Abstract
Objective
Electronic health record (EHR)-based phenotyping infers whether a patient has a disease based on the information in his or her EHR. A human-annotated training set with gold-standard disease status labels is usually required to build an algorithm for phenotyping based on a set of predictive features. The time intensiveness of annotation and feature curation severely limits the ability to achieve high-throughput phenotyping. While previous studies have successfully automated feature curation, annotation remains a major bottleneck. In this paper, we present PheNorm, a phenotyping algorithm that does not require expert-labeled samples for training.
Methods
The most predictive features, such as the number of International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes or mentions of the target phenotype, are normalized to resemble a normal mixture distribution with high area under the receiver operating curve (AUC) for prediction. The transformed features are then denoised and combined into a score for accurate disease classification.
Results
We validated the accuracy of PheNorm with 4 phenotypes: coronary artery disease, rheumatoid arthritis, Crohn’s disease, and ulcerative colitis. The AUCs of the PheNorm score reached 0.90, 0.94, 0.95, and 0.94 for the 4 phenotypes, respectively, which were comparable to the accuracy of supervised algorithms trained with sample sizes of 100–300, with no statistically significant difference.
Conclusion
The accuracy of the PheNorm algorithms is on par with algorithms trained with annotated samples. PheNorm fully automates the generation of accurate phenotyping algorithms and demonstrates the capacity for EHR-driven annotations to scale to the next level – phenotypic big data.
Keywords: high-throughput phenotyping, phenotypic big data, electronic health records, precision medicine
INTRODUCTION
New health problems, medications, and regimens emerge on a daily basis. To understand their clinical impact in a timely manner, large amounts of accurate genotypic and phenotypic data must be readily available for research in a cost-effective manner. The advent of high-throughput gene sequencing technologies has reduced the cost of obtaining genomic data exponentially, from 2.7 billion USD for the first human genome (the Human Genome Project)1 down to 1000 USD per genome in 2016. Currently, million-people-scale sequencing projects are under way to generate genomic data for research.2 However, amassing phenotypic data remains a challenge,3 as it traditionally takes human effort to record the phenotypes of patients.
To overcome the scarcity of phenotypic data, genomic and other medical studies have begun to extract phenotypic information from electronic health records (EHRs) to augment existing biorepositories or quickly create new ones.4,5 Notable efforts include the i2b2 effort led by Harvard University and Partners HealthCare,6–16 the BioVU effort led by Vanderbilt University,17 and the multicenter eMERGE Network.18–20 Typically, EHR-based phenotyping extracts features from the patient’s EHR and assembles them into a classification rule (called a “phenotyping algorithm”) to infer whether the patient has a target phenotype. The features often involve the patient’s demographic information, such as age and sex; codified information, such as diagnosis, medication, lab, and procedure codes (codified features); and information extracted from the narrative notes via natural language processing (NLP features). It has been demonstrated that studies utilizing such data-driven phenotypes can reproduce previously established results based on phenotypic data obtained by traditional means21,22 and can drive novel studies, such as genome-wide and phenome-wide association studies.19,23 However, the current approach to algorithm development relies on tremendous domain expert participation and takes many months to complete.
