Abstract
SH2 domain-containing inositol 5-phosphatase 2 (SHIP2) binds with the Y1356-phosphorylated hepatocyte growth factor (HGF) receptor, c-MET, through its SH2 domain, which is essential for the role of SHIP2 in HGF-induced cell scattering and cell spreading. Previously, the experimental structure of the SH2 domain from SHIP2 (SHIP2-SH2) had not been reported, and its interaction with the Y1356-phosphorylated c-MET had not been investigated from a structural point of view. In this study, the solution structure of SHIP2-SH2 was determined by NMR spectroscopy, where it was found to adopt a typical SH2-domain fold that contains a positively-charged pocket for binding to phosphotyrosine (pY). The interaction between SHIP2-SH2 and a pY-containing peptide from c-MET (Y1356 phosphorylated) was investigated through NMR titrations. The results showed that the binding affinity of SHIP2-SH2 with the phosphopeptide is at low micromolar level, and the binding interface consists of the positively-charged pocket and its surrounding regions. Furthermore, R28, S49 and R70 were identified as key residues for the binding and may directly interact with the pY. Taken together, these findings provide structural insights into the binding of SHIP2-SH2 with the Y1356-phosphorylated c-MET, and lay a foundation for further studies of the interactions between SHIP2-SH2 and its various binding partners.
Keywords: INPPL1, HGFR, SH2 domain, solution NMR structure, tyrosine phosphorylation
1. Introduction
The Src homology 2 (SH2) domain is the central module of the phosphotyrosine (pY) signaling network that mediates phosphotyrosine-dependent protein-protein interactions [1]. More than a hundred SH2 domains are present in mammals and they share a conserved structure that consists of an antiparallel β-sheet flanked by two α-helices [2]. The β-sheet along with the surrounding loops and one of the helices makes up a ligand binding region that typically includes two pockets, one of which is positively charged for binding the pY residue (pY-pocket). The second pocket associates with residues adjacent to the pY in the ligand, and typically determines the binding specificity of the SH2 domain [3, 4]. This binding specificity is important for the accuracy of signal transduction, protein complex assembly and subcellular localization of SH2 proteins [5].
The SH2 domain-containing inositol 5-phosphatase 2 (SHIP2) is a phosphatidylinositol (PI) phosphatase that plays an essential role in cell proliferation, adhesion, migration, metabolism, and immune system function [6–11]. Perturbations of SHIP2 function have been implicated in cancers, diabetes, obesity, opsismodysplasia, and Alzheimer’s disease [12–16]. SHIP2 consists structurally of an N-terminal SH2 domain, a catalytic 5-phosphatase domain, a proline-rich region, and a C-terminal sterile alpha motif (SAM) domain [17]. The 5-phosphatase domain hydrolyzes the 5-phosphate of PI-3,4,5-trisphosphate (PI(3,4,5)P3) to produce PI-3,4-bisphosphate (PI(3,4)P2), and is required for the role of SHIP2 in modulating the levels of PI(3,4,5)P3 and PI(3,4)P2 in cells [18–20]. However, the function of SHIP2 also requires its interactions with various other proteins. The interactions of SHIP2 with several pY-containing proteins such as the p130CAS adapter protein, the immunoreceptor FcγRIIb and the hepatocyte growth factor (HGF) receptor known as c-MET, are mediated by the SHIP2 SH2 domain (SHIP2-SH2) [21–23]. These SH2-mediated interactions either connect SHIP2 function to a specific signaling pathway, or localize SHIP2 to specific subcellular sites, both of which are essential for SHIP2 cellular function [21–24].
The interaction of SHIP2 with c-MET is crucial for HGF-induced lamellipoadia formation and cell spreading [23]. c-MET is a single-pass transmembrane receptor involved in cellular survival, proliferation, invasion, and migration [25]. The binding of its physiological ligand, HGF, leads to c-MET activation and the phosphorylation of multiple residues including Y1003, Y1234, Y1235, Y1349 and Y1356 in the c-MET intracellular domain. This phosphorylation then facilitates the interactions of c-MET with a number of SH2-containing adaptor proteins such as SHIP2, PI3K, GRB2 and GAB1, and subsequently induces relevant cellular responses (reviewed in [26]). SHIP2 specifically binds to phosphorylated Y1356 of c-MET through its SH2 domain [23]. However, until now, the detailed molecular mechanism of this binding had not been determined.
The structures of the 5-phosphatase domain and the SAM domain of SHIP2 have been well studied [27–30]. However, although a 3D model of SHIP2 SH2 domain was described previously [31], the experimental structure had not yet been reported. In this study, we determined the solution structure of SHIP2-SH2 domain using NMR spectroscopy. It adopts a canonical SH2 fold and contains a potential pY-pocket on the surface. Subsequently, its interaction with a pY-containing peptide derived from Y1356-phosphorylated c-MET was investigated using NMR titration experiments. The binding interface was determined to be comprised of the potential pY-pocket and its surrounding region, and the dissociation constant (KD) was at low micromolar level. Furthermore, through alanine scanning mutagenesis followed by a fluorescence polarization (FP) binding assays, R28, S49 and R70 of SHIP2-SH2 were identified as key residues for binding to the Y1356-phosphorylated c-MET peptide.
2. Materials and Methods
2.1. Protein expression and purification
The DNA fragment encoding human SHIP2-SH2 domain (Uniprot ID: O15357, residues 20 to 117) was cloned into an NESG-modified pET15 vector including an N-terminal His-tag (MGHHHHHHSHM), and subsequently transformed into Escherichia coli BL21 (DE3) cells for protein expression. The cells harboring the plasmid were cultured in M9 minimal medium at 37 °C until OD600 reached 0.8, and then 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce protein expression at 17 °C overnight. Cell cultures were harvested by centrifugation and lysed by sonication. The supernatant containing SHIP2-SH2 was purified using an ÄKTAxpress™ (GE Healthcare) equipped with a Ni-affinity column (HisTrap IMAC HP™ column, 5 ml) followed by a gel filtration column (HiLoad 26/60 Superdex 75). The purified protein was concentrated to a final concentration of 0.58 mM for structure determination in the NMR buffer containing 10 % v/v D2O, 20 mM Tris, 100 mM NaCl, 10 mM DTT, and 0.02 % NaN3 at pH 7.5. The samples of S27A, R28A, R47A D48A, S49A, E50A, T68A, Y69A, R70A, Q82A, and S84A mutants were prepared using a similar method to that described above.
