Abstract
As a step towards development of a high-resolution ion mobility mass spectrometer using the orbitrap mass analyzer platform, we describe herein a novel reverse-entry ion source (REIS) coupled to the higher-energy C-trap dissociation (HCD) cell of an orbitrap mass spectrometer with extended mass range. Development of the REIS is a first step in the development of a drift tube ion mobility-orbitrap MS. The REIS approach retains the functionality of the commercial instrument ion source which permits the uninterrupted use of the instrument during development as well as performance comparisons between the two ion sources. Ubiquitin (8.5 kDa) and lipid binding to the ammonia transport channel (AmtB, 126 kDa) protein complex were used as model soluble and membrane proteins, respectively, to evaluate the performance of the REIS instrument. Mass resolution obtained with the REIS is comparable to that obtained using the commercial ion source. The charge state distributions for ubiquitin and AmtB obtained on the REIS are in agreement with previous studies which suggests the REIS - Orbitrap EMR retains native structure in the gas phase.
Graphical abstract
Introduction
Native Ion Mobility-Mass Spectrometry (IM-MS) has rapidly cemented a crucial role in the biophysical characterization of proteins over the last three decades[1–6]; however, the impact of this technique has been hindered by the lack of mobility (RIM) and mass resolutions (Rm/z). Coupling IM to MS facilitates the studies of protein conformational preferences while also improving MS spectral quality and analytical merits[7, 8]. In the last 10 years, a surge of innovation in MS technology has been spurred in large part by the development of orbitrap mass analyzers, a robust and affordable benchtop MS[9, 10]. Orbitrap mass analyzers rely on electric field-based trapping to measure the transients of trapped ions for mass determination. Coupling an achieved Rm/z = 240,000 with a mass range spanning 40,000 m/z, these instruments offer excellent performance for the analysis of small molecules up to large proteins and protein complexes[11]. Continuous improvements in ion injection and trapping efficiencies leverages the capabilities of these instruments to achieve performance similar to other trapping-based mass analyzers such as Fourier Transform Ion Cyclotron Resonance (FTICR) instruments at a greatly reduced cost[12].
Although orbitrap instruments have dramatically improved performance with regards to Rm/z over other benchtop MS, the option for a factory-designed high-resolution IM cell coupled to an orbitrap mass analyzer does not currently exist. Several groups have modified orbitrap platforms, however, the modifications generally eliminate the ability to utilize the instrument in its intended configuration[13–15]. Unlike other modifications to orbitraps reported to date, the completion of the instrument described here preserves the commercial ion source during development. Ultimately, higher RIM and Rm/z will provide the necessary detail to study larger, more complex protein assemblies and their interactions. The development of novel instrumentation to achieve these advances remains at the forefront of our developmental efforts.
Herein, we focus on the development and performance characteristics of a novel interface through which IMS or other tandem MS strategies can be coupled to the orbitrap mass analyzers without the need to significantly modify the commercial instrument. The performance of this reverse-entry ion source (REIS) was characterized through the analysis of ubiquitin, the trimeric transmembrane ammonia transport channel (AmtB) complex as well as the products of lipid binding to AmtB. Mass spectra obtained on the novel REIS were compared to spectra obtained using the commercial ion optics as well as a Waters Synapt G1 IM-MS instrument. The data and instrumental developments described herein set the stage for future work wherein IMS can be coupled to an orbitrap mass analyzer. This coupling will enable more targeted structural studies to be performed using an unprecedented combination of ion mobility and mass spectral resolution which has yet to be realized by both commercial and “homebuilt” instrumentation.
Experimental
Sample Preparation
Ubiquitin (bovine erythrocytes, >90% purity) was purchased from Sigma Aldrich (St. Louis, MO). A 5.0 μM stock solution in deionized water (18 MΩ) with 0.1% formic acid was prepared and used without further purification. Ammonia transport channel (AmtB) was expressed and purified as described previously [16]. AmtB was diluted in 200 mM ammonium acetate and 0.5% C8E4. Lipid samples of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and 1′,3′-bis [1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol (TMCDL) were purchased from Avanti Polar Lipids Inc. (Alabama, USA). Lipids stocks were prepared in 200 mM ammonium acetate and 0.5% C8E4 and used without further purification.
