Abstract
Sphingolipids comprise a diverse family of lipids that perform multiple functions in both structure of cellular membranes and intra- and inter-cellular signaling. The diversity of this family is generated by an array of enzymes that produce individual classes and molecular species of family members and enzymes which catabolize those lipids for recycling pathways. However, all of these lipids begin their lives with a single step, the condensation of an amino acid, almost always serine, and a fatty acyl-CoA, almost always the 16-carbon, saturated fatty acid, palmitate. The enzyme complex that accomplishes this condensation is serine palmitoyltransferase (SPT), a membrane-bound component of the endoplasmic reticulum. This places SPT in the unique position of regulating the production of the entire sphingolipid pool. Understanding how SPT activity is regulated is currently a central focus in the field of sphingolipid biology. In this review we examine the regulation of SPT activity by a set of small, membrane-bound proteins of the endoplasmic reticulum, the Orms (in yeast) and ORMDLs (in vertebrates). We discuss what is known about how these proteins act as homeostatic regulators by monitoring cellular levels of sphingolipid, but also how the Orms/ORMDLs regulate SPT in response to other stimuli. Finally, we discuss the intriguing connection between one of the mammalian ORMDL isoforms, ORMDL3, and the pervasive pulmonary disease, asthma, in humans.
Keywords: Orm, ORMDL, serine palmitoyltransferase, sphingolipid biosynthesis, asthma
1. Introduction
As outlined in the other articles in this issue, sphingolipids are a diverse family of lipids with an enormous impact on cell and organismal biology. Clearly, the regulation of the production of these lipids is essential for the proper functioning of cells and tissues, and dysregulation of their synthesis can have catastrophic effects (Harrison, Dunn, and Campopiano 2018; Kolter and Sandhoff 2006; Marcus et al. 2006; Maceyka and Spiegel 2014; Obeid et al. 1993; Miller et al. 2014; Sasset et al. 2016; Volpert et al. 2017; Ogretmen 2018; Olsen and Faergeman 2017; Espaillat, Kew, and Obeid 2017; Shamseddine, Airola, and Hannun 2015).
The panoply of sphingolipids is constructed by myriad modifications of the basic building block of all lipids in this class, the sphingosine base. The base itself can take several forms, varying in the number of carbons, the degree of unsaturation, and the addition, or not, of hydroxyl groups, plus the phosphorylation of the C1 hydroxyl to form sphingoid base phosphates. However, all sphingoid bases contain the magic nitrogen at carbon 2. Modifications of the base generate the more complex sphingolipids through acylation of the C2 nitrogen with a wide variety of fatty acids (making ceramides) and the addition of groups to the C1 hydroxyl: phosphorylcholine for sphingomyelin, carbohydrate for glycosphingolipids, and phosphate for ceramide-1-phosphate. Once produced, there are enzymatic pathways that can reverse all of these modifications, allowing for a dynamic balance between sphingolipid family members. The sphingoid base therefore finds itself shuttled back and forth amongst the numerous sphingolipid intermediates.
A striking and unique feature of this family of lipids, is that both entry and exit from the biosynthetic pathway is tightly restricted (Bourquin, Capitani, and Grutter 2011). There is only one way in and one way out. That is to say, there is one enzyme that produces the sphingoid backbone, and thus introduces it into the metabolic network, and only one enzyme that can degrade the backbone and remove it from the network. The enzyme responsible for the way out is sphingosine-1-phosphate (S1P) lyase which, as the name denotes, utilizes only sphingosine-1-phosphate or dihydrosphingosine-1-phosphate as its substrate. S1P lyase produces an ethanolamine phosphate and a long chain aldehyde (saturated and unsaturated), both of which can be utilized for glycerolipid metabolism. The exit from the metabolic network of sphingolipids therefore depends on the sphingosine kinases to phosphorylate the sphingoid base. The biochemistry and biology of sphingosine-1-phosphate lyase and the sphingosine kinases have been addressed in several excellent recent reviews and will not be further addressed here (Aguilar and Saba 2012; Liu et al. 2012; Sanllehi et al. 2016; Hatoum et al. 2017; Pyne, Adams, and Pyne 2016).
Here we focus on the only metabolic route into the sphingolipid network and therefore the step that, in concert with S1P lyase, has the weighty task of determining the overall steady state levels of sphingolipids in the cell. The initiating step in sphingolipid biosynthesis is accomplished by serine palmitoyltransferase, (SPT). This enzyme condenses an amino acid, almost always serine, with a fatty acyl-CoA, almost always palmitate (16:0), to produce 3-ketodihydrosphingosine. With that step accomplished, sphingolipid biosynthesis is off to the races. SPT is the initiating and rate-limiting step in this pathway and like any well-behaved biochemical pathway, this rate limiting step is highly regulated. It is this regulation that we will address in this review.
Much of this review is devoted to the Orms/ORMDLs, small membrane-bound proteins that are central regulators of SPT activity. This laboratory (as well as others) have focused on the biochemical aspects of this regulation, both in yeast (Orms) and mammalian systems (ORMDLs), and that will be one aspect of this review. In addition, a number of laboratories have shown that ORMDL/SPT regulation of sphingolipid metabolism has considerable physiological and pathophysiological consequences in both higher and lower eukaryotes and these aspects will also be addressed.
2. Serine Palmitoyltransferase: gateway for de novo sphingolipid biosynthesis
The basic building block of all sphingolipids is the sphingoid base, also known as a long chain base (LCB), which was first discovered in brain tissue (Thudichum 1884). As mentioned above, formation of the precursor to all sphingoid bases, 3-keodihydrosphingosine (3-KDS), is catalyzed by the enzyme, serine palmitoyltransferase (SPT), which condenses L-serine with an Acyl-CoA substrate, and is the rate-limiting step in de novo sphingolipid biosynthesis (Figure 1) ((Merrill, Nixon, and Williams 1985) reviewed (Hanada 2003)). The production of 3-KDS by serine palmitoyltransferase is highly conserved across multiple species even though the downstream enzymes contributing to the overall sphingolipid profile for individual species has diverged. More complex sphingolipids have a fatty acid attached to the C2 amine via N-linked acylation as well as modification of the C1 hydroxyl of the sphingoid base, thus creating a vast array of sphingolipid species (Merrill 2011; Pruett et al. 2008). Complexity and diversity in sphingolipid composition is brought about by variations in the chain length of the fatty acids utilized to acylate the sphingoid base in subsequent enzymatic steps, as well as the variety of head groups that can be attached to the C1 hydroxyl, such as sugars, phosphate, phosphoinositol, and phosphocholine (Merrill 2011; Pruett et al. 2008). The multitude of combinations possible, combined with the rapid turnover of sphingolipid intermediates at each step along the way, results in a complexity of sphingolipid profiles that differs across species. This rapid conversion of intermediates, both biosynthetically and via degradative pathways, yields distinct pools of sphingolipids that are spatially and temporally controlled (reviewed in (Hanada 2003; Harrison, Dunn, and Campopiano 2018)).
Figure 1.
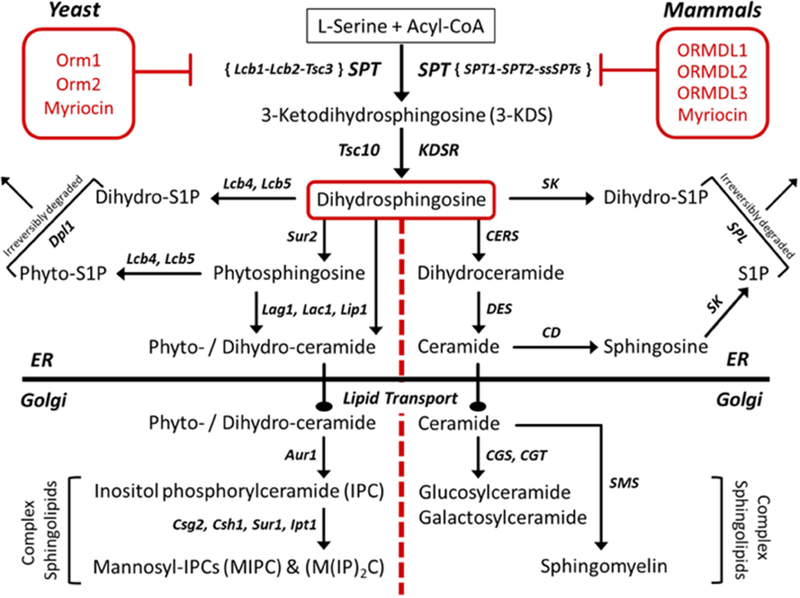
Overview of sphingolipid biosynthesis in yeast and mammals.
Serine palmitoyltransferase is an ER-resident, membrane-bound protein that requires multiple subunits in order to be catalytically active (Hanada 2003). In yeast, these subunits include long chain base 1 (Lcb1), long chain base 2 (Lcb2), and temperature sensitive csg2∆ suppressor 3 (Tsc3) proteins. Lcb1 and Lcb2 compose the minimal complex required for SPT activity in yeast, and while Tsc3 is considered to be nonessential, it has been shown to be needed for optimal SPT activity with tsc3∆ mutants having a 30-fold decrease in SPT activity (Gable et al. 2000). A mammalian orthologue of Tsc3 has not been identified to date. In mammals, there are multiple large subunits of SPT, SPTLC1, 2 and 3, with SPTLC1 pairing with either SPTLC2 or SPTLC3 to form an active, heterodimeric complex (Hanada 2003; Hornemann, Wei, and von 2007). In addition to these large subunits, recent reports have identified small activating subunit proteins in mammals, dubbed ssSPTa and ssSPTb (Han et al. 2009). These subunits have been shown to confer acyl-CoA specificity and increase basal SPT activity nearly 100-fold. Similar subunits have recently been identified and characterized to be SPT enhancers in Aribidopsis (Kimberlin et al. 2013) and were shown to be differentially regulated in various plant tissues. Additionally, these ssSPT proteins were found to interact directly with Orm proteins in Aribidopsis (Li et al. 2016). This exciting discovery will be discussed in more detail below under ORMDL regulation as it suggests a novel way in which SPT activity in mammals might be regulated in an ORMDL-dependent manner.
As discussed above, sphingolipids are essential components of cellular membranes as well as important signaling molecules with diverse and sometimes opposing roles in determining cell fate (reviewed in (Hannun and Obeid 2002; Harrison, Dunn, and Campopiano 2018)). Flux through this pathway is tightly controlled to maintain the proper intracellular balance of each sphingolipid intermediate. As SPT catalyzes the first committed step for all sphingoid bases, regulation of this enzyme has been studied extensively. It has long been known that serine palmitoyltransferase activity can be modulated when exogenous sphingolipids are added to cells (Mandon et al. 1991). Surprisingly, the complete picture of molecular mechanisms governing the regulation of SPT is still not known, especially in higher organisms. Recent advances in the field have led to multiple novel candidates, each of which has the potential to connect de novo sphingolipid biosynthesis to a wide array of cellular processes, such as membrane biogenesis, nutrient uptake, cell cycle control, and differentiation. The following sections detail what is known to date about one family of proteins recently identified to negatively control SPT activity, the Orm/ORMDL proteins.
3. Orm/ORMDL proteins: discovery and homology
The full-length cDNA for human ORMDL1 (ORM1-like) was discovered during a retinal library screen of the RP26 locus on chromosome 2q31, which was a region of the human genome lacking gene assignments at that time (Hjelmqvist et al. 2002). The authors used a 647bp probe which was designed based on an expressed sequence tag (EST) in the retina and homologous to multiple open reading frames (ORFs) across several species. The yeast sequence corresponding to a similar ORF had already been identified and named ORM1 (Borsting et al. 1997), hence the human gene discovered by Hjelmquist et al. was named following the HUGO Gene Nomenclature guidelines as ORM1-like or ORMDL. Using the cDNA for ORMDL1, Hjelmquist et al. searched the genome for human homologues leading to the discovery of ORMDL2 (located on chromosome 12q13) and ORMDL3 (located on chromosome 17q21). This gene family is highly conserved across a multitude of genomes with orthologues being reported in yeast, plants, microsporidia, urochordates, invertebrates and vertebrates (Hjelmqvist et al. 2002; Paulenda and Draber 2016).