In most settings, a “supervised” machine learning method is employed to estimate a statistical model that outputs the probability or classification of the target phenotype based on a number of input features and a training dataset consisting of a few hundred patients with “gold-standard” phenotype labels. The features are curated and engineered by a panel of clinicians, informaticians, statisticians, and computer scientists, while the labels are obtained by experts via laborious manual chart review. Feature curation and sample annotation, requiring up to months of human effort, are thus the rate-limiting factors in phenotyping, due to the heavy human input required. Research is now under way to automate algorithm development to fully leverage the efficiency of EHR-based research and achieve so-called high-throughput phenotyping.24,25
Toward the goal of automating feature curation, some studies have attempted to use all available codified and NLP features potentially predictive of the phenotype of interest for algorithm estimation.26–29 Though this approach eliminates the feature selection step entirely, it tends to be suboptimal when the number of codified and NLP features is substantially larger than the size of the labeled training set. Supervised algorithms trained with a large number of noisy features and a small number of labeled examples can suffer from significant overfitting and instability, leading to suboptimal out-of-sample performance.30 While overfitting can be reduced with estimation procedures that penalize model complexity (penalized regression procedures are a common choice31–33) a price is paid in terms of sampling variability that reduces the out-of-sample accuracy; the more unnecessary complexity, the heavier the price. It is therefore essential to use only informative features in the model. Wright et al. mined codified EHR data to look for possible associations between problems, medications, and lab tests, which can potentially be used for automated feature selection. Unfortunately, their remaining modeling steps involved a large amount of manual screening and design.34,35 More recently, fully automated feature selection has been achieved with satisfying results. The automated feature extraction for phenotyping (AFEP) method36 calls on online sources for the knowledge originally provided by domain experts by scanning articles concerning target phenotypes on Wikipedia and Medscape to extract relevant medical concepts as candidate NLP features. Informative features are then identified by analyzing the co-occurrence patterns of the features in the EHR database. The performance of the automatically selected features is comparable to those designed by experts. The surrogate-assisted feature extraction (SAFE) method37 improved upon AFEP, and was able to cut the feature set down to 10–30 highly informative features that outperform the AFEP features significantly for classifying disease phenotypes when the number of training samples is small.
Despite the success in automated feature curation, sample annotation remains a major obstacle to achieving high-throughput phenotyping. In addition to feature selection, feature refinement or selective prioritization of representative samples for annotation can work to reduce the sample size needed for the training.37–39 However, manual annotation must be entirely removed from the algorithm development process to truly achieve scalable phenotyping. It is therefore desirable to move toward learning associations between the features and the target phenotype without using any gold-standard labels to guide the learning. A feasible approach is to rely on “silver-standard labels” automatically generated from the EHR in place of human-annotated labels for training, such as counts of relevant billing codes or NLP mentions, which are strong but imperfect predictors of true disease status. This approach is known as “distant supervision” in the machine learning community and was employed by Yu et al.37 with SAFE for feature selection but not for model estimation. Recently, Agarwal et al.29 proposed XPRESS to use the silver-standard labels directly for training the phenotyping algorithm in order to completely eliminate the human annotation step. However, the simplistic choice of their silver-standard labels – an indicator of whether the phenotype is positively mentioned in the narrative notes – leads to suboptimal performance of their algorithm when compared to supervised counterparts.
This paper presents PheNorm, a completely annotation-free 2-step classification method for phenotyping involving an initial normalization step of highly predictive features of the target phenotype followed by a denoising step to leverage additional information contained in the remaining candidate features. We validate the performance of our method with 4 phenotypes from the Partners HealthCare EHR and compare the area under the receiver operating curve (AUC) of the PheNorm score to that of the supervised algorithms trained with gold-standard labels.
METHODS
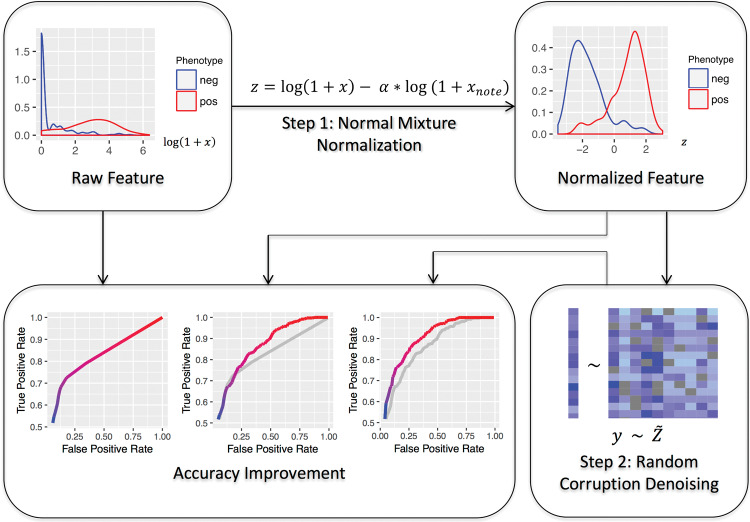
The 2 steps of the PheNorm procedure are outlined in Figure 1. The first step transforms a highly predictive feature of the target phenotype, such as the number of International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes of the target phenotype in the patient records, to resemble a 2-component normal mixture distribution with high accuracy for prediction. The second step involves self-regression with dropout to denoise the transformed feature based on additional candidate features, similarly transformed, to further improve the prediction. The output is a linear combination of all the transformed features.
