Abstract
Background:
Disrupted in schizophrenia 1 (DISC1) has been implicated in a number of psychiatric diseases along with neurodevelopmental phenotypes such as the proliferation and differentiation of neural progenitor cells. While there has been significant effort directed towards understanding the function of DISC1 through the determination of its protein-protein interactions within an in-vitro setting, endogenous interactions involving DISC1 within a cell-type specific setting relevant to neural development remain unclear.
Methods:
Using CRISPR/Cas9 genome engineering technology, we inserted an endogenous 3×-FLAG tag at the C-terminus of the canonical DISC1 gene in human induced pluripotent stem cells. We further differentiated these cells and used affinity purification in order to determine protein-protein interactions involving DISC1 in iPSC-derived neural progenitor cells and astrocytes.
Results:
We were able to determine 150 novel cell-type specific proteins present in DISC1 endogenous interactomes. The DISC1 interactomes can be clustered into several sub-complexes that suggest novel DISC1 cell-specific functions. In addition, the DISC1 interactome in iPSC-derived neural progenitor cells associates in a connected network containing proteins found to harbor de-novo mutations in patients affected by schizophrenia and contains a subset of novel interactions that are known to harbor syndromic mutations in neurodevelopmental disorders.
Conclusions:
Endogenous DISC1 interactomes within iPSC-derived human neural progenitor cells and astrocytes are able to provide context to DISC1 function in a cell-type specific setting relevant to neural development and enables the integration of psychiatric disease risk factors within a set of defined molecular functions.
Keywords: neural development, DISC1, proteomics, CRISPR, schizophrenia, neurodevelopmental disorders
Introduction
Since its discovery, Disrupted in schizophrenia 1 (DISC1) has been implicated in contributing to multiple psychiatric disorders (1–11) and functional studies have shown DISC1 to play critical roles in several cell-types throughout brain development (12, 13). Induced pluripotent stem cells (iPSCs) with clinically-relevant mutations in DISC1 have been shown to exhibit neurodevelopmental phenotypes following differentiation into neural progenitor cells (hNPCs) such as abnormalities in proliferation and altered cell fate (14–16). The proliferation of primary NPCs has also been shown to be affected by astrocytes over-expressing a dominant negative form of DISC1 (17). DISC1 does not have an enzymatic function and has been considered a scaffold protein (18), with its function usually characterized in association with modulation of activity and/or localization of its binding partners (19–21). However, the identification of endogenous DISC1 protein interactions has not been straightforward.
The large-scale identification of DISC1 protein interactions has been largely composed of in-vitro systems such as yeast two hybrid based assays (22–28). These studies have identified dozens of proteins with the potential to directly interact with DISC1 in in-vivo interactions. In addition, a relatively low number of interactions have also been observed in low-throughput studies using recombinant expression systems (7, 19, 20). However, these assays usually do not account for tissue or cell specificity and the relative stoichiometry between interacting partners. The investigation of endogenous DISC1 protein-protein interactions has been limited to low-throughput assays (29, 30).
In order to determine DISC1 protein complexes at endogenous expression levels in cell types representing early brain development and to eliminate non-specific interactions, we inserted a 3×-FLAG (31) coding sequence at the C-terminal end of the endogenous DISC1 gene using CRISPR/Cas9 genome engineering in human iPSCs. DISC1FLAG iPSCs, were differentiated into hNPCs and astrocytes and endogenous DISC1 binding partners were determined by immunoisolation of DISC1-FLAG followed by HPLC-MS/MS in cell type-specific settings. We identified 165 non-redundant proteins in DISC1 protein complexes associated to different subcellular compartments. We confirm the involvement of DISC1 in centrosomal dynamics (32) and mRNA transport (33) along with novel cell-type specific functions such as regulation of transcription in hNPCs and cytoskeleton processes in astrocytes. We also show that DISC1 is expressed at sub-stoichiometric levels when compared to its binding partners. This indicates DISC1 might associate with a discrete subset of its interacting molecules involved in specific molecular functions. Finally, with DISC1 being implicated in a variety of psychiatric disorders, we also show a novel set of protein interactors contributing to complex brain disorders such as schizophrenia and intellectual disability in protein interaction networks identified in hNPCs but not astrocytes or recombinant immunopurification experiments.
Methods and Materials
Cell culture and neural differentiation
The control 03231 iPSC line was generated from a lymphoblastoid cell line derived from a healthy 56 year-old male (NINDS repository, ND03231) as previously described (34). For gene-editing, guide RNAs were cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 and the homologous recombination donor vector was constructed using the pFETCH_Donor plasmid (35, 36). Following nucleofection (Lonza, Basel, Switzerland) and puromycin selection, individual colonies were genotyped to confirm insertion of the 3×-FLAG sequencing in the DISC1 gene. Predicted off-target regions were identified using CRISPOR (37), PCR amplified, and Sanger sequenced. Subsequent differentiation of iPSCs was carried out as previously described with slight modifications (38–41). Full details are available in Supplementary Methods.
Biochemistry
Immunoprecipitation, western blotting, and mass spectrometry were carried out as previously described (42, 43). All primary and secondary antibody antibodies used along with their dilutions can be found in Supplemental Table S16. Full details are available in supplementary methods.
