Abstract
As the sites of communication between neurons, synapses depend upon precisely regulated protein-protein interactions to support neurotransmitter release and reception. Moreover, neuronal synapses typically exist great distances (i.e. up to meters) away from cell bodies, which are the sources of new proteins and the major sites of protein degradation via lysosomes. Thus, synapses are uniquely sensitive to disruptions in proteostasis, and depend upon carefully orchestrated degradative mechanisms for the clearance of dysfunctional proteins [1–3]. One of the primary cellular degradative pathways is macroautophagy, hereafter referred to as ‘autophagy’. Although it has only recently become a focus of research in synaptic biology, emerging studies indicate that autophagy has essential functions at the synapse throughout an organism’s lifetime. This review will discuss recent findings about the roles of synaptic autophagy, as well as some of the questions and issues to be considered in this field moving forward.
Keywords: synapse, autophagy
Autophagy in synapse formation and pruning
Studies in genetic organisms indicate the importance of autophagy and autophagic machinery in early synaptic development. For instance, impairment of autophagy via the knockdown of ATG1, -2, -6, or -18 was found to reduce the number and size of presynaptic terminals at the neuromuscular junction (NMJ) in Drosophila melanogaster [4]. More recently, Stavoe and Colon-Ramos showed that autophagy is required cell-autonomously for presynapse formation in AIY neurons of C. elegans [5]. The authors found that autophagosome biogenesis occurred at developing AIY presynaptic boutons. They then observed that 18 distinct components from every stage of the autophagy pathway – initiation (UNC-15, ATG-13), nucleation (ATG-9), elongation (LGG-1, LGG-3, ATG-3, ATG-4), and retrieval (ATG-2, ATG-9, ATG-18)– were required for the clustering of synaptic vesicle (SV) proteins at nascent presynaptic sites during AIY neuron development [5]. However, these components were not required for SV clustering in other C. elegans neuron types [5], suggesting a cell-specific role for autophagic machinery in presynaptic development. These findings highlight the importance of autophagy for synaptogenesis, and the need for future studies to clarify its roles in different cell types and organisms.
In the mammalian brain, synapse formation is followed by a period of synapse refinement, and autophagy has been shown to play an important role in this process. For example, Tang and colleagues found that autophagy is required for the developmental pruning of dendritic spines, and that this pruning is disrupted in individuals with autism spectrum disorder (ASD)[6]. Intriguingly, the authors found a statistically significant increase in phosphorylated mTOR (p-mTOR) in the brains of ASD patients compared to age-matched controls [6]. Since the activation of mTOR inhibits autophagy, an increase in mTOR activity typically indicates a reduction of autophagy [7]. Indeed, ASD patients exhibited significantly lower levels of LC3-II, a marker for autophagosomes, throughout childhood and adolescence [6]. Moreover, Tang et al. observed that mTOR hyperactivation in mice led to ASD-like behaviors and spine pruning deficits in cortical projection neurons, indicating that autophagy is essential for synaptic pruning, and that defects in autophagy contribute to ASD phenotypes [6].
Overall, these studies show that autophagic machinery is vital for both synapse formation and pruning during development. However, many questions remain. For example, does the autophagic machinery regulate SV clustering via protein degradation or another mechanism? Are other aspects of synapse formation also regulated by this machinery? And most intriguingly, how are autophagy components temporally and spatially regulated to facilitate SV clustering in certain contexts, and synapse pruning or SV degradation (see below) in others? Answers to these questions will lead to a better understanding of how autophagy functions throughout neurodevelopment to facilitate synapse formation and elimination.