The 8-residue phosphopeptide “N-A-T-pY-V-N-V-K” (residues 1353–1360 of c-MET with Y1256 phosphorylated) and its unphosphorylated form “N-A-T-Y-V-N-V-K”, with or without the FAM labeling at N-terminus were synthesized by GL Biochem Ltd. (Shanghai). The purities of the peptides were determined to be above 98% through high performance liquid chromatography (HPLC) analysis and their molecular weights were confirmed through electrospray ionization mass spectrometry (ESI-MS).
2.2. Chemical shift assignment and structure calculation
The SHIP2-SH2 NMR data for structure calculations were collected on a Varian Inova 600 MHz spectrometer and a Bruker Avance III 850 MHz spectrometer at 298 K, and included 2D 1H-15N HSQC and 1H-13C HSQC, 3D HNCA, HNCO, HN(CO)CA, HNCACB, CBCA(CO)NH, (H)CC(CO)NH-TOCSY, H(CC)(CO)NH-TOCSY, H(C)CH-COSY, H(C)CH-TOCSY, two 13C-edited NOESY-HSQC optimized for either aliphatic or aromatic carbons, and an 15N-edited NOESY HSQC on the U-15N, 13C-labeled (NC) sample. A 3D (H)CCH-TOCSY and 4D 13C- 13C-HMQC-NOESY-HMQC was collected on the NC sample in D2O, and a 2D constant time 1H- 13C HSQC (CT-HSQC) was collected on the U- 15N, 5% 13C isotopically-labeled (NC5) sample. The mixing time for all NOESY spectra was 70 ms. Backbone and side-chain resonances were automatically assigned using the PINE server from NMRFAM [32], and subsequently validated and corrected manually. Stereospecific assignments of isopropyl methyl groups of Val and Leu residues were based on analysis of the CT-HSQC spectrum collected on the NC5 sample [33]. Assignments were 100% complete for backbone HN cross peaks and 98.5% complete for all 1H, 13C and 15N chemical shifts. Chemical shift assignments were deposited in the BioMagResDB (BMRB) under the accession number 19749, and a 1H-15N labeled HSQC is shown in Figure S1.
NOE-based inter-proton distance constraints for SHIP2-SH2 were obtained automatically using CYANA 3.0. Input for CYANA included chemical shift assignments, NOESY peak lists from NOESY spectra with peak intensities, the constraints for backbone phi (φ) and psi (ψ) torsion angle derived from backbone chemical shifts using the TALOS+ software [34]. Manual and iterative refinements of NOESY peak picking lists were guided with NMR RPF quality to assess “goodness of fit” between NOESY peak lists and calculated structures [35]. At last step of the iterative calculation process, hydrogen bond constraints for backbone NH and CO distances were introduced based on the proximity of potential donors and receptors in earlier structure calculations. The 20 structures with lowest energy calculated by CYANA 3.0 were further refined using restrained molecular dynamics in explicit water with CNS 1.2 [36] and the PARAM19 force field, based on the NOE-derived distance constraints, hydrogen-bond constraints, and TALOS-derived dihedral angle constraints. The final ensemble of 20 structures and all restraints used in the structure calculations were deposited in the Protein Data Bank (PDB ID: 2MK2). Structural statistics and global structure quality factors computed using PSVS version 1.4 [37] are shown in Table 1.
Table 1.
Structural statistics for human SHIP2-SH2 domaina
| Conformationally restricting constraints b | |
|---|---|
| Distance constraints | |
| Total | 1187 |
| Intra-residue (i=j) | 277 |
| Sequential (|i-j|=1) | 293 |
| Medium-range (1<|i–j|<5) | 146 |
| Long-range (|i–j|≥5) | 471 |
| Hydrogen bond constraints Long-range (|i–j|≥5)/total |
20/46 |
| Dihedral angle constraints | 108 |
| Residue constraint violations b | |
| Average number of distance violations per structure | |
| 0.1–0.2Å | 6.6 |
| 0.2–0.5 Å | 2.05 |
| >0.5Å | 0 |
| Average RMS distance violation/constraint (Å) | 0.02 |
| Maximum distance violation (Å) | 0.38 |
| Average number of dihedral angle violations per structure | |
| 1–10° | 10.65 |
| >10° | 0 |
| Average RMS dihedral angle violation/constraint (degree) | 1.07 |
| Maximum dihedral angle violation (degree) | 8.8 |
| RMSD from average coordinates b,c | |
| Backbone /Heavy atoms (Å) | 0.7/1.3 |
| Ramachandran plot statistics b,c | |
| Most favored/Allowed regions (%) | 96.7/3 |
| Disallowed regions (%) | 0.2 |
| Global quality scores(raw/Z-score) b | |
| Verify3D | 0.37/ −1.44 |
| Prosall | 0.45/−0.83 |
| Procheck(phi-psi) c | −0.37/−1.14 |
| Procheck(all) c | −0.30/−1.77 |
| Molprobity clash | 13.48/−0.79 |
| RPF Scores d | |
| Recall/Precision | 0.98/0.90 |
| F-measure/ DP-score | 0.94/0.77 |
Structural statistics were computed for the ensemble of 20 deposited structures.
Calculated using the PSVS 1.4 program. Residues (20–117) were analyzed.
Ordered residues ranges (with the sum of φ and ψ order parameters > 1.8): 28–38, 42–49, 55–70, 78–82, 87–104, 113–115.
RPF scores reflected the goodness-of-fit of the final ensemble of structures including disordered residues to the NMR data.