Protein sequence alignment of Orm/ORMDL orthologues across multiple species reveals the conserved nature of this gene family and implies a highly conserved function for this family of proteins (Hjelmqvist et al. 2002; Paulenda and Draber 2016). At the amino acid level, there are multiple residues with a strong degree of conservation amongst all species. These ultra-conserved residues align in a way that forms the basis for putative conserved motifs, which likely play a key functional role for this protein family. Sequence analysis at the protein level of the three ORMDL proteins found in humans reveals a high level of identity shared between the isoforms. Human ORMDL1 shares 83% identity with ORMDL2 and 84% identity with ORMDL (Hjelmqvist et al. 2002). This level of homology hints at a gene duplication event in the human genome as the source of the three ORMDL paralogs. Even more striking is how much identity there is between the ORMDL proteins in humans compared to the ORMDL proteins found in other vertebrates. For example, human ORMDL1 shares 99% identity with mouse ORMDL1, with 97% identity reported for both ORMDL2 and ORMDL3 at the protein level. This high conservation between species also suggests that the three isoforms have distinct, as yet unidentified, functions. In yeast, there are two ORM genes, ORM1 (located on chromosome VII) and ORM2 (located on chromosome XII), which encode proteins sharing approximately 70% identity with each other (Hjelmqvist et al. 2002). In contrast to the high degree of sequence identity shared among the vertebrate ORMDLs, when vertebrate ORMDL protein sequences are compared with sequences of the Orm proteins found in yeast or plants, there is only 30–40% similarity (Hjelmqvist et al. 2002; Paulenda and Draber 2016). Some of these differences will be discussed in the sections to follow on Orm/ORMDL regulation. In humans, all three ORMDL genes encode proteins consisting of 153 amino acids with only slight variations in the sequence between the isoforms. In yeast, the resulting proteins are considerably larger with Orm1 being 222 amino acids in length and Orm2 having 216 amino acids. The Orm/ORMDL proteins have been localized to the endoplasmic reticulum (ER) (Araki et al. 2008; Hjelmqvist et al. 2002; Li et al. 2016; Wang et al. 2015). A variety of techniques including fluorescently-tagged recombinant proteins, colocalization of endogenous proteins in multiple cell lines and subcellular fractionation have been used to support this observation. Prediction algorithms such as TopPred, TMPred, and HMMTOP, assign two, three or four possible transmembrane domains (TMDs) for the Orm/ORMDL proteins based on hydrophobicity. Topology of human ORMDL3 was probed using a fluorescence-based protease protection assay (Cantero-Recasens et al. 2010) and that of yeast Orm2 was probed using the insertion of glycosylation cassettes at key locations in the protein (Kimberlin et al. 2016). These studies revealed that both the N and C termini of Orm/ORMDL proteins are located on the cytosolic face of the ER. This orientation of the terminal regions of Orm/ORMDL proteins supports an even number of TMDs. Our lab has recently utilized substituted cysteine accessibility to provide a detailed analysis of the membrane topology of the mammalian ORMDL1 isoform (submitted manuscript). This analysis confirms existence of four TMDs with the N and C terminal regions facing the cytosol and sets the stage for future work to identify key domains of ORMDL proteins required for interacting with SPT and possibly defining the “lipid sensing” domain of ORMDL responsible for the homeostatic feedback regulation of SPT when levels of cellular sphingolipids are perturbed (Siow and Wattenberg 2012).
The endoplasmic reticulum is also the subcellular location of proteins recently identified to have protein-protein interactions with Orm/ORMDL. Breslow et al., combined immunoprecipitation studies with mass spectrometry to identify the Orm1/2 interacting proteins in yeast (Breslow et al. 2010). Direct binding to Orm1 and Orm2 was seen with Lcb1, Lcb2, and Tsc3, the subunits of yeast SPT. In addition, Orm proteins were found to co-immunoprecipitate Sac1, a phosphatidylinositol-phosphate phosphatase in yeast, as well as Orm proteins themselves which indicates a certain degree of self-association. As described above, SPT is an essential enzyme that catalyzes the first step in sphingolipid biosynthesis and in yeast results in the production of long chain base (LCB) intermediates. As covered in more depth below, the direct association of the Orm proteins with SPT in yeast is consistent with a role for the Orms/ORMDLs in the regulation of SPT activity. Tsc3 is an SPT-regulating protein that has been shown to increase activity of the SPT heterodimer (Gable et al. 2000). Sac1 is a phosphoinositide phosphatase and the activity of Sac1 has been shown to be essential for the production of complex sphingolipids (Brice, Alford, and Cowart 2009). The work done by Breslow and co-workers led to creation of the acronym “SPOTS” (Serine Palmitoyltransferase-Orm1/2-Tsc3-Sac1) complex as a way of identifying this group of interacting proteins (Breslow et al. 2010; Walther 2010). Identification of these key interactions also supported the hypothesis that Orm/ORMDL proteins were intricately involved in the sphingolipid biosynthetic pathway.
4. Orm/ORMDL proteins: inhibitory function revealed
For many years following their discovery in 2002 (Hjelmqvist et al. 2002), the function of the ORMDL proteins was not identified. Even though a growth defect and sensitivity to toxins was initially reported in yeast when both ORM genes were deleted (Hjelmqvist et al. 2002), it was not until 2010 that the first solid evidence for a role in sphingolipid homeostasis was reported for the Orm/ORMDL proteins (Breslow et al. 2010; Han et al. 2010). Taking distinctly different approaches, likely at the same time and unbeknownst to each other, members of the Weissman and Chang laboratories began unraveling the function(s) of this novel family of highly conserved genes. Both labs chose yeast as their model system, an organism known to be a powerful tool for gene study, in part due to the relatively small genome size and ease of gene manipulation. While the Weissman group was primarily focused on the role of Orm/ORMDL proteins in the regulation of sphingolipid biosynthesis, the Chang lab was studying this gene family in the context of membrane biogenesis and protein quality control of the endoplasmic reticulum. Both groups made seminal discoveries towards advancing our understanding of the physiological role(s) for the Orm/ORMDL proteins. In the sections below, we will first discuss what is known about regulation of Orms in yeast then we will cover the research that is emerging in regards to regulation of mammalian ORMDLs. Thus far, the regulatory mechanisms for yeast Orms and mammalian ORMDLs appear to be vastly different from each other.
4.1. Orm proteins as homeostatic regulators of sphingolipids in yeast
The Weissman lab started their quest by conducting an E-MAP study in yeast (Breslow et al. 2010). This type of gene ontology analysis can reveal “function” based on genetic interactions that differ between the profiles generated in the presence or absence of the gene in question. Specifically, when the authors deleted the ORM2 gene, the resulting E-MAP profile was opposite from that seen when LCB1 and LCB2 (major SPT subunits in yeast) were deleted. A result like this can be interpreted as the two genes (ORM2 and LCB1/2 in this case) having opposing cellular roles. This led the authors to measure the steady state levels of the long chain bases (sphingolipids) in yeast cells lacking ORM1/2. Lipidomic profiles revealed that when ORM1/2 were deleted, sphingolipids were increased compared to wild type cells. Conversely, when either of the Orm proteins were overexpressed, steady state levels of sphingolipids and ceramides were decreased. These results were recapitulated in HeLa cells where a 3-fold increase in ceramide levels was seen following siRNA-mediated depletion of all three ORMDL isoforms, lending further evidence for a highly conserved function of these proteins (Breslow et al. 2010). Strengthening the emerging role of Orm/ORMDL proteins in sphingolipid biosynthesis, Breslow et al. established a direct interaction of Orm1/2 with proteins known to play a role in the sphingolipid pathway. As detailed in Section 3 above, qualifying these specific protein-protein interactions of Orm1/2 led to what is now known as the “SPOTS” complex (Walther 2010; Breslow et al. 2010)
Breslow et al. (2010) then combined deletion of ORM1/2 in their yeast model with treatment with myriocin, a specific and irreversible inhibitor of SPT (Figure 1). From previous work it was known that loss of certain genes, like LCB2 or SAC1, rendered cells either more or less sensitive to myriocin treatment (Breslow et al. 2008). This degree of myriocin sensitivity can often be attributed to the status of the cellular levels of long chain bases (LCBs). If LCB levels are elevated (as in ∆sac1 mutants), cells are able to cope with the loss of SPT activity that results from myriocin treatment and are deemed “myriocin resistant”. If, however, the cellular levels of LCBs are low (as in ∆lcb2 mutants) prior to myriocin treatment, cells aren’t able to handle the loss of SPT activity and are deemed “myriocin sensitive”. Breslow and co-workers were surprised to find that even though cells lacking ORM1/2 had increased levels of LCBs (a phenotype shared by ∆sac1 mutants), they did not exhibit an altered response to myriocin treatment when compared to wild type cells. This suggested that myriocin treatment of cells was causing some type of inactivation of the Orm proteins thereby removing them from the cell survival picture during myriocin treatment.
Upon further exploration, the Weissman lab was able to demonstrate that myriocin treatment led to phosphorylation of the Orm proteins on distinct residues of the N terminus and in a dose-dependent manner (Breslow et al. 2010). As the concentration of myriocin increased, the number of phosphorylated sites on Orm1/2 increased as evidenced by multiple phospho-Orm bands using a phospho-affinity gel system. They went on to identify the myriocin-induced phosphorylated residues using mass spectrometry (see Table 1 and Section 5.1 below). Mutation of these residues resulted in a decrease in cellular LCB levels and increased the sensitivity to myriocin. These results confirmed that Orm1/2 are negative regulators of SPT activity and indicated that phosphorylation of the Orm proteins blocks their ability to inhibit SPT and therefore relieves the constitutive inhibition of SPT activity. Reducing the ability of the Orm proteins to inhibit SPT allows the cells to make sufficient LCBs to grow while under myriocin treatment.