Figure 1.
Workflow of PheNorm. Top left: density plot (after logarithm transformation) of a highly predictive feature (illustrated here using the ICD-9-CM count of ulcerative colitis from a Partners HealthCare EHR datamart), denoted by x, in patients who do (the right curve) and do not (the left curve) have the phenotype. Top right: Density plot of the ICD-9-CM count after the normal mixture transformation using the total number of notes in the patient’s EHR, denoted by xnote. The densities of the phenotype positive and negative patients are approximately normally distributed, and the 2 populations are separated to a large degree. Bottom right: The transformed feature is denoised by self-regression of the transformed feature, denoted by y, onto the entire transformed and randomly corrupted feature set, denoted by Z˜, with dropout. The transformed features are then combined into a prediction formula for disease status classification based on the estimated regression coefficient. Bottom left: The receiver operating characteristic (ROC) curve of the feature or score in each step, with AUC growing steadily (gray curves are copies of the ROC curves from the previous steps).
Raw feature preparation
The input for the PheNorm algorithm consists of unlabeled data on a set of potentially informative features, either automatically curated or designed by experts. For the purpose of illustration in a high-throughput phenotyping scenario, we use SAFE37 to automatically curate features (listed in the Supplementary Material). Briefly, online articles about the target phenotype from publicly available knowledge sources, such as Wikipedia and Medscape, are scanned with NLP software to extract medical concepts recorded in the Unified Medical Language System.40 These concepts are potentially related to the target phenotype. Then, narrative notes in the EHR database are processed with NLP software, which identifies mentions of the above medical concepts. We include only positive mentions, ie, mentions confirming the presence of a condition, the performance of a procedure, the prescription of a medication, etc., in all analyses. The patient-level counts of these concept mentions are assembled as candidate feature data. The SAFE procedure selects a subset of the candidate features via frequency control and repeated fitting of sparse logistic regressions to predict silver-standard labels created from combinations of ICD-9-CM diagnosis codes and NLP counts of the target phenotype. Features predictive of the silver-standard labels are deemed as informative features for further algorithm training. The NLP analyses used for processing the notes are provided in many out-of-the-box software tools,41–45 and some hospitals and research institutions have their own NLP implementations.
Throughout, we denote by and the counts of ICD-9-CM codes and free-text positive mentions of the target phenotype, respectively. In addition, we let , be the counts of positive mentions of all remaining concepts selected by SAFE and be the count of narrative notes for the patient. The input data for the PheNorm algorithm is then the patient-level data on (. We also include a simple derived feature, .
Normal mixture normalization
Since and are expected to be fairly predictive of the underlying phenotype, we expect them to follow mixture distributions: the counts of phenotype-positive patients cluster around the upper end and the counts of phenotype-negative patients cluster around the lower end. However, , , and tend to be higher for patients with more health care utilization regardless of their true underlying phenotype status. Therefore, we propose to normalize each of these 3 main features by based on our observation that, for , , or , with appropriate choice of , the distribution of the normalized count
approximately follows a normal mixture distribution (see top right of Figure 1) with, where is the true phenotype status. The optimal is chosen as the minimizer of the difference between the empirical distribution of and its normal mixture approximation,
where is the empirical cumulative distribution function (CDF) of under , is the CDF of the standard normal distribution, representing is the mixing proportion, and , , and (shared) are parameters of the 2 components of the normal mixture. For a given , we can find the maximum likelihood estimates of , , , and with the expectation-maximization (EM) algorithm,46,47 and then the distribution divergence is calculated by plugging in these estimated parameters. To overcome overfitting and increase stability, we use bootstrap resampling to repeatedly calculate with increasing from 0 to 1 and record where the divergence starts to increase, and we obtain the final estimate of as the average of those recorded points. Our numerical studies demonstrate that the normalized achieves a much higher AUC than the original , for * = ICD, NLP, and ICDNLP.
