to control implant-associated infections because they can lead to failed implants and thus repeated surgeries, prolonged antibiotic therapy, and increased morbidity (Stoodley et al. 2008). Bacteria present in biofilms are less sensitive to antibiotics due to several factors, including limited penetration of antibiotic agent into biofilm matrix, slow growth rate of biofilm bacteria, high bacterial density and changes in gene expression (Saginur et al. 2006).
Staphylococci, mostly Staphylococcus epidermidis and S. aureus, are the predominant bacteria associated with implant-associated infections (Adam, Baillie and Douglas 2002; Fey and Olson 2010). The ability of the bacteria to adhere to the implant and form a biofilm is related to the pathogenicity (Saginur et al. 2006). Some strains of S. epidermidis can increase the bacteria's ability to form biofilms by secreting extracellular polymeric substances (EPS) that consist of polysaccharides, proteins and teichoid acid (Adam, Baillie and Douglas 2002). Polysaccharide intercellular adhesin (PIA) and capsular polysaccharide adhesion have been identified as two products of S. epidermidis that contribute heavily to biofilm formation in combination with different proteins (O'Gara and Humphreys 2001). Once biofilms and the matrix are established, it becomes difficult to eradicate pathogenic microorganisms via the host's immune response and treatment with antibiotics is often performed unsuccessfully (Grant and Hung 2013).
The fungus Candida albicans has also been associated with implant-associated infections (O'Gara and Humphreys 2001). However, C. albicans tends to cause more virulent infections than bacteria and can cause serious damage to host tissue and patient health (Adam, Baillie and Douglas 2002). Candida species are some of the most prevalent causes of nosocomial bloodstream infections (Klotz et al. 2007). In the case of urinary catheter biofilms, even patients using urinary catheters on a short-term basis of 7 days or shorter can develop a biofilm infection involving this fungal species. A study showed that all of patients who used catheters on a long-term basis of more than 28 days were expected to develop a Candida biofilm infection (Donlan 2001).
The microorganisms S. epidermidis and C. albicans are both present in humans as commensal organisms and they have often been shown to coexist in biofilms as opportunistic pathogens in immuno-compromised patients (Lin et al. 2013). This combination makes polymicrobial biofilms particularly resistant to common antimicrobial therapies. Specifically, the multikingdom fungal–bacterial biofilm may be more resistant to antimicrobial treatment in the presence of significant EPS production (Davies 2003). It was shown by Adam, Baillie and Douglas (2002) that increased virulence of C. albicans can occur in the presence of an EPS overproducing S. epidermidis strain, when compared to a combination fungal–bacterial infection involving a mutant S. epidermidis strain that exhibited limited EPS production (Adam, Baillie and Douglas 2002). This increased resistance was likely due to the presence of the thick EPS layer protecting the biofilm organisms from penetration of drugs (Adam, Baillie and Douglas 2002; Veses and Gow 2009). Candida albicans can exist in three morphological forms: yeast, hyphal and pseudo-hyphal forms (Veses and Gow 2009). The yeast form is dominant in the commensal state, while the filamentous hyphae are usually associated with a pathogenic state (Sudbery 2011). Additionally, C. albicans yeast cannot enter human cells actively, whereas C. albicans hyphae can penetrate and become opportunistic within the infected area (Sudbery 2011). These varying fungal morphologies are important for the degree of virulence of the biofilm infection.
It is valuable to study the interaction between C. albicans and EPS-producing strains of S. epidermidis because their interactions are involved in biofilm drug resistance and EPS production is important for staphylococcal pathogenicity. A research experiment by Adam, Baillie and Douglas (2002) showed that in a multikingdom biofilm, an EPS-producing wild-type strain (RP62A) of S. epidermidis blocks penetration of fluconazole, an antifungal agent, and has no effect on the metabolic activity (Adam, Baillie and Douglas 2002). In addition, the matrix viscosity of C. albicans protects the non-EPS-producing mutant (M7) of S. epidermidis from vancomycin, an antibiotic (Adam, Baillie and Douglas 2002).