Autophagic cargo at the synapse
Mitochondria
The term ‘mitophagy’ was coined by Rodriguez-Enriquez and colleagues in 2006 to describe the selective autophagy of mitochondria [8]. Since then, mitophagy has been shown to be essential for the removal of damaged mitochondria in all eukaryotic cell types, including neurons [9, 10]. Mitochondria are critical sources of ATP at synapses, and indeed several studies indicate that deficient mitophagy leads to synaptic dysfunction [11, 12]. Recent studies implicate the Parkinson’s disease (PD)-related proteins PTEN-induced putative kinase (PINK1) and Parkin, an E3 ubiquitin ligase, as critical mediators of mitophagy in axons and presynaptic terminals of hippocampal neurons [13]. These proteins were previously shown to work in concert to target mitochondria for degradation, as PINK1 localizes to the mitochondrial membrane and upon phosphorylation recruits Parkin to damaged mitochondria [14–16]. Parkin in turn ubiquitinates mitochondrial membrane proteins to promote recruitment of the autophagic machinery [17, 18]. However, more recent studies call into question whether the PINK1/Parkin pathway is essential for triggering the removal of damaged mitochondria from axons and presynaptic boutons. For example, Sung and colleagues found that Parkin deficiency in Drosophila led to the accumulation of abnormal mitochondria in cell bodies, rather than in axons or presynaptic boutons as would be expected if PINK1/Parkin are required for initiating the clearance and retrograde transport of these organelles [19]. Moreover, a study by Lin et al. showed that the removal of stressed mitochondria from axons of hippocampal neurons required the retrograde motor protein syntaphilin but not Parkin [20], suggesting a Parkin-independent mechanism for detecting mitochondrial dysfunction and mediating its retrograde transport prior to mitophagy. Additional evidence from Lee et al. shows that basal mitophagy is unaffected in Pink1 or parkin null Drosophila, although these flies exhibit locomotor defects and dopaminergic neuron loss, demonstrating that PINK1 and Parkin are not essential for in vivo mitophagy [21].
Synaptic Vesicles
Autophagy has also been found to mediate the degradation of synaptic vesicles (SVs). Notably, Hernandez and colleagues found that in dopaminergic neurons, stimulating autophagy with rapamycin decreased SV number and evoked dopamine release, while blocking autophagy via ATG-7 knockout increased dopamine release [22]. These findings suggest that, at least in dopaminergic neurons, induction of autophagy can stimulate SV degradation, while loss of this pathway leads to increased SV release and/or number. However, other studies in Drosophila and mammalian hippocampal neurons, including our own, suggest that the endolysosomal degradative pathway mediates basal and activity-dependent turnover of SV proteins [23–25]. A synthesis of these studies suggests that although activation of autophagy can stimulate SV degradation and regulate neurotransmitter release under some conditions, autophagy may not be the primary mechanism of SV protein degradation. It is also worth noting that the autophagy and endolysosomal pathways are linked, as autophagosomes fuse with late endosomes prior to lysosomal degradation [26–28]. Thus, it can be difficult to parse the roles of the autophagy and endolysosomal pathways in the degradation of specific cargoes, and in the case of synaptic vesicle degradation it is possible that the two pathways work synergistically.
Regardless of the specific substrates, considerable evidence suggests that presynaptic autophagy is tightly regulated and, distinct from basal autophagy, activated only under certain conditions [29]. Indeed, several recent studies have begun to identify the molecular machinery that activates and inhibits autophagy in presynaptic terminals. For example, Okerlund and colleagues report that the active zone cytomatrix proteins Bassoon and Piccolo are negative regulators of presynaptic autophagy in hippocampal neurons [30]. Following up on previous findings that Bassoon/Piccolo double knockdown (DKD) triggered the progressive loss of SV pools by stimulating degradative pathways [31], the authors observed an accumulation of LC3-positive and autophagosome-like vesicles in presynaptic DKD boutons [30]. They subsequently demonstrated that Piccolo and Bassoon regulate autophagy via their suppression of ATG-5, an E3 ubiquitin ligase-like enzyme essential for coupling LC3 to the membrane of nascent autophagosomes. Both active zone proteins directly interact with ATG-5, and its knockdown rescues the increase in LC3-positive structures and concurrent decrease in SV pools seen in DKD neurons [30]. These data indicate that Bassoon/Piccolo act to restrain presynaptic autophagy, and that dysregulated autophagy due to the loss of these proteins stimulates the degradation of SVs. However, it is not yet clear whether autophagy is specifically regulated by Bassoon/Piccolo, or induced by a broader neuronal stress response following the loss of one or both of these critical synaptic structural determinants. More work is needed to clarify the roles of these proteins in regulating presynaptic autophagy and other degradative pathways (e.g. endolysosomal, ubiquitin-proteasome), as well as the involvement of autophagy in presynaptic vesicle degradation under basal conditions.