Instrumentation
The homebuilt ion source used for these experiments utilized a radiofrequency (RF) ion funnel consisting of 33 electrodes electrically coupled through a series of capacitors and resistors[17]. Ions enter the funnel through a heated capillary set between 80–95 °C. A differential pumping region separates a RF-only octupole ion guide built in-house. A dual channel Ardara RF Generator was used to apply RF potentials to both the ion funnel and octupole. This homebuilt source was coupled to the higher-energy C-trap dissociation (HCD) cell of a Thermo Exactive Plus with Extended Mass Range where the HCD endcap was removed and replaced with the octupole. Collision energy (CE) in the HCD cell was set between 10–156 V, maximum injection time was set to 200 ms, trapping gas pressure was set between 1–4 au, orbitrap resolution of 70,000 or 140,000 was selected, and each spectrum was collected using 10 microscans and 100 averaging. Samples were introduced to the MS using gold coated capillaries prepared in-house using borosilicate glass capillaries.
Temperature control for the lipid binding studies was achieved using a similar heated source device similar to what has been described recently[16]. Briefly, a temperature-controlled air circulator was used to heat an enclosed chamber around the ion source. Samples were placed inside a glass capillary, allowed to equilibrate at a given temperature, and subsequently analyzed. All experiments were carried out at 29 °C.
Data Processing
AmtB lipid binding deconvolution and peak picking were performed using Unidec software with the following settings: m/z range of 6500 to 11500, 2.0 smoothing binned every 0.25, charge range of 12 to 19, mass sampling every 2.0 Da, peak FWHM of 0.8, peak detection range of 2.0 Da, and a peak detection threshold of 0.05[18]. The resultant zero-charge mass spectra were then assigned using Python scripts written in-house. Charge state distribution values were calculated using Unidec software.
Results and Discussion
Reverse Entry Ion Source
Native MS requires a great deal of care in sample preparation, ion creation, and manipulation, specifically for biophysical studies[6, 19–23]. In this work, a modified “reverse entry” ion source (REIS) was designed to create a “cold” ion source interfaced to the higher-energy collisional dissociation (HCD) cell of the orbitrap. This design was chosen to retain the full functionality of the orbitrap and its commercial source while simultaneously being able to design and evaluate ion optics using the REIS.
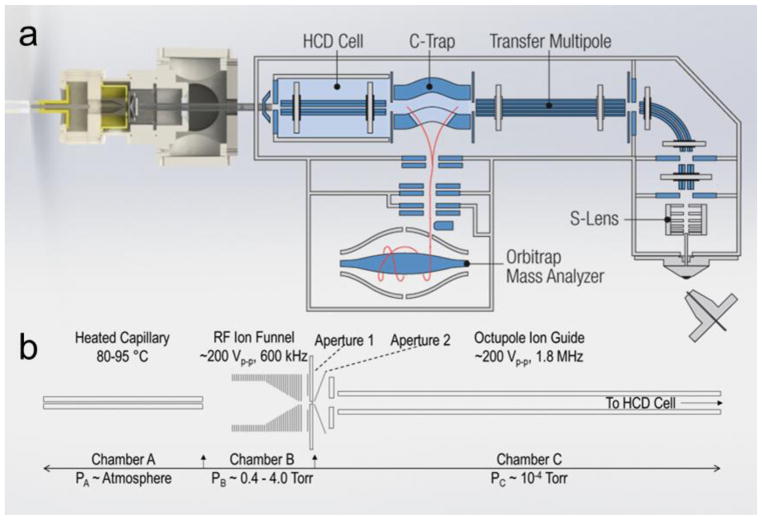
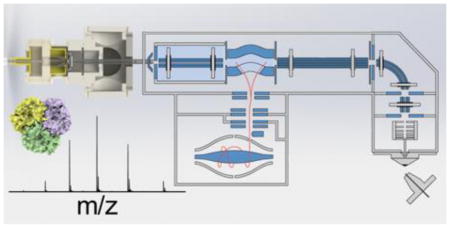