Table 1:
Regulation of Orm1/2 in yeast
| Trigger/Condition | Orm Status | Sphingolipids | Phosphosites | Kinase | Phenotype / Pathway | References |
|---|---|---|---|---|---|---|
| Heat stress | ↑ P-Orm2 | ↑ LCBs (rapid, transient) |
(Orm2) S46, S47, S48 |
Ypk1 | Pkh-Ypk1-PP2A axis | (Sun et al. 2012) |
| ER stress (DTT & tunicamycin) |
↑ Orm2 mRNA ↑ Orm2 protein ↑ P-Orm1 |
↓ LCBs | (Orm1) S29, S32, S34, S35, S36 |
Npr1 | Ca2+/calcineurin - dependent TORC1-Npr1 axis ↑ UPR |
(Gururaj, Federman, and Chang 2013; Han et al.2010; Liu et al. 2012) |
| Nutrient loss (Rapamycin) Nitrogen starvation |
↑ P-Orm1 ↑ P-Orm2 |
↓ LCBs ↑ IPCs ↑ MIPCs |
(Orm1) S29, S32, S34, S35/36 (Orm2) S9, S15, T18, S22, S29, S31 |
Npr1 |
TORC1-Npr1 axis |
(Liu et al. 2012; Shimobayashi et al. 2013) |
| Exogenous lipids (PHS, DHS, C2 ceramide) |
↓ P-Orm1 ↓ P-Orm2 |
↓ LCBs | --------------- | --------- | Feedback regulation via ↓ SPT activity |
(Gururaj, Federman, and Chang 2013; Sun et al. 2012) |
| Myriocin | ↑ P-Orm1 ↑ P-Orm2 |
↓ LCBs | (Orm1) S51, S52, S53 (Orm2) S46, S47, S48 |
Ypk1 (T662) |
TORC2-Ypk1 axis | (Breslow et al. 2010; Gururaj, Federman, and Chang 2013) |
| Aureobasidin A | ↑ P-Orm1 ↑ P-Orm2 |
↓ IPCs ↓ MIPCs |
--------------- | --------- | ↑ SPT activity | (Gururaj, Federman, and Chang 2013) |
| Cyclohexamide (activates TORC1) |
↓ P-Orm1 ↓ P-Orm2 |
↓ LCBs | --------------- | --------- | TORC1 | (Shimobayashi et al. 2013) |
| Increased Orm 2 protein | ↑ P-Orm1 | --------------- | --------------- | --------- | ↑ SPT activity | (Han et al. 2010; Liu et al. 2012) |
| ∆Orm1 ∆Orm2 | ---------------------- | ↑ LCBs ↑ Ceramide ↑ PHS ↓ Ceramide ↓ IPCs ↓ MIPCs ↑ MIP2C |
--------------- | --------- | Growth defect ↑ SPT activity ↑ UPR ↑ PHS sensitivity ↑ Toxin sensitivity ↑ FB1 sensitivity Disrupted phospholipids ER-Golgi transport delay Impaired heat response |
(Gururaj, Federman, and Chang 2013; Han et al. 2010; Hjelmqvist et al. 2002; Liu et al. 2012; Roelants et al. 2011; Shimobayashi et al. 2013; Breslow et al. 2010) |
| ∆ypk1 | ↓ P-Orm1 ↓ P-Orm2 |
↓ LCBs ↓ DHS ↓ PHS |
--------------- | --------- | ↑↑ Myriocin sensitivity Growth defect |
(Gururaj, Federman, and Chang 2013; Liu et al. 2012; Roelants et al. 2011) |
| ∆tsc3 (SPOTS complex) | ↑ P-Orm1 ↑ P-Orm2 |
↓ LCBs | --------------- | --------- | ↑ UPR ↓ SPT activity |
(Gable et al. 2000; Gururaj, Federman, and Chang 2013) |
| ∆sac1 (SPOTS complex) | ↑ P-Orm1 | ↑ LCBs ↑ Ceramide ↓ IPCs ↓ MIPCs |
--------------- | --------- | KO w/∆Orm1/2 is lethal Myriocin resistant |
(Breslow et al. 2010; Brice, Alford, and Cowart 2009; Liu et al. 2012) |
| ∆isc1 | ↑ Orm2 protein ↓ P-Orms |
↑ IPCs ↑ MIPCs |
--------------- | --------- | ↑ UPR | (Gururaj, Federman, and Chang 2013; Shimobayashi et al. 2013) |
| ∆sit4 (PP2A subunit) | ↓ P-Orm1 | ↓ LCBs ↓ Ceramide |
--------------- | --------- | (Liu et al. 2012) | |
| ∆cdc55 (PP2A subunit) | ↓ P-Orm2 | ↑ LCBs | --------------- | --------- | Myriocin resistant | (Sun et al. 2012) |
| ∆swe1 (checkpoint kin) | ↓ P-Orm1 ↓ P-Orm2 |
↓ LCBs ↓ DHS ↓↓ PHS (C20) |
--------------- | --------- | ↑↑ Myriocin sensitivity KO w/ ∆tsc3 is lethal |
(Chauhan et al. 2017) |
Abbreviations: LCB, long chain base; DHS, dihydrosphingosine; PHS, Phytosphingosine; IPC, inositol phosphorylceramide; MIPC, mannosyl-IPC; UPR, unfolded protein response; SPT, serine palmitoyltransferase; FB1, fumonisin B1; TORC1/2, target of rapamycin complexes 1 and 2; ∆, genetic deletion
4.2. Orm proteins integrate protein quality control at the ER with sphingolipid homeostasis
Simultaneous to the work reported by Breslow et al., members of the Chang Lab reported that depleting yeast cells of both ORM genes resulted in a slow growth phenotype and increased sensitivity to toxins known to induce an unfolded protein response (UPR) in yeast (Han et al. 2010) (Table 1). They were able to rescue both of these phenotypic differences by overexpressing just one of the Orm proteins, implying overlapping function for the two Orm isoforms in yeast. Additionally, the authors established that loss of ORM1/2 resulted in basal levels of UPR that were higher than levels seen in wild-type cells, essentially rendering ∆orm1∆orm2 cells more sensitive to toxins known to induce a UPR response, such as DTT or tunicamycin. The ∆orm1∆orm2 cells accumulated unfolded or misfolded proteins and upon further investigation, the authors were able to connect this accumulation to an ER-to-Golgi transport delay in these cells. Interestingly, Araki et al. reported that siRNA-mediated knockdown of all three ORMDL isoforms in human cervical carcinoma cells resulted in failure of nicastrin to mature (Araki et al. 2008). Nicastrin is a protein that requires ER-to-Golgi transport in order to become fully glycosylated and complete the maturation process. Once mature, nicastrin can then interact with the other proteins in the gamma secretase complex which is responsible for the proteolytic cleavage of several proteins. These results suggest that the loss of the ORMDL proteins in the human system could also be causing defects in ER-to-Golgi transport and warrants further investigation.
In agreement with Breslow et al., the ∆orm1∆orm2 mutants made by the Chang lab had increased levels of phytosphingosine (approximately 5-fold) and were sensitive to the addition of exogenous sphingolipids (Han et al. 2010). The authors showed that sensitivity to exogenous lipids could be overcome if they overexpressed a lipid transporter, Rcb1, known to efflux LCBs from yeast cells. Additionally, if ∆orm1∆orm2 mutants were treated with low doses of myriocin to inhibit SPT activity, which would also lower endogenous LCB levels, they were able to overcome the ER-to-Golgi transport delay, the slow growth phenotype and the sensitivity to UPR-inducing toxins. Han and coworkers then conducted a genetic screen to search for suppressors of the phenotypic changes seen in ∆orm1∆orm2 mutants. This led to discovery of Lcb1 and Lcb2 binding to Orm1 and Orm2, which is in complete agreement with the results of (Breslow et al. 2010). In contrast to the increased levels of ceramides reported by Breslow et al. when they deleted both ORM genes in their system, Han et al. reported decreased ceramide levels in the double knockout cells. This may be due to yeast strain differences. It is possible, for example, that in the yeast strain used by the Chang lab excess ceramides are rapidly de-acylated to produce LCBs. Taken together, results from both the Weissman and Chang laboratories confirmed the role of Orm1/2 proteins as negative regulators of sphingolipid biosynthesis in yeast.
5. Yeast Orm proteins: multiple regulatory pathways emerging
5.1. Phosphorylation of Orm proteins in yeast by Ypk1
Building on their discovery that Orm proteins in yeast were phosphorylated in response to myriocin (Breslow et al. 2010), members of the Weissman lab set out to find the kinase(s) responsible for this. There were several pieces of evidence pointing to Ypk1 as being the target kinase to pursue, including previous results showing that loss of YPK1 caused cells to be hyper-sensitive to myriocin treatment and that Ypk1 overexpression rendered cells resistant to myriocin (Roelants, Torrance, and Thorner 2004; Tanoue et al. 2005). Additionally, a previous study had established Ypk1 as a critical component of the kinase network regulating the balance between sphingolipid pools and aminophospholipids on the outer leaflet of the plasma membrane. This evidence was further strengthened when the authors found that each of the serine residues phosphorylated on Orm1/2 aligned with an optimal phosphoacceptor motif for Ypk1, namely the motif [–R-x-R-x-x-S/T- (Hydrophobic residue)–] (Roelants et al. 2011). Through a series of elegant experiments, the authors confirmed that Ypk1 was indeed the kinase responsible for phosphorylation of Orm1/2 on distinct serine residues upon sphingolipid depletion. The authors went on to show that sphingolipid depletion (i.e. myriocin treatment) led to the TORC2-dependent phosphorylation of Ypk1 at threonine 662, thus activating this kinase which led to increased phosphorylation of Orm1/2 at predicted Ypk1 motifs. Once phosphorylated by Ypk1, Orm1/2 inhibition of SPT was blunted and SPT activity was essentially released from this constitutive Orm-dependent inhibition. This then allowed sphingolipid biosynthesis to increase sufficiently for cells to survive under the pressure of myriocin treatment.
The work done by Roelants et al. established Ypk1-dependent phosphorylation of Orm1/2 to occur on specific serine residues, namely S51, S52 and S53 on Orm1 along with S46, S47 and S48 on Orm2 (Roelants et al. 2011) (Table 1). Additionally, for Orm2, these specific serine residues were also found to be phosphorylated in response to heat stress in a Ypk1-Pkh1/2 axis-dependent fashion (Sun et al. 2012). Yet both of these proteins contain at least 17 serine and threonine residues within the first 80 amino acids of the N-terminus, some of which were previously demonstrated to also be phosphorylated (Breslow et al. 2010). This observation has led to a flurry of recent studies that are beginning to link the phosphorylation status of Orm proteins to distinct biological outcomes and signaling pathways in yeast, such as heat stress (Sun et al. 2012), ER stress (Gururaj, Federman, and Chang 2013; Han et al. 2010; Liu et al. 2012), and nutrient loss (Liu et al. 2012; Shimobayashi et al. 2013) (Table 1).
5.2. Phosphorylation of Orm proteins in yeast by Npr1
In addition to the established role of Ypk1 in phosphorylating Orm proteins, recent work by several groups has linked the Npr1-TORC1 axis to Orm regulation (Gururaj, Federman, and Chang 2013; Liu et al. 2012; Shimobayashi et al. 2013). Specifically, Npr1 (nitrogen permease reactivator 1), a TOR-regulated kinase, was shown to phosphorylate Orm1 at residues S29, S32, S34, S35/36 (Liu et al. 2012; Shimobayashi et al. 2013) and Orm2 at residues S9, S15, T18, S22, S29 and S31 (Shimobayashi et al. 2013). These residues are distinctly different than the ones utilized by Ypk1 (as detailed above), thereby affording another component to be added to the expanding regulatory network for control of sphingolipid levels in an Orm-dependent manner. While phosphorylation of Orm1/2 by Npr1 was not directly connected to changes in SPT activity by these authors, the conditions under which these residues were phosphorylated (i.e. ER stress, nutrient loss) resulted in overall decreases in the cellular levels of LCBs.
TOR (target of rapamycin) is a highly conserved serine/threonine kinase that exists in a complex of proteins and regulates cell growth under a variety of conditions (Loewith et al. 2002; Wullschleger, Loewith, and Hall 2006). TOR signaling in yeast has two major arms, TOR complex 1 (TORC1) and TOR complex 2 (TORC2), each of which is structurally and functionally distinct (Wedaman et al. 2003). The work by Liu et al. and Shimobayashi et al. establishes a direct link between TORC1 signaling and sphingolipid biosynthesis via the phosphorylation of Orm1/2 proteins by Npr1 (Liu et al. 2012; Shimobayashi et al. 2013). This TORC1-dependent regulation of Orm1/2 results from nutrient deprivation, as mimicked by rapamycin treatment and nitrogen starvation, which in turn leads to accumulation of complex sphingolipids as a possible means of dealing with nutrient loss. On the other hand, the work by Breslow et al. and Roelants et al., establishes a direct link between TORC2 signaling and sphingolipid biosynthesis via the phosphorylation of Orm1/2 by Ypk1 (Breslow et al. 2010; Roelants et al. 2011). This TORC2-dependent regulation of Orm1/2 is responsive to sphingolipid limitation, as seen with myriocin treatment, and results in the derepression of SPT and increases in cellular LCBs (Roelants et al. 2011).