2.3. NMR titration
NMR titration experiments were carried out on a Bruker Avance III 600 MHz instrument at 298 K. A sample containing 0.2 mM 15N-labeled SHIP2-SH2 in NMR buffer was titrated with unlabeled c-MET peptide with or without Y1356 phosphorylation. Peptide stock solutions in the NMR buffer at a peptide concentration of 7.73 mM were added stepwise with a sample dilution less than 10% at a time, and allowed to equilibrate for over 1 h before each NMR experiment. 1H-15N HSQC spectra for the samples with SHIP2-SH2:peptide molar ratios of 1:0, 1:0.25, 1:0.5, 1:0.75, 1:1, 1:2, and 1:4 were collected, respectively. Chemical shift perturbations (CSPs) were calculated using the following equation: Δδ = [(ΔδHN)2 + (ΔδΝ/5)2]1/2, where ΔδHN and ΔδN are the chemical shift changes in the proton and nitrogen dimensions, respectively [38]. Intensity reductions and CSPs were considered significant if they were greater than 60% or 0.15 ppm, respectively. The titration data was further fitted using the equation for fast chemical exchange [38] with the “Nonlinear Curve Fit” function in Origin 2017 to determine the dissociation constant (KD) of each selected residue. A global fitting of thirteen residues was carried out to determine an average KD, in order to represent the overall binding affinity.
2.4. Fluorescence Polarization
FAM-labeled c-MET peptide with Y1356 phosphorylated was dissolved in a buffer containing 20 mM Tris, 100 mM NaCl, 10 mM DTT, and 0.02 % NaN3, at pH 7.5. The peptide concentration was kept at 50 nM, while SHIP2-SH2 was serially diluted with the concentrations ranging from micromolar to nanomolar, and then mixed with the peptide. Fluorescence polarization (FP) was measured using a Cytation™ 3 cell imaging multi-mode reader (BioTek) by recording the excitation at 485 nm and emission at 528 nm for each binding reaction between 50 nM peptide and indicated concentration of SHIP2-SH2. Each binding reaction was repeated three times, and the polarization values, P, were averaged (given in units of mP). In each parallel test, the polarization value of the free peptide (Pmin) and the maximal polarization value saturated with SHIP2-SH2 (Pmax) were recorded. The normalized FP value (Y) was yielded using the following equation: Y = (P - Pmin) / (Pmax – Pmin). Then, the Y was plotted against the protein concentration (X, in unit of μM). The resulted binding curves were fit to the following equation to yield KD values.
3. Results and Discussion
3.1. Solution Structure of SHIP2-SH2
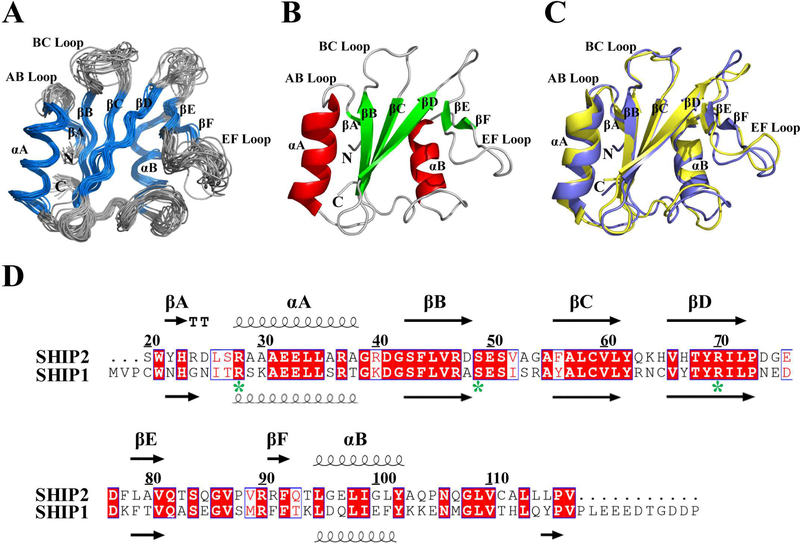
The recombinant SHIP2-SH2 domain with an N-terminal His-tag has a molecular weight of 12.2 kDa. Analytical static light scattering measurement and gel-filtration chromatography indicated that it existed as a monomer under our conditions (Figure S2). The ensemble of 20 solution structures of the SHIP2-SH2 domain (20–117) determined through NMR spectroscopy is shown in Figure 1A and has a RMSD of 0.7 Å for backbone atoms and 1.3 Å for heavy atoms in the ordered regions. All secondary structural elements were well defined as follows: βA (22–23), αA (27–39), βB (43–48), βC (56–61), βD (66–72), βE (79–81), βF (91–92) and αB (94–103) (Figure 1B). The overall structure of SHIP2-SH2 adopts a canonical SH2 fold that consists of a central β-sheet flanked by two α-helices. Three long antiparallel β-strands (βB, βC and βD) compose the central β-sheet, while βE and βF combine with the C-terminal part of βD to form another small β-sheet.
Figure 1.
NMR structure of SHIP2-SH2. (A) Superposition of Cα traces of 20 lowest energy conformers of SHIP2-SH2 determined by NMR spectroscopy, with α-helix and β-strand colored in blue, and loop regions in gray. The disordered N-terminal His-tag is not shown for clarity. (B) Cartoon representation of SHIP2-SH2 structure with the lowest overall energy wherein α-helix, β-strand and loop regions are colored in red, green and gray, respectively. (C) Structural comparison of SHIP2-SH2 (violet) and SHIP1-SH2 (yellow, PDB ID: 2YSX). (D) Sequence alignment of SH2 domains from SHIP2 (20–117) and SHIP1 (1–112). Alignment was rendered using ESPript16 with default settings for the similarity calculations. Identical (white letters filled with red color) and similar (red letters with blue box) amino acids are denoted. All secondary structure elements are labeled on the top for SHIP2-SH2 and bottom for SHIP1-SH2. Three key residues for c-MET binding are labeled with green stars.
SHIP1, the closest homologous protein to SHIP2, is similar to SHIP2 in terms of sequence, domain architecture, PI(3,4,5)P3 degradation activity and cellular function [39]. The SH2 domains of SHIP1 and SHIP2 have similar substrate specificity but different binding kinetics [40]. When the SHIP2-SH2 structure is compared with that of SHIP1-SH2 from the PDB database (PDB ID: 2YSX), it was found that they were highly similar with an RMSD of 1.4 Å for 90 backbone Cα atoms (Figure 1C), consistent with their similar functions. Nevertheless, residues R91 and F92 of SHIP2 form a small β-strand (βF) while the counterparts of SHIP1 do not, possibly because R91 aligns with a phenylalanine at the corresponding position in SHIP1 (Figure 1D).