Data analysis
Mass spectrometry data analysis was carried out as previously described (42, 43). The datasets generated and/or analyzed during the current study have been deposited to the ProteomeXchange Consortium via the Proteomics IDEntifications (PRIDE) partner repository under the dataset identifier PXD007103 (44). Full details for all analyses are available in supplementary methods.
Results
Generation of DISC1 FLAG TAG Lines
In order to detect DISC1 interactions and avoid non-specific interactions, we utilized the CRISPR-Cas9 genome engineering system to insert a 3×-FLAG sequence (31) at the C-terminal region of the canonical DISC1 sequence in a previously characterized control iPSC line derived from a healthy 56-year-old male with no known psychiatric disorders (34) (Figure 1A, Figure S1A). It should be noted that the particular iPSC line used in this study carries the common missense mutation of R264Q (rs3738401). However, this is the most common missense mutation in DISC1 with an allele frequency of 29.23% (45) and recent studies have shown no link between this mutation and schizophrenia (46, 47). Thus, it is highly unlikely that this common missense mutation will have an effect in the protein interactions reported. PCR-based genotyping of individual iPSC colonies was able to show the successful homozygous insertion of the 3×-FLAG tag (designated DISC1FLAG) which was confirmed via Sanger sequencing (Figure 1B and 1C). The top predicted off-target sequences of the guide-RNA used for gene editing were also sequenced to confirm no undesired mutagenesis occurred within these regions (Supplemental Table S1). The newly derived DISC1FLAG iPSC line maintained expression of pluripotency markers such as OCT4 and SSEA4 (Figure 1D, Figure S1B and S1C).
Figure 1. Generation of DISC1FLAG human neural progenitor cell lines.
(A) Gene targeting scheme illustrating the insertion of a 3×-FLAG sequence at the C-terminal region of DISC in induced pluripotent stem cells using the CRISPR/Cas9 genome engineering System. The site of double strand break induction is annotated by the red triangles. (B) Agarose gel showing PCR-based screening for the insertion of the 3×-FLAG tag sequence. (C) Sanger sequencing conformation of the 3×-FLAG tag sequence. (D) DISC1FLAG iPSCs express markers of pluripotency such as OCT4 and SSEA4. (E) DISC1FLAG iPSC-derived neural progenitors express the neural progenitor markers SOX2 and NESTIN. (F) DISC1FLAG Astrocytes express the astrocyte marker S100β. (G) Immunoprecipitation followed by immunoblot of DISC1 using anti-FLAG antibodies in WT and DISC1FLAG hNPCs. (H) Immunoprecipitation followed by immunoblot of DISC1 using anti-FLAG antibodies in WT and DISC1FLAG Astrocytes. All bands corresponding to DISC1 isoforms were absent in WT hNPC controls and include a shift of 3.92 kDa in their molecular weight due to the insertion of the 3×-FLAG sequence. Scale bars in (D), (E), and (F) = 100 μm.
We then differentiated the DISC1FLAG iPSCs along with the WT parental iPSCs into hNPCs using a dual SMAD inhibition based differentiation scheme (38, 39). The iPSC-derived NPCs were positive for the NPC markers PAX6, SOX2, and NESTIN in both the DISC1FLAG and WT cell lines (Figure 1E, Figure S1D and S1E). Further, due to the ability of DISC1 to influence gliogenesis and proliferation of NPCs through astrocytes (17, 48), hNPCs were then differentiated into S100β/GFAP+ astrocytes over a 30 day period (41) and were confirmed to be multipotent. (Figure 1F, Figure S1F–H).
In order to visualize DISC1 hNPCs and astrocytes, we performed immunoisolation of DISC1-FLAG followed by immunoblotting with anti-FLAG antibodies (Figure 1G–H). Previous studies have shown over 50 different mRNA transcript splice variants of DISC1 in the human brain (49) and numerous molecular weights of DISC1 have been observed in western blot assays (50). Western blot analysis shows both cell types express a FLAG-DISC1 isoform corresponding to the commonly reported long isoform of 98 kDa (isoform L, NCBI reference sequence NP001158009.1) (50). We also observed additional bands between 100 and 75 kDa which may correspond to the smaller predicted molecular weight isoform (isoform a, NCBI reference sequence NP_001158012.1). Thus, the insertion of the 3×-FLAG tag labeled multiple isoforms that encompass the canonical C-terminal region of DISC1.
