Abstract
There are many resources available to mycobacterial researchers, including culture collections around the world that distribute biomaterials to the general scientific community, genomic and clinical databases, and powerful bioinformatics tools. However, many of these resources may be unknown to the research community. This review article aims to summarize and publicize many of these resources, thus strengthening the quality and reproducibility of mycobacterial research by providing the scientific community access to authenticated and quality-controlled biomaterials and a wealth of information, analytical tools and research opportunities.
Keywords: Mycobacterium, Culture Collection, repository, biomaterials, reagents, database, strain bank, strain, isolate, resource
This review aims to raise awareness among mycobacterial researchers of the many global resources available, including culture collections distributing mycobacterial biomaterials as well as other valuable resources such as databases and computational/bioinformatics tools.
INTRODUCTION
Mycobacterium is a bacterial genus characterized generally as aerobic, nonmotile and acid fast in microscopic staining methods. They have a unique hydrophobic cell wall rich in mycolic acids/mycolates. The genus is comprised of at least 180 species, but this number frequently changes due to the description of new species and reclassification of others. The genus includes a diverse collection of organisms ranging from relatively fast growers to truly slow growers, environmental to pathogenic, biosafety level 1 (BSL-1) to BSL-3, free-living to obligate intracellular parasites, psychrophilic to thermophilic, etc. Mycobacteria often thrive in unexpected, artificial environments, such as in stagnant or slowly moving water inside water heaters, ice machines or water pipes. Occasionally, exposure to these sources results in human infections. Despite being comprised of such a wide variety of organisms, the mycobacteria pose major challenges in the laboratory to differentiate between species. Phenotypically, mycobacteria frequently show intraspecies variability, making a confident differential identification difficult. Even using molecular techniques, species differentiation can prove difficult. Use of single gene sequences (such as 16S, hsp65 or rpoB) provides limited discriminatory power. Although combining multiple gene sequences increases the discriminatory power, many species may not be sufficiently defined and thus pose difficulties for definitive identification in diagnostic microbiology laboratories. Thus, the use of whole genome sequences (WGS) has increased dramatically and is becoming a prevalent tool for species identification and differentiation.
Mycobacteria are among the most prevalent etiological agents of human disease and are often divided into four groups: Mycobacterium tuberculosis (MTB) complex, M. leprae, M. ulcerans and non-tuberculous mycobacteria (NTM). MTB ranks near the top of the list of pathogens causing human mortality. Researchers working with MTB face several complicating factors, such as working under BSL-3 conditions due to its ability to easily produce infectious aerosols, slow growth rates, a thick cell wall that makes the extraction of nucleic acids and cellular fractions difficult, complex metabolism, limited access to clinical isolates from diverse geographical sites, as well as a lack of complete understanding of most mechanisms of drug resistance, virulence and pathogenesis. Researchers working with NTM also face some of these same challenges, in addition to having fewer isolates available for research, more
limited research funding and a much more limited knowledge base.
Fortunately, the mycobacterial research community is relatively small, and researchers know and support each other through collaborative ties. This leads to constant exchange of materials and information to expand their research and has been key to creating a fertile ground where research thrives. Unfortunately, in some instances the biomaterials shared between collaborators lack appropriate controls and qualifications, resulting in failure, mistaken results or inaccurate interpretations.
Culture collections (CCs) are repositories of biomaterials that acquire, authenticate, preserve, produce, develop and distribute biological materials. CCs often have a website and/or a catalog to allow communication with the user. This interface allows the users to learn which items are available, provides a description of the items and provides the user with an order management system. It should be a responsibility of the CC to ensure that the biological material is packed and shipped under the appropriate conditions, while complying with regulatory and biosafety requirements. The CC should be able to produce documentation indicating the steps they followed to confirm purity, viability, identification and a detailed description of the product to be delivered. Some collections provide their users with recommendations on how to handle, store and use the material distributed. The lack of a regulated quality system for obtaining, verifying and disseminating information regarding these features is what separates the CCs from individual researchers that hold and share their biological material with their peers. Availability of biomaterial and metadata via CCs allows multiple researchers to test the same material, making comparison and correlation of results between researchers more reliable. CCs allow access to qualified, quality-assured biomaterial to a wider range of researchers.
The main objective of this manuscript is to provide the mycobacterial research community with a brief description of mycobacterial biomaterial available from several CCs, as well as other resources to support scientific research. We aim to raise awareness among researchers that CCs are essential to support research and can enhance the quality and reproducibility of their research. Furthermore, CCs stimulate researchers not only to request their material, but also to receive and qualify biological material from researchers to support their scientific efforts, and make their research material accessible to the broader research community and hence returning public investment.
CULTURE COLLECTIONS
A wide variety of global CCs exist, some of which specialize in mycobacterial reagents and some of which are general CCs that have significant mycobacterial reagent holdings. Some of these CCs which distribute biomaterials to the general scientific community are described in detail below and are summarized in Table 1. For a list of most of the mycobacterial items described in the text, see Table S1 (Supporting Information).
Table 1.
Summary of publicly available culture collections containing or specializing in mycobacterial reagents.
| Mycobacterial reagents | ||||
|---|---|---|---|---|
| Name | Location | Approximate number (type strains) | Biosafety level (or equivalent) | Website |
| American Type Culture Collection (ATCC) | Manassas, Virginia, USA | 1300 (100) | BSL-1, -2, -3 | https://www.atcc.org |
| BEI Resources | Manassas, Virginia, USA | 2300 (21) | BSL-1, -2, -3 | https://www.beiresources.org |
| Belgian Co-ordinated Collections of Microorganisms/Institute of Tropical Medicine (BCCM/ITM) | Antwerp, Belgium | 850 (40) | BSL-1, -2, -3 | http://bccm.belspo.be |
| Biological Resource Centre of Institut Pasteur/Collection of Institut Pasteur (CRBIP/CIP) | Paris, France | 659 (159) | BSL-1, -2, -3 | https://www.pasteur.fr/en/public-health/crbip |
| Foundation for Innovative New Diagnostics (FIND) | USA; France | 800 | BSL-1, -2, -3 | https://www.finddx.org/specimen-banks/ |
| Korean Collection for Type Cultures (KCTC) | Jeollabuk-do, Republic of Korea | 82 (67) | RG 1, 2 | http://kctc.kribb.re.kr/English/ekctc.aspx |
| Leibniz-Institut, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) | Braunschweig, Germany | 424 (173) | BSL-1, -2 | https://www.dsmz.de |
| National Collection of Type Cultures (NCTC) | Porton Down, Salisbury, UK | 134 (44) | BSL-2, -3 | https://www.phe-culturecollections.org.uk/collections/nctc.aspx |
| National Jewish Health Colorado Cystic Fibrosis Research & Development Program (CF RDP) | Denver, Colorado, USA | 1,000 | BSL-1, -2 | https://www.nationaljewish.org/cocfrdp |
| NITE Biological Resource Center (NBRC) | Chiba, Japan | 32 (4) | BSL-1, -2 | http://www.nite.go.jp/en/nbrc/index.html |
| RIKEN BioResource Center/Japan Collection of Microorganisms (RIKEN BRC/JCM) | Tsukuba, Ibaraki, Japan | 171 | RG 1, 2 | http://jcm.brc.riken.jp |
BSL: Biosafety Level; RG: Risk Group.