For the study of the interactions between C. albicans and S. epidermidis biofilms, the nematode Caenorhabditis elegans can effectively be used as a model host organism for infection. Caenorhabditis elegans are 1 mm long hermaphroditic invertebrates found in soil and they make a useful model for studying host–pathogen interactions because their genome has been completely sequenced, they have a rapid generation time and a transparent cuticle, and they can be easily maintained (Kurz and Ewbank 2000). Caenorhabditis elegans has been used as a model system to study virulence factors of pathogenic bacteria and host defenses of the nematode (Kurz and Ewbank 2000). Nematodes are present in soil environments and feed on bacteria; therefore, it has been suggested that they have developed defensive immune responses against pathogens (Alegado et al. 2003). Pathogens that gain entrance into the nematode cuticle can proliferate in the intestine of the nematode and cause an infection (Alegado et al. 2003). The innate immune response of the nematode can be equated to the immune response of humans after exposure to pathogens prior to onset of the adaptive immune system components (0–72 h after exposure).
The biofilm infection within the nematode involves the bacteria colonizing within the gut, then disrupting the lining of the gut, which leads to organ destruction and death (Marsh and May 2012). Antibacterial factors, lysozymes and antimicrobial molecules, such as lectins, are involved in the immunity of C. elegans against invasions (Marsh and May 2012). For example, the exopolysaccharide PIA is produced when S. epidermidis colonizes the gut of C. elegans (Begun et al. 2007). Disruption of PIA synthesis greatly reduces the ability of the bacteria to infect and kill C. elegans (Begun et al. 2007). Additionally, PIA-producing S. epidermidis is more virulent towards C. elegans than non-PIA-producing S. epidermidis suggesting that the exopolysaccharides produced during biofilm production increased the virulence of the pathogen by protecting it from the immune defenses of its host (Begun et al. 2007). An increased understanding of the interactions between the fungi and bacteria will allow for modeling of these interactions in vivo within C. elegans.
The hypothesis of this research was that the interactions between C. albicans hyphae and a clinical S. epidermidis strain, which produced EPS at an enhanced rate, would decrease the survival rate of C. elegans due to increased virulence of the biofilm. It was presumed that bacteria attach to the hyphae, and during active tissue penetration, the bacteria can gain access into the nematode tissue. The commensal S. epidermidis bacteria have the ability to become pathogenic because they would be present in a new area within its host. The host–pathogen interaction between the EPS-producing strains of S. epidermidis would then differ from the co-culture exposure. When nematodes consumed the EPS-producing wild-type S. epidermidis, it was hypothesized that the EPS layer of the bacteria could act as a shield towards the nematode immune system, which would prevent it from detecting the pathogen, thus causing increased incidence of fatality. Conversely, if nematodes were exposed to the EPS-deficient S. epidermidis mutant, the survival rate was expected to increase. Without a layer of EPS surrounding the bacteria, it was hypothesized that the nematodes’ innate immune system would recognize the pathogens as foreign bodies and thereby initiate defensive actions, thus preserving the life of the host organism.
MATERIAL AND METHODS
Experimental organisms
In this study, the following bacterial and fungal strains were used: (1) Staphylococcus epidermidis (ATCC strain #155, strain isolated from an infected skin lesion); (2) S. epidermidis RP62A (ATCC strain #35 984) is a EPS-producing wild-type strain (Hussain et al. 1997); (3) S. epidermidis M7 is a EPS-negative mutant strain obtained through chemical mutagenesis of S. epidermidis RP62A with mitomycin C (Hussain et al. 1997). All S. epidermidis species were propagated from freezer stocks in Luria Broth (LB) medium overnight at 37°C. (4) Escherichia coli OP50 was used as a food source for Caenorhabditis elegans. Escherichia coli OP50 was plated on 60 mm nematode growth media (NGM) plates and stored at room temperature for up to 2–3 weeks; (5) Candida albicans strain SC5314 was used. Propagation was obtained by growing C. albicans from a freezer stock in Yeast Extract-Peptone-Dextrose (YPD) medium overnight at 37°C. The multikingdom cultures of C. albicans and S. epidermidis were prepared by growing the microorganisms separately in YPD and LB media, respectively, at 37°C. A 50:50 mix of each of the cultures was made to obtain a co-culture (Peters et al. 2010).
The nematode strain used for all experiments was C. elegans glp-4; sek-1 and was obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota (MN, USA). They were propagated on E. coli OP50 seeded NGM plates at 15°C. The nematodes are sterile above this temperature and will enter a dauer stage below this temperature (WormBook 2017).