Random corruption denoising
The normalized main ICD-9-CM and NLP count features do not leverage information from other features, such as counts of competing diagnoses and medication prescriptions, which provide additional characterization of the presence of the target phenotype. In fact, some of these remaining features have been shown to possess predictive values beyond the main features in supervised algorithms trained with gold-standard labels.7–11,13–16 We thus wish to utilize the additional information contained in the entire candidate feature set to further refine phenotype definition, but in the setting of not using any gold-standard labels.
To this end, we propose to aggregate the information in the entire candidate feature set with denoising self-regression via dropout training, a popular training method in deep learning to control overfitting.48–50 Let be a data matrix whose columns are the transformed features using the above normal mixture transformation ( is calculated for each feature separately) and whose rows represent patients randomly sampled from the EHR database. To obtain a stable result from the dropout training, we recommend that has at least 105 rows. When the EHR cohort size is smaller than 105, we can use bootstrap to sample 105 out of with replacement. We randomly corrupt and obtain , with
where is the mean of the jth column of , and are independent and identically distributed Bernoulli random variables, with . Our empirical results suggest that a dropout rate of 20–30% works well. We let the response be one of the strong predictors in the uncorrupted , that is, , , or . Then we predict with using ordinary least squares regression and obtain the regression coefficient vector , for , , or , depending on whether , , or . Since the column in that corresponds to has been corrupted, cannot be predicted by a single feature. Instead, the regression will utilize the underlying associations among all the features to recover the lost information of . Since the recovered information must be supported by the evidence in other features, the regression is essentially a denoising process. We obtain the final PheNorm score for each patient as an inner product of the patient’s normalized feature vector and the coefficient vector
where in is the uncorrupted version of the normalized features.
Majority vote for robustness
Without labels, it is unclear which of , , or performs the best. Thus, we use a voting scheme to combine the 3 scores for robustness. By approximating ( ICD, NLP, or ICDNLP) as a normal mixture distribution, we classify a patient’s phenotype status as according to whether the posterior probability of phenotype positive given is >0.5. Let be the majority vote of . We let the final score be:
Data and metrics for evaluation
To evaluate the performance of the PheNorm algorithm, we used Partners HealthCare EHR datamarts constructed for phenotyping rheumatoid arthritis (RA), Crohn’s disease (CD), ulcerative colitis (UC), and coronary artery disease (CAD).7,8,13 We aimed to develop algorithms for classifying these 4 phenotypes: CAD, RA, CD, and UC. The RA datamart included the records of 46 568 patients who had at least one ICD-9-CM code of 714.x (Rheumatoid arthritis and other inflammatory polyarthropathies) or had been tested for anticyclic citrullinated peptide. The RA status was annotated by domain experts for a random sample of 435 patients. From the RA datamart, 4446 patients were predicted to have RA by Liao et al.,7 and of those, 758 patients who had at least one ICD-9-CM code or a free-text mention of CAD were reviewed for the CAD phenotype. Since PheNorm relies heavily on clinical notes, the records of 17 of 758 patients that did not have a note were removed from the samples. The datamart for inflammatory bowel diseases included 34 033 patients who had at least one ICD-9-CM code of 555.x (Regional enteritis) or 556.x (Ulcerative enterocolitis). For UC and CD, respectively, 600 patients randomly sampled from those with at least one corresponding ICD-9-CM code were reviewed for the target phenotype. The prevalence for CAD, RA, CD, and UC were estimated at 40.1%, 22.5%, 67.5%, and 63.0%, respectively.
