Abstract
Protein-Observed Fluorine NMR Spectroscopy (PrOF NMR) is an emerging technique for screening and characterizing small molecule-protein interactions. The choice of which amino acid to label for PrOF NMR can be critical for analysis. Here we report the first use of a protein containing two different fluoroaromatic amino acids for NMR studies. Using the KIX domain of the CBP/p300 as a model system, we examine ligand binding of several small molecules elaborated from our previous fragment screen and identify a new ligand binding site distinct from those used by native transcription factors. This site was further supported by computational modeling (FTMap and Schrödinger) and 1H-15N HSQC/HMQC NMR spectroscopy. Metabolic labelling with multiple fluorinated amino acids provides useful probes for further studying ligand binding and has led to new insight for allosterically regulating transcription-factor protein interactions with small molecules.
Keywords: biomolecular NMR, fluorine, protein-protein interactions, fragments, allostery
Introduction
Small-molecule inhibition of protein-protein interactions (PPIs), remains a challenging area of research.[1] Due to their large surface areas, multiple low-affinity binding partners in the case of transcription factors, and allosteric networks, PPIs are notoriously difficult to target with small molecules.[2] Transcription factor-PPIs often have multiple native binding partners which in some cases, bind to different sites on a protein surface.[3] These sites may be allosterically coupled. Multiple binding sites can make it difficult to identify exactly where a small molecule is interacting with a protein. For example, the KIX domain of the CREB Binding Protein (CBP/p300) interacts with over a dozen different transcription factors through two allosterically coupled sites.[4]
In a recent characterization of KIX, Pomerantz et al. found that 3-fluorotyrosine incorporation at all five tyrosine sites for Protein-Observed Fluorine NMR spectroscopy (PrOF NMR) was sufficient for distinguishing the binding footprints of three native ligands from the transcriptional activation domains of c-Myb, MLL, and CREB.[5] These proteins interact with the KIX domain through two distinct binding sites. Allosteric effects on the protein were further shown to be different between c-Myb and CREB despite their similar binding surfaces on KIX.[4c] Screening of 508 small molecules against this fluorinated protein by PrOF NMR revealed several molecules which perturbed the resonance for Y631. Although this result is consistent with binding near the second transcription factor site engaged by MLL,[6] due to only a single amino acid being labeled in the binding site, it was not definitely confirmed if the binding site was orthosteric with MLL or outside but in close proximity.
Although enriched at protein-protein interfaces, aromatic amino acids occur at low frequency in proteins.[7] Thus this sparse labeling strategy may not be sufficient to elucidate the protein binding site of small molecule ligands such as the case above. The presence of a single tyrosine, Y631, within the KIX-MLL binding site limits the amount of structural information that can be inferred and increases the likelihood of false negatives in screening. However, there is a phenylalanine (F612) on KIX present in the MLL binding site near Y631, which could also serve as an additional probe for PrOF NMR if simultaneously fluorine labeled (Figure 1). Here we report on such an approach using KIX as a model system for dual lableing.
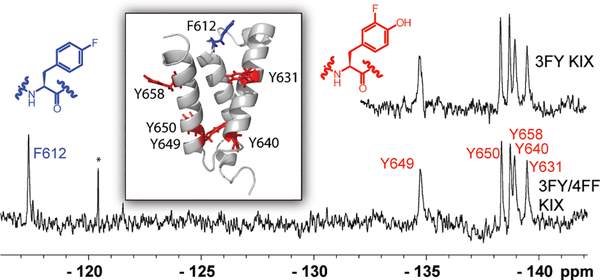
Figure 1:
PrOF NMR comparison of 3FY KIX and 3FY/4FF KIX. Top Right: PrOF NMR spectrum of 3FY KIX. Bottom: PrOF NMR spectrum of 3FY/4FF KIX. Inset: Solution structure of KIX with tyrosines in red and phenylalanine in blue. (PDB ID: 1KDX) * denotes an impurity that others have identified to be fluoride.[12]
Fluorinated aromatic amino acid labeling is challenging due to bacteria growth inhibition from monofluorinated aromatic amino acids under protein expression conditions.[8] Alternatively, para-CF3-phenylalanine incorporated into proteins by the Mehl and Pielak labs using amber suppression is much less toxic to bacteria, but only a single site is labeled.[9] Additional studies with fluorine-labeled proteins have been reviewed by Arntson et al.[10] Budisa and co-workers were the first to show the ability to incorporate multiple non-natural amino acids into proteins via sequence selective methods using auxotrophic bacteria for 4-fluoroproline, 6-fluorotryptophan, and 4-fluorophenylalanine.[8b] In this study, fluorine labeling was high but still resulted in a heterogeneously labeled sample, which may have a significant effect on the resulting NMR spectrum. As such, a 19F NMR study on a multi-labeled protein has yet to be reported. To address this need, we explored a dual-labeling strategy using both 3-fluorotyrosine (3FY) and 4-fluorophenylalanine (4FF) as a test case for characterizing native peptides sequences and newly discovered small molecules for the KIX domain for future chemical probe development. We report on an improved small molecule and data supporting a new binding site distinct from native transcription factor binding sites for regulating KIX PPIs. Results from this study provide insights into new approaches for using fluorine-labeled proteins to characterize protein-ligand interactions by PrOF NMR.