Experimental protocol
Survival assays were carried out in a sterile environment with the use of sterile techniques. All assays had three to nine replicates. Candida albicans yeast survival assays were adapted from Pukkila-Worley et al. (2009). Briefly, nematodes were placed onto a plate containing C. albicans for several hours and then washed with M9 media. The cleaned nematodes were added to a 6-well plate and then were individually classified as alive or dead based on observable response to prodding with a worm pick. Staphylococcus epidermidis and E. coli OP50 survival assays were performed under similar conditions as C. albicans yeast. Experiments performed with C. albicans hyphae were adapted from a protocol used in the study by Breger et al. (2007) and adjusted for the applied experimental conditions in this study as briefly explained here. To complete the C. albicans hyphae infection assays, C. elegans were placed on a lawn of C. albicans where they consumed the yeast for 2 h. After 2 h, the nematodes were added to M9 media. Dead nematodes, determined by a lack of response to external stimuli, were counted and removed from the culture. Live nematodes were counted allowed to grow for 2 days to allow for hyphal penetration into the body; after 2 days, the number of live and dead nematodes were counted.
In vivo biofilm formation in Caenorhabditis elegans
Cultures were inoculated overnight at 37°C in 6-well plates containing 3 mL Roswell Park Memorial Institute media (RPMI) with 5% fetal bovine serum (FBS) in order to develop biofilms. Once the biofilms were established in the well plates, sterile nematodes were transferred into the liquid and allowed to feed for 4 h. Then, a subsample of the nematodes was washed four times with M9 buffer and subsequently washed on a vacuum filter to clear them of excess E. coli. From the filter, the nematodes were washed into M9 medium and pipetted into an assay plate containing a RPMI + 5% FBS and M9 buffer solution.
Assessing in vivo biofilm formation in Caenorhabditis elegans
The nematodes were observed and quantified after 24 and 48 h following exposure to the biofilm. Counts of live and dead nematodes were recorded. Dead nematodes were determined if they do not respond to the stimulation of the nematode pick and they were subsequently removed from the assay plate and imaged under a fluorescent microscope using LIVE/DEAD BacLight stain (Thermo Fisher Scientific) or PNA-FISH probes (OpGen). LIVE/DEAD stain contains two stains: SYTO9 which stains live cells green and propidium iodide which stains dead cells red. This stain can easily identify whether the organisms inside the nematode are alive or dead.
RESULTS
The effects of exposure to opportunistic pathogens Staphylococcus epidermidis and Candida albicans on the nematode survival rate were assessed after the nematodes had been feeding on biofilms for 4 h. To account for any effects of Escherichia coli, a control assay was performed, in which the nematodes fed on GFP-labeled E. coli OP50. The average nematode survival rates after 24 and 48 h were 85% and 72%, respectively (Figs 1 and 2). To determine if coexistence of S. epidermidis and C. albicans in a biofilm increased the virulence, nematodes were fed on biofilms grown from single inoculated or dual inoculated cultures. Dual inoculated cultures contained S. epidermidis and C. albicans present in its hyphael form. The filamentous hyphae make C. albicans a more virulent pathogen, once inside its host. However, there were not any observations of nematode–hyphal interactions in these nematode assays. Survival assays based on C. albicans hyphae showed a 63% nematode survival rate after 48 h (P < 0.05), but S. epidermidis and C. albicans yeast alone did not show significant results. The clinical strain of S. epidermidis combined with C. albicans hyphae, however, showed a significant decrease in the survival rate of 67% (P < 0.05) after 24 h (Fig. 1). The average survival rate of the clinical S. epidermidis strain was 86% compared to 81.5% from C. albicans yeast. When the two organisms were combined, the survival rate was 67% after 24 h and 47% after 48 h, respectively (Figs 1 and 2).
Figure 1.