BEI Resources (https://www.beiresources.org)
BEI Resources was established by the National Institute of Allergy and Infectious Diseases (NIAID) to provide reagents, tools and information for studying category A, B, and C priority pathogens, emerging infectious disease agents, non-pathogenic microbes and other microbiological materials of relevance to the research community. BEI Resources acquires, authenticates and produces reagents that scientists need to carry out basic research and develop improved diagnostic tests, vaccines and therapies. BEI Resources has been managed under contract by the American Type Culture Collection (ATCC®) since 2003. BEI Resources reagents are shared free of charge to registered individuals and organizations doing research on emerging infections and other relevant areas of interest related to microbiology. Registrants must demonstrate they work in an established institution with facilities and safety programs appropriate for the materials requested. BEI Resources produces and distributes materials following standardized procedures and quality control (QC) standards to provide users with assurance regarding item identity, purity, viability and characterization. The catalog of more than 13 000 organisms and related reagents includes bacteria, viruses, parasitic protists, pathogenic fungi, parasitic worms, rickettsiales, arthropod vectors, host and vector cell lines, and toxins. BEI Resources produces and distributes materials from well-known resources and projects such as the Human Microbiome Project, the Network of Antibiotic Resistant Staphylococcus aureus (NARSA), Malaria Research and Reference Reagent Resource Center (MR4), the Kilbourne Archive of reassortants or mutants of Influenza viruses, the Filariasis Research Reagent Resource Center (FR3) and the Schistosome Related Reagent Repository (SR3), among others. Among the materials requested through BEI Resources, mycobacterial reagents account for 20% of the items distributed annually.
The BEI Resources mycobacterial reagent resource comprises ≈2300 reagents including laboratory strains, clinical isolates of mycobacterial pathogens (e.g. H37Rv, H37Ra, HN878, CDC1551, Erdman), a growing number of NTM species including several type strains, gamma-irradiated whole cells, cell lysate and cell fractions, nucleic acids, antibodies and antisera, protein reference standards, protein expression clones, peptide and DNA microarrays, knockout and entry clone libraries, and leprosy research reagents.
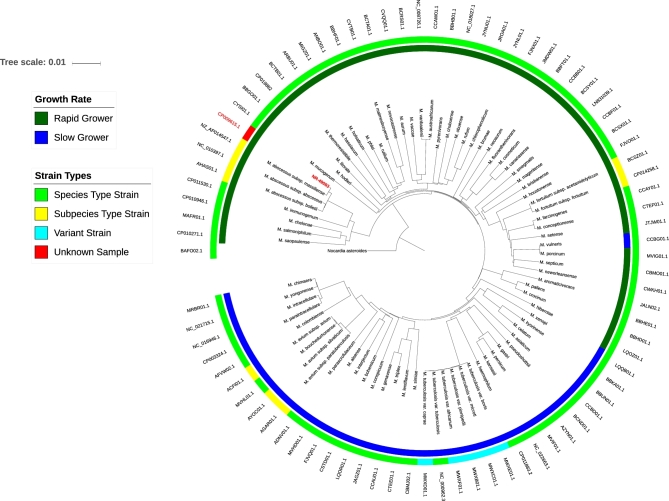
BEI Resources is using WGS as part of the production process for deposited Mycobacterium (mostly NTM) strains, thus obviating the identification issues commonly observed when relying only on routine phenotypic (biochemical tests) and genotypic (16S, hsp65 and/or rpoB sequence similarities) methods. WGS are compared to a database of type strain genomes from most mycobacterial species (to date, 173 species and subspecies) to confirm ID down to the species and/or subspecies level. This enables a reliable and reproducible identification of the strain, providing users with validated and characterized material. A real-world example of BEI Resources’ use of phylogenomic comparison is shown in Fig. 1. The item NR-49093 was originally deposited incorrectly as M. xenopi. However, upon comparison to the genome sequences of Mycobacterium type strains, it became abundantly clear that NR-49093 (shown in red) actually represents a strain of M. abscessus subsp. massiliense. Thus, this analysis enabled not only the determination of the correct species for the item but also its accurate identification down to the subspecies level.
Figure 1.
An example phylogenomic tree used to identify unknown Mycobacterium samples to the species- and/or subspecies-level. Tree was constructed as previously described (Riojas et al.2018). Each leaf on the tree corresponds to the test sample (in red) or to the reference genomes of the species and/or subspecies type strains, or in the case of the M. tuberculosis complex, the variants (Riojas et al.2018). The outer ring is comprised of the accession numbers for the WGS of each strain.
BEI Resources distributes MTB clinical isolates of diverse geographical origins, allowing researchers to access isolates from regions that impose strict export regulations. For example, sets of multidrug-resistant (MDR) clinical isolates of MTB from South Africa and Belarus (including both from MDR and extensively drug-resistant (XDR) which have detailed clinical information and susceptibility profiles) are available. BEI Resources is among the few distributors of M. leprae reagents to the scientific community. These materials include cellular fractions, inactivated cells, proteins, antigens and antibodies. Furthermore, distribution of inactivated MTB items (downgraded to BSL-2 or BSL-1) widens the number of researchers capable of performing research into MTB. The top five mycobacterial reagents distributed by BEI Resources are purified lipoarabinomannan (LAM), whole cell lysate, culture filtrate proteins, monoclonal anti-MTB LAM (CS-35), recombinant protein reference standard ESAT-6 and monoclonal anti-MTB LAM (CS-40).
ATCC: American Type Culture Collection (https://www.atcc.org)
With over 80 000 microbes in the collection, ATCC is a premier global biological materials resource and standards organization. For 90 years, ATCC has been providing cultures that represent the benchmark standard for performance in authentication and identification assays. ATCC cultures are released only after passing a rigorous QC process that confirms identity, viability and purity. Identity is always confirmed by at least two methods, including phenotypic and genetic. ATCC retains extensive records of each lot produced and provides quality documentation and growth instructions to customers. In addition to the off-the-shelf items, ATCC also offers custom solutions including custom configurations of our cultures for standards, controls, cell banks, cell and microbial banking, cell and microbial authentication, controls and derivatives, custom nucleic acids and CRISPR/Cas9 genome editing. ATCC is an ISO 9001:2008 certified, ISO 13485:2003 certified, ISO 17025:2005 and ISO Guide 34:2009 accredited organization.
The Mycobacterium collection at ATCC has over 1300 reagents including strains, phages and purified DNAs. The collection represents the broad diversity of the Mycobacterium genus, with isolates from humans, soil, water, livestock, birds, reptiles, plants and fish. The species and strains were collected from around the world, allowing for the investigation of geographic diversity within some species. All ATCC Mycobacterium are phenotypically and genotypically tested to confirm their species.
Over 100 of the strains in the ATCC Mycobacterium collection are the species type strain. The type strain designation is given to the strain of a species that is the first to be described and published and can be used to provide a benchmark standard for the species identification via phenotypic assays. ATCC has obtained the WGS of most of the type strains available from the collection. The MTB type strain, H37Rv, is offered in the ATCC catalog as ATCC 27294 (see Table 2). To accommodate researchers who do not have a BSL-3 facility but still wish to work with ATCC 27294, DNA is available that has been downgraded to a BSL-1 (ATCC 27294 D-2) after testing to confirm no viable material is present. In addition to the type strains, ATCC has BCG strains deposited by the Trudeau Mycobacterial Collection.
Table 2.