Incorporation of the 3×-FLAG tag does not alter expression or function of DISC1
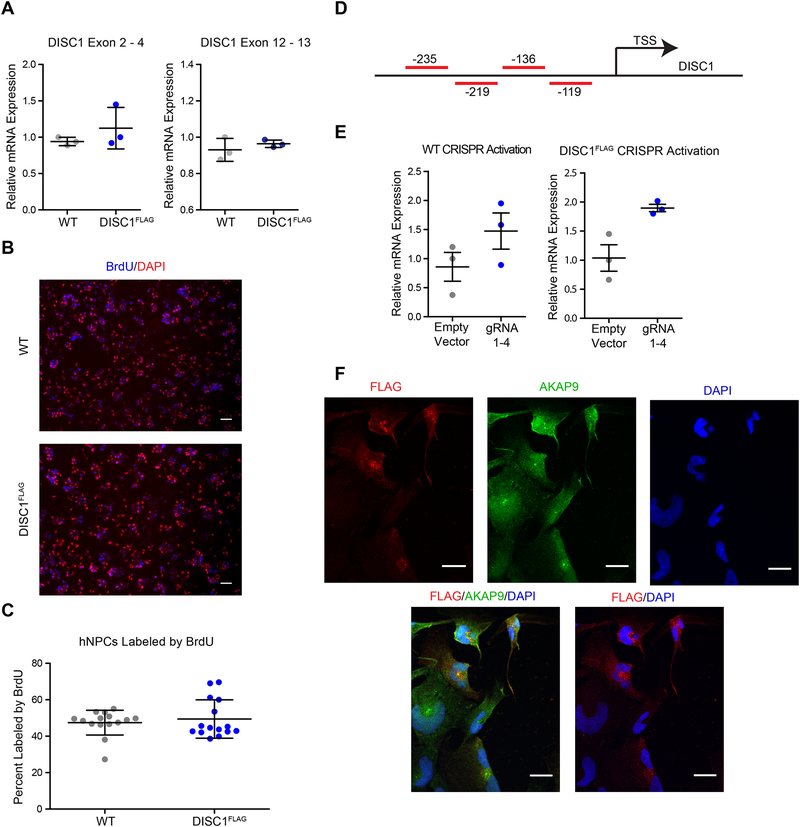
Decreased expression of DISC1 has been observed in whole blood from patients affected by schizophrenia (21, 51) and has been reported to modulate neural progenitor proliferation (12, 13). Therefore, we wanted to verify that the insertion of the 3×-FLAG tag did not alter DISC1 expression or hNPC proliferation. Quantification of DISC1 mRNA levels via qPCR using primers that were specific to either a large majority of the reported DISC1 isoforms (exons 2–4) or only those isoforms containing the conical C-terminal sequence (exons 12–13) (Figure S2A) was able to show no significant difference in DISC1 expression between WT and DISC1FLAG hNPC lines regardless of the primer set used for qPCR analysis (Figure 2A). In addition, quantification of BrdU incorportation was able to show no significant difference in proliferation between DISC1FLAG and parental hNPCs (Fig. 2B and C).
Figure 2. Insertion of 3×-FLAG at the C-terminus of DISC1 does not alter expression or function of DISC1.
(A) DISC1 expression levels showed no significant difference between DISC1FLAG hNPCs and the WT hNPC line as measured by qPCR (n = 3 biological replicates). (B) Representative immunofluorescence images of BrdU incorporation in WT and DISC1FLAG hNPCs. (C) Quantification of BrdU incorporation in both WT and DISC1FLAG hNPC cells lines shows no significant difference in proliferation (n = 3 biological replicates with 5 random fields imaged and quantified for each). (D) Schematic illustrating the position of guide RNAs used to upregulate expression of DISC1 relative to the transcription start site of DISC1. (E) qPCR demonstrating up-regulation of DISC1 following CRISPR activation. (F) Immunoflorescence following upregulation of DISC1 using the CRISPR activation system in WT and DISC1FLAG hNPCs. Images display co-staining of DAPI and the centrosomal marker, AKAP9, along with FLAG (DISC1). Data in panels (A) (C), and (E) are plotted as mean ± SD. Scale bars in (B) = 100 μm. Scale bars in (F) = 20 μm.
In addition to effects on expression and proliferation, we wanted to ensure the 3×-FLAG tag did not alter the localization of DISC1. Multiple studies have determined that DISC1 is localized to the centrosome (23, 32, 52, 53) and the perinuclear region (53). These studies have indicated that the C-terminal region of DISC1 is essential for its subcellular localization as the lack of the C-terminal region can disrupt localization and cause DISC1 to be diffusely spread throughout the cytoplasm (52, 53). However, we found that traditional immunofluorescence methods using an anti-FLAG antibody were unable to visualize the endogenous tagged-DISC1 (Figure S2B). This may be a result of the relatively low expression of DISC1 as it has been previously reported that DISC1 is a low-abundance protein in mouse brain (54) and we were unable to detect endogenous DISC1 in total cell lysate without the use of immunoisolation enrichment assays (data not shown).
Due to its low abundance, we utilized the CRISPR-VPR activation system to upregulate the expression of DISC1 in hNPCs for the sole purpose of visualizing DISC1-FLAG (55). Here, we employed four single-guide RNAs (sgRNAs) spread throughout the first 250 BP of the DISC1 promoter region (Figure 2D). With an approximate 34% transfection efficiency, this resulted in an approximate 2-fold increase in gene expression and allowed us to detect FLAG-DISC1 localization in hNPCs (Figure 2E and F, Figure SC–D). Co-staining with the centrosomal marker and DISC1 interactor, AKAP9 (22), along with DAPI, was able to show DISC1 localization at both the centrosome and perinuclear region in hNPCS coinciding with previous reports (23, 32, 56) (Figure 2F). This suggests that the 3×-FLAG tag does not impair DISC1 localization.
Identification of Endogenous DISC1 Interactomes
We co-immunopurified endogenous DISC1 and its binding partners in hNPCs using an anti-FLAG-M2 antibody (57) attached to magnetic beads and performed sequential elutions using an anti-3×-FLAG peptide followed by a final release of all bound protein via boiling of the magnetic beads. Each step was subject to immunoblotting in order to measure the efficiency, yield, and specificity of DISC1 protein elution (Figure 3A, right panel). We observed that more than 90% of DISC1 could be released within a single elution. This was followed by identification of DISC1 interactors via high performance liquid chromatography (HPLC) coupled to tandem mass spectrometry (HPLC-MS/MS).