Additional insights about the molecular regulation of presynaptic autophagy come from two recent studies in the Verstreken lab [32, 33]. Soukup and colleagues showed that autophagic factors are recruited to presynaptic boutons by the endocytic protein EndophilinA (EndoA) following its phosphorylation by LRRK2, a Parkinson’s disease-related kinase [32]. Vanhauwaert et al. found that synaptojanin, a presynaptic lipid phosphatase also important for SV endocytosis, is similarly crucial for autophagy. In Drosophila synapses and neurites of iPSC-derived neurons with mutant synaptojanin, nascent autophagosomes accumulated Atg18a on their membranes and could not mature into functional autophagosomes [33]. Moreover, Parkinson’s disease-associated synaptojanin mutations caused dopaminergic neuron loss in Drosophila [33], linking the dysregulation of presynaptic autophagy to neurodegenerative disease, and highlighting the importance of this process for nervous system health.
Postsynaptic AMPA receptors
Across the synaptic cleft, autophagy also mediates the degradation of postsynaptic cargo, including neurotransmitter receptors. Some of the first evidence for this came from Rowland and colleagues, who showed that GABAA receptors colocalize with autophagosome markers at the C. elegans motor neuron/body wall junction in the absence of neuronal activity, and that post-synaptic GABAA activity is attenuated in an autophagy-dependent manner [34]. Interestingly, the authors also found that this autophagic degradation was selective, as GABAA but not acetylcholine receptors localized to autophagosomes [34]. Subsequently, Matsuda et al. observed that hippocampal and cerebellar AMPA receptors were mistargeted to axonal autophagosomes in the absence of the trafficking adaptor complex AP-4, instead of being delivered to the dendritic plasma membrane [35]. Although this study did not demonstrate autophagy of AMPA receptors in dendrites, a later study reported more direct evidence of AMPA receptor degradation at synapses through the autophagy-lysosomal pathway. Specifically, Shehata and colleagues found that high potassium stimulation or chemical stimuli to induce long-term depression (LTD) briefly increased autophagosome numbers in dendritic shafts and spines, an effect that was partially NMDA receptor-dependent [36]. Moreover, the levels of AMPA receptor subunit GluA1 decreased following chemical LTD, and this degradation was partially mitigated by autophagy inhibitors. These results indicate that postsynaptic autophagy is regulated by neuronal activity, and at least partially responsible for the degradation of AMPA receptors following LTD [36], suggesting a possible role for autophagy in synaptic plasticity mechanisms. However, given the preponderance of evidence demonstrating that AMPA receptors are degraded through the endolysosomal pathway [37–40] and ubiquitin proteasome system (UPS) [41, 42], it will be important to clarify whether autophagy works in parallel with these other pathways, or is stimulated only during specific circumstances, e.g. induction of LTD. Despite many lingering questions, it is clear that autophagy mediates the degradation of important synaptic substrates (Fig. 1), and that future studies are needed to disentangle and clarify the many roles it plays on both sides of the synaptic cleft.
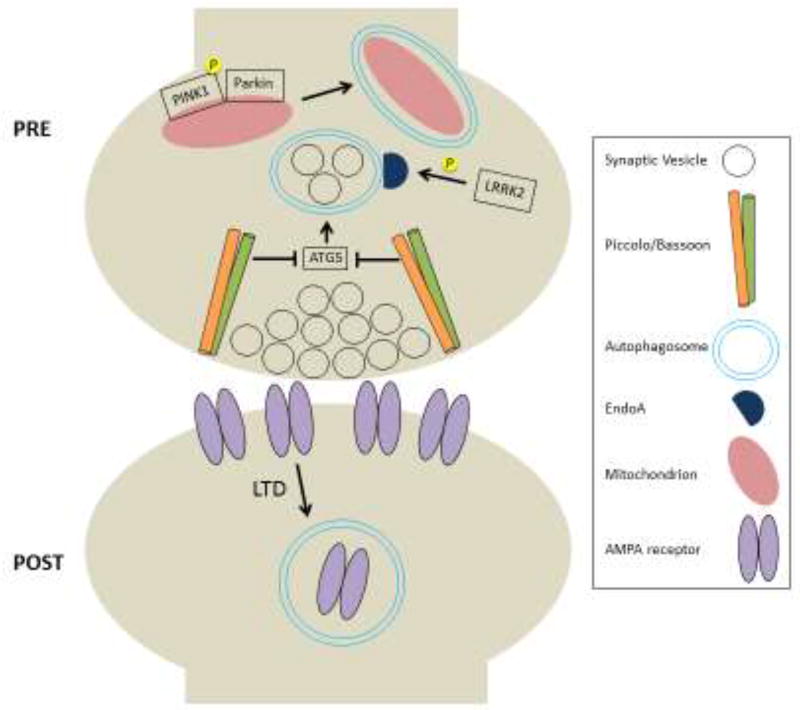
Figure 1. Proposed regulators and cargo of synaptic autophagy.