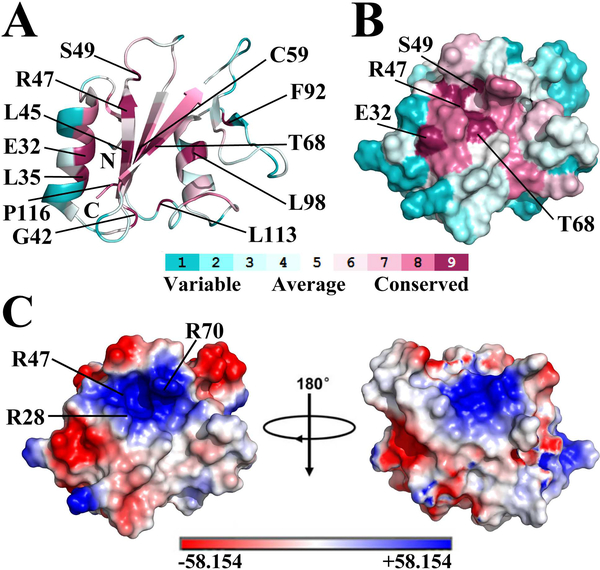
ConSurf [41] analysis of SHIP2-SH2 identified twelve highly conserved residues including E32, L35, G42, L45, R47, S49, C59, T68, F92, L98, L113 and P116 (conservative level 9, Figure 2A). Most of these residues are located in the secondary structure regions with the exception of S49, G42, L113 and P116. The pY-pocket of a typical SH2 domain is made up of several conserved residues that are located in the BC loop, N-terminal part of αA, and the central β-sheet [2]. The corresponding region of SHIP2-SH2 also forms a pocket on the protein surface that includes the three conserved residues R47, S49 and T68 (Figure 2B), implicating that it may be the pY-pocket of SHIP2-SH2. Further, electrostatic surface potential analysis showed that this probable pY-pocket is positively charged (Figure 2C), consistent with this function. The side-chain of R47 (conservative level 9) along with those of R28 and R70 (conservative level 8) around the pocket contribute the positive charges that are expected to be important for phosphotyrosine binding.
Figure 2.
The potential pY-pocket of SHIP2-SH2. (A) Cartoon representation of SHIP2-SH2 structure with conservative level for each residue analyzed using the ConSurf Server. Residue colors represent the conservative levels, ranging from cyan (variable) to purple (conserved). Twelve most conservative residues (conservative level 9) are labeled. ConSurf analysis was performed using the first structure of SHIP2-SH2 (PDB ID 2MK2) as a query for homologue search with search algorithm of HMMER. 150 homologous sequences were collected from UNIREF90 database for conservation score calculation with an E-value cut-off of 0.0001, maximal and minimal sequence identity of 95% and 35%, and HMMER iteration number of 1. (B) Molecular surface representation of SHIP2-SH2 structure (same orientation as in A) with conservative level for each residue. Only the most conservative residues on protein surface are labeled. (C) Molecular surface representation of SHIP2-SH2 structure with surface electrostatic potential analysis. Positively-charged residues around the potential pY-pocket are indicated.
3.2. The binding interface of SHIP2-SH2 for Y1356-phosphorylated c-MET
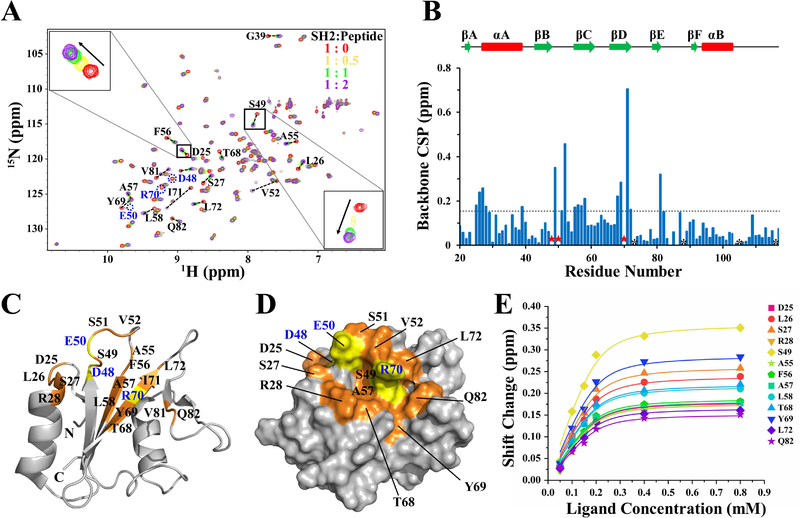
Based on the solution structure and chemical shift assignments of SHIP2-SH2, the interaction of SHIP2-SH2 with Y1356-phosphorylated c-MET was subsequently investigated. NMR titrations of 15N-labeled SHIP2-SH2 with unlabeled c-MET peptides (1353–1360) with and without Y1356 phosphorylation were carried out. 2D 1H-15N HSQC spectra for the mixtures at a series of SHIP2-SH2:peptide ratios (1:0, 1:0.25, 1:0.5, 1:0.75, 1:1, 1:2, and 1:4) were collected, respectively. Titration with Y1356-unphosphorylated c-MET peptide did not lead to any significant chemical shift perturbations (CSPs) of the SHIP2-SH2 amide protons (Figure S3). In contrast, titration with Y1356-phosphorylated c-MET peptide resulted in many significant CSPs (Figure 3A), indicating that phosphorylation of Y1356 is required for binding, consistent with the previous functional studies [23]. Upon increasing the ratio of SHIP2-SH2:peptide from 1:0 to 1:0.5 and then to 1:1 for the Y1356-phosphorylated c-MET peptide, most peaks from SHIP2-SH2 that had CSPs shifted successively from the unbound to completely-bound positions, indicating that the population weighted averaging of chemical shifts, which is characteristic of fast time-scale chemical exchange (Figure 3A). Further titrations that increased the ratio to 1:2 and then to 1:4, resulted in only minor additional changes in peak positions, indicating that the SH2-peptide binding was basically saturated by a ratio of 1:1. There were also a few peaks showing line broadening and decreased intensities upon titration, representing intermediate chemical exchange behavior (Figure 3A).