What is now emerging in yeast is an intricate and complex system of regulating de novo sphingolipid biosynthesis through manipulation of the Orm proteins, with possible connections to ER to Golgi protein transport (Han et al. 2010), phospholipid utilization and synthesis (Han et al. 2010), nutrient uptake (Shimobayashi et al. 2013), Ca2+/calcineurin signaling (Gururaj, Federman, and Chang 2013), cell cycle control (Chauhan et al. 2017), and membrane remodeling/biogenesis (Liu et al. 2012). This ability of Orm proteins to be responsive to multiple regulatory pathways underscores the important role that Orm1/2 play in maintaining tight control over cellular sphingolipid levels. A complete picture of the molecular mechanisms connecting Orm-dependent regulation of sphingolipid biosynthesis to a wide variety of cellular processes has yet to emerge but is currently an active area of research. Ongoing efforts in multiple labs should soon provide answers to key questions regarding Orm-dependent regulation of sphingolipids, such as what are the specific lipids being sensed by this system, how are sphingolipid levels connected to lipid transport, and why have multiple isoforms of the Orm/ORMDL proteins been maintained across multiple species, just to name a few.
6. Mammalian ORMDL proteins: Distinct mechanisms of SPT regulation
Above we have discussed the regulation of SPT by the Orms in yeast. Here we continue that discussion with the somewhat distinct regulation found in mammalian cells. While the focus of this review is on the ORMDLs, recent studies have indicated that there is at least one additional regulator of SPT in mammals, the Nogo-B protein (Cantalupo et al. 2015). We touch on that regulation below.
6.1. The mammalian ORMDL proteins are negative regulators of SPT that respond to cellular levels of sphingolipid
In 1991 Mandon et al. sought to test whether SPT activity was homeostatically regulated by examining the response of SPT activity to exogenously added sphingosine in primary cultured neurons (Mandon et al. 1991). Indeed they reported a very profound inhibition of SPT by incubation of cultures with sphingosine. Because sphingosine added to cells is rapidly metabolized to other sphingolipids, they could not determine exactly which sphingolipid(s) triggered this inhibition. Regardless, these experiments established the critical point that there was some type of system which could sense levels of an as yet unspecified sphingolipid to inhibit SPT, a classic homeostatic response. It was not until the reports of Orm regulation of SPT by the Weissman and Chang groups that the mechanism that mediated this response gained a molecular foothold (Section 4 of this review). As detailed above, the Weissman group reported that deletion of the ORMDLs increased cellular ceramide levels in mammalian cells (Breslow et al. 2010). Subsequently our group looked directly at the sphingolipid de novo biosynthetic pathway by utilizing labeled serine to track de novo synthesis of sphingolipids (Siow and Wattenberg 2012). To elevate cellular sphingolipid levels we incubated cells with a ceramide in which the N-acyl group was a 6-carbon fatty acid (termed here C6- ceramide), and therefore water-soluble making it easily delivered to and taken up by cells in culture. Parallel to the observations of Mandon et al., we saw that de novo synthesis of ceramide was strongly inhibited by incubating cells with C6-ceramide. Extending this observation, we found that siRNA depletion of the ORMDLs had two notable effects on de novo sphingolipid biosynthesis. Under control conditions, in the absence of C6-ceramide treatment, de novo sphingolipid biosynthesis was elevated by ORMDL depletion. This suggests that the ORMDLs are constitutively inhibiting SPT. Just as importantly, ORMDL depletion completely abolished the ability of C6-ceramide to inhibit sphingolipid biosynthesis. Therefore, as in yeast, the mammalian ORMDLs mediate the homeostatic regulation of SPT. The control of de novo biosynthesis is mirrored in the effect of ORMDL depletion on steady-state levels of sphingolipids in which it is observed that ORMDL knockdown elevates sphingolipid levels in A549 cells, particularly ceramides (Oyeniran et al. 2015). Returning to the question of which sphingolipid(s) trigger the ORMDL-mediated inhibition of SPT, we recapitulated the experiment of Mandon et al. by treating cells with sphingosine. We observed, as they did, that de novo synthesis of sphingolipid was strongly inhibited and, not surprisingly, that ORMDL depletion blocked this inhibition. To test whether the sphingosine inhibition of SPT was due to sphingosine itself, rather than a downstream metabolite, we treated cells with Fumonisin B1 (FB1), a potent and specific inhibitor of the ceramide synthases. In these experiments, FB1 blocked the ability of sphingosine to inhibit SPT. Therefore sphingosine, when added exogenously to cells in culture, must be converted to ceramide in order to result in subsequent inhibition of SPT. This demonstrates that ceramide, or a downstream sphingolipid metabolite, is the lipid sensed by the ORMDL/SPT system. Identifying this lipid is important because it is an indication of which lipid is critical to control in order to preserve cell function.
6.2. Why are there three ORMDL Isoforms?
The three mammalian ORMDL isoforms, ORMDL1-3, are highly homologous, as discussed in Section 3. Expression analysis for human adult and fetal tissues revealed ubiquitous expression patterns for all three ORMDL genes (Hjelmqvist et al. 2002). Similar homogeneity of isoform expression, both spatially and temporally, with some minor isoform specificity, has been reported for mice (Araki et al. 2008), as well as in the plant species Arabidopsis (Kimberlin et al. 2016) and rice (Chueasiri et al. 2014). These studies have failed to demonstrate a profound developmental or tissue-specific expression of the ORMDLs/Orms at the RNA level. Just as importantly, at a basic functional level, the three isoforms appear to be redundant. This conclusion is drawn from experiments in which we depleted, with siRNA, each of the ORMDL isoforms individually and in all possible combinations (Siow, Sunkara, Dunn, et al. 2015; Siow, Sunkara, Morris, et al. 2015). Only when all three ORMDL isoforms were knocked down did we observe a loss of SPT regulation either when measuring constitutive inhibition of SPT by the ORMDLs, or when measuring ORMDL- dependent inhibition of SPT by exogenously added C6-ceramide. These experiments demonstrated that at least in HeLa cells, any single ORMDL isoform can do the trick.
So why are there three ORMDL isoforms in vertebrates? Given the conservation of these three isoforms, it seems very unlikely that they are truly redundant. One possibility is that individual isoforms respond to different sphingolipids or to different cellular levels of sphingolipid. This would be one mechanism to tune the levels and composition of the sphingolipids present in a tissue or condition-specific manner. Another possibility is that, although the RNA expression of the isoforms is relatively uniform, the levels of the proteins themselves are differentially regulated. As outlined below, under some circumstances the levels of ORMDL proteins are regulated post-transcriptionally, most probably by a degradative mechanism. In this context, although the RNA levels of the ORMDL isoforms appears uniform, the protein levels within specific tissues or cellular compartments may be quite different. However, measuring the protein levels of each individual isoform is technically challenging. The production of isoform-specific antibodies is precluded by the high level of homology between the three isoforms at the protein sequence level. The sequence differences between the isoforms are primarily single residue differences dispersed throughout the sequence or are found in non-antigenic hydrophobic residues in the putative transmembrane segments (Hjelmqvist et al. 2002; Paulenda and Draber 2016). Therefore, the individual protein levels of each isoform in different tissues, developmental stages, and/or under specific physiological conditions cannot be determined by immunoblots or immunohistochemistry at this time. The field awaits a robust proteomics approach that can be used to routinely assess the protein levels of the three isoforms.
6.3. The molecular mechanism underlying ORMDL-dependent homeostatic regulation of SPT
As in yeast, the mammalian ORMDLs form a stable complex with SPT as measured by immunoprecipitation (Kiefer et al. 2015; Siow, Sunkara, Dunn, et al. 2015). In our hands the level of association of the ORMDLs with SPT is the same in both high and low sphingolipid conditions (Siow, Sunkara, Dunn, et al. 2015). This is a key finding when considering the mechanism by which the ORMDLs differentially regulate SPT depending on cellular sphingolipid content. These results indicate that regulation is accomplished by a change in the conformation of the ORMDL/SPT complex from non-inhibitory (under low sphingolipid conditions) to inhibitory (under high sphingolipid conditions), rather than by a regulated association of ORMDL with SPT or by changes in expression levels of either of these proteins (Figure 2). Thus, to establish the molecular mechanism of regulation by the ORMDL proteins, we should focus on how sphingolipids change the conformation of the ORMDL/SPT complex.
Figure 2.
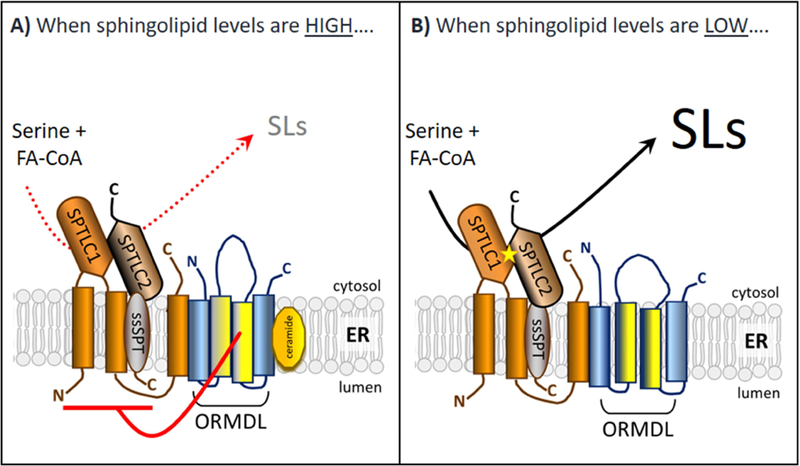
Working model depicting the homeostatic regulation of SPT by the ORMDL protiens in response to changes in sphingolipid levels in mammalian system.
As covered in detail in Section 5, the major sphingolipid-responsive regulation of the Orms in yeast is mediated by phosphorylation of the Orm proteins on specific residues. This is not true of the mammalian ORMDLs. Mammalian ORMDLs lack the stretch of amino acids found in the amino terminus of the yeast Orms that contains the Orm phosphorylation sites (Hjelmqvist et al. 2002; Paulenda and Draber 2016). This is, in fact, the major structural difference between the ORMDLs and the Orms and strongly suggests that the ORMDLs utilize a distinct regulatory mechanism. Moreover, direct phosphate labeling experiments of the ORMDLs have failed to demonstrate any phosphorylation under conditions known to produce ORMDL-dependent regulation of SPT activity (unpublished data from this laboratory). Therefore, we must look beyond ORMDL phosphorylation to discover the mechanism of regulation. An important clue comes from experiments using cells in which the plasma membrane was selectively permeabilized to release the soluble cytoplasmic proteins and co-factors as well as ATP (Siow and Wattenberg 2012). Remarkably, these permeabilized cells are fully responsive to C6 ceramide with respect to inhibition of SPT.
This regulation requires the ORMDLs, establishing that the permeabilized cells recapitulate the regulatory system of intact cells (Siow and Wattenberg 2012). This observation narrows the potential molecular mechanisms of regulation considerably. Considering that the permeabilized cells are depleted of cytosolic proteins, cofactors, and ATP, we can rule out most post-translational modifications of either SPT or the ORMDLs, including phosphorylation. Because permeabilized cells lack many components required for protein translation, this experiment also confirms that changes in protein expression levels cannot explain sphingolipid regulation of the ORMDL/SPT complex. It is also notable that the permeabilized cells, in which further metabolism of ceramide within the sphingolipid network should be virtually non-existent, still exhibited C6-ceramide-induced inhibition of de novo sphingolipid biosynthesis. This strongly suggests that ceramide is the sphingolipid to which this system is responsive. What then are potential models of the molecular basis of regulation? We favor a model where ceramide directly binds to the ORMDL/SPT complex to accomplish the change in conformation that inhibits SPT activity (Figure 2). These experiments do not rule out that there might be a separate ceramide sensor that binds to the SPT/ORMDL complex to trigger SPT inhibition. There are, of course, other possibilities. Ceramide is known to be a potent modifier of the physical properties of lipid membranes (Castro, Prieto, and Silva 2014; Castro et al. 2009). It is an attractive concept that the ORMDL/SPT system may monitor the biophysical properties of the endoplasmic reticulum to maintain a lipid composition that is optimal for function of that membrane. Clearly there is work to be done to sort out these possibilities.