Results and Discussion
Dual Labeling of KIX
To obtain a protein labeled with two different fluorinated amino acids, 3FY and 4FF, we selected the DL39(DE3) cell line which is auxotrophic for both tyrosine and phenylalanine.[11] We have obtained yields as high as 70 mg/L of singly-labeled KIX with 3FY with this cell line, and thus chose it for this study.[5-6] Previous expressions for 4FF-labeled KIX resulted in highly variable yields, possibly due to varying conditions, including expression and recovery time (the time after the media change for bacteria to recover and residual unlabeled amino acid to be depleted). Optimization experiments identified ideal conditions for expression of 4FF KIX: media containing 10 μM phenylalanine, 160 μM 4FF, and a 30 min recovery time after the defined media change, resulting in 80–88% fluorine labeling (Table S1).
The first successful expression of a dual-labeled 3FY/4FF KIX was achieved using the conditions established for 4FF KIX above, with the addition of 400 μM 3FY and removal of tyrosine from the defined media. Fluorine incorporation (determined via ESI-TOF, see Supporting Information) of 70–85% was achieved with yields of 3FY/4FF KIX ranging from 14–22 mg/L, with fluorine incorporation ranging from 70–85%. By leaving phenylalanine out of expressions and a 30–40 min recovery time, fluorine incorporation of 95% can reliably be obtained but with reduced protein yield to 3–7 mg/L. High labeling is important to avoid a heterogenous sample; therefore, all subseqent studies have used this highly-labeled protein (Figure 1, S1).
Although this strategy results in lower yields than the singly-labeled constructs, dual-labeled proteins provide some advantages over the single-labeled constructs, such as our previously reported 4FF-KIX.[5] Two advantages of dual-labeling are the incorporation of additional probes to more fully characterize a binding site to validate ligand binding and to serve a control for protein folding. While KIX has five tyrosines, it only has one tryptophan and one phenylalanine. With a single fluorinated amino acid in a binding site, it can be challenging to determine if a chemical shift perturbation is due to a direct ligand-binding event, an allostreic effect, or to some other change (e.g. protein degradation or non-specific binding). The presence of multiple fluorine resonances can provide improved controls to mitigate these challenges.
Secondary Structure and Function Validation
Once protein expression conditions were established, we first sought to test the possibility of any significant structural or functional perturbation from fluorine incorporation. In prior reports, fluorescence polarization (FP) binding studies and circular dichroism (CD) investigations of thermal stability and secondary structure revealed only a modest perturbation of structure and binding from 3FY incorporation.[5] Far-UV CD spectra showed all three protein constructs to be α-helical based on molar ellipticity at 222 nm, with the dual-labeled protein with 11% lower molar ellipticity (Figure S2 A, D). Similar to 3FY-labeling, the dual-labeled protein did show a remarkable thermal stabilization of 7 degrees relative to the unlabeled protein, although a shallower rate of unfolding was observed consistent with less cooperative folding. The lower molar ellipticity, but higher thermal stability, may result from an altered extinction coefficient of one of the fluorinated tyrosine residues used to determine the protein concentration. However, such a difference in protein concentration could not be used to explain the enhanced stability, for which the origin of this effect is still unclear. Wang et al. demonstrated that a covalent ligand was able to stabilize KIX by 15–18 degrees leading to the first and only x-ray structure of this protein.[13] We speculate that fluorine labeling may serve the same purpose.
Because we labeled F612 which is in the MLL binding site, we also compared binding of the unlabeled, 3FY-labeled, and 3FY/4FF dual-labeled-KIX protein to MLL by FP. We saw approximately a 2-fold difference in binding affinity across samples (Figure S2 C, D, Kd = 3.8 ± 0.1 μM, 1.7 ±0.1 μM, and 1.8 ± 0.1 μM, respectively). Together these results, consistent with prior studies, confirm only a modest perturbation from fluorine labeling to global secondary structure and ligand binding, as well as a remarkable increase in stability.
The fluorine nucleus is highly sensitive to small changes in chemical environment and thus is also a sensitive reporter of subtle conformational effects.[14] In the first PrOF NMR experiment on this new construct, we compared the chemical shift of the resonances of 3FY-labeled KIX protein to the 95% 3FY/4FF-labeled protein (Figure 1). In both cases, the 3FY resonances were very similar (0.04 ppm average difference) between the two spectra indicating a minimal change in chemical environment in the folded protein, despite the slightly lower molar ellipticity from CD. To date, this data is the first example of a dual-labeled protein analyzed by 19F NMR. The single 4FF resonance found at −117.3 ppm is also in a similar region as previously reported in the singly-labeled protein.[5] The large chemical shift dispersion between the two sets of resonances (~20 ppm between 3FY and 4FF) indicates distinct windows for fluorinated aromatic amino acid resonances. Furthermore, 5-fluorotryptophan resonances typically originate near −125 ppm,[15] allowing sufficient chemical shift space for future triple-labeling experiments.