Here, the novel REIS was constructed by coupling an ion source via an RF-only octupole ion guide to the HCD cell of the instrument as depicted in Figure 1. Ions were introduced into a heated capillary using nanoflow ESI (n-ESI) where they are subsequently focused using an RF ion funnel[17]. Ions exiting the funnel pass through a skimmer region before being focused through a ring electrode and finally enter an RF-only octupole ion guide[24]. The ions entering via REIS are indiscriminately guided and trapped in the HCD cell. The HCD cell retains its functionality: ions can be stored and transmitted to the C-Trap as well as heated or fragmented before mass analysis. Although this configuration retains most of the functionality of the orbitrap, the interface eliminated the ability to use automatic gain control (AGC) to limit space charge effects in the orbitrap. AGC issues can be addressed, however, with manual control of the “maximum injection time” which allows for the tuning of the number of ions loaded into the orbitrap for mass analysis.
Figure 1.
(a) Solidworks rendering of the reverse entry ion source and configuration of the instrument. (b) Starting from left to right, ions enter the instrument through an 11.4 cm, 800 μm i.d. heated capillary and are radially focused by a 5.08 cm in length RF ion funnel. The RF ion funnel consists of 33 electrodes electrically coupled through a series of capacitors and resistors, wherein the first 15 electrodes have inner diameters of 2.54 cm and the last 18 electrodes linearly taper to 2 mm. Ions are then guided through an RF-only octupole ion guide (rod diameter of 1.59 mm and 4.76 mm inscribed diameter) prior to being loaded into the HCD Cell of the Exactive Plus EMR.
Retention of Mass Performance
Retaining both the native-like structure of ions and the high mass resolution afforded by orbitrap MS were the most important factors in the design of the REIS. Mass spectral resolution (Rm/z) can be defined by the ratio of mass to charge (m/z) to its peak width at half maximum (FWHM) as describe in Equation 1 below. Rm/z is not only instrument dependent but also analyte specific.
| (1) |
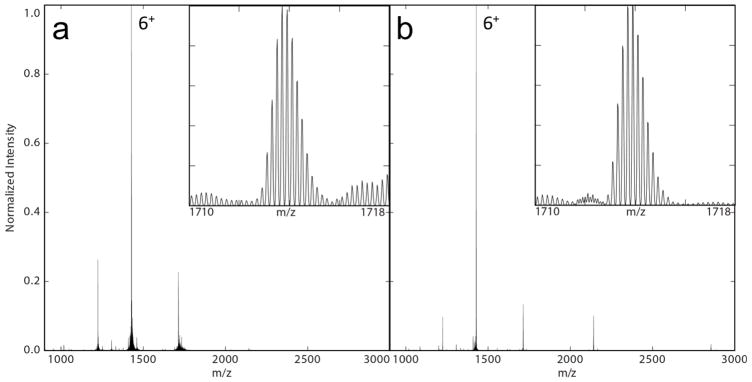
To evaluate the performance of the modified source, identical samples of ubiquitin were analyzed using both the commercial ion source of the orbitrap and the REIS. As shown in Figure 2, ubiquitin was isotopically resolved for each observed charge state using both ion sources giving nearly identical maximum resolutions of 30.2k and 30.9k for the commercial and REIS, respectively.
Figure 2.
Mass spectra of ubiquitin acquired on the (a) commercial ion source and (b) REIS. Shown in the insets are the 5+ charge state giving isotopic resolution from both ion sources. Note the lower charge states observed using the REIS giving rise to the 3+.