A list of selected Mycobacterium type strains available at ATCC.
| Mycobacterium species | Type strain | Source |
|---|---|---|
| Mycobacterium abscessus subsp. abscessus | ATCC 19977 | Human infection (knee) |
| Mycobacterium avium subsp. avium | ATCC 25291 | Liver of diseased hen, Denmark |
| Mycobacterium chelonae subsp. chelonae | ATCC 35752 | Tortoise, England, UK |
| Mycobacterium fortuitum subsp. fortuitum | ATCC 6841 | Cold abscess, Human, Brazil |
| Mycobacterium kansasii | ATCC 12478 | Inguinal Sinus, Human, USA |
| Mycobacterium marinum | ATCC 927 | Fish, USA |
| Mycobacterium septicum | ATCC 700731 | Venous catheter, 2-year-old cancer patient, Australia |
| Mycobacterium simiae | ATCC 25275 | Macaca mulatta, Hungary |
| Mycobacterium tuberculosis var. tuberculosis | ATCC 27294 | H37Rv: derived from H37, Lung, Human, New York, USA |
| Mycobacterium tuberculosis var. bovis | ATCC 19210 | Lymph node, 6 month heifer, Iowa, USA |
| Mycobacterium tuberculosis var. microti | ATCC 19422 | Field vole, England, UK |
| Mycobacterium tuberculosis var. pinnipedii | ATCC BAA-688 | Lung, seal, Australia |
| Mycobacterium ulcerans | ATCC 19423 | Leg ulcer, Human, Australia |
| Mycobacterium xenopi | ATCC 19250 | Adult, female toad, England, UK |
ATCC continues to actively collect new species and clinically relevant strains of all species of Mycobacterium. The type strains available from the collection have been used to revise taxonomy of mycobacterial species. The most recent example is the reclassification of the MTB complex as M. tuberculosis (Riojas et al.2018). Similar studies to reclassify other species are in progress.
ATCC is currently beginning the production under a research grant of a library in Escherichia coli of ≈4000 plasmids from the laboratory of Prof. William Jacobs, which can be used to obtain knockouts in each non-essential gene of MTB. This grant aims also at making available the corresponding ≈4000 phage lysates to allow researchers to excise the genes directly in the MTB strain of interest.
BCCM/ITM: Belgian Co-ordinated Collections of Microorganisms/Institute of Tropical Medicine (http://bccm.belspo.be)
The Mycobacteria Strain Collection of the Belgian Co-ordinated Collections of Microorganisms hosted by the Institute of Tropical Medicine in Antwerp, Belgium (BCCM/ITM) is one of the largest and most diverse collections of well-documented mycobacteria worldwide. BCCM aims to be a solution partner for providing services of quality in microbial and genetic resources for academia and industry. The consortium operates under an ISO 9001:2015 certified quality management system and ensures secured back-up storage of biomaterial, which can be searched through online catalogs. It hosts over 72 000 bacteria, diatoms, yeasts and molds, as well as 200 000 plasmids and DNA libraries.
The BCCM/ITM mycobacterial collection contains over 850 strains from human, animal and environmental origin, from over 90 different species/subspecies, about half of which belong to the MTB complex. The collection has 40 different type strains, including the widely used MTB type strain H37Rv. All strains are available as viable freeze-dried bacilli, heat-inactivated bacterial suspensions or upon request as fresh cultures or purified genomic DNA. Each item is supplied with a conformity certificate containing information on the strain's geographical and biological origin, date of isolation, history and other relevant information.
The diversity of drug-resistant MTB strains in the collection is unique, both in terms of drug-resistance profiles and phylogeographic origin. Strains resistant to first-line drugs are best represented among the 229 clinical isolates from the former WHO-TDR TB-Strain Bank (Vincent et al.2012), integrated in BCCM/ITM since 2013 and consulted by multiple diagnostic tool developers (Helb et al.2010). In addition, BCCM/ITM has incorporated clinical isolates and in vitro-selected strains resistant to fluoroquinolones, second-line injectables, pyrazinamide and new or repurposed drugs like bedaquiline and clofazimine. These strains have documented phenotypic resistance profiles, with the majority having detailed minimal inhibitory concentrations (MIC) determined for one or more drugs or drug classes, as well as genotypic resistance documented by targeted sequencing and/or WGS. This collection is about to release to its catalog more than 8200 transposon insertion mutants from a library of the M. bovis BCG Pasteur 1721 strain (Master et al.2008; Vandewalle et al.2015).
Building on the strong expertise of the host laboratory, BCCM/ITM hosts the world's largest and most diverse collection of M. ulcerans strains, the causative agent of the third-most important mycobacterial disease, Buruli ulcer. The genomes of these strains were recently described, including analysis of M. ulcerans across Africa (Vandelannoote et al.2017).
BCCM/ITM also contains over 400 strains from other mycobacterial species including the most common potential pathogens such as M. avium, M. kansasii, M. intracellulare, M. xenopi and M. marinum, important rapid growers like M. abscessus, some fastidious species like M. genavense and M. haemophilum, as well as widespread, generally non-pathogenic species like M. gordonae and M. nonchromogenicum. These NTM strains have 16S rRNA gene and rpoB gene sequences available, and many also have additional targets, e.g. the hsp65 gene or IS1245 or IS901 for M. avium strains, analyzed to allow characterization to the subspecies or subtype level. Genomes from these NTM type strains recently contributed to the elucidation of phylogenetic relationships within the Mycobacterium genus (Tortoli et al.2017).
RIKEN BRC/JCM: RIKEN BioResource Center/Japan Collection of Microorganisms (http://jcm.brc.riken.jp)
The Microbe Division of the RIKEN BioResource Center (RIKEN BRC) has been collecting, preserving and distributing cultured microbial strains as one of the world's leading CCs since it was established as the Japan Collection of Microorganisms (JCM) in 1981. JCM aims to contribute to scientific communities by maintaining and distributing high-quality microbial resources useful for general microbial studies and various research fields, particularly in health and environmental science. JCM has participated in the National BioResource Project supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan as the core facility for ‘General Microbes’ (http://www.nbrp.jp).
As of November 2017, JCM maintains ∼25 600 strains including ∼11 700 strains of bacteria, 530 strains of archaea and 5200 strains of fungi. Approximately 3500 to 4000 strains are annually distributed to domestic and international researchers. Genomic DNA samples of some strains are also distributed in cooperation with RIKEN BRC-DNA Bank. The mycobacterial collection is regarded as a part of the JCM general collection and mainly comprised of type strains. At the time of writing, 171 strains of 151 species and subspecies of the genus Mycobacterium are available, limited to those classified in Risk Group 1 or 2. Information of the available strains is opened to the public through the JCM On-line Catalogue Database (http://jcm.brc.riken.jp/en/catalogue_e).
JCM is working to continuously improve its function as a microbial resource center, to exploit new microbial resources, to describe novel microbial taxa and to develop the methods for investigating and handling extremophiles, uncultured microorganisms and microbial communities. JCM has acquired ISO 9001:2015 certification for its quality management system to maintain and improve the quality of its service. In order to promote the use of the microbial strains, JCM regularly issues mail news and gives training courses for both basic and advanced microbial techniques.
DSMZ: Leibniz-Institut DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, https://www.dsmz.de)
The Leibniz-Institut DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) is one of the largest biological resource centers of the world, with over 50 000 items. All biological materials accepted in the DSMZ collection are subject to extensive QC and physiological and molecular characterization. In addition, DSMZ provides an extensive documentation and detailed diagnostic information on the biological materials available from their catalog. The collection has 424 BSL-1 and BSL-2 mycobacterial items, including 173 type strains.