Binding Interaction Characterization
With both sets of fluorinated resonances clearly resolved, we sought to characterize the binding interactions of native and synthetic ligands. The solution NMR structure of MLL binding to KIX (PDB ID: 2AGH) indicates close interactions with the MLL transcriptional activation domain and KIX at both F612 and Y631. PrOF NMR experiments with the dual labeled KIX are able to capture these interactions, where both resonances yield large perturbations (Δδ = 1.1–1.2 ppm) upon increasing MLL concentration in the slow exchange regime (Figure 2, S3).
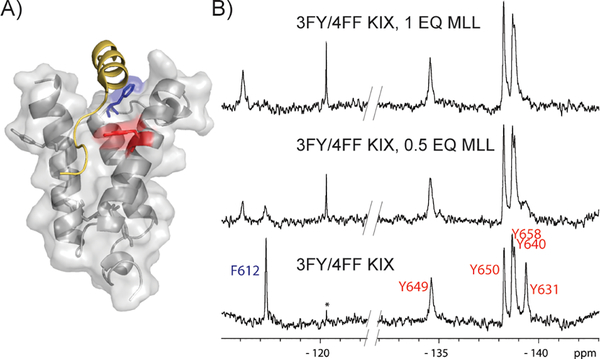
Figure 2:
Evaluation of MLL binding to 3FY/4FF KIX A) Solution structure (PDB ID: 2AGH) showing the native MLL peptide (gold) near Y631 (red) and F612 (blue). B) PrOF NMR results from MLL binding demonstrating slow chemical exchange binding kinetics. The concentration of 3FY/4FF KIX in the samples was 51 μM. Slashes in the baseline denote edited spectra to focus on the fluorine resonances. Full spectra are shown in Figure S3. * denotes an impurity that others have identified to be fluoride.[12]
A second transcription factor, CREB, binds a second site on the KIX surface but is known to enhance MLL binding through an allosteric network.[16] We previously showed that the 3FY resonances (Y658, Y650, and Y649) which mapped to the binding site were affected as well as Y631 in the MLL binding site. Independent experiments with 4FF-labeled KIX showed no effects on the F612 resonance.[5] Because the singly-labeled KIX and dual-labeled KIX are different and could be presenting slightly different binding surfaces, we characterized this binding-induced allosteric effect again with dual-labeled KIX (Figure S4). In this experiment, Y631 was similarly perturbed and F612 was unaffected, supporting an allosteric network that affects Y631, but does not involve F612 within the binding site.
Few small molecules have been reported to bind to the KIX domain. Naphthol AS-E phosphate is one of the most well characterized PPI inhibitors of KIX-transcription factor interactions between both c-Myb and CREB.[17] Our studies and those by Best et al. using 1H-15N HSQC, argue against binding at the CREB or c-MYB site, despite the similarity of the phosphate group in the inhibitor and phosphoserine 133 of CREB which binds to KIX, in contrast to the binding-site hypothesis proposed by Uttarkar et al.[17b] Similar to the experiments with the native peptides, we sought to investigate whether dual-labeled KIX had an altered structure or function which could mask the effects of small-molecule interactions in or near the MLL binding site. However, through a titration experiment of the daul-labeled protein with Naphthol-AS-E phosphate we confirmed that Y631, as previously shown, is the sole residue that exhibits a large dose dependent effect yielding a Kd of 160 ± 30 μM, and supports the existence of an alternate small molecule binding site distinct from those used by the native transcription factors (Figures 3, S5).