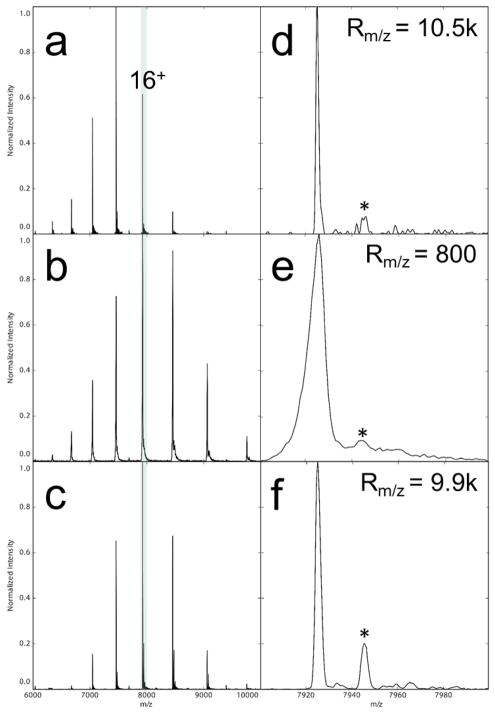
Additionally, a more challenging membrane protein, AmtB (126 kDa), was analyzed on both the commercial (Figure 3 a and d) and REIS (Figure 3 c and f) ion sources. A maximum resolution of ~10.5k and 9.9 k was achieved for the commercial source and REIS, respectively, on an identical sample of AmtB. We attribute the small difference in resolution to the retention of small molecules non-covalently adducted to the complex using the REIS leading to increased heterogeneity in the observed ion. The commercial source and REIS on the orbitrap yielded an Rm/z that was over 12-fold higher than the Rm/z = 800 achieved by the Waters Synapt G1 (Figure 3 e). These results demonstrate that the addition of the REIS did not adversely affect mass performance of the orbitrap EMR instrument. This finding is a crucial step towards the addition of IMS to the orbitrap platform.
Figure 3.
Mass spectra collected from the unmodified orbitrap (a,d), Waters Synapt G1 (b,e), and the REIS (c,e). The full mass spectra are shown on the left, and the 16+ charge state is expanded on the right. The asterisks denote the adducted C8E4 detergent molecule bound to the AmtB in all three cases which was impossible to strip from the protein complex without dissociation of the complex itself.
Retention of Native-like Charge States
Native MS refers to the analysis of solution phase protein structure in the gas phase by using biologically relevant solution conditions and careful MS tuning parameters. It is becoming increasingly important for the study of non-covalent interactions and structures of proteins[25–27]. Typically, native-like structures are confirmed using IMS by matching experimentally measured collisional cross sections (CCS) to those determined from crystal structures[4]. Native structures have also been characterized in MS by their retention of low charge states as the average charge state is related to the solvent accessible surface area [28, 29]. Folded tertiary structures do not typically display all their basic residues to the surface of the droplet in the ESI process and therefore cannot be protonated [30–32]. Notably, native MS can be performed by using biological pH buffers and salts amenable to MS experiments [23, 28, 33, 34]. Furthermore, recent studies have suggested the ability to retain native structures using MS for the analyses of membrane proteins through the addition of detergents and buffers [26, 35, 36]. Detergent molecules surround membrane proteins in the form of micelles, acting as a substitute for a lipid membrane, to maintain their solubility [37]. During early stages of analysis in the gas phase, these detergent molecules can be removed in collision cells; however, careful tuning is necessary to prevent unfolding or activation of the protein upon removal of these detergents.
Although this instrument is not currently equipped with IMS, the charge state distributions of ubiquitin and AmtB were compared to previously published standards for native MS to evaluate whether native-like ions were preserved with REIS. The Clemmer group reported the retention of native compact conformers for the same dominant charge states observed in our experiment [38, 39]. In the current configuration and solution conditions, the 3+ charge state was observed and is lower than reported by our group and others, suggesting the REIS is a “cold” source [6, 40, 41].