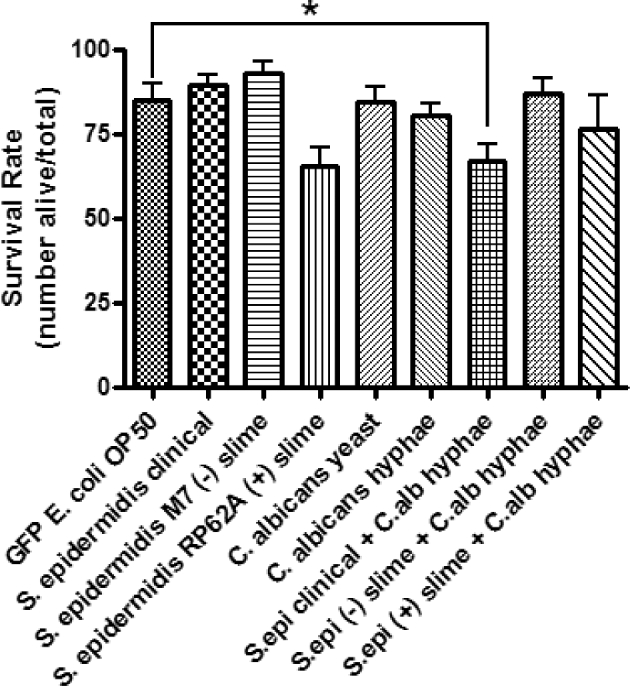
Single and dual inoculation assays of C. elegans after 24 h following exposure to treatments. Caenorhabditis elegans from an L3, L4 population recently fed on an E. coli OP50 lawn, were washed with M9 buffer prior to feeding to avoid carry over. The living and dead nematodes were counted after 24 h. Error bars represent SEM; n = 3 for all treatments. Statistical analysis showed P = 0.022 for S. epidermidis clinical + C. albicans hyphae compared to the control.
Figure 2.
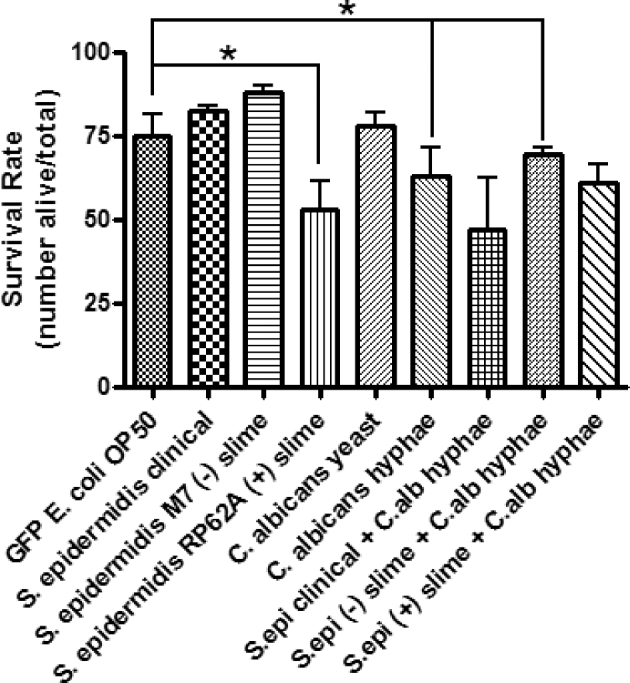
Single and dual inoculation assays of C. elegans after 48 h following exposure to treatments. Caenorhabditis elegans from an L3, L4 population recently starved on an E. coli OP50 lawn, were washed with M9 buffer prior to feeding to avoid carry over. The living and dead nematodes were counted after 48 h. Error bars represent SEM; n = 3 for all treatments. Statistical analysis showed P = 0.027 for S. epidermidis (+) slime, P = 0.032 for C. albicans hyphae, and P = 0.046 for S. epidermidis (–) slime + C. albicans hyphae compared to the control.