Figure 3. Identification of the endogenous DISC1 Interactome in hNPCs and astrocytes.
(A) Right Panel - Western blot following Immunoprecipation and elution of DISC1 from DISC1FLAG and WT hNPCs using an anti-3×-FLAG peptide. Comparing elutions shows that a majority of DISC1 is eluted within the first elution. Lane 1 = first elution, Lane 2 = second elution, Lane 3 = boiling of magnetic beads post elutions. Left Panel - DISC1 Interactome in hNPCs as characterized via immunoprecipitation followed by mass spectrometry or western blotting. Criteria for inclusion is indicated by colored border of nodes. (B) Western blots in hNPCs showing conformation of proteins identified via immunoprecipitation followed by mass spectrometry or previously identified protein-protein interactions involving DISC1. (C) DISC1 Interactome in astrocytes as characterized via immunoprecipitation followed by mass spectrometry or western blotting. (D) Western blots in astrocytes showing conformation of proteins identified via immunoprecipitation followed by mass spectrometry or previously identified protein-protein interactions involving DISC1. (E) Gene ontology analysis of the identified DISC1 interactome in hNPCs. (F) Gene ontology analysis of the identified DISC1 interactome in Astrocytes.
DISC1 hNPC-interactors were considered if they were identified with multiple unique peptides in duplicate assays and no peptides in controls (36 interactors). However, because of the low expression level of DISC1, this stringent criteria might impair the identification of true DISC1 interactors. Thus, we also considered interactors with different levels of confidence bringing the number of interactors identified via mass spectrometry to a total of 107 interactors (Figure 3A left panel, Supplemental Table S2). In order to confirm DISC1 interactions and the validity of our approach, we performed DISC1 immunoisolation followed by western blot analysis from DISC1 interactors that we identified with different levels of confidence (Figure 3B). Moreover, we were also able to confirm several previously reported DISC1 protein interactors including TNIK, PDE4DIP, CEP170, and AKAP9 bringing the total number of interactors to 112 (Figure 3A–B).
Inference of hNPC DISC1 localization through its interactome indicates DISC1 associates with proteins in different subcellular structures such as the nucleus, centrosome, endoplasmic reticulum (ER), and the cytoplasm. Gene ontology (GO) enrichment analysis of DISC1 interactors provides further evidence for this as terms such as “cytosol”, “ER”, and “nucleoplasm” are significantly enriched (P < 0.05 for all, Bonferroni corrected) (Figure 3E, Supplemental Table S3). This corresponds with previous reports of DISC at the ER and nucleus in neuronal systems (58, 59).
We then identified DISC1-interacting partners in astrocytes differentiated from hNPCs. Here, we were able to identify 51 interactors that were categorized according the various confidence levels as was completed for hNPCs (Figure 3C, Supplemental Table S4). Western blot analysis of DISC1 interactors was able to confirm a number of the identified interactors including NDE1, GRIPAP1, and GSN. In addition, we were again able to confirm reported DISC1 interactors including centrosomal proteins LIS1 and NDEL1, bringing the total number of interactors to 56 (Figure 3D). This Indicates that a core DISC1 function, such as modulation of centrosomal dynamics, is conserved in astrocytes. The astrocyte interactome also shows an increased number of cytoskeletal proteins which is corroborated through GO enrichment analysis of DISC1 astrocyte interactors. Here, terms such as “actin filament binding,” “focal adhesion,” and “actin cytoskeleton” are significantly enriched (P < 0.05 for all, Bonferroni corrected) (Figure 3F, Supplemental Table S5), indicating that DISC1 may also play a role in astrocyte cytoskeletal organization and therefore, astrocyte vesicle mobility and trafficking.
While the enrichment of cytoskeletal proteins may indicate a cell type-specific function or localization of DISC1 in astrocytes, “Poly(A) RNA binding” is significantly enriched in both cell types (P < 0.05 for all, Bonferroni corrected) (Figure 3E–F, Supplemental Table S3, S5). A recent report characterized DISC1 as an RNA binding protein and showed its involvement in RNA transport in adult hippocampal dendrites (33), suggesting a common regulatory function in hNPCs, astrocytes, and neurons. This is exemplified by the association of DISC1 with RNA binding proteins, including members of the nuclear pore complex (e.g NUP210, NUP98, and RANGAP1) in hNPCs, along with ALYREF which is involved in nuclear mRNA transport and is a common DISC1 interactor in both hNPCs and astrocytes.
We identified multiple unique DISC1 peptides accounting for 21.66% and 34.13% coverage of the DISC1 canonical (L) isoform in hNPCs and astrocytes, respectively (Supplemental Table S6). This relatively low coverage, along with the inability to detect DISC1 above background with traditional immunofluorescence or immunoblotting assays, is in agreement with low expression levels as previously reported (54). Characterization of the expression of DISC1 in hNPCs and astrocytes relative to the identified DISC1 interactors using publicly available data that directly compares the expression of genes in both iPSC-derived hNPCs and astrocytes (41) was able to show over 90% of DISC1-interacting proteins have higher expression levels than DISC1 in both cell types (Supplemental Figure S3, Supplemental Table S7). While transcript levels may not always be directly correlated to protein levels, our MS results are in agreement with DISC1 low expression levels in hNPCs and astrocytes. This suggests that for a particular interactor, only a small fraction of the overall protein levels might be interacting with DISC1 at any given time, indicating DISC1 may interact at substoichiometric ratios.