Figure 3.
The binding interface and affinity of SHIP2-SH2 for Y1356-phosphorylated c-MET revealed by NMR titration. (A) Overlay of 2D 1H-15N HSQC spectra of the apo SHIP2-SH2 (red) and the mixtures of SHIP2-SH2 with Y1356-phosphorylated c-MET peptide at the ratio of 1:0.5 (orange), 1:1 (green), 1:2 (purple). Residue D25 and S49 are zoomed in respectively. For clarity, only four points of the titration are shown. (B) The chemical shift perturbation (CSP) of each residue during NMR titrations were calculated and shown with the secondary elements on top. The residues that had significant CSPs but were untraceable in 1H-15N HSQC spectra during the titration are labeled with red stars. Prolines are labeled with black asterisks. The broken line represents the cutoff level (CSP ≥ 0.15 ppm) for significant perturbations. (C) The residues affected significantly by the titrations are mapped onto a cartoon representation of the SHIP2-SH2 structure. Residues with CSPs greater than the cutoff level are highlighted in orange with black text, and the residues that disappeared during the titration are highlighted in yellow with blue text. The peak intensities for R16, S84 and G86 were poor, and therefore their CSP values could not be definitely determined. (D) Molecular surface representation of SHIP2-SH2 structure with residue and text colors the same as those in (C) is show. Only the residues on the protein surface are labeled. (E) Global fitting of the NMR titration data for thirteen residues. The selected residues were all suitable for fitting to the fast-exchange equation.
The CSP values for each residue of SHIP2-SH2 during titration with Y1356-phosphorylated c-MET peptide were calculated, and the residues with CSP values greater than 0.15 ppm along with the residues that disappeared during the titration were mapped onto the structure of SHIP2-SH2 (Figure 3B-D). These residues were located in AB loop (D25 and L26), αA (S27 and R28), βB (D48), BC loop (S49-V52 and A55), βC (F56-L58), βD (T68-L72), βE (V81), and EF loop (Q82). The majority of these residues were located in the central β-sheet, the BC loops and N-terminus of αA, and make up the putative pY binding pocket (Figure 3C and3D). Residues R28, S49, A57, T68 and R70 whose side-chains are involved in forming the surface of the pY-pocket are conserved residues with ConSurf scores of 8 or 9, while S27, D48, E50 and V52 surrounding the pY-pocket are more variable. In the canonical model of SH2-ligand binding, the pY-pocket the SH2 domains interact with the pY mainly through hydrogen bonds and electrostatic interactions. Therefore, based on the NMR titration data, we predict that S27, R28, D48, S49, E50, T68 and R70 are shifted because of direct-binding interactions with pY. In addition, other residues that were distant from the pY-pocket, including D25, L26, S51, A55, F56, L72, Y69 and Q82, also had significant CSPs. These may be involved in the contact with the residues adjacent to pY1356 in the c-MET peptide. Overall, it could clearly be seen that the region with significant CSPs contained a central pY-pocket along with a larger binding interface that could accommodate the extended peptide (Figure 3D). Due to the conservation of the binding-interface residues between SHIP2-SH2 and SHIP1-SH2, and the fact that SHIP1-SH2 also binds to Y1356-phosphorylated c-MET [42], we expect that SHIP2-SH2 and SHIP1-SH2 will share a similar binding interface for c-MET.
3.3. The binding affinity of SHIP2-SH2 for Y1356-phosphorylated c-MET
To evaluate the binding affinity of SHIP2-SH2 for the Y1356-phosphorylated c-MET peptide, the average dissociation constant (KD) was estimated through a global fitting of the following thirteen residues with significant CSPs: D25, L26, S27, R28, S49, A55, F56, A57, L58, T68, Y69, L72 and Q82 (Figure 3E). Consistent with the result that the binding was largely completed at a SHIP2-SH2:peptide ratio of 1:1, the titration curves fit well to a 1:1 binding model and gave an average KD of 16.5 μM. Meanwhile, the binding affinity was also evaluated through fluorescence polarization (FP) assays using FAM-labeled peptides (Figure 4), with the resulted KD (7.4 μM, Table 2) similar to that from the NMR titration data. In agreement with the fitting of the NMR titration data, the FP data also fit well to a 1:1 binding model. Both NMR titration and FP assays measured affinity at the low micromolar level. However, the difference in the measured KD values could be caused by the differences in protein and ligand concentrations that were used for the measurements. In order to achieve good signal to noise in the NMR experiments, the concentration of 15N-labelled SHIP-SH2 was 0.2 mM, higher than the determined KD values, while the concentration of FAM-labeled peptides used in FP assays for fluorescence measurement was 50 nM. The higher concentration of 15N-labelled SHIP-SH2 may lead to an overestimation of the KD determined by NMR titrations. The measured KD of SHIP2-SH2 for Y1356-phosphorylated c-MET peptide was about 5 to 11-fold higher than that found for the ITIM motif in FcγRIIB (KD =1.5 μM) determined previously through surface plasmon resonance (SPR) analysis [40]. The lower binding affinity of SHIP2-SH2 for Y1356-phosphorylated c-MET may be due to non-optimal binding interactions for residues following the pY. It is notable that the c-MET sequence following the pY (pYVNV) does not match the previously determined the consensus sequence (pY [S/Y] [L/Y/M] [L/M/I/V]) recognized by SHIP2-SH2 at pY+1 and pY+2 sites (the neighboring first and second residues C-terminal to the pY) [40].
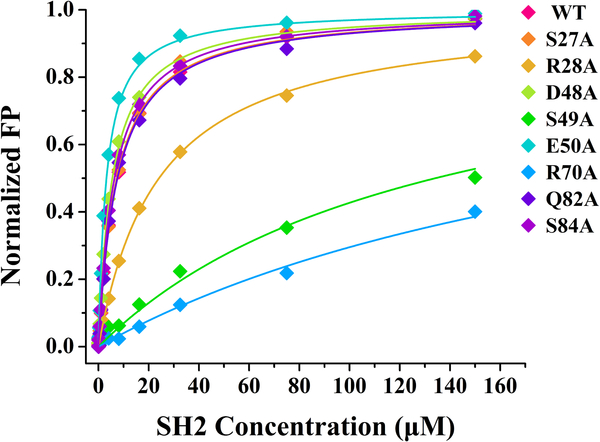
Figure 4.