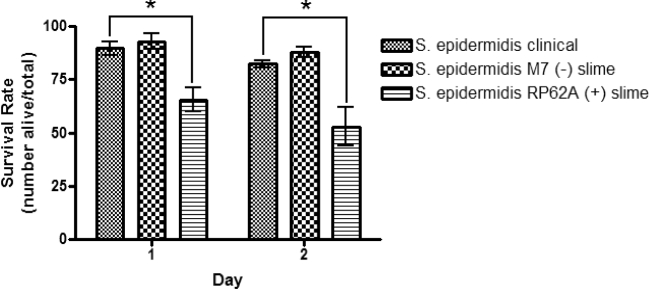
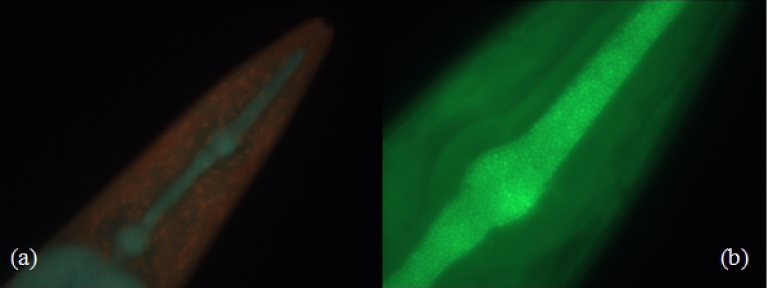
The effects of EPS on the virulence of S. epidermidis were tested. EPS-producing S. epidermidis could use the EPS to deter the nematode's innate immune system. As hypothesized, it was shown that non-EPS-producing S. epidermidis did not provide the bacterial cells with protection against the nematode's immune response thus making it more susceptible to clearance. The average survival rate for the nematodes that were exposed to the non-EPS-producing S. epidermidis was 90.4% after 48 h (Fig. 3). Conversely, the EPS-producing S. epidermidis assay showed a significant decrease in survival of the nematode (Figs 1 and 2). After 24 h, an average survival rate of 65.5% was observed (P < 0.05) and 52.8% after 48 h (P < 0.05) (Figs 1 and 2, Table 1). Visualization of the bacterial biofilms within the gut of the nematode was captured using fluorescence microscopy and Live/Dead BacLight differential staining. Live, intact bacterial cells stained green, while dead and compromised bacterial cells stained red. A biofilm established from the clinical S. epidermidis strain was seen colonizing inside the gut of a dead nematode (Fig. 4).
Figure 3.
Single inoculation survival assay of C. elegans fed on S. epidermidis clinical strain (ATCC #155) and laboratory strain (ATCC #35984) individually for 4 h. Caenorhabditis elegans from an L3, L4 population recently fed on an E. coli OP50 lawn, were washed with M9 buffer prior to feeding to avoid carry over. The living and dead nematodes were counted after 24 and 48 h, respectively. Error bars represent SEM. n = 6 for S. epidermidis clinical and S. epidermidis (+) slime; n = 3 for S. epidermidis (–) slime. Statistical analysis showed **P < 0.01 for S. epidermidis RP62A (+) slime after 24 h and ***P < 0.001 after 48 h compared to the clinical S. epidermidis strain.
Table 1.
Comparison of survival assays to control assays.
| Assay | P value (24 h) | P value (48 h) | Conclusion |
|---|---|---|---|
| GFP E. coli OP50 vs. S. epidermidis clinical | 0.58 | 0.26 | Nematode survival rate was not affected by clinical strain of S. epidermidis after 48 h. |
| GFP E. coli OP50 vs. S. epidermidis M7 (–) EPS | 0.49 | 0.92 | Nematode survival rate was not affected by non-slime producing strain of S. epidermidis after 48 h. |
| GFP E. coli OP50 vs. S. epidermidis RP62A (+) EPS | 0.06 | 0.03 | Nematode survival rate was only significantly affected by slime-producing strain of S. epidermidis after 48 h, but not so after 24 h. |
| GFP E. coli OP50 vs. C. albicans yeast | 0.94 | 0.73 | Nematode survival rate was not affected by C. albicans yeast after 48 h. |
| GFP E. coli OP50 vs. C. albicans hyphae | 0.07 | 0.03 | Nematode survival rate was affected by C. albicans hyphae after 48 h, but not after 24 h. |
| GFP E. coli OP50 vs. S. epidermidis clinical + C. albicans hyphae | 0.02 | 0.08 | The clinical strain of S. epidermidis combined with C. albicans hyphae showed a significant decrease in nematode survival after 24 h. But the decrease was not significant after 48 h. |
| GFP E. coli OP50 vs. S. epidermidis M7 (–) EPS + C. albicans hyphae | 0.62 | 0.04 | After 48 h, nematode survival was affected by the non-slime producing S. epidermidis when combined with C. albicans hyphae. |
| GFP E. coli OP50 vs. S. epidermidis RP62A (+) EPS + C. albicans hyphae | 0.23 | 0.070 | Nematode survival rate was not affected by the slime-producing strain of S. epidermidis when combined with C. albicans hyphae. |
| S. epidermidis clinical vs. S. epidermidis M7 (–) EPS | 0.22 | 0.20 | The non-slime producing strain of S. epidermidis did not show any significant decrease in nematode survival after 48 h, when compared to the clinical strain. |
| S. epidermidis clinical vs. S. epidermidis RP62A (+) EPS | 0.01 | 0.03 | The slime-producing strain of S. epidermidis was shown to decrease the survival rate of the nematode after 24 and 48 h, compared to the clinical strain. |
P values obtained using a paired t-test in order to assess statistical significance of survival rates. n refers to the number of replicates compared to n = 6 for the E. coli OP50 control.