The Endogenous DISC1 Interactome and Previously Reported Protein-Protein Interactions
With this being the first report of endogenous DISC1 protein-protein interactions in iPSC-derived hNPCs and astrocytes, we wanted to compare our data with what has been previously reported. Here, we curated a list of previously reported DISC1 interactions (Supplemental Table S8) (Methods) and were able to identify a total of 9 proteins in hNPCs that we confirmed either by mass spectrometry or via immunoprecipitation followed by western blot and 9 proteins in astrocytes (Supplemental Table S10). We also observed an additional 7 and 3 proteins derived from yeast two-hybrid assays present in at least one sample in hNPCs and astrocytes, respectively (Supplemental Table S10).
Since a large number of protein interactors in hNPCs and astrocytes were novel, we hypothesized that this may be due to cell-type specific interactions and DISC1 stoichiometry ratios. To test this, we performed immunoprecipitation of the full-length recombinant DISC1 in adult mouse cortex. This is comparable to previous studies that have characterized DISC1 interactions with respect to both non-stoichiometric conditions and developmental context. As opposed to the 20 – 30% coverage obtained under endogenous conditions, we obtained close to 100% using the recombinant protein. Under these conditions, we identified 148 proteins in DISC1 immunoprecipitation experiments and absent in corresponding controls, several of which were verified via western blot (Supplemental Figure S4 A–B, Supplemental Table S9). As opposed to 9 proteins identified in NPCs or astrocytes, we identified a total of 42 previously reported interactions using recombinant DISC1 in a tissue similar to what has been used previously (Supplemental Table S10).
We also found a number of related proteins to previously reported interactions in both structure and function (Supplemental Table S11). For example, while the nuclear pore protein, NUP160, was previously characterized via yeast-two hybrid and detected in the DISC1 recombinant IP, we identified NUP210 and NUP98 as interactors of DISC1 in hNPCs. This highlights the role cellular context plays in determining protein interactions.
Clustering of DISC1 Interactors and Psychiatric Disease
Next, we asked if the endogenous DISC1 interactomes could cluster in subsets of protein interactions that can indicate both protein functions and complexes that DISC1 may associate with. Here, we extracted reported protein interactions for each DISC1 interactor identified in our screens and clustered them in a hNPC- or astrocyte-DISC1 protein interaction network. We identified a number of molecular functions and cellular locations where DISC1 interactors associated in interconnected clusters of protein interactions. Both hNPCs and astrocyte interactomes contain the centrosomal interactome including TNIK, PDE4DIP, and CEP170 along with proteins that clustered into functions such as RNA binding and transport (Figure 4A, Supplemental Table S12). hNPCs interactions also clustered in the peri-nuclear region including the nuclear pore and nuclear lamina complexes. In contrast, DISC1 interactors identified in astrocytes clustered into complexes involving cytoskeletal processes such as actin-related proteins or the spectrin complex (Figure 4A, Supplemental Table S12). While clusters of proteins involved in centrosome dynamics have been previously described, a majority of the clusters uncovered are novel and include proteins characterized as contributing to psychiatric disorders such as GATAD2B and FOXP1 which were identified in hNPCs (60, 61).
Figure 4. Clustering and Enrichment of DISC1 Interactome in Psychiatric Disease Risk Factors.
(A) Clustering of the endogenous DISC1 hNPC and astrocyte interactomes based on protein-protein interactions derived from public databases. (B) Enrichment of the DISC1 hNPC interactome in proteins found to have de novo mutations in patients affected by schizophrenia (SCZ), intellectual disability (ID), autism spectrum disorder (ASD), congenital heart disease (CHD), along with all controls listed in denovo-db. Variant classes are split between missense mutations, protein truncating variants (PTV), nonsynonomous mutations (includes missense and protein truncating variants), and synonomous mutations. Error bars represent the 95% confidence interval and nominal p-values are displayed in bold for each test. (C) Heatmap representing proteins implicated in contributing to neurological disorders via OMIM along with proteins containing nonsynonomous de novo mutations in SCZ, ASD, or ID when found in two or more gene lists.
DISC1 has been associated with numerous psychiatric disorders (9, 11, 62, 63). However, the lack of association to schizophrenia through both genome wide association (GWAS) and exome sequencing studies has brought the applicability of DISC1 to psychiatric disorders into question (64, 65). Irrespective of this, proteins may be able to regulate disease-relevant pathways through the modulation of protein interactions and function in protein networks where disease risk factors are associated, as the case has been made for DISC1 (20). Therefore, we tested the hNPC, astrocyte, and recombinant DISC1 interactomes for enrichment in proteins affected by de novo mutations, separated by mutation class, in Schizophrenia (SCZ), Autism Spectrum Disorder (ASD), Intellectual Disability (ID), Congenital Heart Disease (CHD), and an aggregate of all controls in denovo-db (66) (Figure 4C, Supplemental Table S13). For both the astrocyte and recombinant DISC1 interactomes, there was no statistically significant enrichment in any category tested. However, in the hNPC DISC1 interactome, we did find enrichment of nonsynonymous mutations in SCZ following correcting for multiple comparisons (nominal p-value = 1.84 × 10−3, Bonferroni threshold p < 2.40 × 10−3). ID also presented enrichment in nonsynonymous mutations, but did not pass the Bonferroni threshold (nominal p-value = 3.34 × 10−3) (Figure 5C, Supplemental Table S13). While the SCZ enrichment was driven by a diverse set of proteins, the non-significant enrichment in ID was driven by proteins that were found to have recurrent mutations including FOXP1, GATAD2B, and USP7 (Figure 5C-D, Supplemental Table S13). This Suggests that DISC1 function within hNPCs may contribute more to the risk of disease when compared to DISC1 function in astrocytes or in the adult cortex.