For each phenotype, we used PheNorm to generate a score for each patient using , , and and calculated the AUC as a metric of accuracy. The corruption rate in the denoising step was set to 0.3. For sensitivity analysis, we also trained PheNorm using a corruption rate of 0.2 and using features selected from AFEP in the denoising step. As benchmarks, we trained supervised algorithms with randomly sampled gold-standard labels of patients for , , and using the SAFE and AFEP features, and used the remaining samples to estimate the out-of-sample AUC. The algorithms were obtained from fitting adaptive elastic-net penalized logistic regression models.32,33 The penalty parameters were chosen by the Bayesian information criterion.51 We repeated the supervised training process 30 times and report the average AUC. We also trained XPRESS algorithms as proposed in Agarwal et al.29 with silver-standard labels defined as if and if . As suggested in the paper, we manually validated the dictionary for the target phenotype to ensure that there was no ambiguity. Except for CAD, we sampled 750 patients from and , respectively, and trained a logistic regression model with L1 penalty. The optimal tuning parameter was selected with 5-fold cross-validation. For CAD, since the population was defined as patients from the identified RA patients who had at least an ICD-9-CM code or a free-text mention of CAD, the entire population had only 741 patients. We therefore used all patients for training. The features for the regression included all the SAFE-selected NLP features, the note count, and expert-curated codified features, including related diagnosis, prescription, and procedure codes, as well as demographic information such as age and sex (listed in Supplementary Material). In addition to XPRESS, we also experimented with the Anchor method,52 which was originally developed for annotating single visit notes and relies on expert-curated filters to define positive labels with high positive predictive value. Here we adapted the method for the multivisit scenario, using as the filter to identify positive labels and removing from the predictors due to the conditional independence requirement.
We used bootstrap to estimate the standard errors of the difference in the AUC estimates from comparing different algorithms and to obtain the P-value for testing whether the difference is zero.
RESULTS
The out-of-sample AUC estimates for various algorithms are shown in Table 1. Recall that the PheNorm algorithm involves 2 main steps: (1) normalization and (2) denoising via dropout regression. Comparing the AUC of the ICD-9-CM codes before and after normalization, we found that the normalization step substantially improved the accuracy of the codes, with average improvement in AUC of around 0.04 across phenotypes. The denoising step further improved the AUC with varying degrees of magnitude depending on the phenotype and the feature; it substantially improved the AUC of the normalized ICD-9-CM count, but was not as critical for the NLP or ICD + NLP count as it was for the ICD-9-CM. Comparing the PheNorm algorithm applied to the different features, it appears that using the ICD + NLP count gave the most robust results across phenotypes, and the score based on majority voting achieved similar accuracy.
Table 1.
AUCs of the raw feature , the normalized feature , the PheNorm scores using SAFE feature for denoising with a dropout rate of 0.3, , the supervised algorithms trained with SAFE features with N = 100, 200, or 300 labels, as well as the XPRESS and Anchor algorithms.
| CAD | RA | CD | UC | ||
|---|---|---|---|---|---|
| 0.844 | 0.868 | 0.824 | 0.812 | Comparison is with the previous step; asterisk indicates positive increment at the significance level of 0.05. | |
| 0.840 | 0.898 | 0.906 | 0.904 | ||
| 0.899 | 0.937 | 0.945 | 0.933 | ||
| 100 labels | Comparison is with ; asterisk indicates difference at the significance level of 0.05. | ||||
| 200 labels | |||||
| 300 labels | |||||
| XPRESS | |||||
| Anchor | |||||
Comparisons are shown in the form , where the superscript is the increment in AUC and the subscript is the standard error of the difference in AUC.