6.4. Regulation of mammalian ORMDL protein levels
In the preceding section we discussed the mechanisms by which the ORMDL/SPT system responds to acute changes in sphingolipid levels to regulate sphingolipid biosynthesis. However, evidence is emerging to indicate that there is another layer of control at work whereby changes in ORMDL protein levels become regulatory. It is notable that initial experiments did not support the concept that increased ORMDL levels would be a regulatory mechanism. Initially, it was expected that since the ORMDLs are negative regulators of SPT, increased expression of ORMDL protein would further inhibit de novo sphingolipid biosynthesis. Surprisingly, this was not the case when studied in cultured cells. In fact, we and others observed just the opposite result. Overexpression of human ORMDL3 increased SPT activity and ceramide levels in A549, HeLa and RAW264.7 cells (Oyeniran et al. 2015; Siow, Sunkara, Dunn, et al. 2015). This increase in SPT activity is puzzling and may be explained by the artificial nature of the overexpression. However, while the increase in SPT activity is mysterious, the lack of inhibition is clearly explained by the observation that in these cells, at endogenous levels of SPT subunit proteins, there is sufficient ORMDL expression to maximize levels of the association with SPT. For this reason, additional ORMDL expression does not force an increase in levels of the ORMDL/SPT complex (Siow, Sunkara, Dunn, et al. 2015). We confirmed that endogenous levels of SPT were limiting for complex formation by demonstrating that when SPT was overexpressed above endogenous levels, we did observe that increasing the level of ORMDL protein expression resulted in inhibiting de novo sphingolipid biosynthesis, as expected. Under these conditions there is not sufficient endogenous ORMDL protein to satisfy the requirements of the overexpressed SPT and expressing more ORMDL protein promotes formation of ORMDL/SPT complexes, and therefore inhibition, of the extra SPT. These experiments establish that the stoichiometric ratio of SPT to ORMDL determines whether changes in ORMDL levels will control SPT activity and this ratio may be depend on cell type and physiological conditions. For example, Kiefer et al. demonstrated that under specific conditions, when HEK293 cells are treated with palmitate, overexpression of all three ORMDL isoforms was needed for maximal inhibition of SPT activity (Kiefer et al. 2015). Taken together, these results underscore the importance of monitoring both SPT and ORMDL expression levels in order to completely understand how changes in levels of either protein will affect sphingolipid biosynthesis. Evidence is emerging that ORMDL protein levels are indeed regulated under specific conditions. As outlined below, we are now beginning to understand that the acute versus long term control of SPT activity by ORMDL is accomplished by distinct mechanisms.
Gupta et al. have uncovered an important regulation of ORMDL protein levels that depends on activity of SPT (Gupta et al. 2015). In Hek293 cells they increased SPT levels by ectopic expression and observed a remarkable increase in ORMDL protein levels. This was not accompanied by an increase in ORMDL message, indicating that the increase in ORMDL protein was at the level of increased translation or decreased degradation. They favor the latter mechanism. Strikingly, this was not solely due to the protein/protein interaction between SPT and ORMDL, which might have been thought to stabilize ORMDL against degradation. They found instead that the increase in ORMDL protein levels was only observed with enzymatically active SPT. Overexpression of a catalytically inactive SPT did not lead to increased ORMDL levels, nor was there increased ORMDL protein when wild-type SPT protein was overexpressed then subsequently inhibited pharmacologically. These experiments point to a fascinating mechanism that detects SPT enzymatic activity and subsequently regulates ORMDL protein levels to modulate that increased activity.
Another case in which ORMDL protein levels appear to be regulated is in the inflammatory response in certain cells. Spiegel and coworkers sought to understand if sphingolipid signaling during an inflammatory response might be conferred by changes in de novo sphingolipid biosynthesis (Cai et al. 2016). They examined ORMDL levels in HepG2 liver cells treated with the pro-inflammatory cytokines Interleukin-1 (IL-1) and Oncostatin M (OSM). This treatment induced a profound decrease in ORMDL levels. This loss of ORMDL would be expected to stimulate SPT activity and indeed they observed an increase in cellular sphingolipid levels. Importantly, this treatment did not affect ORMDL1-3 message at the RNA level. Therefore the decrease in ORMDL protein levels was post-transcriptional, either via an increase in ORMDL turnover or a decrease in ORMDL translation. This group then extended their observations to an in vivo model of inflammation. Turpentine treatment in mice causes tissue destruction at the site of injection resulting in release of pro-inflammatory cytokines (Cai et al. 2016). This treatment induced a decrease of ORMDL protein without decreases in ORMDL mRNA levels. The mechanism behind this loss of ORMDL has yet to be clarified. An attractive possibility is that this loss of ORMDL is mediated by regulated degradation. It remains to be determined if this is the case and, if so, which of the known mechanisms of regulated protein degradation are involved. Considering that the ORMDLs are membrane proteins of the ER, the likely candidate mechanisms include endoplasmic reticulum associated protein degradation (ERAD, (Huang et al. 2018)) or organelle-specific autophagy (Anding and Baehrecke 2017).
The decrease of ORMDL protein levels induced by inflammatory signals in HepG2 cells stands in stark contrast to an increase in ORMDL levels found under allergic and inflammatory conditions in airway epithelia and other cell types implicated in the asthmatic response (Oyeniran et al. 2015; Ha et al. 2013; Liu et al. 2017; Miller et al. 2012; Wang et al. 2017). For clarity, these results are covered in more detail below in the section on ORMDL and asthma. However it should be noted that initial evidence indicates these increases are driven by gene expression rather than post-transcriptional mechanisms. Suffice it to say that the mechanisms which control sphingolipid biosynthesis in different physiological and pathophysiological states are clearly complex and involve regulating ORMDL protein content and interactions with SPT at multiple levels.
An example of a pathophysiological regulation of ORMDL protein levels is the response of macrophages to cholesterol loading (Wang et al. 2015). Gulshan and colleagues were exploring the mechanism behind the known increase in sphingomyelin levels in atherosclerotic plaques. They hypothesized that macrophages, when loaded with cholesterol, might have increased sphingolipid biosynthesis. Their supposition was correct. SPT activity in macrophages was increased following free cholesterol loading. To explain the increases in SPT activity, they probed ORMDL protein levels and found that levels of ORMDL protein were depressed by cholesterol loading, presumably decreasing the ORMDL-dependent inhibition of SPT, thus explaining the increased SPT activity. These authors then asked what mechanism might reduce ORMDL protein levels with cholesterol loading. To address this they turned to HEK293 cells, a more tractable system for ectopic expression of proteins. They used these cells to express GFP-tagged ORMDLs and track their localization under conditions of cholesterol loading. They observed that increased levels of free cholesterol led to translocation of the GFP-tagged ORMDL1 and ORMDL3 constructs out of the ER membrane and into punctate cytosolic structures. Moreover, inhibition of autophagy blocked the decrease in ORMDL levels. Together these data indicate that increased cholesterol levels in cells leads to selective autophagic degradation of the ORMDLs. The mechanism by which cholesterol achieves this effect is still to be determined. Cholesterol, like ceramides, has an important role in bilayer physical properties, and ceramides are known to interact with cholesterol (Castro, Prieto, and Silva 2014; Castro et al. 2009). This tantalizing physical connection between cholesterol and sphingolipids in mammalian systems has yet to be fleshed out although strong associations have been established in yeast (Guan et al. 2009). In yeast, deletion studies of the enzymes involved in sterol biosynthesis often cause perturbations in sphingolipid levels and vice versa which indicates coordinate regulation between these two biosynthetic pathways (Guan et al. 2009). The proposed involvement of cholesterol adds yet another mechanism by which control of SPT activity by ORMDLs could be achieved.
6.5. Regulation of mammalian SPT by Nogo-B
Recent studies done by Di Lorenzo and her co-workers suggests that the ORMDLs are not the only regulators of SPT activity (Cantalupo et al. 2015). These workers were studying Nogo-B, a membrane protein of the ER that is found primarily in the vasculature, and observed that knockdown of Nogo-B induced hypotension and further tracked that to an increase in the vasodilator, nitric oxide. In an apparent leap of logic, they decided to determine whether changes in sphingolipid levels could explain the increase in nitric oxide. Indeed, they found that levels of ceramide were increased, and that pharmacologically inhibiting that increase reversed the effects seen with Nogo-B knockdown. Prompted by this finding they looked for, and found, a direct protein/protein interaction between Nogo-B and SPT subunits, going on to demonstrate that Nogo B is an inhibitor of SPT, much like the ORMDL proteins. This may be an interaction specific to the vasculature, but more work needs to be done to establish whether this is a more wide-spread phenomenon. An interesting aspect of this work is that Nogo-B is a member of the reticulon family, members of which shape the structure of the endoplasmic reticulum. The interaction between Nogo-B and SPT raises a number of important questions. First, is there an interaction between Nogo-B and the ORMDLs? One possibility is that Nogo-B exerts its effects directly through ORMDL. Second, are the membrane-shaping properties of Nogo-B intrinsic to its regulatory function? Third, why is there an additional (beyond ORMDL) control of SPT activity needed in the vasculature? Is this one mechanism through which the homeostatic control of SPT activity conferred by the ORMDLs is over-ridden in order to achieve other physiological ends? If so, the area of control over SPT activity may be in its infancy and raises the possibility that there are a number of SPT regulators that work with, or alongside, the ORMDLs.
7. ORMDL and Asthma
Inserting the search terms “ORMDL3” and “Asthma” in to PubMed returns over 140 citations, of which almost 30 are reviews. The interest in this connection stems from a 2007 genome wide association study in which ORMDL3-linked polymorphisms were very strongly associated with elevated risk for childhood asthma (Moffatt et al. 2007). Below we review this association, what it may mean and how ORMDL3 may play a role in the development of this pervasive disease.
7.1. Genome-wide association studies: genetic evidence for a link between ORMDL3 and asthma
Beginning with a landmark genome-wide association study (GWAS) in 2007, ORMDL3 has received intense scrutiny as an important element in the risk for asthma. That study, by Moffat and colleagues (Moffatt et al. 2007) identified single nucleotide polymorphisms (SNPs) that were strongly associated with risk for childhood asthma. The strongest association was with a region on chromosome 17, the 17q21 locus, which is proximal to the gene for ORMDL3. It is important to note here that this region also includes the gene for Gasdermin B. There have been a large number of subsequent studies that have confirmed and extended the observation of a correlation between haplotype at the 17q21 locus and the risk for asthma. These studies have been the subject of several excellent reviews (Das, Miller, and Broide 2017; Ober and Yao 2011; Ono, Worgall, and Worgall 2014) and meta-analyses (Qu et al. 2018; Zhao et al. 2015) and will not be further reviewed here in detail.
7.2. What do the GWAS studies tell us about the role of ORMDL3 in asthma?
Asthma is a multi-component disease, involving the immune and inflammatory systems, the lung epithelium, and the smooth muscle in the airways. A significant complication is that factors can influence the immune component early in life, when individuals are sensitized to allergens or other environmental factors, or much later, during the expression of symptoms, such as airway hypersensitivity (Stein et al. 2018). This complexity is reflected in the degree to which the risk alleles elevate risk for asthma. The association of the risk-alleles in the 17q21 locus is strong, with p values below 0.001 in most cases. However while the increased risk is significant, it is somewhat moderate, with the increased risk, as expressed as an Odds ratio, of around 1.5, depending on the haplotype. The risk alleles are highly prevalent in many populations. One of the most highly cited SNPs, rs7216389, has an almost equal representation by the risk and non-risk genotypes in European populations. Clearly, the expression of the elevated risk of asthma imparted by the genetic effect at this locus is highly dependent on other genetic influences as well as strong environmental components. This presents a considerable challenge in understanding the role of these genetic effects at a mechanistic level.