CRBIP/CIP: Biological Resource Centre of Institut Pasteur/Collection of Institut Pasteur (https://www.pasteur.fr/en/public-health/crbip)
The Biological Resource Centre of Institut Pasteur (CRBIP) harmonizes the management of collections on the Institut Pasteur campus, to enrich collections in an organized manner (e.g. with strains from collections of research laboratories), to ensure that the strains or products stored are characterized as thoroughly as possible, to improve conservation conditions (in two forms, at two different locations) and to ensure the distribution of samples according to health and environmental safety norms in accordance with the laws and regulations in force. It is organized to improve the access to microbial material and in particular to bacterial resources, in order to enhance exchange within Europe and non-European countries, to facilitate cross-disciplinary experimental work and to innovate with new better-performing methodologies such as collaborations between private and public laboratories. It has globally been certified ISO 9001:2000 since 2004 then NF S96-900 since 2009.
The CRBIP holds the Collection of Institut Pasteur (CIP), which contains more than 12 000 bacterial strains and almost 130 plasmids. The mission of the CIP is to maintain and enrich the collection of bacteria, primarily through collaborations with scientists from the Institut Pasteur and other scientists by the deposit of strains from French and foreign researchers. The CIP disseminates information about distributed strains, including properties, preservation and identification. It also develops a research activity on identification, taxonomy and strain preservation, between others. It distributes its biological resources in France and abroad following safety standards for health and the environment in compliance with regulations and laws (Best practices, Nagoya Protocol, etc.) and ensuring maximum traceability. The mycobacterial collection at CRBIP/CIP has ≈650 strains, from which 159 are type strains. The collection includes BSL-1, BSL-2 and BSL-3 items, from which many are clinical isolates obtained in different research projects carried out at the Institut Pasteur.
NBRC: National Institute of Technology and Evaluation Biological Resource Center (http://www.nite.go.jp/en/nbrc/cultures/index.html)
NBRC, National Institute of Technology and Evaluation (NITE) Biological Resource Center, which is the part of the NITE, is one of the largest microbial CCs in Japan. At the time of writing, NBRC maintains more than 32 000 strains including bacteria, fungi, yeast, archaea, microalgae and bacteriophages, and distributes ∼9000 strains per year to domestic and international researchers. The mycobacterial collection in NBRC is comprised of 32 NTM items. Strains preserved at NBRC are limited to those classified as BSL-1 and BSL-2. The collection has four type strains: M. aquiterrae, M. chlorophenolicum, M. fortuitum and M. vaccae. As clinical isolates, the collection offers several isolates of M. abscessus, M. chelonae, M. fortuitum, M. intracellulare and M. kansasii, which were isolated from Japanese patients. The mycobacterial strains are preserved as liquid-dried cultures in glass ampules. These items are released after rigorous QC tests including viability, purity checks and confirmation of the 16S rRNA gene sequences. NBRC strains and their information can be searched using the NBRC Culture Catalogue Search (http://www.nbrc.nite.go.jp/NBRC2/NBRCDispSearchServlet?lang=en).
National Jewish Health Colorado Cystic Fibrosis Research & Development Program( https://www.nationaljewish.org/cocfrdp)
National Jewish Health, one of the leading respiratory hospitals in the USA, has established and maintains a clinical NTM isolate repository that provides isolates to researchers on request and after committee review. This NTM isolate repository is primarily focused on archiving NTM strains that have been isolated from cystic fibrosis (CF) patients across the country for WGS, speciation, antimicrobial susceptibility analysis and subsequent research. The CC was initiated in 2015, through efforts of the CF Foundation-sponsored Colorado CF Research and Development Program (RDP) which oversees the NTM isolate repository, genome sequencing effort, and isolate distribution. NTM infections have become an increasing concern in the CF population (Martiniano, Davidson and Nick 2017) due to their high prevalence (Adjemian, Olivier and Prevots 2014) and suggested potential for transmission within CF populations (Bryant et al.2016).
The Colorado CF RDP at National Jewish Health actively accepts, archives and analyzes CF-associated NTM isolates from across the USA to better understand the species spectrum and genomic diversity of NTM infecting the CF population. Next-generation genomic sequencing and phylogenomic analysis are utilized to compare isolates within M. abscessus and its subspecies, the M. avium complex and other species to better understand the genomic diversity of clinically significant NTM species, and to identify occurrences of patient-to-patient transmission, shared sources of environmental acquisition, or to rule out such shared environmental acquisition. These efforts are essential to identify potential outbreaks, instances of nosocomial transmission, and to limit their occurrence and spread. Genomic sequencing efforts greatly expand our understanding of the phylogenetic diversity of clinically significant NTM species (Davidson et al.2014).
Currently, this collection has over 1000 NTM isolates, with several hundred isolates added each year, including members of the species M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, M. abscessus subsp. massiliense, M. avium, M. chimaera and M. intracellulare. Isolates can be requested from the culture and biorepository core of the Colorado CF RDP. Requests can be made by species or other characteristic, and many isolates have associated metadata including WGS information, phylogenomic analysis and antimicrobial susceptibility information. The collection also contains isolates from subjects enrolled in the CF Foundation-sponsored diagnostic PRospective Evaluation of nontuberculous mycobacterial Disease In CysTic Fibrosis (PREDICT: NCT02073409) and treatment Prospective Algorithm for the TrEatment of NTM in CF (PATIENCE: NCT02419989) trials. For these isolates, longitudinal collection of clonal isolates over time, pre- and post-treatment may be available, as well as detailed information concerning the virulence of the isolate, clinical features of the patient and response to treatment.
KCTC: Korean Collection for Type Cultures (http://kctc.kribb.re.kr)
The Korean Collection for Type Cultures (KCTC) was established in 1985 and obtained official approval from the Ministry of Science and Technology as a gene bank node. KCTC has three functions: (1) to collect, preserve and distribute biological resources; (2) to develop core technologies to characterize, produce and distribute valuable bioresources; and (3) to establish local and international networks with biological resource providers, and support the information to be provided to users of the collection. KCTC performs basic and applied scientific research to better characterize and produce novel microbial strains, performs microbiome research (mostly focused on anaerobic strains), and develops new processes to improve handling and preservation of fastidious strains. KCTC offers its users training courses on topics such as microbial handling and preservation as well as chemotaxonomic and phylogenetic analysis of novel microbes.
As of October 2017, KCTC maintains over 30 900 items including ∼12 600 bacteria, 230 archaea, 7000 fungi and yeast, 5600 patent strains, 1600 microalgae, 780 plant cell lines and 180 animal cell lines. To date, KCTC's mycobacterial collection (limited to Risk Groups 1 and 2) consists of 82 items, 67 of which are type strains. The information of each strain is available in the KCTC website.