In addition to proteins that have been implicated through exome-sequencing studies, we also focused on proteins known to be syndromic for neurodevelopmental disorders as classified by the Online Mendelian Inheritance in Man (OMIM) database (Figure 5D). Here, DISC1 interactors included the previously described DISC1 interacting kinases CIT (microcephaly) and TNIK (ID) along with NDE1 (Microhydranencephaly) and PAFAH1B1 (Lissencephaly). Novel interactors that are also known to by syndromic include the transcription factors FOXP1 (ID and ASD) and GATAD2B (ID), USP7 (ID and ASD), PGAP1 (ID), SGSH (Mucopolysaccharidosis type IIIA), and EMC1 (Developmental Delay). This is in accordance with our previous findings, showing that the interactomes of molecules involved in developmental disorders, such as TNIK (67), show an enrichment in de novo mutations in a variety of complex brain disorders (43).
Mutations in DISC1 Interactors Regulate Shared Cellular Functions
Proteins associated in protein interaction networks are usually involved in the regulation of shared cellular processes and mutations in genes encoding for these proteins are more likely to be associated to the same disease (68–70). We observed that DISC1 mutations alter the expression levels of many novel DISC1 interactors, with a significant overlap in differentially expressed genes found in iPSC-derived neurons from patients with a frameshift mutation that affects the isoforms selectively tagged here (14) (see Supplementary Text S1). In addition, with DISC1 interacting with several centrosomal proteins in hNPCs, we hypothesized that similarly to DISC1, they might also affect hNPC proliferation. One of the proteins, the TRAF and Nck interacting Kinase (TNIK), has also been involved in a number of psychiatric disorders and intellectual disability (67, 71–73). This suggests that TNIK mutations might also be associated with alterations in cellular proliferation in hNPCs. Thus, we introduced a truncating mutation at the Citron homology domain (CNH) domain of TNIK, which resulted in TNIK knockout cell line (Supplemental Figure S4A). Compared to WT cell lines, we found a marked decrease of proliferation in TNIK KO hNPCs, as measured by a 40% decrease in phospho-histone H3 positive cells (p < 0.001, Student’s t Test) (Supplemental Figure S4B–C). Interestingly, it has been reported that DISC1 may inhibit TNIK function in mature neurons in vitro (74). However, our results indicate a synergistic interaction of DISC1 and TNIK where both molecules are needed to regulate hNPC proliferation, highlighting the importance of the identification of cell-specific protein networks
Discussion
Through the combination of genome-engineering and mass-spectrometry, we are able to report the first endogenous interactome of DISC1 in human iPSC-derived NPCs and astrocytes. While we ensured that there were no transcriptional changes at the mRNA level due to the insertion of the endogenous tag, we should note the unlikely possibility of an endogenous tag interfering with protein expression levels, distribution, or protein interactions. Nonetheless, the novel interactions identified here can be categorized into numerous functional modules that are in line with previously reported functions of DISC1 and expand upon them. For example, DISC1 is associated with a number of proteins that are known to reside at the centrosome including AKAP9, CEP170, NDEL1, LIS1. Our results are able to include molecules such as PDE4DIP into the DISC1/centrosome protein interaction network in both hNPCs and astrocytes.
Using the DISC1 interactions determined in hNPCs, astrocytes, the recombinant protein, or all previously reported interactions, we were able to show that only the DISC1 hNPC interactome is enriched in nonsynonymous mutations as a whole identified in sporadic cases of SCZ. Determining DISC1 interactors in hNPCs is also able to provide context to neurodevelopmental phenotypes observed in previous studies. For example, clinically relevant mutations in DISC1 created via genome engineering in iPSCs were shown to result in differential sup-populations of neural progenitors following differentiation (16). Here, we show that DISC1 binds to nuclear pore proteins such as NUP210 and NUP98, which have both been shown to influence neural differentiation (75, 76). While relatively little is known concerning the function of DISC1 in astrocytes, it has been shown to influence gliogenesis of primary NPCs (48), indicating a common function of influencing differentiation potential. Here, the common functional module of regulatory RNA-binding proteins between hNPCs and astrocytes may play a role in this process.
While DISC1 has been regularly described as a scaffold protein, the expression of DISC1 appears to be extremely low which is corroborated through both transcriptomic and proteomic data. Mass spectrometry data derived from public repositories such as PeptideAtlas do not contain any DISC1 peptides observed in any neural cell types or brain tissue as a whole (77), Thus, the sub-stoichiometric characteristics of DISC1 protein interactions suggests that DISC1 binds and affects only a specific subset of its interactors at any given time.