In the presynaptic terminal, autophagosome formation is promoted by EndophilinA (EndoA) following its phosphorylation by LRRK2. Conversely, the presynaptic structural proteins Bassoon and Piccolo inhibit autophagic degradation of synaptic vesicles via negative regulation of ATG-5. Mitophagy is induced when PINK1 is phosphorylated and recruits Parkin to damaged mitochondria. Postsynaptically, AMPA receptors are degraded via autophagy following induction of LTD.
Outstanding questions about synaptic autophagy
Given the distinct molecular compositions and features of the juxtaposed pre- and postsynaptic compartments, one important but unresolved question is whether pre and post-synaptic autophagosomes have unique characteristics and/or regulatory mechanisms. A recent study by Maday and colleagues supports this idea, as the authors find that autophagosomes originating in axons of hippocampal neurons exhibit distinct characteristics from those originating in the somatodendritic region [43]. The former appear overwhelmingly ‘mature,’ i.e. positive for the late endosome/lysosome marker LAMP1, once they enter the somatodendritic compartment following their retrograde axonal transport, while the latter are less mature and less mobile overall [43]. These findings suggest that neuronal and/or synaptic compartmentalization creates molecularly distinct pools of autophagosomes that exhibit different dynamic behavior and functional properties. Additional work will be needed to determine whether these autophagosomes are indeed molecularly distinct and/or responsive to compartment-specific regulatory stimuli.
This possibility of distinct spatially and conditionally regulated autophagosomes dovetails with mounting evidence that there are multiple classes of LC3-positive structures. Although LC3 is considered a specific and reliable marker of autophagosomes, it is worth noting that LC3 was first identified in rat neurons as the light chain of microtubule-associated proteins MAP1a and MAP1b, components of the neuronal cytoskeleton [44]. Its role in autophagy was recognized only later by Kabeya and colleagues [45]. In keeping with the potentially versatile role of LC3, Kononenko et al. reported that a subset of LC3-positive structures undergoing retrograde transport from presynaptic terminals exhibited colocalization with the endocytic adaptor AP-2 and with BDNF-activated TrkB receptors, which are traditionally found in signaling endosomes [46]. One interpretation of these data is that presynaptic autophagosomes can merge with signaling endosomes, or that these retrograde cargoes are co-transported. Another possibility is that LC3 associates with multiple vesicle types, including autophagosomes, signaling endosomes, and potentially other structures. Interestingly, it was observed by Waites, Okerlund, and colleagues in the Garner lab that Bassoon/Piccolo DKD boutons, shown to be LC3-immunopositive, contained multiple heterogeneous membrane structures, most of which did not resemble classical autophagosomes [30, 31](Fig. 2). It is also worth noting that the other mammalian Atg8 homologs, namely GABARAP and GATE-16, likewise exhibit many functions at the synapse, some unrelated to autophagosome formation. GABARAP in particular has been shown to be involved in the trafficking of GABA-A receptors to the neuronal plasma membrane [47], and has numerous binding partners that are involved in a diversity of dynamic processes underlying synapse formation, maintenance, and plasticity [48]. Together, these studies indicate how much there still is to learn about protein trafficking and degradative pathways at the synapse, and caution against assuming that ‘traditional’ autophagy markers such as LC3 always represent autophagosomes in highly specialized cell types such as neurons. In the future, it will be important to use the most stringent possible criteria for identifying putative autophagosomes. Techniques like correlative light and electron microscopy, and proteomics-based identification of unique molecular markers for pre- and/or postsynaptic autophagosomes will help to expand our repertoire for characterizing and investigating autophagy at the synapse.