Figure 4.
Caenorhabditis elegans fed on S. epidermidis clinical strain (ATCC #155) stained with Live/Dead BacLight differential stain. (a) Dual filter overlay used to show both live and dead cells (×40), whereas (b) shows only live cells (×100). Green fluorescence shows live cells and red fluorescence shows dead cells. Arrows show S. epidermidis colonized in the pharynx and intestine of the dead nematode.
DISCUSSION
Live/Dead staining confirmed that the ingested microorganisms were able to colonize inside the intestine of the nematode. Therefore, it was assumed that a biofilm mode of growth comparable to a biofilm infection was the cause of death for the nematodes. Living cells were observed with fluorescence microscopy, suggesting that they were able to survive within the nematode. As predicted, the survival rate of co-infected nematodes was decreased compared to exposure by the pathogens individually. However, individual infection with Candida albicans hyphae showed a significant decrease in nematode survival. This shows that morphology is a factor in fungal virulence and that hyphae are the more virulent form of C. albicans, as opposed to yeast cells.
As previously stated, the presence of EPS increased the pathogenicity of staphylococci in part by shielding the bacterium from immune responses. Caenorhabditis elegans, instead of possessing neutrophils and other immune cells to fight infections, uses only the innate immune system to fight pathogens (Ermolaeva and Schumacher 2014). Their innate immune system involves a cascade of ligands and receptors. However, the exact pathway is not clearly understood at this point and not all of the components of this cascade have been identified (Ermolaeva and Schumacher 2014). It is known that at the end of the pathway, a group of effectors is activated that will actively work against the pathogen aiming at eliminating the infection (Ermolaeva and Schumacher 2014). These effectors include C-type lectins, which adhere carbohydrates to the surface of bacteria; CUB domain containing factors, which adhere extracellular proteins to bacteria; and antimicrobial peptides, whose exact function in the immune system is not clear (Ermolaeva and Schumacher 2014). Although C. elegans do not have a cell-based innate immune system, the nematode is used frequently as a model for the human immune system because of the nematodes’ susceptibility to many human pathogens. The similarities between the pathogen defense systems of C. elegans and humans allow for comparisons to be made between the results of experiments that involve either organism (Marsh and May 2012). For example, Staphylococcus epidermidis EPS has been found to negatively affect the rate of phagocytosis and destruction of the bacteria by neutrophils, which are human immune cells, indicating that the EPS and its components are key for maintaining the strength of an S. epidermidis biofilm infection.
Strains of S. epidermidis that produce higher amounts of EPS are closely linked to the presence of a biofilm infection that can be found on intravenous catheters (Mateo et al. 2008). Using optical density, Mateo et al. examined 66 separate isolates of S. epidermidis taken from intravascular catheters and healthy skin, where coagulase-negative staphylococci were detected. Each isolate was compared with the optical density of S. epidermidis strain ATCC 35983, which is known to profusely produce EPS. Therefore, the isolates that were closest in optical density to the positive control strain were classified as excessive EPS producers. Forty-two per cent of the S. epidermidis isolates taken from the catheters exhibited characteristics of strong EPS production, compared with 0% of the non-EPS producing S. epidermidis isolates. Of the isolates taken from healthy skin, 35.7% were non-EPS producing and 11.5% were strong-EPS producers (Mateo et al. 2008). The increased prevalence of S. epidermidis on patient catheters compared to their prevalence on healthy tissue led to the conclusion that EPS was an impactful virulence factor for these bacteria and thus created a challenge for organism's immune systems working to eliminate these bacteria from their infected host.