Our results are also able to provide the first confirmation via immunoprecipitation followed by western for DISC1 interactors identified in Y2H screens including AKAP9, CEP170, and GRIPAP1. However, comparison of the endogenous interactome of DISC1 in iPSC-derived hNPCs and astrocytes to the multiple DISC1 yeast-two hybrid screens is able to show that a vast majority of the interactions detected here were not apparent in those studies. While protein interactions derived from affinity purification may include secondary or tertiary interactions compared to binary interactions identified via yeast-two hybrid assays, these assays do not take context or stoichiometric ratios into account. In contrast to the endogenous interactions, when we performed immunopurification of recombinant DISC1 in a non-stoichiometric fashion and in an experimental setting resembling what has been previously published, we found an abundance of previously reported interactions. This underscores the importance of defining endogenous protein-protein interactions within a particular cellular context of interest with the correct stoichiometric ratios between interacting proteins.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Child Health and Human Development (MH104603-01A1; to M.P.C). J.K.I is a New York Stem Cell Foundation-Roberton Investigator. We would like to thank Daniel Howrigan (Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.) for providing guidance in the enrichment analysis of proteins found to harbor denovo mutations in patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
J.K.I. is a co-founder of Acurastem Inc. All other authors report no biomedical financial interests or potential conflicts of interests.
References
- 1.Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, et al. (2003): Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Human molecular genetics. 12:3151–3159. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, et al. (2006): Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Human molecular genetics. 15:3024–3033. [DOI] [PubMed] [Google Scholar]
- 3.Hennah W, Thomson P, Peltonen L, Porteous D (2006): Genes and schizophrenia: beyond schizophrenia: the role of DISC1 in major mental illness. Schizophrenia bulletin. 32:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porteous DJ, Thomson P, Brandon NJ, Millar JK (2006): The genetics and biology of DISC1--an emerging role in psychosis and cognition. Biological psychiatry. 60:123–131. [DOI] [PubMed] [Google Scholar]
- 5.Mackie S, Millar JK, Porteous DJ (2007): Role of DISC1 in neural development and schizophrenia. Current opinion in neurobiology. 17:95–102. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, et al. (2010): The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Molecular psychiatry.1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandon NJ, Sawa A (2011): Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nature reviews Neuroscience. 12:707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porteous DJ, Millar JK, Brandon NJ, Sawa A (2011): DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends Mol Med. 17:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. (2008): Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 13:187–196. [DOI] [PubMed] [Google Scholar]
- 10.Millar JK, Christie S, Semple CA, Porteous DJ (2000): Chromosomal location and genomic structure of the human translin-associated factor × gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics. 67:69–77. [DOI] [PubMed] [Google Scholar]
- 11.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. (2000): Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human molecular genetics. 9:1415–1423. [DOI] [PubMed] [Google Scholar]
- 12.Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, et al. (2011): DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, et al. (2009): Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 136:1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. (2014): Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 515:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murai K, Sun G, Ye P, Tian E, Yang S, Cui Q, et al. (2016): The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat Commun. 7:10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA, et al. (2015): Genomic DISC1 Disruption in hiPSCs Alters Wnt Signaling and Neural Cell Fate. Cell Rep. 12:1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terrillion CE, Abazyan B, Yang Z, Crawford J, Shevelkin AV, Jouroukhin Y, et al. (2017): DISC1 in Astrocytes Influences Adult Neurogenesis and Hippocampus-Dependent Behaviors in Mice. Neuropsychopharmacology. 42:2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yerabham AS, Weiergraber OH, Bradshaw NJ, Korth C (2013): Revisiting disrupted-in-schizophrenia 1 as a scaffold protein. Biol Chem. 394:1425–1437. [DOI] [PubMed] [Google Scholar]
- 19.Tomppo L, Hennah W, Lahermo P, Loukola A, Tuulio-Henriksson A, Suvisaari J, et al. (2009): Association between genes of Disrupted in schizophrenia 1 (DISC1) interactors and schizophrenia supports the role of the DISC1 pathway in the etiology of major mental illnesses. Biological psychiatry. 65:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradshaw NJ, Porteous DJ (2012): DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 62:1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampino A, Walker RM, Torrance HS, Anderson SM, Fazio L, Di Giorgio A, et al. (2014): Expression of DISC1-interactome members correlates with cognitive phenotypes related to schizophrenia. PloS one. 9:e99892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, et al. (2007): Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Molecular psychiatry. 12:74–86. [DOI] [PubMed] [Google Scholar]
- 23.Morris JA, Kandpal G, Ma L, Austin CP (2003): DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Human molecular genetics. 12:1591–1608. [DOI] [PubMed] [Google Scholar]
- 24.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, et al. (2003): Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 100:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, et al. (2010): Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc Natl Acad Sci U S A. 107:17785–17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, et al. (2003): Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 8:685–694. [DOI] [PubMed] [Google Scholar]
- 27.Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, et al. (2014): Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 5:3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar JK, Christie S, Porteous DJ (2003): Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochemical and biophysical research communications. 311:1019–1025. [DOI] [PubMed] [Google Scholar]
- 29.Ishizuka K, Chen J, Taya S, Li W, Millar JK, Xu Y, et al. (2007): Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Molecular psychiatry. 12:897–899. [DOI] [PubMed] [Google Scholar]
- 30.Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, et al. (2008): A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 105:7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terpe K (2003): Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 60:523–533. [DOI] [PubMed] [Google Scholar]