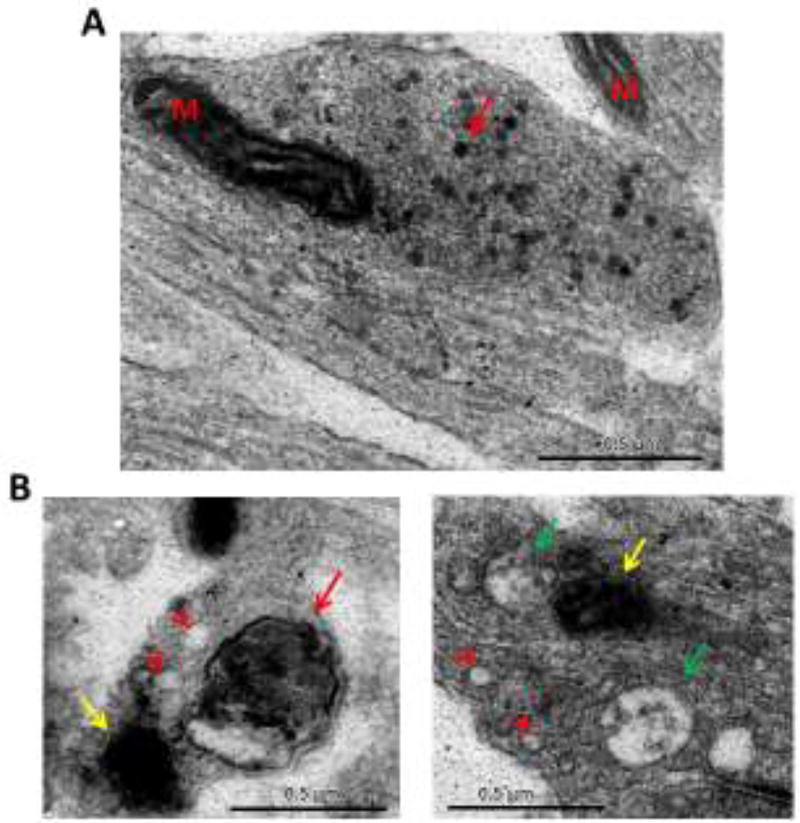
Figure 2. Diverse membrane structures observed at Bassoon/Piccolo DKD synapses.
A) Electron micrograph of a presynaptic terminal expressing VAMP2-HRP to label synaptic vesicles (red arrow). ‘M’ denotes mitochondria. B) Examples of structures found at DKD synapses, shown to have increased autophagy and LC3-positive vesicles. These structures include vacuoles (arrowheads), putative double-membrane autophagosome (red arrow), multivesicular bodies (green arrows), as well as structures that are difficult to characterize due to electron-dense VAMP2-HRP labeling (yellow arrows).
Overall, many questions remain to be answered in the subfield of synaptic autophagy, from identifying cargoes and regulatory mechanisms to deciphering the complex interactions between autophagy and other degradative pathways, and finally understanding how these processes shape neuronal proteostasis and neurotransmission over an organism’s lifetime.
Acknowledgments
This work was funded by NIH/NINDS grant number NS080967 to C. Waites.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bezprozvanny I, Hiesinger PR. The synaptic maintenance problem: membrane recycling, Ca2+ homeostasis and late onset degeneration. Mol Neurodegener. 2013;8:23. doi: 10.1186/1750-1326-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D, et al. Membrane trafficking in neuronal maintenance and degeneration. Cell Mol Life Sci. 2013;70(16):2919–34. doi: 10.1007/s00018-012-1201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YC, Lauwers E, Verstreken P. Presynaptic protein homeostasis and neuronal function. Curr Opin Genet Dev. 2017;44:38–46. doi: 10.1016/j.gde.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187(1):71–9. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stavoe AK, et al. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev Cell. 2016;38(2):171–85. doi: 10.1016/j.devcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang G, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83(5):1131–43. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Enriquez S, et al. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2(1):39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393(7):547–64. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Um JH, Yun J. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017;50(6):299–307. doi: 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebrahimi-Fakhari D, et al. Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell Rep. 2016;17(4):1053–1070. doi: 10.1016/j.celrep.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haddad DM, et al. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell. 2013;50(6):831–43. doi: 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Ashrafi G, et al. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206(5):655–70. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189(2):211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107(1):378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouton-Liger F, et al. PINK1/Parkin-Dependent Mitochondrial Surveillance: From Pleiotropy to Parkinson's Disease. Front Mol Neurosci. 2017;10:120. doi: 10.3389/fnmol.2017.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung H, et al. Compartmentalized Regulation of Parkin-Mediated Mitochondrial Quality Control in the Drosophila Nervous System In Vivo. J Neurosci. 2016;36(28):7375–91. doi: 10.1523/JNEUROSCI.0633-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin MY, et al. Releasing Syntaphilin Removes Stressed Mitochondria from Axons Independent of Mitophagy under Pathophysiological Conditions. Neuron. 2017;94(3):595–610. e6. doi: 10.1016/j.neuron.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JJ, et al. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin; but minimally affected by loss of Pink1 or parkin. The Journal of Cell Biology. 2018;217(5):1613. doi: 10.1083/jcb.201801044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez D, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74(2):277–84. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uytterhoeven V, et al. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 2011;145(1):117–32. doi: 10.1016/j.cell.