The PheNorm algorithms using ICD + NLP count achieved an AUC comparable to that of the corresponding supervised algorithms when 100 labels were used for CAD and 300 labels were used for RA and CD. The AUC of the PheNorm score of UC appeared to be lower than those of the supervised algorithms, but an AUC of 0.935 is acceptably high when comparing across phenotypes. None of the supervised algorithms attained an AUC significantly higher than the unsupervised . The AUCs from the XPRESS and Anchor methods were significantly lower than that of PheNorm. In addition, as reported in the Supplementary Material, the performance of PheNorm was not sensitive to the choice of the corruption rate or feature set.
DISCUSSION
Maturation of high-throughput phenotyping technology is key to enabling phenomics – the next big challenge for the study of precision medicine.53 However, scalable phenotyping relies on the ability to generate an accurate algorithm without intense involvement of clinical experts. Existing automated feature selection methods, including AFEP and SAFE, serve as a step toward automated phenotyping, but ultimately rely on expert-annotated labels to train supervised phenotyping algorithms with the selected features. PheNorm exploits the underlying distribution and association between features to aggregate them into a score for disease status classification without any gold-standard labels.
Though the training of the XPRESS algorithms does not require gold-standard labels either, our analysis indicates that the resulting algorithms have low accuracy. The suboptimal performance is due to the construction of the silver-standard labels as dichotomized versions of one of the model features, , and hence the AUC of the XPRESS algorithms essentially approximates the AUC of . It is also important to note that the original implementation of XPRESS used tens of thousands of features without preselection for training,29 leading to further overfitting and decreased out-of-sample performance.
Using as the anchor filter potentially automates the Anchor method and adapts it for patient-level multivisit phenotyping. However, this leads to algorithms with suboptimal performance. It is unclear whether alternative anchors would yield more accurate algorithms or how to choose better anchor filters that satisfy the conditional independence requirement in the multivisit setting.
The results from our numerical studies indicate that PheNorm achieves the same accuracy as supervised algorithms based on training set sizes between 100 and 300, depending on the phenotype. Current large-scale phenotyping efforts (eg, 10 phenotypes at a time) rarely have the bandwidth to offer more than 200 gold-standard labels for training for each disease, thus illustrating the potential of our method to streamline phenotyping without compromising the accuracy of a supervised approach. Additionally, our results demonstrate that the normalization step () always significantly improves prediction performance, while the subsequent denoising step contributes in varying degrees. Denoising appears to be critically important for ICD-9–based algorithms, but contributes minimally to ICD + NLP–based algorithms. This suggests that the effectiveness of the denoising step is inversely related to the predictiveness of the normalized feature. In practice, one may wonder if the denoising step is still necessary, since is typically highly accurate. We would argue that such a step is still potentially beneficial, particularly in settings where the ICD-9-CM code provides a poor characterization of the desired phenotype or the NLP software fails to accurately capture the description of the phenotype. In this case, would benefit from the additional information in the related features offered by denoising.
Our experiments also show that and perform differently across the phenotypes, with the former better for CAD and the latter better for RA, CD, and UC. It is important to note that is not necessarily a tradeoff between and , although and its accuracy may surpass both. Though consistently performed well for the 4 phenotypes, it is difficult to determine which score to use in practice without gold-standard labels for validation. Our empirical studies indicate that is a good hedging policy, as it consistently achieved accuracy close to the best performing one.
The PheNorm score can be converted to a predicted probability of having the disease phenotype using the EM algorithm. If the goal of the phenotyping is to link the phenotype to genomic data, one can directly use the predicted probability as a continuous trait and perform association analysis by fitting a quasi-binomial model. In fact, one could gain power by leveraging the predicted probability, as compared to converting the probability to a binary trait.54 When a small number of labels are available for validation, one can use these labels to estimate the receiver operating characteristic (ROC) curve and then select a threshold value optimizing the tradeoff between positive predictive value and sensitivity.14
Though our results demonstrate the ability of PheNorm to provide accurate phenotyping for 4 different diseases in the absence of gold-standard labels, further work is needed to understand the performance of our method across a diverse range of phenotypes, particularly for phenotypes that have more subtle definitions, in which case a combination of PheNorm and handcrafted rules or regular expressions might be effective. Additionally, while PheNorm eliminates the annotation typically required for algorithm estimation, labeled examples are still needed to evaluate the algorithm’s accuracy. Future research is thus warranted in unsupervised approaches to estimating the ROC parameters where the statistical inference is particularly challenging.