7.3. Connecting the risk alleles in 17q21 to asthma: Expression levels of ORMDL3 and Gasdermin B
The SNPs associated with increased risk for asthma lie in a region that is adjacent to the genes for both ORMDL3 and Gasdermin B, but are far outside of the coding regions for both. The effects of the genotype in this region therefore must affect expression of these genes, either by influencing enhancer or silencing sequences or by altering chromatin structure. Unfortunately, to date, no clear picture has emerged as to which of these mechanisms may be operating (Berlivet et al. 2012; Verlaan et al. 2009) although a change in CpG island methylation has been observed (Kothari et al. 2018). In principal either ORMDL3 or Gasdermin B or both may be the relevant protein that mediates the genetic effects at this locus. Determining which gene (or both) are affected by the genetic elements in 17q21 is challenging. The samples must come from clinical studies (mice do not share these SNPs) and so are confined to analysis of peripheral blood, which may not be the relevant tissue. Moreover there will other genetic and environmental influences on expression of these proteins that will further complicate the analysis.
Nevertheless in the original study by Moffat and co-workers, expression levels of these proteins were determined in peripheral blood leukocytes from a large cohort. This established a strong correlation between the risk alleles and elevated expression of ORMDL3, but not Gasdermin B, at the mRNA level (Moffatt et al. 2007). This observation sparked the investigations of the role of ORMDL3 in asthma, outlined below. However subsequent studies have yielded more ambiguous results. Several have demonstrated a correlation between risk alleles and an increased expression of Gasdermin B as well as ORMDL3 (Kothari et al. 2018; Carreras-Sureda et al. 2013). The role of elevated Gasdermin B expression in asthma has not received the attention that ORMDL3 has attracted, however a recent study suggests that Gasdermin B may also play a role in the asthmatic phenotype (Das et al. 2016). In fact quite early after the original observation by Moffat and co-workers it was proposed that it was the genetic influence in the entire 17q21 region that may result in increased asthma risk rather than an effect on any individual gene (Wjst 2008).
7.4. ORMDL levels are induced by inflammatory and allergic stimulation in the airway
One potential mechanism underlying the influence of SNPs on ORMDL3 expression is that these effects are magnified by a stimulated ORMDL expression in all genotypes under allergic or inflammatory conditions. Indeed an increase in ORMDL expression has been reported for viral infection (Liu et al. 2017; Wang et al. 2017), by inflammatory cytokines in eosinophils (Ha et al. 2013), lipopolysaccharide in macrophages (Miller et al. 2012; Oyeniran et al. 2015), and in lung tissue in house dust mite and ovalbumin challenged mouse models (Miller et al. 2012; Oyeniran et al. 2015). These elevations are specific for ORMDL3 and not the other ORMDL isoforms at the RNA level. At least in the case rhinovirus infection, the risk genotype at the rs7216389 demonstrates an increased expression of ORMDL3 relative to the non-risk genotype (Schmiedel et al. 2016).
7.5. ORMDL3 levels impact the asthmatic phenotype in animal models of the disease
As noted above, an increase in ORMDL3 expression has been correlated with the risk alleles for asthma (Moffatt et al. 2007). Consequently a number of investigators turned their attention to the mechanism by which elevated expression of ORMDL3 could influence the asthmatic phenotype.Workers have approached this through two major strands of research. One approach is to directly test the effects of artificially elevated ORMDL3 levels in murine models of asthma. A second avenue is to investigate the effects of elevated ORMDL expression on cellular physiology and to leverage those findings into animal models of the disease.
The effect of overexpression of ORMDL3 has been tested in several murine models of asthma. Miller and co-workers produced a mouse line globally overexpressing ORMDL3 and tested the effect of this overexpression in an ovalbumin model of allergic asthma (Miller et al. 2014). These workers found that ORMDL3 overexpression markedly promoted airway remodeling, resulting in increased airway smooth muscle, collagen deposition, and mucus secretion. Moreover there were increases in the inflammatory marker TGF-beta and the metalloprotease ADAM8 and an increase in lung macrophages, eosinophils, and neutrophils. They also observed an increase in serum IgE, the immunoglobulin long associated with asthma. This phenotype strongly suggests that, even prior to antigen challenge, ORMDL3 overexpression primes these animals for an asthmatic response. The animals were challenged with ovalbumin, a traditional (but not uncontroversial) model of allergic asthma. After challenge they observed a marked increase, relative to wildtype animals, in airway hyper-responsiveness to methacholine, a standard measure of the asthmatic phenotype. Importantly they noted that ORMDL3 overexpression exacerbated the ovalbumin-induced elevation of the asthma-associated cytokines IL-4 and IL-13 and enhanced lung-infiltration of eosinophils. Taken together, these data indicate that high levels of ORMDL3 expression pre-dispose these animals to an asthmatic phenotype. These results were derived from a globally overexpressing ORMDL3 model, leaving unresolved in which cell types and tissues overexpression of ORMDL3 was having its effects. To address this issue this group, in a subsequent publication (Chen et al. 2018) used irradiation to ablate the hematopoietic lineage, and reconstituted the ORMDL3 overexpressing mice with wild-type bone marrow cells. This would distinguish effects of immune cells from those of other tissues, such as in the lung itself. They observed that the phenotype of the ORMDL3 overexpressing mice reconstituted with wild-type bone marrow was identical with the whole animal overexpressors, although they lacked the lung inflammation that they had observed earlier. This strongly suggests that the effects of ORMDL3 overexpression was directly on the lung tissue, including the lung epithelium and the underlying smooth muscle, rather than the immune/inflammatory component of their model.
While the ovalbumin-sensitized model of asthma has been a traditional approach in the field, there has been a shift to antigens that are more relevant to the human disease. Loser and colleagues have utilized the fungus Alternaria, an established asthma-associated allergen, to sensitize mice and test the role of ORMDL3 (Loser et al. 2017). In contrast to the studies of Miller and co-workers, who looked at the effect of overexpressing ORMDL3, this group produced a global knockout of ORMDL3. Their results were consistent with an important role for ORMDL3 for the asthmatic phenotype. Alternaria-challenged, ORMDL3 depleted mice had reduced airway hyper responsiveness as compared to the wild-type mice. There was no change, however, in the in allergic markers, such as IL-4 and −13 and eosinophilia. This suggests that ORMDL3 expression has a direct effect on the airway tissue. To test the tissue specificity of the ORMDL3 effect, these workers re-introduced ORMDL3 into the knockout animals by instilling adenovirus driving ORMDL3 expression into the lung. They found that this reconstituted the wild-type response to Alternaria challenge. They did note that they did not see an effect of ORMDL3 knockout in an alternative asthma model which uses house dust-mite challenge as the allergen. This suggests that the involvement of ORMDL3 may be allergen-dependent. This is further emphasized by a recent manuscript from Miller and colleagues in which the effect of an airway epithelium-specific ORMDL knockout was tested in the ovalbumin-challenge model (Miller et al. 2017). In that model these investigators found an increased sensitivity with the loss of ORMDL3, the opposite result to that found with Alternaria. These opposing results may be the result of the different allergens used or may reflect the different genetic models constructed. Currently this dichotomy has yet to be definitively explained.
7.6. Sphingolipids as mediators of the effect of elevated ORMDL3 in asthma
As outlined above, the major cellular role of the ORMDL proteins is to regulate sphingolipid biosynthesis as sphingolipid-dependent inhibitors of the initiating enzyme in the pathway, serine palmitoyltransferase (SPT). It would be reasonable to conclude that as an inhibitor of SPT, elevating ORMDL3 would inhibit sphingolipid synthesis (although as discussed below, this supposition may be questionable.) Worgall and colleagues therefore tested whether reduced sphingolipid biosynthesis exacerbates the asthmatic phenotype (Worgall et al. 2013). They utilized both pharmacological and genetic approaches in the intact animal. The pharmacological approach used myriocin, a specific inhibitor of SPT. To genetically reduce SPT activity, these workers produced a heterozygous knockout of one of the SPT subunits, as a homozygous knockout was embryonic lethal. Both approaches yielded an increase in airway hypersensitivity to methacholine challenge without inducing an inflammatory reaction. Allergen sensitivity was not tested in this model. However these results are consistent with reduced sphingolipid levels mediating the effects of elevated ORMDL3 expression in asthma.
A different picture emerges from the study by Oyeniran and co-workers (Oyeniran et al. 2015). This study utilized the house dust-mite (HDM) model of allergic asthma. These workers found that HDM challenge led to a profound increase in ORMDL protein levels in the lung. It should be noted that because of the high homology between ORMDL isoforms, antibodies cannot distinguish between them. This elevation of ORMDL was accompanied by an increase in lung ceramide. This was not totally unexpected, despite the known role of ORMDLs as inhibitors of SPT. Previous work has shown that in many cell types, elevation of ORMDLs does not further inhibit SPT. Why not? It is the stoichiometry between ORMDL and SPT that is important. In many cell types it appears that there is sufficient endogenous ORMDL to regulate SPT and increasing those levels has no effect (Siow, Sunkara, Dunn, et al. 2015). Indeed, Oyeniran and co-workers found that overexpression of ORMDL3 in A549 lung adenocarcinoma cells increases ceramide levels. These investigators therefore surmised that an ORMDL-dependent elevation of ceramide might underlie the increased risk for asthma. To test this, they inhibited ceramide synthesis with myriocin and the ceramide synthase inhibitor Fumonisin B1, and tested their effects in the HDM model. Consistent with their hypothesis, these inhibitors reduced airway hyper responsiveness.
7.7. Non-sphingolipid related effects of ORMDL3
The emphasis on ORMDL3 effects of sphingolipid metabolism in asthma is reasonable given the known roles of sphingolipids as signaling molecules. However other potential ORMDL3 regulatory roles should also be considered. ORMDL3 overexpression in airway epithelia increases levels of the ER calcium transporter SERCA-2B (Chen et al. 2018; Miller et al. 2014) and stimulates one arm of the unfolded protein response (Miller et al. 2012; Cantero-Recasens et al. 2010). The effect of ORMDL3 overexpression on SERCA-2B is consistent with the observation that resting calcium levels are elevated in ORMDL3 overexpressing cells (Cantero-Recasens et al. 2010). These effects have been replicated in animals with global ORMDL3 overexpression (Chen et al. 2018; Miller et al. 2014). The effects of ORMDL3 on calcium levels may be particularly relevant for the hyper responsive phenomenon. Chen and co-workers found that ORMDL3 overexpression in cultured smooth muscle increased contractility in response to acetylcholine and in the airway of globally ORMDL3 overexpressing animals, there was increased calcium oscillations in response to methacholine that accompany increased airway restriction. Direct tests of the role of these changes in asthma and the degree to which they are dependent on ORMDL regulation of sphingolipid biosynthesis have yet to be performed.
8. Future Directions
In the preceding sections we have summarized what is known to date about the function and mechanisms of action of the Orms (in yeast) and ORMDLs (in mammals) in controlling sphingolipid biosynthesis. As with any system, the deeper we dive, the more questions emerge.