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 24.Sheehan P, et al. Activity-Dependent Degradation of Synaptic Vesicle Proteins Requires Rab35 and the ESCRT Pathway. J Neurosci. 2016;36(33):8668–86. doi: 10.1523/JNEUROSCI.0725-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandes AC, et al. Reduced synaptic vesicle protein degradation at lysosomes curbs TBC1D24/sky-induced neurodegeneration. J Cell Biol. 2014;207(4):453–62. doi: 10.1083/jcb.201406026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg TO, et al. Isolation and Characterization of Rat Liver Amphisomes: Evidence For Fusion Of Autophagosomes With Both Early And Late Endosomes. Journal of Biological Chemistry. 1998;273(34):21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 27.Liou W, et al. The Autophagic and Endocytic Pathways Converge at the Nascent Autophagic Vacuoles. The Journal of Cell Biology. 1997;136(1):61. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death And Differentiation. 2008;16:70. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 29.Vijayan V, Verstreken P. Autophagy in the presynaptic compartment in health and disease. J Cell Biol. 2017;216(7):1895–1906. doi: 10.1083/jcb.201611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okerlund ND, et al. Bassoon Controls Presynaptic Autophagy through Atg5. Neuron. 2017;93(4):897–913. e7. doi: 10.1016/j.neuron.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 31.Waites CL, et al. Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. The EMBO journal. 2013;32(7):954–69. doi: 10.1038/emboj.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukup SF, et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 2016;92(4):829–844. doi: 10.1016/j.neuron.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 33.Vanhauwaert R, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017;36(10):1392–1411. doi: 10.15252/embj.201695773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland AM, et al. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26(6):1711–20. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda S, et al. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57(5):730–45. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Shehata M, et al. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci. 2012;32(30):10413–22. doi: 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28(2):511–25. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 38.Esteves da Silva M, et al. Positioning of AMPA Receptor-Containing Endosomes Regulates Synapse Architecture. Cell Rep. 2015;13(5):933–43. doi: 10.1016/j.celrep.2015.09.062. [DOI] [PubMed] [Google Scholar]
- 39.Zheng N, et al. Synaptic activity regulates AMPA receptor trafficking through different recycling pathways. Elife. 2015;4 doi: 10.7554/eLife.06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(49):16718–29. doi: 10.1523/JNEUROSCI.3686-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira JS, et al. GluN2B-Containing NMDA Receptors Regulate AMPA Receptor Traffic through Anchoring of the Synaptic Proteasome. J Neurosci. 2015;35(22):8462–79. doi: 10.1523/JNEUROSCI.3567-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goo MS, Scudder SL, Patrick GN. Ubiquitin-dependent trafficking and turnover of ionotropic glutamate receptors. Front Mol Neurosci. 2015;8:60. doi: 10.3389/fnmol.2015.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maday S, Holzbaur EL. Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci. 2016;36(22):5933–45. doi: 10.1523/JNEUROSCI.4401-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269(15):11492–7. [PubMed] [Google Scholar]
- 45.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kononenko NL, et al. Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat Commun. 2017;8:14819. doi: 10.1038/ncomms14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leil TA, et al. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. The Journal of Neuroscience. 2004;24(50):11429. doi: 10.1523/JNEUROSCI.3355-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohrlüder J, Schwarten M, Willbold D. Structure and potential function of γ - aminobutyrate type A receptor - associated protein. The FEBS Journal. 2009;276(18):4989–5005. doi: 10.1111/j.1742-4658.2009.07207.x. [DOI] [PubMed] [Google Scholar]