Further evidence of native MS can be seen in Figure 3. Note that AmtB retains a Gaussian charge state distribution centered at 16+ using the REIS, which is similar to that obtained on the Synapt G1, where IMS data have previously shown the retention of a single, compact conformer for the low charge states centered at 16+[16]. Average charge states of AmtB were found to be 15.93 and 15.88 for the G1 and REIS, respectively. While charge state distributions are not a direct reflection of protein structure, lower charge states are indicative of native-like structures [42]. To further evaluate and tune the REIS, IMS would provide an optimal platform to validate and tune for native-like conditions on the orbitrap EMR instrument.
Importance and Illustration of Mass Resolution
Continual development in instrumentation over the past four decades has pushed mass resolution and resolving powers to heights that were unimaginable when MS was initially developed. The full potential of the REIS-orbitrap instrument could be realized for investigating heterogeneous lipid binding events to membrane proteins, such as for AmtB with Rm/z = 10,000. It has recently been shown by Patrick et al. that the binding of two different types of phospholipids to AmtB can be resolved using the Synapt G1 [43]. One of the major instrumental limitations cited in this work was a lack of mass resolution that limited the ability to distinguish between lipids of different type with similar molecular weights, which was overcome by using a non-natural lipid.
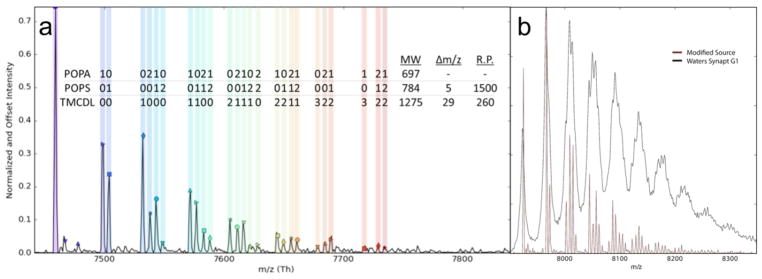
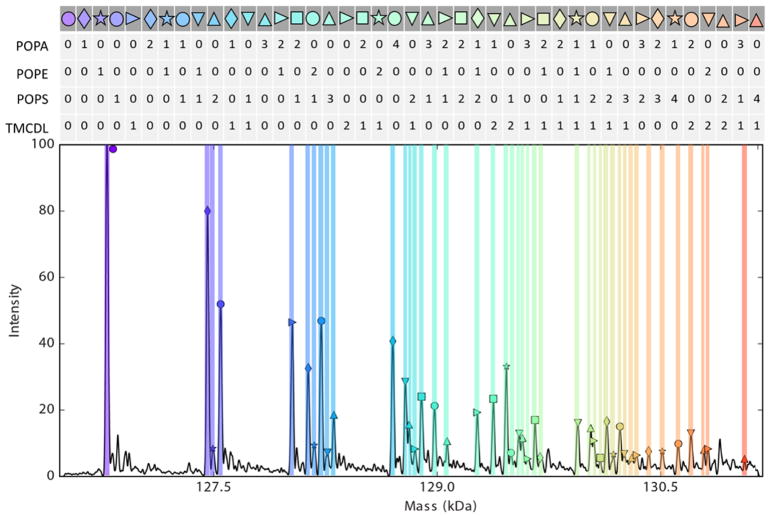
AmtB was mixed with three different natural phospholipids to demonstrate the utility of the REIS-orbitrap platform and its ability to overcome the issues described above. Figure 4 a shows a mass spectrum of AmtB in the presence of a lipid mixture containing POPA (697 Da), POPS (784 Da), and TMCDL (1285 Da) collected on the REIS-orbitrap and the Waters Synapt G1 instruments (Figure 4 b). With a Rm/z of ~500 using the Synapt G1, the determination of lipid binding events for the three different lipids was not resolved owing to the overlap of several peaks representing lipid bound species with similar masses. In contrast, up to 26 individual lipid binding events were, for the most part, baseline-resolved when the same analysis was performed on the REIS-orbitrap. In Figure 4, each colored band represents a different combination of lipids bound; the number of POPA, POPS, and TMCDL bound are shown above the respective peak. Binding of lipids with identical headgroups that differ in lipid tail composition to a small membrane protein using an orbitrap instrument has previously been shown by Gault et al. [44]. However, the presented mass spectrum represents the first example of fully resolved heterogeneous lipid binding events of three unique lipids. Here, we were able to baseline resolve and observe up to five individual lipids bound in total to AmtB that are composed of mixtures of all three lipids as highlighted in Figure 4. Clearly, at elevated m/z, lower resolution instruments do not allow for the same level of detailed analyses as higher resolution instruments. These limitations preclude such instruments from performing such complex experiments on large complexes.