FIND (Foundation for Innovative New Diagnostics) Specimen Bank (https://www.finddx.org/specimen-banks)
FIND is an international non-profit organization that enables the development and delivery of much-needed diagnostic tests for poverty-related diseases, including TB. FIND acts as a bridge between experts in technology development, policy and clinical care, reducing barriers to innovation and implementation of diagnostic solutions in low- and middle-income countries. FIND manages a high-quality biorepository of well-characterized MTB specimens—including its own collection and one for the World Health Organization—for use by researchers, developers and manufacturers of TB diagnostics. Providing access to these specimens supports the development and evaluation of new and existing tools to improve TB diagnosis. Sample types range from sputum to urine, plasma and serum. All specimens are collected at qualified clinics under protocols approved by institutional review boards (IRB) with informed consent from patients meeting the inclusion criteria for sample collection. Since 2015, FIND has added strains and DNA from patients with MDR-TB and pre-XDR-TB. Specimens from MDR/XDR-TB patients are collected from globally representative sites in South America, South East Asia, Africa and Eastern Europe. These specimens (sputum, plasma, serum, PAXgene tubes) are matched with strain isolates and DNA preparations. Characterization includes whole genome sequencing, phenotypic testing against first- and second-line drugs and MICs. Approximately 800 strains are currently available in FIND's specimen bank.
NCTC: National Collection of Type Cultures (https://www.phe-culturecollections.org.uk/collections/nctc.aspx)
The National Collection of Type Cultures (NCTC) is one of four CCs that make up Public Health England's (PHE) Biological Resource Centre. NCTC specializes in the curation and distribution of bacterial strains of clinical and veterinary importance. The collection includes nearly 6000 bacterial strains from 900 distinct species in at least 83 different families.
NCTC currently provides 134 strains of mycobacteria, including 44 type strains. Of these strains, 27 are BSL-3, including numerous strains of MTB. Mycobacteria represent some of the oldest strains in the collection with NCTC 358, the type strain of M. duvalii, being one of the first to be deposited when NCTC was first established in 1920. Historical strains such as these are proving to be of increasing importance in understanding microbial evolution.
NCTC is a dynamic collection, as new strains are added every week to ensure that the collection remains scientifically relevant. The collection is co-located with PHE's National Mycobacterium Reference Laboratory, which provides the ability to quickly and efficiently accession new strains as they emerge. At the time of writing, there are currently 22 new mycobacterial deposits undergoing QC testing in the process of being added to the NCTC catalogue. Many of these are clinical isolates that represent MTB clades of global significance.
NCTC, in collaboration with the Sanger Institute and Pacific Biosciences, is making freely available raw and annotated long-read WGS data for 3000 bacteria through the NCTC 3000 Project (https://www.phe-culturecollections.org.uk/collections/nctc-3000-project.aspx). NCTC 3000 aims to produce a website that will bring together the complete strain information of type and reference bacteria and viruses of global public health importance. The project is nearing completion, with 98% of the 900 type strains in the collection processed. To date, WGS data are available via the NCTC website for 59 mycobacterial strains (a subset of which are shown in Table 3), with the remainder anticipated to be uploaded in the coming months. For all NCTC Mycobacterium strains, DNA is available as an inactivated product that is safe to use within BSL-1 facilities.
Table 3.
A list of selected Mycobacterium genomes available through the NCTC 3000 Project.
RESOURCES
In addition to CCs that distribute physical biomaterials to the scientific community, there also exist numerous databases and tools relevant to mycobacterial researchers. Some of these resources are described in detail below and are summarized in Table 4.
Table 4.
Selected resources supporting mycobacterial research.
| Name | Hosted By | Function | Website |
|---|---|---|---|
| Advanced Analytics Tool for TB Portals' Data | National Institute of Allergy and Infectious Diseases (NIAID) | Virtual patient cohorts using real clinical data and metadata | https://depot.tbportals.niaid.nih.gov |
| Genomic Centers for Infectious Diseases (GCID) | National Institute of Allergy and Infectious Diseases (NIAID) | Provide insights into the biology of microbes, their role in pathogenesis and their interactions with the host, including the microbiome | https://www.niaid.nih.gov/research/genomic-centers-infectious-diseases |
| Global Catalogue of Microorganisms (GCM) | World Federation of Culture Collections (WFCC) | Dissemination of information related to culture collection holdings | http://gcm.wdcm.org |
| PATRIC Bioinformatics Resource Center (BRC) | National Institute of Allergy and Infectious Diseases (NIAID) | Integrated data and tools for bacterial infectious disease research | https://www.patricbrc.org |
| Preclinical Therapeutic Development Services | National Institute of Allergy and Infectious Diseases (NIAID) | Preclinical services to support development of products intended to cure, mitigate, diagnose or treat disease caused by a pathogen or certain toxins | https://www.niaid.nih.gov/research/therapeutic-development-services |
| Relational Sequencing for Tuberculosis (ReSeqTB) | Critical Path Institute (C-Path) | Regulatory-grade TB drug-resistance knowledgebase based on NGS data | https://platform.reseqtb.org/ |
| SITVITWEB | Institut Pasteur de la Guadeloupe | MTB molecular markers (spoligotypes, MIRU, and VNTR markers) database | http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE |
| SpolSimilaritySearch | Institut Pasteur de la Guadeloupe | Similarity search algorithm of spoligotype patterns in the SITVIT2 proprietary database | http://www.pasteur-guadeloupe.fr:8081/SpolSimilaritySearch |
| Virtual Strain Bank | Foundation for Innovative New Diagnostics (FIND) | Online tool to find and select TB strains based on user-defined criteria | https://strainbank.finddx.org |
GCM: Global Catalogue of Microorganisms (http://gcm.wdcm.org)
The World Federation of Culture Collections (WFCC) Global Catalogue of Microorganisms (GCM) is a robust, reliable and user-friendly system to help CCs to manage, disseminate and share the information related to their holdings (Wu et al.2017). Since its launch in 2012, GCM has received and published information on 402 778 strains and other holdings (plasmids and antibodies) deposited in 117 collections from 46 countries and regions.
GCM provides data retrieval, analysis and visualization system of microbial resources, and also serves as a knowledge base hosting taxonomy, phenotype and -omics data, together with scientific papers, patents and other outputs linked with its catalogue information.
ReSeqTB: Relational Sequencing for Tuberculosis (https://platform.reseqtb.org)
The Relational Sequencing for Tuberculosis (ReSeqTB) platform provides diagnostic developers, health care professionals and clinical laboratories a regulatory-grade tuberculosis (TB) drug-resistance knowledgebase based on next generation sequencing (NGS) data that has been standardized and curated with WHO-endorsed, culture-based drug susceptibility tests (Schito and Dolinger 2015; Starks et al.2015). This public initiative is a collaborative effort of the Bill & Melinda Gates Foundation (BMGF), Critical Path Institute (C-Path), Foundation for Innovative New Diagnostics (FIND), World Health Organization (WHO), US Centers for Disease Control and Prevention (CDC), the StopTB Partnership New Diagnostics Working Group (NDWG) and NIAID. The knowledgebase includes an MTB-validated Unified Variant Pipeline (UVP) to standardize WGS analysis (Personal communication, Matthew Ezewudo) and a targeted sequencing module that is still in development. Identified variants are clinically graded using evidence-based medicine approaches based on statistical evaluation combined with expert review. We have recently applied this approach to grade mutations identified in one of the largest systematic reviews of genotype/phenotype associations for both first and second-line TB drugs (Miotto et al.2017) and used the resulting confidence-graded list of mutations to interpret global sequencing-based drug resistance surveillance data (Zignol et al.2018).