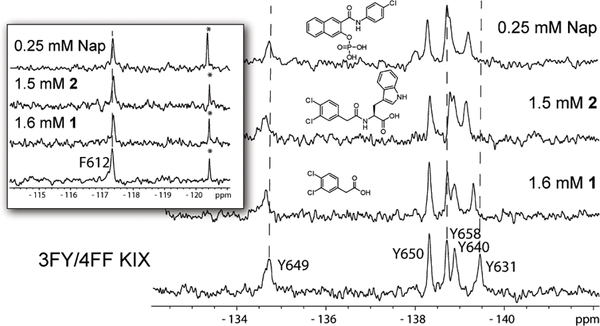
Figure 3:
Small molecule binding to 3FY/4FF KIX. Y631 shows significant perturbations while F612 (inset) shows no significant perturbations to chemical shift upon small molecule binding. The concentration 3FY/4FF KIX in the samples was 35 μM. * denotes an impurity that others have identified to be fluoride.[12]
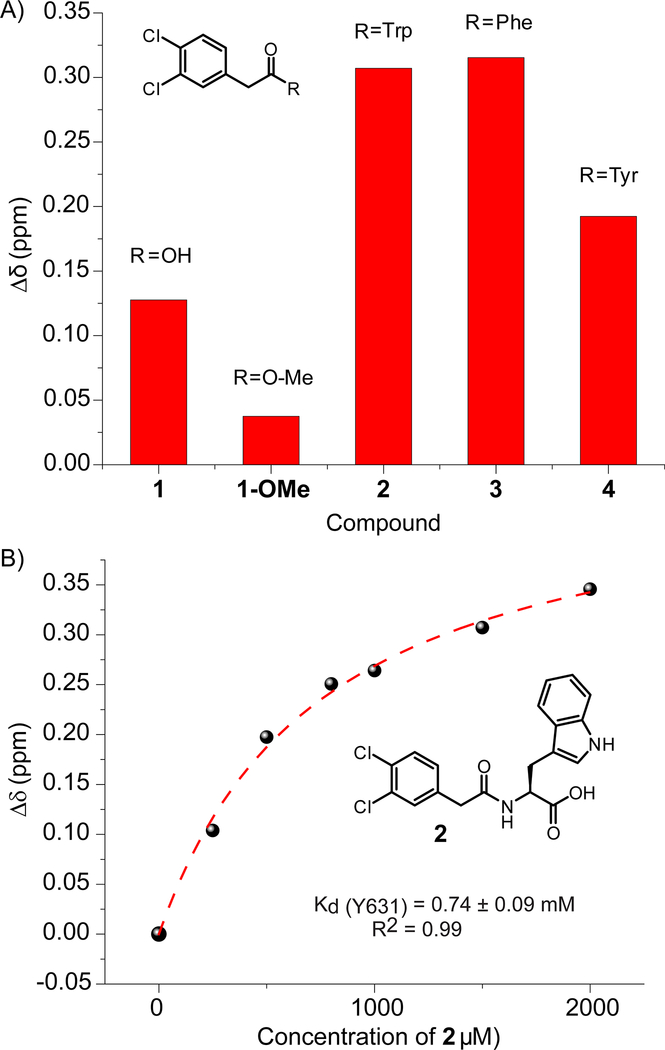
The above results led us to test small molecule 1, discovered in our previously reported fragment screen, which perturbed Y631 and at higher concentrations exhibited a non 1:1 interaction.[6] We speculated that this interaction was caused by two molecules fitting within the MLL site. However, addition of 1 at increasing concentration also did not affect F612. (Figure 3) Either 1, due to its small size, was too far away from F612, or it bound near but outside the MLL binding site similar to Naphthol AS-E-phosphate. To this end, we designed several more extended molecules through acylation with amino acids tryptophan, phenylalanine, and tyrosine (molecules 2, 3, 4, Figure 4A). Structure-activity relationships studies showed the carboxylic acid of 1 to be important for binding, as noted by the methyl ester of 1 being unable to affect a large change in chemical shift in the PrOF NMR spectrum (Figure 4A, S6). Therefore, 2, 3, and 4 were synthesized to maintain the carboxylic acid. Titration of 2, against 3FY KIX indicated binding near the MLL site, and a saturating binding isotherm, consistent with 1:1 binding was observed, yielding a Kd of 740 ± 90 μM (Figure 4B, S7). Compounds 3 and 4 did not improve the affinity with 3FY KIX relative to the parent compound, 1 (Figure S8, S9). Testing 2 with the 3FY/4FF labeled protein, again perturbed Y631 near the MLL binding site but did not affect F612 (Figure 3, S10). Comparing the NMR spectra from 2 binding to both 3FY KIX and 3FY/4FF KIX indicated similar binding modes though a slightly weaker affinity (Figure S10, S11 A,B). Together these data support 1, 2, and Naphthol AS-E phosphate interacting at a similar binding site, distinct from native transcription factors. We further note, that not all small molecules engage this site. As we previously showed a small molecule, 1–10, could also perturb both fluorinated resonance for Y631 and F612 of single labeled constructs.[5] A subsequent crystal structure demonstrated this molecule to indeed bind within the MLL site.[13]
Figure 4:
PrOF NMR data from synthesized analogs. A) Comparison of Y631 chemical shift perturbations in the presence of 1666 μM 1, 5 mM 1-OMe, and 1500 μM 2, 3, and 4. B) Y631 Binding isotherm for 2 demonstrating a slightly improved binding affinity for 3FY KIX.
Investigation of New Binding Site
This PrOF NMR data led us to reevaluate the data that initially pointed towards the conclusion that 1 and its related analogs bound to KIX in the MLL site. We turned to the FTMap server to investigate possible binding sites in KIX.[18] This computational solvent mapping server analyzes protein structures and identifies binding hotspots through the use of solvent molecules as probes. As expected, FTMap identified the MLL and pKID binding sites with clusters of its probe solvent molecules. However, additional clusters were found distal to these binding sites on an outer face of the protein between the α1 and α2 helices yet still near Y631, the residue most perturbed in the PrOF NMR experiments (Figure 5B). Further, these clusters were situated in close proximity to amino acid residues perturbed by Naphthol AS-E phosphate in previous 1H-15N HSQC experiments by Best et al.[17a] These clusters were also situated close to the residues perturbed by 1 in our prior 1H-15N HSQC validation studies.[6] Clusters were consistently found in similar locations within the protein across four KIX PDB structures (1KDX, 2AGH, 2LXT, 2LXS), lending more weight to this potential, previously unreported ligand binding site. Figure S12).