CONCLUSION
In this paper, we introduce PheNorm for training accurate phenotyping algorithms without using gold-standard labels. In our road map to high-throughput phenotyping, we have decomposed the task into automated feature curation and algorithm training without gold-standard labels. The former goal has been achieved by AFEP and SAFE, and the latter goal has now been achieved by PheNorm. PheNorm is easy to implement, and its accuracy is similar to algorithms trained with gold-standard labels. The bandwidth provided by SAFE + PheNorm can potentially reduce the algorithm development process from months to a few days, providing the valuable phenotypic big data necessary for the study of precision medicine.55
Funding
This work was supported by US National Institutes of Health grants U54-HG007963, U54-LM008748, R01-HL089778, R01-HL127118, F31GM119263-01A1, and K23-DK097142, the Harold and Duval Bowen Fund, and internal funds from Tsinghua University and Partners HealthCare.
Competing Interests
None.
Contributors
All authors made substantial contributions to: conception and design; acquisition, analysis, and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
References
- 1. National Human Genome Research Institute. Human Genome Project Completion: Frequently Asked Questions. www.genome.gov/11006943/Human-Genome-Project-Completion-Frequently-Asked-Questions. Accessed April 112 017. [Google Scholar]
- 2. Gaziano JM, Concato J, Brophy M, et al. . Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–23. [DOI] [PubMed] [Google Scholar]
- 3. Murphy S, Churchill S, Bry L, et al. . Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19:1675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–28. [DOI] [PubMed] [Google Scholar]
- 5. Pathak J, Kho AN, Denny JC. Electronic health records–driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc. 2013;20:e206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy SN, Mendis ME, Berkowitz DA, et al. . Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006;2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 7. Liao KP, Cai T, Gainer V, et al. . Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. 2010;62:1120–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ananthakrishnan AN, Cai T, Savova G, et al. . Improving case definition of Crohn's disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia Z, Secor E, Chibnik LB, et al. . Modeling disease severity in multiple sclerosis using electronic health records. PLoS ONE. 2013;8:e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar V, Liao K, Cheng S-C, et al. . Natural language processing improves phenotypic accuracy in an electronic medical record cohort of type 2 diabetes and cardiovascular disease. J Am Coll Cardiol. 2014;1263:A1359. [Google Scholar]
- 11. Castro VM, Minnier J, Murphy SN, et al. . Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am J Psychiatry. 2014;172:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu S, Kumamaru KK, George E, et al. . Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform. 2014;52:386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao KP, Ananthakrishnan AN, Kumar V, et al. . Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS ONE. 2015;10:e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao KP, Cai T, Savova GK, et al. . Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ. 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castro V, Shen Y, Yu S, et al. . Identification of subjects with polycystic ovary syndrome using electronic health records. Reprod Biol Endocrinol. 2015;13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castro VM, Dligach D, Finan S, et al. . Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology 2017;882:164–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roden D, Pulley J, Basford M, et al. . Development of a Large-Scale De-Identified DNA Biobank to Enable Personalized Medicine. Clin Pharmacol Ther. 2008;84:362–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clayton EW, Smith M, Fullerton SM, et al. . Confronting real time ethical, legal, and social issues in the eMERGE (Electronic Medical Records and Genomics) Consortium. Genet Med Off J Am Coll Med Genet. 2010;12:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kullo IJ, Ding K, Jouni H, et al. . A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5:e13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarty CA, Chisholm RL, Chute CG, et al. . The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ritchie MD, Denny JC, Crawford DC, et al. . Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Denny JC, Ritchie MD, Crawford DC, et al. . Identification of genomic predictors of atrioventricular conduction using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denny JC, Ritchie MD, Basford MA, et al. . PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richesson RL, Sun J, Pathak J, et al. . Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artif Intell Med. 2016;71:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pakhomov SV, Buntrock J, Chute CG. Identification of patients with congestive heart failure using a binary classifier: a case study. In: Proceedings of the ACL 2003 Workshop on Natural Language Processing in Biomedicine, Volume 13. Stroudsburg, PA: Association for Computational Linguistics; 2003: 89–96. [Google Scholar]