In this study, the nematode hosts that ingested EPS-producing S. epidermidis biofilms showed signs of increased infection level compared to non-EPS producing and clinical strains of S. epidermidis. Although the EPS-negative S. epidermidis monoinfection was not as virulent as the infections created by EPS-producing strains, the C. elegans survival rate decreased when EPS-negative bacteria were combined with C. albicans hyphae. Therefore, it is possible that fungal EPS is a contributing factor to the virulence of a biofilm infection. This assertion can be corroborated by Pammi et al. (2013), in whose experiment C. albicans and S. epidermidis biofilm virulence was tested in both in vitro and in vivo in a subcutaneous catheter infection model. Here, the authors observed that a mixed biofilm (polymicrobial) increased the catheter infection. The authors evaluated the S. epidermidis EPS production and found that several genes related to extracellular EPS production were upregulated and concluded that the increase in virulence likely was due to EPS originating from S. epidermidis. Further evidence that fungal EPS encouraged the lethality of an EPS-negative bacterial strain infection can be found in the research of Adam, Baillie and Douglas (2002) and Chen et al. (2015). Conclusions from both of these studies indicate that bacterial and fungal EPS might interfere with the function of antifungal and antibiotic compounds, respectively. However, the studies did not specifically cover whether the EPS without antibiotics/antifungals would impact the virulence of the polymicrobial biofilm.
Originally, based on the virulence of EPS-producing S. epidermidis and C. albicans hyphae was expected that combining the EPS-producing bacteria and hyphae together would decrease the survival of C. elegans and cause a more severe infection. However, this result was not observed. In contrast, the only significant decrease in nematode survival rate was observed for the coinfection assays with the clinical S. epidermidis strain combined with C. albicans hyphae. This result, although not consistent with the originally proposed hypothesis, aligns with other studies that combined C. albicans with pathogenic bacteria to imitate multikingdom infections. Peleg et al. (2008) coinfected C. elegans with C. albicans and varying bacterial species (individually) to evaluate the effects of these coinfections (Peleg et al. 2008). It was found that the highest nematode survival rates occurred, when an infection of the fungus with Gram-negative bacteria was taking place. Increased death rates were recorded when the fungus was paired with Gram-positive bacteria. It appeared that the Gram-positive bacteria Acinetobacter baumannii had the ability to structurally dismantle C. albicans populations within the nematodes. The weakened fungi became less virulent than it would be if alone, and therefore, C. elegans had higher rates of survival (Peleg et al. 2008). In the case of coinfection with C. albicans and the EPS-overproducing S. epidermidis strain in this study, the results showed that the EPS-producing bacteria were not as virulent as the clinical strain, when coexisting with hyphae in biofilms. This can likely be explained by the EPS initially (24 h) interfering with hyphal attachment in the EPS-producing S. epidermis strain within C. elegans, thus severely weakening a key virulence factor of the fungus. This effect did not continue throughout the experiment since the survival rates for the combined assays were not statistically different at 48 h (Fig. 2).
The next step for investigating the interactions between C. albicans and S. epidermidis for forming a biofilm coinfection would be to quantify the effects of hyphal tissue penetration on nematode survival. It is proposed that if C. elegans feed on C. albicans yeast, hyphae will be induced inside the intestine. Upon hyphael activity within the nematode intestine, S. epidermidis would then attach to the hyphae and subsequently penetrate the nematode's cuticle, leading to a biofilm infection.
CONCLUSION
The interaction between Staphylococcus epidermidis and Candida albicans was shown to increase the virulence of a biofilm infection within the nematode host Caenorhabditis elegans. These interactions can occur within human hosts as well, specifically when the bacterium becomes pathogenic upon attachment to the fungus and penetrates the tissue. The commensal S. epidermidis can gain entry into tissue, where it would otherwise not exist. Visualization of hyphal penetration of the C. elegans cuticle and determination of how it affects the survival rate of the nematodes could improve the understanding of multikingdom biofilm interactions. Increasing the knowledge of these interactions can aid in determining how to fight polymicrobial biofilm infections in human hosts. In addition, further research could identify the genomic responses of the biofilm infections modeled in the nematode host model.