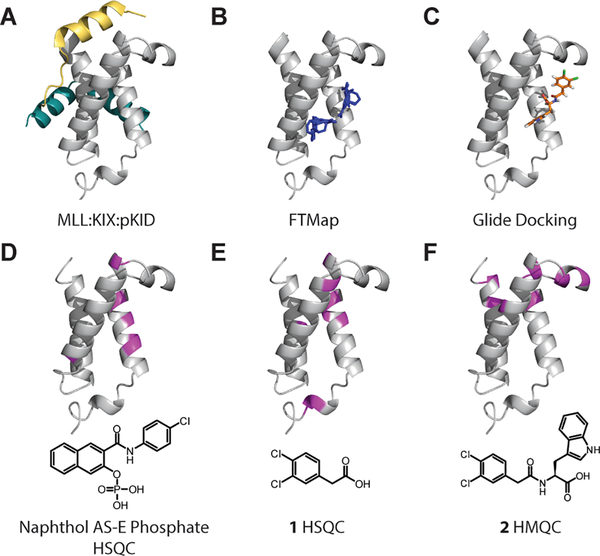
Figure 5:
Structural representations of KIX highlighting binding sites mapped onto 1KDX. A) Ternary complex of MLL (gold), KIX (gray), and pKID (teal). B) Select FTMap results displaying probe molecule clusters (blue) in a third site in the protein (PDBID 1KDX). C) Glide docking pose of 2 (orange) using the Schrödinger Maestro Suite. D, E, F) Chemical shift perturbations from 1H-15N HSQC or HMQC experiments. Perturbations greater than one standard deviation are shown in magenta.
As an additional followup, Glide docking studies in Schrödinger were performed with Naphthol AS-E Phosphate and 2 against the four previously listed KIX PDB structures. Grids large enough to encompass both the MLL binding site and this potentially new binding site were used to provide an unbiased comparative prediction. Using extra precision (XP) docking, two poses for each molecule were generated per PDB structure. Six out of eight poses for Naphthol AS-E phosphate were found to be in the new binding site while two were predicted to be in the MLL site. For 2, all eight poses were found in the new binding site consistent with the clusters predicted by FTMap (Fig 5C, S13).
Reevaluation of the HSQC NMR data from Best et al. (Naphthol AS-E Phosphate)[17a] and Gee et al. (1)[6] in light of the computational data further supported the possibility of this new ligand binding site. The most significant perturbations in the amide backbones for both ligands were found on the α1 helix, outside the MLL binding site. 1H-15N HMQC NMR data on 2 resulted in similar perturbations to both Naphthol AS-E phosphate and 1. (Figure 5D, E, F, 6, S14)
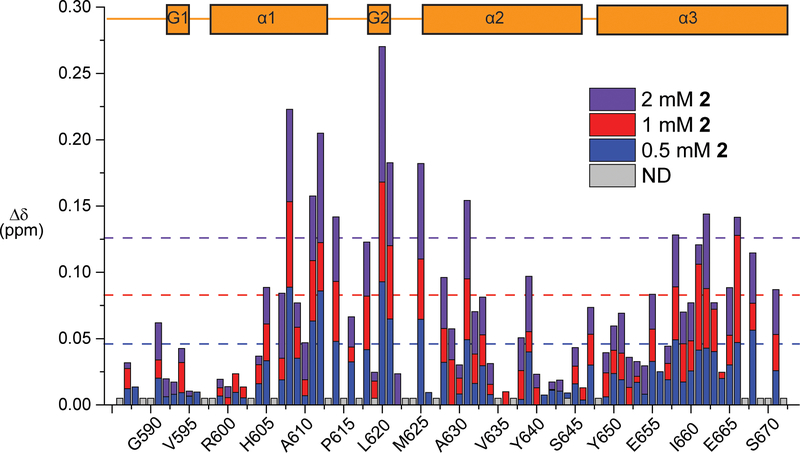
Figure 6:
B) Chemical shift perturbation mapping of KIX residues with 0.5 mM, 1 mM and 2 mM 2. Residues that were not detected or not assigned are shown in gray. Dashed lines represent the average chemical shift perturbation + one standard deviation at the corresponding concentration. Assignments previously obtained by Majmudar et al.[24] In the secondary structure box schematic, G1 and G2 refer to the two 310-helices, while α1, α2, and α3, refer to the three alpha helices comprising the structure
Notably, in contrast to 1, vectors for the chemical shift perturbations in the presence of increasing concentrations of 2 were consistently linear, indicative of 1:1 binding (Figure S15). Surprisingly, one of the most significantly perturbed amide backbones was that of F612. In comparing the HMQC and PrOF NMR results, the 4FF may be sensitive enough to detect stronger binders like the native MLL peptide but not sensitive enough to detect weaker binders that are farther away. Additionally HMQC NMR experiments report on changes in the chemical environment of the amide backbone while PrOF NMR reports on changes in the chemical environment of the fluorine on the side chain, which may be less sensitive to local secondary structure effects. It is also possible that the chemical environment of this solvent-exposed 4FF has not changed sufficiently, relative to the change in chemical environment near the backbone amide due to either direct or distant binding effects.