Key residues of SHIP2-SH2 for binding Y1356-phosphorylated c-MET revealed by alanine scanning mutagenesis and FP assays. The wild-type (WT) and eight mutants of SHIP2-SH2 were titrated into FAM-labeled c-MET peptide with Y1356 phosphorylation, respectively, to obtain the binding curves. The dissociation constants (KD) obtained by the fits are given in Table 2. The fluorescence polarization (mP) values have been normalized for comparison between different mutants of SHIP2-SH2.
Table 2.
The dissociation constants (KD) of wild-type (WT) and eight mutants of SHIP2-SH2 determined by fluorescence polarization assays.
| Number | Protein | KD (μM) |
|---|---|---|
| 1 | WT | 7.4 ± 0.3 |
| 2 | S27A | 7.3 ± 0.3 |
| 3 | R28A | 24.7 ± 1.0 |
| 4 | D48A | 5.5 ± 0.2 |
| 5 | S49A | 134.1 ± 5.6 |
| 6 | E50A | 3.2 ± 0.1 |
| 7 | R70A | 238.6 ± 12.1 |
| 8 | Q82A | 7.5 ± 0.3 |
| 9 | S84A | 6.4 ± 0.2 |
3.4. Key residues of SHIP2-SH2 for binding Y1356-phosphorylated c-MET
The residues with significant CSPs in the binding interface of SHIP2-SH2 could participate in the interaction with Y1356-phosphorylated c-MET directly, but could also only be indirectly affected by the binding, which can be caused by changes in structure and chemical environment. To identify key residues for binding, eleven residues including S27, R28, R47, D48, S49, E50, T68, Y69, R70, Q82 and S84 were selected for alanine scanning mutagenesis combined with FP binding assays, based on their large CSP values, with the exception of S84. S84 was close to Y69 and Q82 in space, but its CSP value could not be definitely determined in the NMR titration. The mutant proteins were prepared using a similar method to that used for wild-type SHIP2-SH2. Unfortunately, three of these mutants, R47A, T68A and Y69A, were not able to be obtained with these methods. Y69A was expressed at similar level to that of the wild type but went into inclusion bodies, while the expression levels of T68A and R47A were extremely low. These results suggest that the three mutations led to remarkable changes of the properties of SHIP2-SH2.
The other eight mutants were successfully expressed and subsequently assayed for their binding with Y1356-phosphorylated c-MET peptide through FP experiments. The results are shown in Figure 4 and the fitted KD values are in Table 2. The mutants S27A, D48A, Q82A and S84A showed KD values close to that of the wild type, indicating that these four mutated residues are not essential for the binding. On the other hand, the mutants R28A, S49A and R70A showed remarkably reduced affinities for Y1356-phosphorylated c-MET peptide, with KD values increasing to 24.7 μM, 134.1 μM and 238.6 μM, respectively, indicating that these three residues are crucial for the binding. S49 is a highly conserved residue, and the hydroxyl side-chain of this serine usually forms hydrogen bond with the pY in other well-studied SH2-ligand interactions [3]. R20 and R70 are important to form the positively-charged surface and may mediate electrostatic interactions. Unexpectedly, the mutant E50A displayed a two-fold increase in binding affinity by compared with the wild type. This may because the mutation from glutamic acid to alanine led to an increased positive charge of BC loop that is located in the pY-pocket.
The crystal structure of GRB2-SH2 in complex with the tetrapeptide Ac-pY-V-N-V corresponding to residues 1356–1359 of c-MET was reported previously (PDB ID: 1FYR) [43]. SHIP2-SH2 and GRB2-SH2 have a sequence similarity of 50%, and their structures are also similar, as revealed by the RMSD of 1.3 Å (52 Cα atoms, including the major region of chain-A and a small part of chain-B of 1FYR) based on backbone structural alignment (Figure S4A and S4B). As shown in the complex structure, GRB2-SH2 binds to the c-MET peptide mainly using the side chains of residues (R67, R86, S88, S90 and S96) in its pY-pocket through polar contacts, while the side chains of F108 and W121 also make hydrophobic contacts with the valine in a pY+1 site (Figure S4B-D). These residues in GRB2 correspond to residues R28, R47, S49, S51, A57, Y69 and Q82 in SHIP2 (Figure S4A), all of which are located in the binding interface determined by NMR titration. The key roles of R28 and S49 have been demonstrated using function assays of their corresponding mutants, while Q82 was not indispensable for the binding (Figure 4). S51 is relatively far away from the pY-pocket in the apo SHIP2-SH2 structure and is not likely to be involved in direct contact with Y1356-phosphorylated c-MET peptide. On the other hand, although the roles of R47 and Y69 in the binding were not successfully evaluated due to protein expression problems, they may also have direct contact with c-MET by comparison with interactions observed in GRB2-SH2:Ac-pY-V-N-V complex [43]. It would be expected that Y69 could be responsible for selectivity of SHIP2-SH2 since it is located at the pY+1 site of c-MET. R47 is a critical arginine within a highly conserved FLVR motif, which has been shown to be important for ligand binding with some other SH2 domains [4]. Meanwhile, the expression problems encountered for the R47A mutant suggests that R47 may be also important for protein stability of SHIP2-SH2. A mutation of R47, R47G, also resulted in a disturbance of SHIP2 cellular function, which failed to localize to focal contacts and was largely absent from the membrane periphery in spreading cells [22].
4. Conclusions
In this study, the solution structure of the SHIP2-SH2 domain was determined using solution NMR spectroscopy. The structure is comprised of two α-helices and six β-strands and adopts a canonical SH2 fold. A positive-charged pY-pocket is formed on the surface of SHIP2-SH2. NMR titrations were used to demonstrate that the pY-pocket and surrounding regions make up the binding interface of SHIP2-SH2 for Y1356-phosphorylated c-MET. NMR titration data and FP assays were used to determine that the binding affinity (KD) was at the low micromolar level. Three residues R28, S49 and R70 in the pY-pocket play key roles in the binding and may directly interact with the pY residue. Taken together, this study reveals the structural basis of SHIP2-SH2 for binding to Y1356-phosphorylated c-MET, and is helpful for a comprehensive understanding of the ligand binding of SHIP2-SH2. This work lays the foundation for the possible design of inhibitors to compete for binding to SHIP2-SH2 as well as its homologs.