The yeast Orms, as outlined in Section 5, are controlled by several phosphorylation systems (Table 1). Yeast, as free-living organisms, are evolved to respond to variations in their environment. The TORC pathways, classic mediators of changes in nutrient status, control at least two phosphorylation cascades that impact on Orm regulation. There are additional responses to various states of cell stress. It will be intriguing to discover how these responses are integrated and if there is a hierarchy. Which responses are most important to yeast survival and how does this regulatory system decide? The ultimate result of regulation of SPT by the Orms are changes in sphingolipid production and content. Ultimately, understanding the physiological impact of these changes under various conditions will illuminate how and when the Orms perform their regulatory function.
We note in Section 6.2 that it remains unclear why there are three ORMDL isoforms and discuss what their individual functions might be. Future studies, both in cultured cells, as well as using genetic manipulation in intact animals, where single isoforms of ORMDL are depleted, will be needed to tease out the individual roles of the three isoforms in mammals.
There is also a fascinating cell biological puzzle to be solved. What and where are the sphingolipid pools that are monitored by this system? The ORMDLs and SPT reside in the ER, although it cannot be ruled out that other organelles harbor these proteins at levels that are not readily visible by immunofluorescence. Is it only levels of sphingolipid in the ER that are monitored, or are sphingolipid levels detected in other organelles, such as the plasma membrane? Sensing of plasma membrane sphingolipid levels by the ER-localized ORMDL/SPT complex could be accomplished, for example, by localization of the ORMDL/SPT complex to contact sites between the ER and plasma membrane. Answering these questions will require a combination of advanced microscopy techniques and organelle-specific manipulation of sphingolipid levels.
The mammalian ORMDLs are regulated quite differently from the yeast Orms. They are not phosphorylated and instead directly respond to changes in sphingolipid levels by an as yet undiscovered mechanism. There are significant technical challenges to discovering the mechanism by which the ORMDL/SPT complex senses and responds to changes in cellular sphingolipid levels. All of the members of this complex are membrane embedded, so techniques that have been applied to analyze soluble enzyme/regulator complexes are not useful. And yet there are key questions to be answered. What is/are the sphingolipid(s) to which this system responds? What is the mechanism of detection? Is there a direct binding interaction or is the system responding to sphingolipid-dependent changes in the lipid environment? These questions will require the development of reconstituted systems in which the components can be analyzed in a more biochemically tractable system. The structural basis of the inhibition of SPT activity by ORMDL is also an open question. The active site of SPT is formed by the contact of the two major subunits of SPT, SPTLC-1 and −2 and the activity of the enzyme is greatly enhanced by the association with the small accessory subunits, ssSPTa and ssSPTb. A complete understanding of the sphingolipid-induced changes in the conformation of the ORMDL/SPT complex that result in SPT regulation will await structural determination of the complex.
The changes that have been noted under various conditions in the protein levels of the ORMDLs (Section 6.4) deserve considerable future scrutiny. Both the mechanisms that trigger these changes (transcriptional, degradation, translation), when and where they happen, and the consequences of this regulatory apparatus all need further study. As in yeast, the ORMDLs appear to function beyond their homeostatic role to control sphingolipid biosynthesis. We are only beginning to understand the mechanisms that control their function and under what circumstances that control operates. While all cells require sphingolipids to function, certain tissues are especially sphingolipid-dependent. These include skin, in which ceramides form the permeability barrier (Meckfessel and Brandt 2014) and in myelination in both the central and peripheral nervous systems (Schmitt, Castelvetri, and Simons 2015). It will be fascinating to explore how the ORMDL control of sphingolipid metabolism impacts on these processes and how manipulation of ORMDL function might be a useful strategy in the many diseases that involve those tissues.
Supplementary Material
Acknowledgements
Abbreviations
- SPT
serine palmitoyltransferase
- KDSR
3-ketodihydrospingosine reductase
- CERS
ceramide synthase
- DES
dihydroceramide desaturase
- CD
ceramidase
- CGS
ceramide glucosyltransferase
- CGT
ceramide galactosyltransferase
- SMS
sphingomyelin synthase
- SPL
sphingosine-1-phospate lyase
- SK
sphingosine kinase
- S1P
sphingosine-1-phosphate
- ER
endoplasmic reticulum
- PM
plasma membrane
- SPTLC1
SPT large subunit 1
- SPTLC2
SPT large subunit 2
- TORC1
target of rapamycin complex 1
- TORC2
target of rapamycin complex 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest Conflicts
The authors of this manuscript have no conflicts of interest to report.
References:
- Aguilar A, and Saba JD. 2012. ‘Truth and consequences of sphingosine-1-phosphate lyase’, Adv. Biol. Regul, 52: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding AL, and Baehrecke EH. 2017. ‘Cleaning House: Selective Autophagy of Organelles’, Dev Cell, 41: 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki W, Takahashi-Sasaki N, Chui DH, Saito S, Takeda K, Shirotani K, Takahashi K, Murayama KS, Kametani F, Shiraishi H, Komano H, and Tabira T. 2008. ‘A family of membrane proteins associated with presenilin expression and gamma-secretase function’, FASEB J, 22: 819–27. [DOI] [PubMed] [Google Scholar]
- Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A, Kwan T, Sinnett D, Pastinen T, and Naumova AK. 2012. ‘Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines’, Hum Genet, 131: 1161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsting C, Hummel R, Schultz ER, Rose TM, Pedersen MB, Knudsen J, and Kristiansen K. 1997. ‘Saccharomyces carlsbergensis contains two functional genes encoding the acyl-CoA binding protein, one similar to the ACB1 gene from S. cerevisiae and one identical to the ACB1 gene from S. monacensis’, Yeast, 13: 1409–21. [DOI] [PubMed] [Google Scholar]
- Bourquin F, Capitani G, and Grutter MG. 2011. ‘PLP-dependent enzymes as entry and exit gates of sphingolipid metabolism’, Protein Sci, 20: 1492–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, and Weissman JS. 2008. ‘A comprehensive strategy enabling high-resolution functional analysis of the yeast genome’, Nat Methods, 5: 711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, and Weissman JS. 2010. ‘Orm family proteins mediate sphingolipid homeostasis’, Nature, 463: 1048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice SE, Alford CW, and Cowart LA. 2009. ‘Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae’, J Biol Chem, 284: 7588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Oyeniran C, Biswas DD, Allegood J, Milstien S, Kordula T, Maceyka M, and Spiegel S. 2016. ‘ORMDL proteins regulate ceramide levels during sterile inflammation’, J Lipid Res, 57: 1412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo A, Zhang Y, Kothiya M, Galvani S, Obinata H, Bucci M, Giordano FJ, Jiang XC, Hla T, and Di Lorenzo A. 2015. ‘Nogo-B regulates endothelial sphingolipid homeostasis to control vascular function and blood pressure’, Nat Med, 21: 1028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, and Vicente R. 2010. ‘The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress’, Hum. Mol. Genet, 19: 111–21. [DOI] [PubMed] [Google Scholar]
- Carreras-Sureda A, Cantero-Recasens G, Rubio-Moscardo F, Kiefer K, Peinelt C, Niemeyer BA, Valverde MA, and Vicente R. 2013. ‘ORMDL3 modulates store-operated calcium entry and lymphocyte activation’, Hum Mol Genet, 22: 519–30. [DOI] [PubMed] [Google Scholar]
- Castro BM, Prieto M, and Silva LC. 2014. ‘Ceramide: a simple sphingolipid with unique biophysical properties’, Prog Lipid Res, 54: 53–67. [DOI] [PubMed] [Google Scholar]
- Castro BM, Silva LC, Fedorov A, de Almeida RF, and Prieto M. 2009. ‘Cholesterol-rich fluid membranes solubilize ceramide domains: implications for the structure and dynamics of mammalian intracellular and plasma membranes’, J. Biol. Chem, 284: 22978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N, Han G, Somashekarappa N, Gable K, Dunn T, and Kohlwein SD. 2017. ‘Regulation of sphingolipid biosynthesis by the morphogenesis checkpoint kinase Swe1’, J Biol Chem, 292: 9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Miller M, Unno H, Rosenthal P, Sanderson MJ, and Broide DH. 2018. ‘Orosomucoid-like 3 (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca(2+) oscillations in asthma’, J Allergy Clin Immunol, 142: 207–18 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueasiri C, Chunthong K, Pitnjam K, Chakhonkaen S, Sangarwut N, Sangsawang K, Suksangpanomrung M, Michaelson LV, Napier JA, and Muangprom A. 2014. ‘Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development’, PLoS One, 9: e106386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Miller M, Beppu AK, Mueller J, McGeough MD, Vuong C, Karta MR, Rosenthal P, Chouiali F, Doherty TA, Kurten RC, Hamid Q, Hoffman HM, and Broide DH. 2016. ‘GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation’, Proc Natl Acad Sci U S A, 113: 13132–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Miller M, and Broide DH. 2017. ‘Chromosome 17q21 Genes ORMDL3 and GSDMB in Asthma and Immune Diseases’, Adv Immunol, 135: 1–52. [DOI] [PubMed] [Google Scholar]
- Espaillat MP, Kew RR, and Obeid LM. 2017. ‘Sphingolipids in neutrophil function and inflammatory responses: Mechanisms and implications for intestinal immunity and inflammation in ulcerative colitis’, Adv Biol Regul, 63: 140–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K, Slife H, Bacikova D, Monaghan E, and Dunn TM. 2000. ‘Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity’, J Biol Chem, 275: 7597–603. [DOI] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K, Wakabayashi H, Ivanova T, Castillon GA, Piccolis M, Abe F, Loewith R, Funato K, Wenk MR, and Riezman H. 2009. ‘Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology’, Mol Biol Cell, 20: 2083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SD, Gable K, Alexaki A, Chandris P, Proia RL, Dunn TM, and Harmon JM. 2015. ‘Expression of the ORMDLS, Modulators of Serine Palmitoyltransferase, Is Regulated by Sphingolipids in Mammalian Cells’, J. Biol. Chem, 290: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj C, Federman RS, and Chang A. 2013. ‘Orm proteins integrate multiple signals to maintain sphingolipid homeostasis’, J. Biol. Chem, 288: 20453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP, and Sriramarao P. 2013. ‘ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48’, Nat Commun, 4: 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RH Jr., Harmon JM, and Dunn TM. 2009. ‘Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities’, Proc. Natl. Acad. Sci. U. S. A, 106: 8186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, and Chang A. 2010. ‘Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control’, Proc. Natl. Acad. Sci. U. S. A, 107: 5851–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K 2003. ‘Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism’, Biochim Biophys Acta, 1632: 16–30. [DOI] [PubMed] [Google Scholar]