FUNDING
This work was funded through the National Science Foundation's STEP Program, the Successful Engineering Education and Development Support (SEEDS) Program, where fellowships during 2016–17 were awarded to Jillian Holt. We also want to thank The Caenorhabditis Genetics Center (CGC) that is supported by the National Institutes of Health—Office of Research Infrastructure Programs (P40 OD010440).
Conflict of interest. None declared.
REFERENCES
- Adam B, Baillie G, Douglas L. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol 2002;51:344–9. [DOI] [PubMed] [Google Scholar]
- Alegado R, Campbell M, Chen W et al. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol 2003;5:435–44. [DOI] [PubMed] [Google Scholar]
- Begun J, Gaiani J, Rohde H et al. Staphylococcal biofilm extrapolysaccharide protects from Caenorhabditis elegans immune defenses. PLoS Pathog 2007;3:526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breger J, Fuchs BB, Aperis G et al. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog 2007;3:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang XY, Huang YC et al. Study on the structure of Candida albicans-Staphylococcus epidermidis mixed species biofilm on polyvinyl chloride biomaterial. Cell Biochem Biophys 2015;73:461–8. [DOI] [PubMed] [Google Scholar]
- Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2003;2:114–22. [DOI] [PubMed] [Google Scholar]
- Donlan R. Biofilms and device-associated infections. Emerg Infect Dis 2001;7:277–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B. Insights from the worm: the C. elegans model for innate immunity. Semin Immunol 2014;26:303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P, Olson M. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol 2010;5:917–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S, Hung D. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 2013;4:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Herrmann M, von Eiff C et al. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 1997;65:519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz S, Chasin B, Powell B et al. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Micr Infec Dis 2007;59:401–6. [DOI] [PubMed] [Google Scholar]
- Kurz C, Ewbank J. Caenorhabditis elegans for the study of host-pathogen interactions. Trends Microbiol 2000;8:142–4. [DOI] [PubMed] [Google Scholar]
- Lin Y, Alsad L, Vogel F et al. Interactions between Candida albicans and Staphylococcus aureus within mixed species biofilms. BIOS 2013;84:30–9. [Google Scholar]
- Marsh E, May R. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microb 2012;78:2075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M, Maestre JR, Aguilar L et al. Strong slime production is a marker of clinical significance in Staphylococcus epidermidis isolated from intravascular catheters. Eur J Clin Microbiol 2008:27:311, doi: 10.1007/s10096-007-0433-y. [DOI] [PubMed] [Google Scholar]
- Mekni M, Bouchami O, Archour W et al. Strong biofilm production but not adhesion virulence factors can discriminate between invasive and commensal Staphylococcus epidermidis strains. APMIS 2012;120:605–11. [DOI] [PubMed] [Google Scholar]
- O'Gara J, Humphreys H. Staphylococcus epidermidis biofilms: importance and implications. J Med Microbiol 2001;50:582–7. [DOI] [PubMed] [Google Scholar]
- Pammi M, Liang R, Hicks J et al. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol 2013;13:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Jabra-Rizk M, Scheper M et al. Microbial interactions and differential protein expression in Staphylococcus aureus-Candia albicans dual-species biofilms. FEMS Immunol Med Micr 2010;59:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A, Tampakakis E, Fuchs B et al. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. P Natl Acad Sci USA 2008;38:14585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Peleg A, Tampakakis E et al. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 2009;8:1750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saginur R, Denis M, Ferris W et al. Multiple combinations bacteridical testing of Staphylococcal biofilms from implant-associated infections. Antimicrob Agents Ch 2006;50:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P, Nistico L, Johnson S et al. Direct demonstration of viable Staphylococcus aureus biofilms in an infected total joint arthroplasty. J Bone Joint Surg 2008;90:1751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. Growth of Candida albicans hyphae. Nat Rev 2011;9:737–48. [DOI] [PubMed] [Google Scholar]
- Veses V, Gow N. Pseudohypha budding patterns of Candida albicans. J Mycol Med 2009;47:268–75. [DOI] [PubMed] [Google Scholar]
- WormBook. The online review of C. elegans biology. 2017, http://www.wormbook.org/ (9 May 2017, date last accessed).