Finally, we performed additional FP experiments to further test the inhibitory effects from ligand binding in this new site. We performed FP competition experiments with the MLL:KIX complex and Napthol-ASE-phosphate due to its higher binding affinity than 2. Naphthol-AS-E Phosphate has already been shown to inhibit the KIX:CREB interaction with a Ki of 90 μM.[17a] Napthol-ASE-phosphate showed incomplete inhibition of MLL binding with an estimated IC50 of 200 ± 10 μM. (Figure S16 A) Additionally, we investigated the inverse phenomenon to determine the impact of Naphthol AS-E phosphate on the affinity of MLL for KIX. With the concentration of Naphthol AS-E phosphate and DMSO being held constant, MLL binding was attenuated by as much as 16 fold. (Figure S16 B) Together with our NMR experiments and computational predictions, these data support a new binding site that only partially overlaps with the MLL:KIX interface or that allosterically affects binding from adjacent to Y631 but outside the MLL binding site.
Conclusion
This dual-labeling strategy augments previously reported PrOF NMR methods for studying ligand-protein interactions[19], small molecule screening[20], as well as protein conformational changes[14, 21] and allostery[22]. Dual-labeling provides the ability to insert additional probes into the protein of interest and report on a new ligand binding mode. Using this novel dual-labeling strategy in conjunction with two different computational methods as well as the evaluation of both old and new HSQC NMR data, the data presented herein supports a new small-molecule binding site in KIX, distal to the two native transcription factor binding sites. Such a new binding site provides an opportunity to positively or negatively allosterically regulate KIX PPIs, such as with Naphthol AS-E phosphate, without competing with endogenous transcription factors. Identifying ligand binding modes in this site will be the subject of future studies., Although this method produces lower protein expression yields making it less ideal for full screens, dual-labeling a protein can serve as a useful tool and extension of PrOF NMR for further studying ligand binding and characterizing their binding sites. The BPTF bromodomain has recently been studied by PrOF NMR for discovering small molecules with anticancer activities.[23] This protein has only has one tryptophan in the bromodomain binding site, but multiple tyrosine residues. Such a protein would be an ideal candidate for future dual labeling experiments.
Experimental Section
Protein Expression and Purification:
Unlabeled and fluorinated proteins were expressed as previously described. See the Supporting Information for full experimental details, including the optimization of the dual labeling procedure.
Circular Dichroism:
CD spectra were acquired using a peltier equipped temperature controlled Jasco J-815 spectropolarimeter. Protein samples were prepared in a 10 mM sodium phosphate, 100 mM sodium chloride buffer at pH 7.2. Unlabeled KIX was prepared at 10.5 μM while 3FY and 3FY/4FF KIX were prepared at 11.5 μM. Molar ellipticity was monitored from 200–260 nm. Samples were collected at 25 °C using a 1 mm cuvette path-length. Reported data is the average of 5 acquired spectra. Thermal stability of unlabeled and fluorine-labeled KIX was measured by following the change in ellipticity at 222 nm from 20 °C to 95 °C. For the thermal melt experiments, the concentration of 3FY/4FF KIX was increased to 35 μM. The Tm was calculated using a sigmoidal fit. Data analysis was performed using Origin 2016. Molar ellipticity was calculated using the following equation:
| (1) |
In equation 1, Ψ is the CD signal in degrees, n is the number of amides in the protein construct, l is the path lenth in centimeters, and c is the concentration of the protein sample in decimoles per cubic centimeter.
Fluorescence Polarization:
Fluorescence polarization experiments were performed on a Tecan Infinte F500. For the validation experiments, a FITC-MLL peptide[5] was used to compare the binding activity of the three different protein constructs. UL and 3FY/4FF KIX were prepared at a starting concentration of 40 μM while 3FY KIX was prepared at a starting concentration of 35 μM. Samples also contained 85 nM tracer (FITC-MLL), 0.01% Triton X-100, with a total DMSO concentration of 0.5%. Excitation and emission wavelengths were 485 nm and 535 nm respectively. Samples were incubated for 30 min at room temperature prior to scanning. Each data point in Figure S2 C is the average of three independent experiments with the standard deviations listed as the error. Data analysis was performed with GraphPad Prism. Kd values were determined as follows
| (2) |
In the above equation, y corresponds to the observed anisotropy, c and b and the minimum and maximum observed entropies, respectively, Kd is the dissociation constant, and a and x are the concentrations of fluorescent peptide and KIX, respectively.