Figure 4.
(a) REIS-orbitrap fully resolves AmtB bound to different combinations of POPA, POPS, and TMCDL. A total of twenty-six distinct lipid bound species were observed with up to five total lipids bound to AmtB. (b) Notably, none of these individual lipids species were resolved on the Waters Synapt G1 instrument.
To further demonstrate the possibilities of REIS-orbitrap instrument resolution, an AmtB sample was prepared in the presence of four natural phospholipids: POPA (697 Da), POPE (718 Da), POPS (784 Da), and TMCDL (1285 Da). Again, lipid binding events were observed and 45 AmtB-lipid species were assigned (Figure 5). However, it is clear the limitations of this instrument’s resolution are becoming realized with this complex mixture of lipids. Though the majority of species were fully resolved, several peaks were seen as shoulders of adjacent peaks. Continual progress in instrument development, protein purification, and lipid modifications will drive us towards these currently “unanswerable” discoveries in the future.
Figure 5.
RESI-orbitrap can resolve complex mixtures of AmtB bound to four different lipid species: POPA, POPE, POPS, and TMCDL. Forty-five lipid species bound to AmtB were observed.
Conclusions
Here, proof-of-concept experiments were performed to illustrate the ability to modify the orbitrap platforms with REIS. Mass spectra for ubiquitin and AmtB illustrate the retention of functionality of the commercial orbitrap instrument. Furthermore, the REIS was shown to be capable of the same mass spectral performance in terms of mass resolution as the unmodified instrument. Comparisons of the modified source to a Waters Synapt G1 demonstrate that the modified instrument is capable of recording native measurements. This development opens the door to future instrument modifications wherein additional gas phase coupled techniques such as IMS can be adapted to the existing Exactive platform architecture.
Acknowledgments
We wish to thank Will Seward of the Chemistry Department Machine Shop for the fabrication of all custom instrumentation, Greg Matthijetz of the Laboratory for Biological Mass Spectrometry for his electronics expertise, Junho Jeon for his design and development of the RF ion funnel, and Alexander Makarov for numerous helpful discussions relating to interfacing drift tube ion mobility to the orbitrap. Funding for this work was provided by the National Science Foundation (CHE-1707675) and National Institutes of Health (DP2GM123486 and GM121751-01A1).