- 32.Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, et al. (2008): DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochemical and biophysical research communications. 377:1091–1096. [DOI] [PubMed] [Google Scholar]
- 33.Tsuboi D, Kuroda K, Tanaka M, Namba T, Iizuka Y, Taya S, et al. (2015): Disrupted-in-schizophrenia 1 regulates transport of ITPR1 mRNA for synaptic plasticity. Nat Neurosci. 18:698–707. [DOI] [PubMed] [Google Scholar]
- 34.Pepper JP, Wang TV, Hennes V, Sun SY, Ichida JK (2016): Human Induced Pluripotent Stem Cell-Derived Motor Neuron Transplant for Neuromuscular Atrophy in a Mouse Model of Sciatic Nerve Injury. JAMA Facial Plast Surg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, et al. (2015): CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res. 25:1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013): Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, et al. (2016): Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, et al. (2017): Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 35:154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L (2009): Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Topol A, Tran NN, Brennand KJ (2015): A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. J Vis Exp. e52495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, et al. (2017): An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Reports. 9:600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson B, Li J, Coba MP (2017): Synaptic GAP and GEF Complexes Cluster Proteins Essential for GTP Signaling. Sci Rep. 7:5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Zhang W, Yang H, Howrigan DP, Wilkinson B, Souaiaia T, et al. (2017): Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. (2016): 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44:D447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. (2016): Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang HY, Liu Y, Yan JW, Hu XL, Zhu DM, Xu XT, et al. (2018): Gene polymorphisms of DISC1 is associated with schizophrenia: Evidence from a meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 81:64–73. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, Ren J, Ye H (2018): Association between variations in the disrupted in schizophrenia 1 gene and schizophrenia: A meta-analysis. Gene. 651:94–99. [DOI] [PubMed] [Google Scholar]
- 48.Wang S, Liang Q, Qiao H, Li H, Shen T, Ji F, et al. (2016): DISC1 regulates astrogenesis in the embryonic brain via modulation of RAS/MEK/ERK signaling through RASSF7. Development. 143:2732–2740. [DOI] [PubMed] [Google Scholar]
- 49.Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. (2009): DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 106:15873–15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ (2011): DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS Chem Neurosci. 2:609–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. (2005): DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 310:1187–1191. [DOI] [PubMed] [Google Scholar]
- 52.Millar JK, James R, Christie S, Porteous DJ (2005): Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol Cell Neurosci. 30:477–484. [DOI] [PubMed] [Google Scholar]
- 53.Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, et al. (2005): A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 7:1167–1178. [DOI] [PubMed] [Google Scholar]
- 54.Kuroda K, Yamada S, Tanaka M, Iizuka M, Yano H, Mori D, et al. (2011): Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum Mol Genet. 20:4666–4683. [DOI] [PubMed] [Google Scholar]
- 55.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, et al. (2015): Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 12:326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandon NJ, Schurov I, Camargo LM, Handford EJ, Duran-Jimeniz B, Hunt P, et al. (2005): Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Molecular and cellular neurosciences. 28:613–624. [DOI] [PubMed] [Google Scholar]
- 57.Brizzard BL, Chubet RG, Vizard DL (1994): Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques. 16:730–735. [PubMed] [Google Scholar]
- 58.Norkett R, Modi S, Birsa N, Atkin TA, Ivankovic D, Pathania M, et al. (2016): DISC1-dependent Regulation of Mitochondrial Dynamics Controls the Morphogenesis of Complex Neuronal Dendrites. J Biol Chem. 291:613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soda T, Frank C, Ishizuka K, Baccarella A, Park YU, Flood Z, et al. (2013): DISC1-ATF4 transcriptional repression complex: dual regulation of the cAMP-PDE4 cascade by DISC1. Mol Psychiatry. 18:898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EM, Castells-Nobau A, et al. (2013): GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet. 50:507–514. [DOI] [PubMed] [Google Scholar]
- 61.Hamdan FF, Daoud H, Rochefort D, Piton A, Gauthier J, Langlois M, et al. (2010): De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 87:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crepel A, Breckpot J, Fryns JP, De la Marche W, Steyaert J, Devriendt K, et al. (2010): DISC1 duplication in two brothers with autism and mild mental retardation. Clin Genet. 77:389–394. [DOI] [PubMed] [Google Scholar]
- 63.Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J, et al. (2014): 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry. 19:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan PF (2013): Questions about DISC1 as a genetic risk factor for schizophrenia. Mol Psychiatry. 18:1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Porteous DJ, Thomson PA, Millar JK, Evans KL, Hennah W, Soares DC, et al. (2014): DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol Psychiatry. 19:141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, F Stessman HA, et al. (2017): denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 45:D804–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anazi S, Shamseldin HE, AlNaqeb D, Abouelhoda M, Monies D, Salih MA, et al. (2016): A null mutation in TNIK defines a novel locus for intellectual disability. Hum Genet. 135:773–778. [DOI] [PubMed] [Google Scholar]
- 68.Carter H, Hofree M, Ideker T (2013): Genotype to phenotype via network analysis. Curr Opin Genet Dev. 23:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL (2007): The human disease network. Proc Natl Acad Sci U S A. 104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, et al. (2015): Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. (2009): A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 35:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. (2009): Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 460:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, et al. (2016): Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 19:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, et al. (2011): The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry. 16:1006–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW (2012): A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 22:446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW (2013): Dynamic association of NUP98 with the human genome. PLoS Genet. 9:e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, et al. (2006): The PeptideAtlas project. Nucleic Acids Res. 34:D655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.