- 27. Carroll RJ, Eyler AE, Denny JC. Naïve electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc. 2011;2011:189. [PMC free article] [PubMed] [Google Scholar]
- 28. Bejan CA, Xia F, Vanderwende L, et al. . Pneumonia identification using statistical feature selection. J Am Med Inform Assoc. 2012;195:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Agarwal V, Podchiyska T, Banda JM, et al. . Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc. 2016;236:1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer; 2009. [Google Scholar]
- 31. Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Methodol. 1996;58:267–88. [Google Scholar]
- 32. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B. 2005;67:301–20. [Google Scholar]
- 33. Zou H, Zhang HH. On the adaptive Elastic-Net with a diverging number of parameters. Ann Stat. 2009;37:1733–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform. 2010;43:891–901. [DOI] [PubMed] [Google Scholar]
- 35. Wright A, Pang J, Feblowitz JC, et al. . A method and knowledge base for automated inference of patient problems from structured data in an electronic medical record. J Am Med Inform Assoc. 2011;18:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu S, Liao KP, Shaw SY, et al. . Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22:993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu S, Chakrabortty A, Liao KP, et al. . Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc. 2017;24:e143–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Carroll RJ, Hinz ERM, et al. . Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc. 2013;20(e2):e253–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiu P-H, Hripcsak G. EHR-based phenotyping: bulk learning and evaluation. J Biomed Inform. 2017;70:35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993;81:170. [PMC free article] [PubMed] [Google Scholar]
- 41. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001;17–21. [PMC free article] [PubMed] [Google Scholar]
- 42. Denny JC, Smithers JD, Miller RA, et al. . “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc. 2003;10:351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. HITEx Manual. www.i2b2.org/software/projects/hitex/hitex_manual.html. Accessed January 14, 2014. [Google Scholar]
- 44. Savova GK, Masanz JJ, Ogren PV, et al. . Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uzuner Ö, South BR, Shen S, et al. . 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;185:552–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. J R Stat Soc Ser B Methodol. 1977;39:1–38. [Google Scholar]
- 47. Wu CFJ. On the Convergence Properties of the EM Algorithm. Ann Stat. 1983;11:95–103. [Google Scholar]
- 48. Vincent P, Larochelle H, Bengio Y, et al. . Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th International Conference on Machine Learning. New York: ACM; 2008: 1096–103. [Google Scholar]
- 49. Wager S, Wang S, Liang PS. Dropout training as adaptive regularization. In: Burges CJC, Bottou L, Welling M, et al., eds. Advances in Neural Information Processing Systems 26. Curran Associates, 2013: 351–59. http://papers.nips.cc/paper/4882-dropout-training-as-adaptive-regularization.pdf. Accessed November 22, 2016. [Google Scholar]
- 50. Srivastava N, Hinton G, Krizhevsky A, et al. . Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58. [Google Scholar]
- 51. Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–64. [Google Scholar]
- 52. Halpern Y, Horng S, Choi Y, et al. . Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc. 2016;23:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–66. [DOI] [PubMed] [Google Scholar]
- 54. Sinnott JA, Dai W, Liao KP, et al. . Improving the power of genetic association tests with imperfect phenotype derived from electronic medical records. Hum Genet. 2014;133:1369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Delude CM. Deep phenotyping: the details of disease. Nature. 2015;527:S14–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.