References
- 1.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 2.von Helden G, Wyttenbach T, Bowers MT. Inclusion of a MALDI ion source in the ion chromatography technique: conformational information on polymer and biomolecular ions. Int J Mass Spectrom Ion Process. 1995;146–147:349–364. [Google Scholar]
- 3.Wittmer D, Chen YH, Luckenbill BK, Hill HH. Electrospray Ionization Ion Mobility Spectrometry. Anal Chem. 1994;66:2348–2355. [Google Scholar]
- 4.Jurneczko E, Barran PE. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst. 2011;136:20–28. doi: 10.1039/c0an00373e. [DOI] [PubMed] [Google Scholar]
- 5.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility–mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen SH, Russell DH. How Closely Related Are Conformations of Protein Ions Sampled by IM-MS to Native Solution Structures? J Am Soc Mass Spectrom. 2015;26:1433–1443. doi: 10.1007/s13361-015-1191-1. [DOI] [PubMed] [Google Scholar]
- 7.Cohen MJ, Karasek FW. Plasma Chromatography™—A New Dimension for Gas Chromatography and Mass Spectrometry. J of Chromatogr Sci. 1970;8:330–337. [Google Scholar]
- 8.Revercomb HE, Mason EA. Theory of plasma chromatography/gaseous electrophoresis. Review Anal Chem. 1975;47:970–983. [Google Scholar]
- 9.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J Mass Spec. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 10.Makarov AA. Mass spectrometer. US5886346A US Grant. 1999
- 11.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 12.Fort KL, van de Waterbeemd M, Boll D, Reinhardt-Szyba M, Belov ME, Sasaki E, Zschoche R, Hilvert D, Makarov AA, Heck AJR. Expanding the structural analysis capabilities on an Orbitrap-based mass spectrometer for large macromolecular complexes. Analyst. 2018;143:100–105. doi: 10.1039/c7an01629h. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim YM, Garimella SVB, Prost SA, Wojcik R, Norheim RV, Baker ES, Rusyn I, Smith RD. Development of an Ion Mobility Spectrometry-Orbitrap Mass Spectrometer Platform. Anal Chem. 2016;88:12152–12160. doi: 10.1021/acs.analchem.6b03027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keelor JD, Zambrzycki S, Li A, Clowers BH, Fernández FM. Atmospheric Pressure Drift Tube Ion Mobility–Orbitrap Mass Spectrometry: Initial Performance Characterization. Anal Chem. 2017;89:11301–11309. doi: 10.1021/acs.analchem.7b01866. [DOI] [PubMed] [Google Scholar]
- 15.Hagan N, Goldberg I, Graichen A, St Jean A, Wu C, Lawrence D, Demirev P. Ion Mobility Spectrometry - High Resolution LTQ–Orbitrap Mass Spectrometry for Analysis of Homemade Explosives. J Am Soc Mass Spectrom. 2017;28:1531–1539. doi: 10.1007/s13361-017-1666-3. [DOI] [PubMed] [Google Scholar]
- 16.Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A. Determining Membrane Protein–Lipid Binding Thermodynamics Using Native Mass Spectrometry. J Am Chem Soc. 2016;138:4346–4349. doi: 10.1021/jacs.6b01771. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer SA, Tang K, Anderson GA, Prior DC, Udseth HR, Smith RD. A novel ion funnel for focusing ions at elevated pressure using electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1813–1817. [Google Scholar]
- 18.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharon M. How far can we go with structural mass spectrometry of protein complexes? J Am Soc Mass Spectrom. 2010;21:487–500. doi: 10.1016/j.jasms.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Sharon M, Robinson CV. The Role of Mass Spectrometry in Structure Elucidation of Dynamic Protein Complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 21.Kitova EN, El-Hawiet A, Schnier PD, Klassen JS. Reliable Determinations of Protein–Ligand Interactions by Direct ESI-MS Measurements. Are We There Yet? J Am Soc Mass Spectrom. 2012;23:431–441. doi: 10.1007/s13361-011-0311-9. [DOI] [PubMed] [Google Scholar]
- 22.Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5:927. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 23.Heuvel RHHvd, Heck AJR. Native protein mass spectrometry: from intact oligomers to functional machineries. Curr Opin Chem Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Tosi P, Fontana G, Longano S, Bassi D. Transport of an ion beam through an octopole guide operating in the R.F.-only mode. Int J Mass Spectrom Ion Process. 1989;93:95–105. [Google Scholar]