For the competition experiments, the FITC-MLL peptide and unlabeled KIX were held constant at 25 nM and 1.3 μM respectively while the concentraiton of Naphthol AS-E phosphate being diluted with a top concentration of500 μM and a final DMSO concentration of 0.5%. Anisotropy values were fit using GraphPad Prism’s log(inhibitor) vs response (four parameters function. Similar conditions were used for the additional direct binding experiments with the exception of Naphthol AS-E phosphate being held constant in each set of experiments (0, 50, 100, 250, and 500 μM) and protein being diluted from a top concentration of 30 μM.
Protein Observed Fluorine (PrOF) NMR:
All NMR spectra were obtained on a Bruker Avance III 500 MHz instrument equipped with a 5 mm Prodigy TCI Cryoprobe (19F S/N 2100:1). Samples containing 30–50 μM 3FY, 4FF, or 3FY/4FF KIX were prepared in NMR buffer as noted above with 5% D2O and 50 μM trifluoroacetic acid (−76.5 ppm) as a reference. T1 relaxation times were calculated using an inversion recovery experiment. Optimized parameters for PrOF NMR experiments included a 0.6 s delay time, a 0.1–0.3 s acquisition time (AQ), and 400–800 scans. The dead time or pre-scan delay was set at 50 μs. Spectral width and offset were 10 ppm and −136 ppm respectively for 3FY KIX. Spectral width and offset were 10 ppm and −115 ppm respectively for 4FF KIX. Both of the aforementioned parameter sets were used when analyzing the 3FY/4FF KIX in addition to an experiment with a spectral width of 26 ppm and offset of −128 ppm for a full spectrum containing the 3FY and 4FF resonances. A line-broadening of 15–20 Hz was applied to all protein spectra upon processing. Spectra were acquired at 294 K)
PrOF NMR Kd values are determined using equation 3 which accounts for receptor depletion.[19b]
| (3) |
In equation 3, S corresponds to the observed change in chemical shift, D is the maximum change in chemical shift, Kd is the dissociation constant of the ligand, L is the ligand concentration, and P is the protein concentration.
2D 1H-15N HSQC NMR Experiments:
Uniformly 15N labeled KIX protein was expressed and purified using M9 minimal media supplemented with [15N] ammonium chloride as described previously.[24] A 30–35 μM solution of 15N-labeled KIX was prepared in a 9:1 H2O:D2O 10 mM phosphate buffer with 100 mM NaCl and 2.5% DMSO at pH 7.2. HSQC experiments were recorded at 27 °C on a Bruker Ascend 850 MHz NMR spectrometer equipped with a 5 mM TCI Cryo probe. Spectra were collected with 0, 0.5, 1, and 2 mM 2. Data was processed using NMRPipe and analyzed using Sparky (UCSF).
Computational Methods:
FTMap Computational Solvent Mapping was provided by Boston University at ftmap.bu.edu. [18] Experiments were performed on the main KIX chain from each of the four following PDB structures : 1KDX, 2AGH, 2LXT, and 2LXS
Glide docking was performed using Schrodinger Software Suit provided through the Minnesota Supercomputing Institute (MSI). The four following KIX PDB structures were used for docking studies: 1KDX, 2AGH, 2LXT, and 2LXS. Each protein structure was prepared using the Protein Preparatization Wizard with the OPLS-2005 force field. The generated grid that was used was 25 Å x 25 Å x 25 Å, encompassing the MLL site and the space predicted by FTMap solvent clusters. Ligands were prepared using the Ligprep module. Docking studies were performed with extra precision (XP). Two poses for each molecule were generated per PDB structure resulting in eight poses for Naphthol AS-E Phosphate and eight poses for 2.
Small Molecule Characterization:
All NMR data were acquired on a Bruker Avance III 500 MHz instrument equipped with a 5 mm Prodigy TCI Cryoprobe (1H S/N 2500:1, 13C S/N 350:1). High resolution mass spectrometry data was acquired on a Bruker BioTOF II ESI/TOF-MS. In the case of 1-OMe, ESI-MS data was not acquirable. In this case, mass spectrometry data was collected on an Agilent GC-MS system (GC: 6890N, MSD: 5975) with a DB-5 column (30 m length, 0.32 mm ID). The sample was ionized at 70 eV. The GC method used the following conditions: 50 °C for 1.5 min, 20 °C/min ramp, then 250 °C for 3.5 min with a split ratio of 50:1.
Supplementary Material
Acknowledgements
We kindly thank Prof. Sandor Vajda for helpful discussions on the applications of FTMap.
This project was supported by the University of Minnesota, the NSF-CAREER Award CHE-1352091, the NIH training grant T32-GM08700, and the UMN Interdisciplinary Doctoral Fellowship (C.T.G.)