- Hannun YA, and Obeid LM. 2002. ‘The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind’, J Biol Chem, 277: 25847–50. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Dunn TM, and Campopiano DJ. 2018. ‘Sphingolipid biosynthesis in man and microbes’, Nat Prod Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum D, Haddadi N, Lin Y, Nassif NT, and McGowan EM. 2017. ‘Mammalian sphingosine kinase (SphK) isoenzymes and isoform expression: challenges for SphK as an oncotarget’, Oncotarget, 8: 36898–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, and Gonzalez-Duarte R. 2002. ‘ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins’, Genome Biol, 3: research0027-research27.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann T, Wei Y, and Eckardstein A von. 2007. ‘Is the mammalian serine palmitoyltransferase a high-molecular-mass complex?’, Biochem. J, 405: 157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, To M, Tran E, Dionisio LTA, Cho HJ, Baney KLM, Pataki CI, and Olzmann JA. 2018. ‘A VCP inhibitor substrate trapping approach (VISTA) enables proteomic profiling of endogenous ERAD substrates’, Mol Biol Cell, 29: 1021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer K, Carreras-Sureda A, Garcia-Lopez R, Rubio-Moscardo F, Casas J, Fabrias G, and Vicente R. 2015. ‘Coordinated Regulation of the Orosomucoid-like Gene Family Expression Controls de Novo Ceramide Synthesis in Mammalian Cells’, J. Biol. Chem, 290: 2822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin AN, Han G, Luttgeharm KD, Chen M, Cahoon RE, Stone JM, Markham JE, Dunn TM, and Cahoon EB. 2016. ‘ORM Expression Alters Sphingolipid Homeostasis and Differentially Affects Ceramide Synthase Activity’, Plant Physiol, 172: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin AN, Majumder S, Han G, Chen M, Cahoon RE, Stone JM, Dunn TM, and Cahoon EB. 2013. ‘Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity’, Plant Cell, 25: 4627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T, and Sandhoff K. 2006. ‘Sphingolipid metabolism diseases’, Biochim. Biophys. Acta, 1758: 2057–79. [DOI] [PubMed] [Google Scholar]
- Kothari PH, Qiu W, Croteau-Chonka DC, Martinez FD, Liu AH, Lemanske RF Jr., Ober C, Krishnan JA, Nicolae DL, Barnes KC, London SJ, Barraza-Villarreal A, White SR, Naureckas ET, Millstein J, Gauderman WJ, Gilliland FD, Carey VJ, Weiss ST, and Raby BA. 2018. ‘Role of local CpG DNA methylation in mediating the 17q21 asthma susceptibility gasdermin B (GSDMB)/ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) expression quantitative trait locus’, J Allergy Clin Immunol, 141: 2282–86.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yin J, Rong C, Li KE, Wu JX, Huang LQ, Zeng HY, Sahu SK, and Yao N. 2016. ‘Orosomucoid Proteins Interact with the Small Subunit of Serine Palmitoyltransferase and Contribute to Sphingolipid Homeostasis and Stress Responses in Arabidopsis’, Plant Cell, 28: 3038–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Huang C, Polu SR, Schneiter R, and Chang A. 2012. ‘Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast’, J. Cell Sci, 125: 2428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Rajamanikham V, Baron M, Patel S, Mathur SK, Schwantes EA, Ober C, Jackson DJ, Gern JE, Lemanske RF Jr., and Smith JA. 2017. ‘Association of ORMDL3 with rhinovirus-induced endoplasmic reticulum stress and type I Interferon responses in human leucocytes’, Clin Exp Allergy, 47: 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, and Hall MN. 2002. ‘Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control’, Mol Cell, 10: 457–68. [DOI] [PubMed] [Google Scholar]
- Loser S, Gregory LG, Zhang Y, Schaefer K, Walker SA, Buckley J, Denney L, Dean CH, Cookson WOC, Moffatt MF, and Lloyd CM. 2017. ‘Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease’, J Allergy Clin Immunol, 139: 1496–507.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, and Spiegel S. 2014. ‘Sphingolipid metabolites in inflammatory disease’, Nature, 510: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon EC, van Echten G, Birk R, Schmidt RR, and Sandhoff K. 1991. ‘Sphingolipid biosynthesis in cultured neurons. Down-regulation of serine palmitoyltransferase by sphingoid bases’, Eur J Biochem, 198: 667–74. [DOI] [PubMed] [Google Scholar]
- Marcus J, Honigbaum S, Shroff S, Honke K, Rosenbluth J, and Dupree JL. 2006. ‘Sulfatide is essential for the maintenance of CNS myelin and axon structure’, Glia, 53: 372–81. [DOI] [PubMed] [Google Scholar]
- Meckfessel MH, and Brandt S. 2014. ‘The structure, function, and importance of ceramides in skin and their use as therapeutic agents in skin-care products’, J. Am. Acad. Dermatol, 71: 177–84. [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr. 2011. ‘Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics’, Chem Rev, 111: 6387–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill AH Jr., Nixon DW, and Williams RD. 1985. ‘Activities of serine palmitoyltransferase (3-ketosphinganine synthase) in microsomes from different rat tissues’, J. Lipid Res, 26: 617–22. [PubMed] [Google Scholar]
- Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, Doherty TA, McGeough MD, Pena CA, Suzukawa M, Niwa M, and Broide DH. 2014. ‘ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma’, Immunol, 192: 3475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, Rosenthal P, Mueller JL, Hoffman HM, Suzukawa M, Niwa M, and Broide DH. 2012. ‘ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6’, Proc Natl Acad Sci S A, 109: 16648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Tam AB, Mueller JL, Rosenthal P, Beppu A, Gordillo R, McGeough MD, Vuong C, Doherty TA, Hoffman HM, Niwa M, and Broide DH. 2017. ‘Cutting Edge: Targeting Epithelial ORMDL3 Increases, Rather than Reduces, Airway Responsiveness and Is Associated with Increased Sphingosine-1-Phosphate’, J Immunol, 198: 3017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, and Cookson WO. 2007. ‘Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma’, Nature, 448: 470–73. [DOI] [PubMed] [Google Scholar]
- Obeid LM, Linardic CM, Karolak LA, and Hannun YA. 1993. ‘Programmed cell death induced by ceramide’, Science, 259: 1769–71. [DOI] [PubMed] [Google Scholar]
- Ober C, and Yao TC. 2011. ‘The genetics of asthma and allergic disease: a 21st century perspective’, Immunol. Rev, 242: 10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B 2018. ‘Sphingolipid metabolism in cancer signalling and therapy’, Nat Rev Cancer, 18: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen ASB, and Faergeman NJ. 2017. ‘Sphingolipids: membrane microdomains in brain development, function and neurological diseases’, Open Biol, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono JG, Worgall TS, and Worgall S. 2014. ‘17q21 locus and ORMDL3: an increased risk for childhood asthma’, Pediatr Res, 75: 165–70. [DOI] [PubMed] [Google Scholar]
- Oyeniran C, Sturgill JL, Hait NC, Huang WC, Avni D, Maceyka M, Newton J, Allegood JC, Montpetit A, Conrad DH, Milstien S, and Spiegel S. 2015. ‘Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice’, J Allergy Clin Immunol, 136: 1035–46.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulenda T, and Draber P. 2016. ‘The role of ORMDL proteins, guardians of cellular sphingolipids, in asthma’, Allergy, 71: 918–30. [DOI] [PubMed] [Google Scholar]
- Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, and Merrill AH Jr. 2008. ‘Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols’, J Lipid Res, 49: 1621–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne S, Adams DR, and Pyne NJ. 2016. ‘Sphingosine 1-phosphate and sphingosine kinases in health and disease: Recent advances’, Prog Lipid Res, 62: 93–106. [DOI] [PubMed] [Google Scholar]
- Qu YL, Ji YR, Zhang LX, Wu CM, Wen BL, Zhang X, Ma C, Wang DM, Zhang YX, and Zhou X. 2018. ‘17q21 locus rs7216389 polymorphism and childhood asthma risk: a meta-analysis’, Minerva Pediatr, 70: 98–102. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, and Thorner J. 2004. ‘Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9’, Microbiology, 150: 3289–304. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, and Thorner J. 2011. ‘Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae’, Proc. Natl. Acad. Sci. U. S. A, 108: 19222–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanllehi P, Abad JL, Casas J, and Delgado A. 2016. ‘Inhibitors of sphingosine-1-phosphate metabolism (sphingosine kinases and sphingosine-1-phosphate lyase)’, Chem Phys Lipids, 197: 69–81. [DOI] [PubMed] [Google Scholar]
- Sasset L, Zhang Y, Dunn TM, and Di Lorenzo A. 2016. ‘Sphingolipid De Novo Biosynthesis: A Rheostat of Cardiovascular Homeostasis’, Trends Endocrinol Metab, 27: 807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel BJ, Seumois G, Samaniego-Castruita D, Cayford J, Schulten V, Chavez L, Ay F, Sette A, Peters B, and Vijayanand P. 2016. ‘17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells’, Nat Commun, 7: 13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Castelvetri LC, and Simons M. 2015. ‘Metabolism and functions of lipids in myelin’, Biochim Biophys Acta, 1851: 999–1005. [DOI] [PubMed] [Google Scholar]
- Shamseddine AA, Airola MV, and Hannun YA. 2015. ‘Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes’, Adv Biol Regul, 57: 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Oppliger W, Moes S, Jeno P, and Hall MN. 2013. ‘TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis’, Mol. Biol. Cell, 24: 870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siow D, Sunkara M, Dunn TM, Morris AJ, and Wattenberg B. 2015. ‘ORMDL/serine palmitoyltransferase stoichiometry determines effects of ORMDL3 expression on sphingolipid biosynthesis’, J Lipid Res, 56: 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siow D, Sunkara M, Morris A, and Wattenberg B. 2015. ‘Regulation of de novo sphingolipid biosynthesis by the ORMDL proteins and sphingosine kinase-1’, Adv Biol Regul, 57: 42–54. [DOI] [PubMed] [Google Scholar]
- Siow DL, and Wattenberg BW. 2012. ‘Mammalian ORMDL Proteins Mediate the Feedback Response in Ceramide Biosynthesis’, J. Biol. Chem, 287: 40198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, Igartua C, Morin A, Washington C 3rd, Nicolae D, Bonnelykke K, and Ober C. 2018. ‘A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle’, J Allergy Clin Immunol [DOI] [PMC free article] [PubMed]
- Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, and Drubin DG. 2012. ‘Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways’, Mol. Biol. Cell, 23: 2388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue D, Kobayashi T, Sun Y, Fujita T, Takematsu H, and Kozutsumi Y. 2005. ‘The requirement for the hydrophobic motif phosphorylation of Ypk1 in yeast differs depending on the downstream events, including endocytosis, cell growth, and resistance to a sphingolipid biosynthesis inhibitor, ISP-1’, Arch Biochem Biophys, 437: 29–41. [DOI] [PubMed] [Google Scholar]
- Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, Hoberman R, Swiatek M, Dias J, Lam KC, Koka V, Harmsen E, Soto-Quiros M, Avila L, Celedon JC, Weiss ST, Dewar K, Sinnett D, Laprise C, Raby BA, Pastinen T, and Naumova AK. 2009. ‘Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease’, Am J Hum Genet, 85: 377–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpert G, Ben-Dor S, Tarcic O, Duan J, Saada A, Merrill AH Jr., Pewzner-Jung Y, and Futerman AH. 2017. ‘Oxidative stress elicited by modifying the ceramide acyl chain length reduces the rate of clathrin-mediated endocytosis’, J Cell Sci, 130: 1486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC 2010. ‘Keeping sphingolipid levels nORMal’, Proc Natl Acad Sci U S A, 107: 5701–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Robinet P, Smith JD, and Gulshan K. 2015. ‘ORMDL orosomucoid-like proteins are degraded by free-cholesterol-loading-induced autophagy’, Proc Natl Acad Sci U S A, 112: 3728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Shu J, Jiang CM, Zhuang LL, Yang WX, Zhang HW, Wang LL, Li L, Chen XQ, Jin R, and Zhou GP. 2017. ‘Mechanisms and roles by which IRF-3 mediates the regulation of ORMDL3 transcription in respiratory syncytial virus infection’, Int J Biochem Cell Biol, 87: 8–17. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S, Yates J 3rd, McCaffery JM, and Powers T. 2003. ‘Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae’, Mol Biol Cell, 14: 1204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wjst M 2008. ‘ORMDL3--guilt by association?’, Clin Exp Allergy, 38: 1579–81. [DOI] [PubMed] [Google Scholar]
- Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, Silver RB, Jiang XC, and Worgall S. 2013. ‘Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity’, Sci. Transl. Med, 5: 186ra67. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, and Hall MN. 2006. ‘TOR signaling in growth and metabolism’, Cell, 124: 471–84. [DOI] [PubMed] [Google Scholar]
- Zhao CN, Fan Y, Huang JJ, Zhang HX, Gao T, Wang C, Wang T, and Hou LF. 2015. ‘The Association of GSDMB and ORMDL3 Gene Polymorphisms With Asthma: A Meta-Analysis’, Allergy Asthma Immunol. Res, 7: 175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