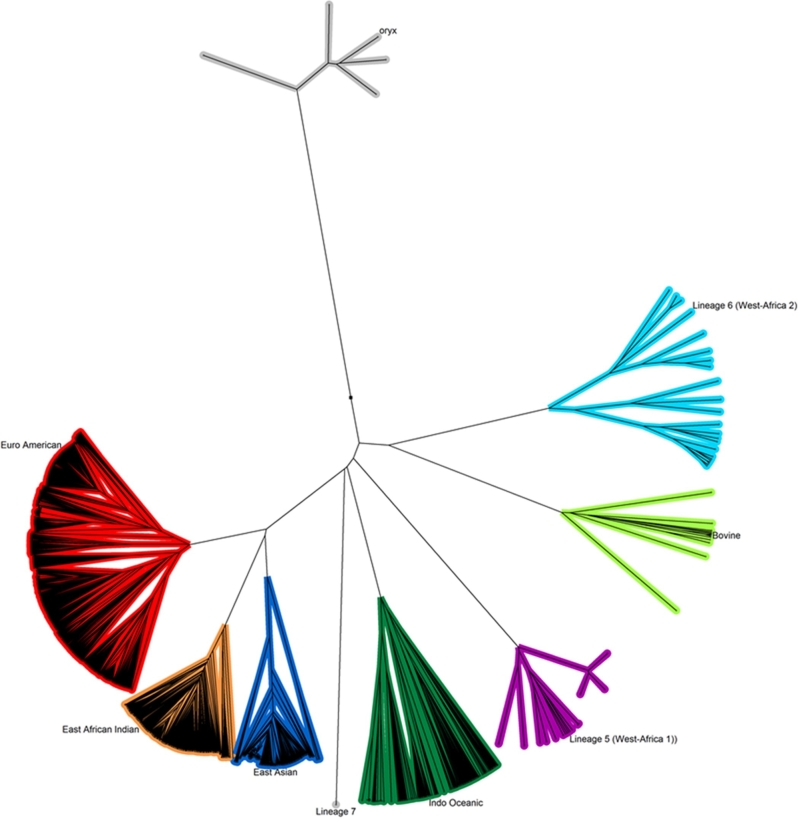
The ReSeqTB knowledgebase features a searchable genomic mutation browser to aid in the identification and analysis of mutations and their correlation with drug resistance and downloadable reports derived from aggregated data tracking 179 genetic loci of interest. As the clinical utility of the ReSeqTB knowledgebase for diagnosis and treatment decisions support is still being developed, the immediate value of the knowledgebase is mostly to support diagnostic developers designing probe-based assays to ensure they incorporate all high confidence mutations and account for mutations not associated with drug resistance. Data contributors own all the data they submit to the platform and have the choice to make it publicly available, embargo it until publication or restrict access to other contributors. The platform averages about 500 new isolates per month and is scalable based on demand. The genetic diversity of publicly available MTB isolates currently in ReSeqTB is shown in the phylogenetic tree (Fig. 2). The aggregate statistics in the downloadable reports are derived from all data in the platform. There are several advantages to contributing TB sequence data to the ReSeqTB platform prior to publication: (1) independent verification and validation of identified variants on a standardized validated pipeline created specifically for TB, (2) comparison of detected variants with other global variants, and (3) early inclusion of data into aggregate test statistics on genotype/phenotype associations which have the potential to facilitate timely patient care, without fear of jeopardizing publication rights.
Figure 2.
Phylogenetic tree based on whole genome multilocus sequence type (WgMLST) data of 3536 publicly available MTB isolates processed through the UVP pipeline in the ReSeqTB knowledgebase. The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) phylogenetic tree was created based on similarity matrix for a cross-section of isolates in the data set drawn from the WGS of the isolates.
ReSeqTB also incorporates standardized clinical data from global collaborations with TB clinicians, trialists and researchers. A vital next step of the ReSeqTB knowledgebase development is the incorporation of high-quality clinical data to correlate detected mutations with clinical outcomes. This effort will be particularly important for improving TB patient management, especially as new TB drugs and regimens become available to more patients globally. The curated, relational ReSeqTB knowledgebase is being designed for controlled global access to promote data sharing efforts and to support clinical interpretation of drug resistance mutations through cloud-based data analytics. In addition, ReSeqTB fills the global need for a centralized curated TB data archive. This will accelerate uptake of culture-free, drug-resistant MTB surveillance programs based on targeted NGS technologies. It is envisioned that the continued development of ReSeqTB will also encourage the uptake and improve feasibility of NGS technologies to support timely high-quality patient management decisions.
SITVIT Databases and other web tools (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE)
Although most of the earlier molecular epidemiological studies on MTB were performed using IS6110-RFLP, this labor-intensive methodology was replaced with alternative PCR-based typing strategies in the last decade (Jagielski et al.2016). Various versions of MTB genotyping databases were developed over last 15 years that allow the comparison of the huge amount of data cumulated worldwide using two widely used methods—spacer-based macroarrays for CRISPR genotyping, namely spoligotyping based on the polymorphism of the direct repeat locus (Kamerbeek et al.1997; Mokrousov and Rastogi 2015) and MIRU-VNTR minisatellites used in 12-, 15- or 24-loci formats (Supply et al.2006). A query in PubMed on 18 December 2017 made for ‘IS6110-RFLP AND tuberculosis’ returned 243 publications versus 1136 for ‘spoligotyping AND tuberculosis’, 943 using the term ‘MIRU OR VNTR OR MLVA OR MIRU-VNTR AND tuberculosis’ and 431 publications looking into the association of last two queries. Indeed, the numerical format of the spoligotype patterns and MIRU-VNTRs facilitates easy storage of the information generated in huge international databases in an organized way. Data handling, interrogation and knowledge-based data mining are further simplified by the development of new web tools allowing the study of MTB epidemiology and compare genotyping information at a global scale. The two most recent versions include SITVITWEB with genotyping data on 62 582 MTB clinical isolates corresponding to 153 countries of patient origin (Demay et al.2012) and SITVIT2 with genotyping data on 111 635 clinical isolates from 169 countries of patient origin (Couvin and Rastogi 2015). Briefly, each spoligotype pattern shared by two or more patients is assigned a spoligotype international type (SIT) number, while shared MIRU patterns are assigned MIRU international type (MIT) numbers, labeled 12-MIT, 15-MIT or 24-MIT depending on the loci formats used (Demay et al.2012; Couvin and Rastogi 2015).
The SITVIT resources are valuable not only to assign MTB genotypic lineages but also for phylogenetic and statistical analysis and geographical distribution of circulating clades worldwide, both at the country level and macrogeographically (based on the defined United Nations subregions; http://unstats.un.org/unsd/methods/m49/m49regin.htm ). They allow searches based both on individual criteria and combined explorations (year, isolation country, country of origin, investigator's name, genotype, genotypic lineage, drug resistance, etc.), making it possible to retrieve genotyping data in conjunction with geographical distribution, drug resistance, demographic and epidemiologic characteristics. One can therefore instantaneously map MTB genotyping data in function of search criteria, enabling the definition of the MTB genetic landscape and its ongoing evolution due to selection pressure exerted by emergence of drug resistance, socioeconomic inequalities and poverty-based migratory trends from high MTB-endemic countries to developed countries (García de Viedma, Mokrousov and Rastogi 2011).