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Arkin MR, Tang Y, Wells JA, Chem. Biol. 2014, 21, 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cesa LC, Mapp AK, Gestwicki JE, Front. Bioeng. Biotechnol. 2015, 3, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee LW, Mapp AK, J. Biol. Chem. 2010, 285, 11033–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Goodman RH, Smolik S, Genes Dev 2000, 14, 1553–1577 [PubMed] [Google Scholar]; b) Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE, J. Biol. Chem 2002, 277, 43168–43174 [DOI] [PubMed] [Google Scholar]; c) Palazzesi F, Barducci A, Tollinger M, Parrinello M, Proc. Natl. Acad. Sci 2013, 110, 14237–14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pomerantz WC, Wang N, Lipinski AK, Wang R, Cierpicki T, Mapp AK, ACS Chem. Biol. 2012, 7, 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gee CT, Koleski EJ, Pomerantz WC, Angew. Chem. Int. Ed. Engl. 2015, 54, 3735–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bogan AA, Thorn KS, J. Mol. Biol. 1998, 280, 1–9. [DOI] [PubMed] [Google Scholar]
- [8].a) Richmond MH, Bacteriol. Rev 1962, 26, 398–420 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Merkel L, Schauer M, Antranikian G, Budisa N, Chembiochem 2010, 11, 1505–1507. [DOI] [PubMed] [Google Scholar]
- [9].Li C, Wang G-F, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RAS, Mehl RA, Pielak GJ, J. Am. Chem. Soc. 2010, 132, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Arntson KE, Pomerantz WCK, J. Med. Chem. 2016, 59, 5158–5171. [DOI] [PubMed] [Google Scholar]
- [11].Frieden C, Hoeltzli SD, Bann JG, Methods Enzymol. 2004, 380, 400–415. [DOI] [PubMed] [Google Scholar]
- [12].Leung EW, Yagi H, Harjani JR, Mulcair MD, Scanlon MJ, Baell JB, Norton RS, Chem. Biol. Drug Des. 2014, 84, 616–625. [DOI] [PubMed] [Google Scholar]
- [13].Wang N, Majmudar CY, Pomerantz WC, Gagnon JK, Sadowsky JD, Meagher JL, Johnson TK, Stuckey JA, Brooks CL 3rd, Wells JA, Mapp AK, J. Am. Chem. Soc. 2013, 135, 3363–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kitevski-LeBlanc JL, Prosser RS, Prog. Nucl. Magn. Reson. Spectrosc. 2012, 62, 1–33. [DOI] [PubMed] [Google Scholar]
- [15].Gee CT, Arntson KE, Urick AK, Mishra NK, Hawk LML, Wisniewski AJ, Pomerantz WCK, Nat. Protoc. 2016, 11, 1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bruschweiler S, Konrat R, Tollinger M, ACS Chem. Biol. 2013, 8, 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M, Proc. Natl. Acad. Sci. U.S.A 2004, 101, 17622–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Uttarkar S, Dukare S, Bopp B, Goblirsch M, Jose J, Klempnauer KH, Mol. Cancer Ther 2015, 14, 1276–1285. [DOI] [PubMed] [Google Scholar]
- [18].a) Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S, Bioinformatics 2009, 25, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S, Proc. Natl. Acad. Sci. USA 2011, 108, 13528–13533 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bohnuud T, Beglov D, Ngan CH, Zerbe B, Hall DR, Brenke R, Vajda S, Frank-Kamenetskii MD, Kozakov D, Nucleic Acids Res. 2012, 40, 7644–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S, Nat. Protoc. 2015, 10, 733–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Matei E, Andre S, Glinschert A, Infantino AS, Oscarson S, Gabius HJ, Gronenborn AM, Chemistry 2013, 19, 5364–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Curtis-Marof R, Doko D, Rowe ML, Richards KL, Williamson RA, Howard MJ, Org. Biomol. Chem. 2014, 12, 3808–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Norton RS, Leung EW, Chandrashekaran IR, MacRaild CA, Molecules 2016, 21, 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li H, Frieden C, Proc. Natl. Acad. Sci. U.S.A 2007, 104, 11993–11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K, Science 2012, 335, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Song L, Teng Q, Phillips RS, Brewer JM, Summers AO, J. Mol. Biol. 2007, 371, 79–92. [DOI] [PubMed] [Google Scholar]
- [23].Urick AK, Hawk LM, Cassel MK, Mishra NK, Liu S, Adhikari N, Zhang W, Dos Santos CO, Hall JL, Pomerantz WC, ACS Chem. Biol. 2015, 10, 2246–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Majmudar CY, Højfeldt JW, Arevang CJ, Pomerantz WC, Gagnon JK, Schultz PJ, Cesa LC, Doss CH, Rowe SP, Vásquez V, Tamayo-Castillo G, Cierpicki T, Brooks CL, Sherman DH, Mapp AK, Angew. Chem. Int. Ed. 2012, 51, 11258–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.