Supplementary Material
Highlights:
SHIP2-SH2 consists of two α-helices and six β-strands adopting a typical SH2 fold
SHIP2-SH2 surface contains a positive-charged pocket for binding phosphotyrosine
The pocket and its surrounding regions are for binding Y1356-phosphorylated c-MET
The SHIP2-SH2 affinity for Y1356-phosphorylated c-MET is at low micromolar level
R28, S49 and R70 of SHIP2-SH2 are crucial for binding Y1356-phosphorylated c-MET
Acknowledgments
This work was supported by funds from National Key R&D Program of China (Grant number 2016YFA051201), the National Natural Sciences Foundation of China (Grant Number: 21575155), the Hundred Talent Program by Chinese Academy of Sciences, the Open-end Funds of Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening (Grant Number: HY201701), and the National Institute of General Medical Sciences of USA (Grant Numbers: U54-GM094597). We thank the scientists in the Rutgers University Protein Production Facility for construct design and sample preparation of human SHIP2-SH2 for NMR structure determination.
Footnotes
Compliance with ethical standards
Conflicts of interest
The authors do not have potential conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD, The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes, Sci Signal, 4 (2011) ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Waksman G, Kumaran S, Lubman O, SH2 domains: role, structure and implications for molecular medicine, Expert Rev Mol Med, 6 (2004) 1–18. [DOI] [PubMed] [Google Scholar]
- [3].Liu BA, Engelmann BW, Nash PD, The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction, FEBS Lett, 586 (2012) 2597–2605. [DOI] [PubMed] [Google Scholar]
- [4].Bradshaw JM, Waksman G, Molecular recognition by SH2 domains, Adv Protein Chem, 61 (2002) 161–210. [DOI] [PubMed] [Google Scholar]
- [5].Pawson T, Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems, Cell, 116 (2004) 191–203. [DOI] [PubMed] [Google Scholar]
- [6].Giuriato S, Blero D, Robaye B, Bruyns C, Payrastre B, Erneux C, SHIP2 overexpression strongly reduces the proliferation rate of K562 erythroleukemia cell line, Biochem Biophys Res Commun, 296 (2002) 106–110. [DOI] [PubMed] [Google Scholar]
- [7].Wang Y, Keogh RJ, Hunter MG, Mitchell CA, Frey RS, Javaid K, Malik AB, Schurmans S, Tridandapani S, Marsh CB, SHIP2 is recruited to the cell membrane upon macrophage colony-stimulating factor (M-CSF) stimulation and regulates M-CSF-induced signaling, J Immunol, 173 (2004) 6820–6830. [DOI] [PubMed] [Google Scholar]
- [8].Sleeman MW, Wortley KE, Lai KM, Gowen LC, Kintner J, Kline WO, Garcia K, Stitt TN, Yancopoulos GD, Wiegand SJ, Glass DJ, Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity, Nat Med, 11 (2005) 199–205. [DOI] [PubMed] [Google Scholar]
- [9].Kato K, Yazawa T, Taki K, Mori K, Wang S, Nishioka T, Hamaguchi T, Itoh T, Takenawa T, Kataoka C, Matsuura Y, Amano M, Murohara T, Kaibuchi K, The inositol 5-phosphatase SHIP2 is an effector of RhoA and is involved in cell polarity and migration, Mol Biol Cell, 23 (2012) 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Clement S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, Sasaki T, Penninger J, Doherty M, Malaisse W, Dumont JE, Le Marchand-Brustel Y, Erneux C, Hue L, Schurmans S, The lipid phosphatase SHIP2 controls insulin sensitivity, Nature, 409 (2001) 92–97. [DOI] [PubMed] [Google Scholar]
- [11].Paternotte N, Zhang J, Vandenbroere I, Backers K, Blero D, Kioka N, Vanderwinden JM, Pirson I, Erneux C, SHIP2 interaction with the cytoskeletal protein Vinexin, FEBS J, 272 (2005) 6052–6066. [DOI] [PubMed] [Google Scholar]
- [12].Marion E, Kaisaki PJ, Pouillon V, Gueydan C, Levy JC, Bodson A, Krzentowski G, Daubresse JC, Mockel J, Behrends J, Servais G, Szpirer C, Kruys V, Gauguier D, Schurmans S, The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man, Diabetes, 51 (2002) 2012–2017. [DOI] [PubMed] [Google Scholar]
- [13].Li B, Krakow D, Nickerson DA, Bamshad MJ, Chang Y, Lachman RS, Yilmaz A, Kayserili H, Cohn DH, Opsismodysplasia resulting from an insertion mutation in the SH2 domain, which destabilizes INPPL1, Am J Med Genet A, 164A (2014) 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kam TI, Park H, Gwon Y, Song S, Kim SH, Moon SW, Jo DG, Jung YK, FcgammaRIIb-SHIP2 axis links Abeta to tau pathology by disrupting phosphoinositide metabolism in Alzheimer’s disease model, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rajadurai CV, Havrylov S, Coelho PP, Ratcliffe CD, Zaoui K, Huang BH, Monast A, Chughtai N, Sangwan V, Gertler FB, Siegel PM, Park M, 5’-Inositol phosphatase SHIP2 recruits Mena to stabilize invadopodia for cancer cell invasion, J Cell Biol, 214 (2016) 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ye Y, Ge YM, Xiao MM, Guo LM, Li Q, Hao JQ, Da J, Hu WL, Zhang XD, Xu J, Zhang LJ, Suppression of SHIP2 contributes to tumorigenesis and proliferation of gastric cancer cells via activation of Akt, J Gastroenterol, 51 (2016) 230–240. [DOI] [PubMed] [Google Scholar]
- [17].Thomas MP, Erneux C, Potter BV, SHIP2: Structure, Function and Inhibition, Chembiochem, 18 (2017) 233–247. [DOI] [PubMed] [Google Scholar]
- [18].Batty IH, van der Kaay J, Gray A, Telfer JF, Dixon MJ, Downes CP, The control of phosphatidylinositol 3,4-bisphosphate concentrations by activation of the Src homology 2 domain containing inositol polyphosphate 5-phosphatase 2, SHIP2, Biochem J, 407 (2007) 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blero D, De Smedt F, Pesesse X, Paternotte N, Moreau C, Payrastre B, Erneux C, The SH2 domain containing inositol 5-phosphatase SHIP2 controls phosphatidylinositol 3,4,5-trisphosphate levels in CHO-IR cells stimulated by insulin, Biochem Biophys Res Commun, 282 (2001) 839–843. [DOI] [PubMed] [Google Scholar]