- 25.Loo JA. Studying Noncovalent Protein Complexes by Electrospray Ionization Mass Spectrometry. Mass Spectrometry Reviews. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson CV, Chung EW, Kragelund BB, Knudsen J, Aplin RT, Poulsen FM, Dobson CM. Probing the Nature of Noncovalent Interactions by Mass Spectrometry. A Study of Protein CoA Ligand Binding and Assembly. J Am Chem Soc. 1996;118:8646–8653. [Google Scholar]
- 28.Benesch JLP, Ruotolo BT. Mass spectrometry: come of age for structural and dynamical biology. Curr Opin Chem Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall Z, Politis A, Robinson Carol V. Structural Modeling of Heteromeric Protein Complexes from Disassembly Pathways and Ion Mobility-Mass Spectrometry. Structure. 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Kaltashov IA, Mohimen A. Estimates of Protein Surface Areas in Solution by Electrospray Ionization Mass Spectrometry. Anal Chem. 2005;77:5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Santambrogio C, Brocca S, Rossetti G, Carloni P, Grandori R. Conformational effects in protein electrospray-ionization mass spectrometry. Mass Spectrom Rev. 2016;35:111–122. doi: 10.1002/mas.21465. [DOI] [PubMed] [Google Scholar]
- 32.Testa L, Brocca S, Grandori R. Charge-Surface Correlation in Electrospray Ionization of Folded and Unfolded Proteins. Anal Chem. 2011;83:6459–6463. doi: 10.1021/ac201740z. [DOI] [PubMed] [Google Scholar]
- 33.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 34.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT. Ion Mobility Analysis of Molecular Dynamics. Annu Rev Phys Chem. 2014;65:175–196. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 35.Mehmood S, Allison TM, Robinson CV. Mass Spectrometry of Protein Complexes: From Origins to Applications. Annu Rev Phys Chem. 2015;66:453–474. doi: 10.1146/annurev-physchem-040214-121732. [DOI] [PubMed] [Google Scholar]
- 36.Laganowsky A, Reading E, Hopper JTS, Robinson CV. Mass Spectrometry of Intact Membrane Protein Complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles Protect Membrane Complexes from Solution to Vacuum. Science. 2008;321:243. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 38.El-baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE. Melting Proteins: Evidence for Multiple Stable Structures upon Thermal Denaturation of Native Ubiquitin from IMS-MS Measurements. J Am Chem Soc. 2017;139:6306–6309. doi: 10.1021/jacs.7b02774. [DOI] [PubMed] [Google Scholar]
- 39.Shi H, Clemmer DE. Evidence for Two New Solution States of Ubiquitin by IMS–MS Analysis. J Phys Chem B. 2014;118:3498–3506. doi: 10.1021/jp4097327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kostyukevich Y, Kononikhin A, Popov I, Nikolaev E. Conformational changes of ubiquitin during electrospray ionization as determined by in-ESI source H/D exchange combined with high-resolution MS and ECD fragmentation. J Am Soc Mass Spectrom. 2014;49:989–994. doi: 10.1002/jms.3409. [DOI] [PubMed] [Google Scholar]
- 41.Shi H, Atlasevich N, Merenbloom SI, Clemmer DE. Solution Dependence of the Collisional Activation of Ubiquitin [M+7H](7+) Ions. J Am Soc Mass Spectrom. 2014;25:2000–2008. doi: 10.1007/s13361-014-0834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall Z, Robinson CV. Do Charge State Signatures Guarantee Protein Conformations? J Am Soc Mass Spectrom. 2012;23:1161–1168. doi: 10.1007/s13361-012-0393-z. [DOI] [PubMed] [Google Scholar]
- 43.Patrick JW, Boone CD, Liu W, Conover GM, Liu Y, Cong X, Laganowsky A. Allostery revealed within lipid binding events to membrane proteins. Proc Natl Acad Sci U S A. Mar 5;108 doi: 10.1073/pnas.1719813115. pii: 201719813. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gault J, Donlan JAC, Liko I, Hopper JTS, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, Wu B, Kleanthous C, Belov M, Damoc E, Makarov A, Robinson CV. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]