The SITVIT2 version has been extensively tested as a proprietary database of the Institut Pasteur de la Guadeloupe (a PubMed query ‘SITVIT2 AND tuberculosis’ yielded 43 publications on 18 December 2017), and a public version will now be released in early 2018. In the meantime, the SpolSimilaritySearch web-tool (http://www.pasteurguadeloupe.fr:8081/SpolSimilaritySearch), which incorporates a similarity search algorithm, allows users to get a complete overview of spoligotype patterns in the SITVIT2 proprietary database (Couvin, Zozio and Rastogi 2017), in particular by looking for spoligotyping patterns with a specific signature worldwide. By allowing retrieval of country-based distribution maps, it is able to highlight shared patterns versus specific and/or confined patterns. Further characterization using other genotyping markers or NGS in the similarity search would undoubtedly allow extra discriminatory power. Nevertheless, the SpolSimilaritySearch web tool ensures that huge data generated using older genotyping methods are not deprecated, allowing instead establishment of useful links between the ‘older’ and ‘newer’ typing methodologies. Among such newer methods, one of the most exciting developments is a computer program named ‘SpoTyping’ which allows in silico determination of MTB spoligotyping patterns with high accuracy from NGS sequence and subsequent lineage determinations by interrogating the SITVIT resources (Xia, Teo and Ong 2016). Among the major advances of SpoTyping are its ability to predict spoligotyping patterns from sequencing experiments with non-uniform read length, its ability to work on different input files and operating systems, and the possibility of generating a graphical report showing the distribution summaries of the metadata corresponding to MTB phylogenetic lineages, years and countries of isolation by automated interrogation of the SITVIT database.
The SITVIT databases and other resources developed by researchers at the Institut Pasteur de la Guadeloupe have largely facilitated a more comprehensive global overview on MTB molecular epidemiology and public health surveillance. Ongoing global mapping of MTB genomes will facilitate the precise determination of exact thresholds of single-nucleotide polymorphism diversity between linked and unlinked isolates promoting the use of genomic epidemiology in MTB prevention and control programs.
FIND Virtual Strain Bank (https://strainbank.finddx.org)
In September 2017, FIND launched the Virtual Strain Bank (VSB), an online platform that allows users to find and select TB strains that can be used for TB research and diagnostic development. The strains can be selected based on several criteria including country, clade, drug resistance, drug sensitivity, mutations and sequencing data availability. The VSB currently lists all strains available at FIND. Other biorepositories may also freely upload information on their own well-characterized strains on this platform as a means to increase awareness and interest. Test developers and researchers can browse the VSB, query specific strains and request them directly from FIND or other biorepositories listed. The VSB aims to become a collaborative platform, enabling broad access to strains from around the world.
NIAID: National Institute of Allergy and Infectious Diseases (https://www.niaid.nih.gov)
NIAID, part of the US National Institutes of Health, is committed to finding new ways to better understand, diagnose, treat and ultimately prevent TB. NIAID supports a comprehensive portfolio of research covering basic, translational and clinical studies to better understand the natural history of TB and the development of drug resistance. NIAID also provides resources and services to investigators worldwide to facilitate biomedical research and help move drugs, vaccines and diagnostics closer to patients.
NIAID has aligned its resources, services and funding opportunities to address critical gaps along the research and product development pathway, from basic scientific research to translational research, preclinical development and clinical evaluation. Because the resources and services NIAID provides change in response to the public health and research community needs, the most up-to-date information on available resources and how to access them is the NIAID research resources website (https://www.niaid.nih.gov/research/resources). Resources include the Bioinformatics Resource Centers (BRCs) (https://www.niaid.nih.gov/research/bioinformatics-resource-centers), which collect, archive, update and integrate a wide variety of research data on infectious disease pathogens and provide this information through user-friendly interfaces and computational analysis tools. The primary BRC for bacterial species—called PATRIC (https://www.patricbrc.org)—currently contains over 100 000 genomes and includes an MTB-specific resource which provides precomputed data including antimicrobial resistance phenotypes. Other genomic-specific resources include the Genomic Centers for Infectious Diseases (GCIDs, https://www.niaid.nih.gov/research/genomic-centers-infectious-diseases), which provide insights into the biology of microbes, their role in pathogenesis and their interactions with the host, including the microbiome, by supporting a diverse set of genomic capabilities, such as NGS and related genomic technologies. The GCID currently focusing on MTB is at the Broad Institute (https://www.broadinstitute.org/scientific-community/science/projects/gscid/genomic-center-infectious-diseases). Three-dimensional structure determination for high-priority TB targets can be conducted as a free service under the Structural Genomics Centers for Infectious Diseases (https://www.niaid.nih.gov/research/structural-genomics-centers), which includes two centers, the Seattle Structural Genomics Center for Infectious Disease (SSGCID, https://www.ssgcid.org) and the Center for Structural Genomics of Infectious Diseases (CSGID, https://csgid.org). An additional TB-specific database that allows the integration of clinical and genomic data is the Advanced Analytics Tool for TB Portals’ Data (https://depot.tbportals.niaid.nih.gov), which encourages users to create virtual patient cohorts using real clinical data and metadata to promote analysis across four types of data.
NIAID resources also provide a range of preclinical Therapeutic Development Services (https://www.niaid.nih.gov/research/therapeutic-development-services) to advance the development of promising vaccine and therapeutic candidates, including Interventional Agents (https://www.niaid.nih.gov/research/interventional-agent-development-services) and Biopharmaceutical Products (https://www.niaid.nih.gov/research/biopharmaceutical-product-development-services) and the In Vitro Assessment for Antimicrobial Activity Program (https://www.niaid.nih.gov/research/vitro-assessment-antimicrobial-activity-resources), which screens products for antimicrobial activity and develops and provides in vitro assays. The ChemDB database (https://chemdb.niaid.nih.gov; https://www.niaid.nih.gov/research/chemdb) contains information extracted from scientific literature on the structure and activity of compounds that have been tested against HIV, opportunistic pathogens and MTB, which is a valuable database resource for TB drug developers. For more advanced vaccine and drug candidates that show promise as interventions for TB, NIAID provides animal model preclinical testing (https://www.niaid.nih.gov/research/pre-clinical-models-infectious-disease), which provides researchers who do not have access to or experience with animal models targeted help in which to study their product. The contractors providing these services have expertise in MTB, as well as access to the appropriate types of animal models, including mice, guinea pigs and non-human primates, for TB drug and vaccine testing. For advanced product development for TB, NIAID also provides contract services to conduct clinical evaluations for TB interventions through the Phase I Clinical Trial Units (https://www.niaid.nih.gov/research/phase-i-clinical-trial-units) and the Vaccine and Treatment and Evaluation Units (VTEUs, https://www.niaid.nih.gov/research/vaccine-treatment-evaluation-units-services). International support for clinical studies is provided under the International Clinical Sciences Support Center (ICSSC, https://www.niaid.nih.gov/research/international-clinical-sciences-support-center; http://icssc.org) and the ClinRegs database (https://www.niaid.nih.gov/research/clinregs; https://clinregs.niaid.nih.gov).
Investigators are encouraged to contact an NIAID Program officer for additional guidance (https://www.niaid.nih.gov/about/respiratory-diseases-branch-contacts).
CONCLUSIONS
The different CCs and resources described in this manuscript clearly indicate that there is a wide range of valuable reagents and tools available. The collections vary in size, processes followed, type of services provided, accessibility, cost, etc. However, each one is able to provide material, manufactured under strict QC standards, to the scientific community. The information tools available provide researchers with a variety of options that can help in understanding the organisms they are working with, while allowing a valid comparison with isolates from other sources.