- [20].Pesesse X, Moreau C, Drayer AL, Woscholski R, Parker P, Erneux C, The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity, FEBS Lett, 437 (1998) 301–303. [DOI] [PubMed] [Google Scholar]
- [21].Bruhns P, Vely F, Malbec O, Fridman WH, Vivier E, Daeron M, Molecular basis of the recruitment of the SH2 domain-containing inositol 5-phosphatases SHIP1 and SHIP2 by fcgamma RIIB, J Biol Chem, 275 (2000) 37357–37364. [DOI] [PubMed] [Google Scholar]
- [22].Prasad N, Topping RS, Decker SJ, SH2-containing inositol 5’-phosphatase SHIP2 associates with the p130(Cas) adapter protein and regulates cellular adhesion and spreading, Mol Cell Biol, 21 (2001) 1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Koch A, Mancini A, El Bounkari O, Tamura T, The SH2-domain-containing inositol 5-phosphatase (SHIP)-2 binds to c-Met directly via tyrosine residue 1356 and involves hepatocyte growth factor (HGF)-induced lamellipodium formation, cell scattering and cell spreading, Oncogene, 24 (2005) 3436–3447. [DOI] [PubMed] [Google Scholar]
- [24].McNulty S, Powell K, Erneux C, Kalman D, The host phosphoinositide 5-phosphatase SHIP2 regulates dissemination of vaccinia virus, J Virol, 85 (2011) 7402–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bradley CA, Salto-Tellez M, Laurent-Puig P, Bardelli A, Rolfo C, Tabernero J, Khawaja HA, Lawler M, Johnston PG, Van Schaeybroeck S, Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges, Nat Rev Clin Oncol, 14 (2017) 562–576. [DOI] [PubMed] [Google Scholar]
- [26].Hass R, Jennek S, Yang Y, Friedrich K, c-Met expression and activity in urogenital cancers - novel aspects of signal transduction and medical implications, Cell Commun Signal, 15 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Le Coq J, Camacho-Artacho M, Velazquez JV, Santiveri CM, Gallego LH, Campos-Olivas R, Dolker N, Lietha D, Structural basis for interdomain communication in SHIP2 providing high phosphatase activity, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tresaugues L, Silvander C, Flodin S, Welin M, Nyman T, Graslund S, Hammarstrom M, Berglund H, Nordlund P, Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases, Structure, 22 (2014) 744–755. [DOI] [PubMed] [Google Scholar]
- [29].Leone M, Cellitti J, Pellecchia M, NMR studies of a heterotypic Sam-Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor, Biochemistry, 47 (2008) 12721–12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mills SJ, Persson C, Cozier G, Thomas MP, Tresaugues L, Erneux C, Riley AM, Nordlund P, Potter BV, A synthetic polyphosphoinositide headgroup surrogate in complex with SHIP2 provides a rationale for drug discovery, ACS Chem Biol, 7 (2012) 822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Saqib U, Siddiqi MI, Insights into structure and function of SHIP2-SH2: homology modeling, docking, and molecular dynamics study, J Chem Biol, 4 (2011) 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bahrami A, Assadi AH, Markley JL, Eghbalnia HR, Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy, PLoS Comput Biol, 5 (2009) e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Neri D, Szyperski T, Otting G, Senn H, Wuthrich K, Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling, Biochemistry, 28 (1989) 7510–7516. [DOI] [PubMed] [Google Scholar]
- [34].Shen Y, Delaglio F, Cornilescu G, Bax A, TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts, J Biomol NMR, 44 (2009) 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang YJ, Powers R, Montelione GT, Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics, J Am Chem Soc, 127 (2005) 1665–1674. [DOI] [PubMed] [Google Scholar]
- [36].Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M, Refinement of protein structures in explicit solvent, Proteins, 50 (2003) 496–506. [DOI] [PubMed] [Google Scholar]
- [37].Bhattacharya A, Tejero R, Montelione GT, Evaluating protein structures determined by structural genomics consortia, Proteins, 66 (2007) 778–795. [DOI] [PubMed] [Google Scholar]
- [38].Williamson MP, Using chemical shift perturbation to characterise ligand binding, Prog Nucl Magn Reson Spectrosc, 73 (2013) 1–16. [DOI] [PubMed] [Google Scholar]
- [39].Ooms LM, Horan KA, Rahman P, Seaton G, Gurung R, Kethesparan DS, Mitchell CA, The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease, Biochem J, 419 (2009) 29–49. [DOI] [PubMed] [Google Scholar]
- [40].Zhang Y, Wavreille AS, Kunys AR, Pei D, The SH2 domains of inositol polyphosphate 5-phosphatases SHIP1 and SHIP2 have similar ligand specificity but different binding kinetics, Biochemistry, 48 (2009) 11075–11083. [DOI] [PubMed] [Google Scholar]
- [41].Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N, ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules, Nucleic Acids Res, 44 (2016) W344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stefan M, Koch A, Mancini A, Mohr A, Weidner KM, Niemann H, Tamura T, Src homology 2-containing inositol 5-phosphatase 1 binds to the multifunctional docking site of c-Met and potentiates hepatocyte growth factor-induced branching tubulogenesis, J Biol Chem, 276 (2001) 3017–3023. [DOI] [PubMed] [Google Scholar]
- [43].Schiering N, Casale E, Caccia P, Giordano P, Battistini C, Dimer formation through domain swapping in the crystal structure of the Grb2-SH2-Ac-pYVNV complex, Biochemistry, 39 (2000) 13376–13382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.