During the preparation of this manuscript, we communicated with numerous institutions that have excellent and wide-ranging collections of mycobacteria (often with thorough characterization and metadata) accumulated over long periods of time. It was unfortunate to learn that sometimes access to their materials is restricted. There are several valid reasons for these restrictions. Often, they receive materials under specific conditions that prevent them from sharing with third parties (e.g. some countries impose strict restrictions for re-distribution of their isolates, they were obtained for specific clinical studies and there is no IRB approval for distribution, etc.) or they are regulated by a defined set of rules that limit their distribution. Furthermore, because these entities are not CCs, for them to share their material may require the establishment of specific scientific projects, official requests and/or to have institutional support from a specific type of entity. It is understandable that in most cases these entities serve a different purpose than supporting scientific research across the broader microbiological community. Many of these repositories were created for specific epidemiological purposes, and they are neither appropriately staffed nor designed to propagate, validate or distribute their material. We urge these institutional collections to consider either facilitating the scientific community's access to their material or depositing their material (or at least some of the most representative items) into an established CC. Depositing this material that is currently unavailable to the broader scientific community into any of the many CCs across the world would help promote increased scientific research and broader collaboration without burdening these institutional collections with the administrative and technical burdens inherent to broader distribution of physical biomaterials.
Similarly, it is also very common for researchers to collect clinical isolates, produce mutants, obtain valuable cellular fractions, antibodies, etc. These items or collections sometimes become well-known thanks to scientific publications, and these researchers manage requests from colleagues on a regular basis. This is a key component in the establishment of fruitful collaborations between scientists, but this poses a burden on the researcher's laboratory staff. Further, the material shared might lack appropriate validation or identification, which may lead to erroneous conclusions, artifacts or irreproducible results. As CCs, we know this for a fact because we receive materials from scientists and other depositors on a regular basis. When these materials are processed following the normal production and QC processes of a CC, an important percentage of them either fail to grow or simply do not pass the expected QC parameters. It is common to discover that the item deposited is not the material expected (Fig. 1), is contaminated or has been mischaracterized. This underscores the value of CCs in sharing material for research. We hope researchers consider depositing their valuable materials with established CCs, which in many cases can provide them with a free-of-charge thorough characterization of their material. As an added value to the depositor, this will ensure open access to research output. This can be achieved not only by sharing their findings and data through open access scientific publications but also by sharing generated biomaterial and metadata through public repositories.
The different CCs described in this manuscript clearly indicate that there is a wide range of valuable biomaterial available. Furthermore, access and use of microbial diversity is subject to new regulations, such as the Nagoya Protocol, covering access and benefit sharing by governing the socioeconomic use of biomaterials. CCs are encouraged to design their policy and practice in line with the protocol, to guarantee safe and legally fit-for-use microbiological material and data. To this end, administrative measures such as Material Accession Agreements and Material Transfer Agreements for acquisition and distribution of material are implemented. Ensuring informed consent from the depositor and country of origin prevents the end user to be confronted with legal issues or claims early on or in the final stage of their research or product development.
The resources available also provide access to large quantity of data and provide a context for the findings of researchers at a global scale. They provide user-friendly tools and access to processes that require different expertise, allowing the researchers to advance the research and give a strong and representative value to their results.
Acknowledgements
The authors wish to thank Dr Katharina Huber, Dr Sabine Gronow and Dr Lorenz Reimer from DSMZ for providing a list of reagents form their collection.
Conflict of interest. None declared.
REFERENCES
- Adjemian J, Olivier KN, Prevots DR. Nontuberculous mycobacteria among patients with cystic fibrosis in the United States: screening practices and environmental risk. Am J Resp Crit Care 2014;190:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JM, Grogono DM, Rodriguez-Rincon D et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous Mycobacterium. Science 2016;354:751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvin D, Rastogi N. Tuberculosis - A global emergency: tools and methods to monitor, understand, and control the epidemic with specific example of the Beijing lineage. Tuberculosis 2015;95(Suppl 1):S177–89. [DOI] [PubMed] [Google Scholar]
- Couvin D, Zozio T, Rastogi N. SpolSimilaritySearch - A web tool to compare and search similarities between spoligotypes of Mycobacterium tuberculosis complex. Tuberculosis 2017;105:49–52. [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hasan NA, Reynolds PR et al. Genome sequencing of Mycobacterium abscessus isolates from patients in the United States and comparisons to globally diverse clinical strains. J Clin Microbiol 2014;52:3573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demay C, Liens B, Burguière T et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 2012;12:755–66. [DOI] [PubMed] [Google Scholar]
- García de Viedma D, Mokrousov I, Rastogi N. Innovations in the molecular epidemiology of tuberculosis. Enferm Infec Micr Cl 2011;29:8–13. [DOI] [PubMed] [Google Scholar]
- Helb D, Jones M, Story E et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 2010;48:229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagielski T, Minias A, van Ingen J et al. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev 2016;29:239–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 1997;35:907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniano SL, Davidson RM, Nick JA. Nontuberculous mycobacteria in cystic fibrosis: updates and the path forward. Pediatr Pulmonol 2017;52:S29–36. [DOI] [PubMed] [Google Scholar]
- Master SS, Rampini SK, Davis AS. et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe 2008;3:224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto P, Tessema B, Tagliani E et al. A standardised method for interpreting the association between mutations and phenotypic drug-resistance in Mycobacterium tuberculosis. Eur Respir J 2017;50:1701354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrousov I, Rastogi N. Spacer-based macroarrays for CRISPR genotyping. Methods Mol Biol 2015;1311:111–31. [DOI] [PubMed] [Google Scholar]
- Riojas MA, McGough KJ, Rider-Riojas CJ et al. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int J Syst Evol Micr 2018;68:324–32. [DOI] [PubMed] [Google Scholar]
- Schito M, Dolinger DL. A collaborative approach for “ReSeq-ing” mycobacterium tuberculosis drug resistance: convergence for drug and diagnostic developers. EBioMedicine 2015;2:1262–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starks AM, Aviles E, Cirillo DM et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis 2015;61(Suppl 3):S141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006;44:4498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E, Fedrizzi T, Meehan CJ et al. The new phylogeny of the genus Mycobacterium: the old and the news. Infect Genet Evol 2017;56:19–25. [DOI] [PubMed] [Google Scholar]
- Vandelannoote K, Meehan CJ, Eddyani M et al. Multiple introductions and recent spread of the emerging human pathogen Mycobacterium ulcerans across Africa. Genome Biol Evol 2017;9:414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle K, Festjens N, Plets E et al. Characterization of genome-wide ordered sequence-tagged Mycobacterium mutant libraries by Cartesian Pooling-Coordinate Sequencing. Nat Commun 2015;6:7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent V, Rigouts L, Nduwamahoro E et al. The TDR Tuberculosis Strain Bank: a resource for basic science, tool development and diagnostic services. Int J Tuberc Lung D 2012;16:24–31. [DOI] [PubMed] [Google Scholar]
- Wu L, Sun Q, Desmeth P et al. World data centre for microorganisms: an information infrastructure to explore and utilize preserved microbial strains worldwide. Nucleic Acids Res 2017;45:D611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E, Teo YY, Ong RT. SpoTyping: fast and accurate in silico Mycobacterium spoligotyping from sequence reads. Genome Med 2016;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zignol M, Cabibbe A, Dean A et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis 2018;S1473-3099(18)30073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]