Abstract
Recent studies point to the evolution of drug resistance in lung cancer as being centered, at least in part, on the upregulation of various genes involved in controlling efflux or drug inactivation. Among the most important of these genes is Nuclear Factor Erythroid 2-Related Factor (NRF2), considered the master regulator of 100–200 target genes involved in cellular responses to oxidative and/or electrophilic stress. With increased focus on the development of combinatorial approaches for cancer treatment, we utilized CRISPR/Cas9 to disable the NRF2 gene in lung cancer cells by disrupting the NRF2 nuclear export signal (NES) domain; phenotypically, the protein is largely blocked from transiting into the nucleus after translation. In tissue culture, cells with this gene knockout were found to have a reduced proliferation phenotype and are more sensitive to chemotherapeutic agents, such as cisplatin and carboplatin. These observations were confirmed in xenograft mouse models wherein the homozygous knockout cells proliferate at a slower rate than the wild-type cells, even in the absence of drug treatment. Tumor growth was arrested for a period of 16 days, with a dramatic decrease in tumor volume being observed in samples receiving the combined action of CRISPR-directed gene editing and chemotherapy.
Keywords: CRISPR, Cas9, gene editing, NRF2, lung cancer, chemoresistance, NSCLC
Introduction
Lung cancer is the leading cause of cancer mortality in the United States, accounting for more than 1 in 4 cancer deaths. It kills more people than breast, prostate, and colon cancer combined;1 yet, despite these grim statistics, there are reasons to be optimistic about the potential to reduce mortality. Advances in treatment have shown promise, and emerging targeted treatments (see Hirsch et al.2) for various forms of lung cancer will soon be made more widely available. Some of these therapies include the use of endothelial growth factor receptor (EGFR) monoclonal antibodies and vascular EGFR inhibitors.2, 3, 4 EGFR tyrosine kinase inhibitors continue to be a superior choice as first-line treatment in patients with EGFR mutation-positive non-small-cell lung cancer (NSCLC).5, 6, 7, 8 Despite these positive results, however, EGFR mutations account for approximately 17% of the driver mutations in lung adenocarcinoma. The other 85% of the mutations reside in genes such as K-RAS, ALK, HER2, or in unknown genes, demonstrating the need to design combinatorial strategies even when some specific mutations in target genes are known.
To this end, immunotherapy is also becoming part of cancer treatment plans, but transformative clinical benefit is often limited to the patients containing infiltrated T cells or biomarkers of a specific type.9, 10 As such, combinatorial strategies for immunotherapy are now being clinically evaluated in a similar fashion to other strategies for tyrosine kinase inhibitors. Chemotherapy remains an important option in the treatment of lung cancer, but issues involving efficacy and toxicity can become problematic with extended care. In most cases, resistance to a variety of chemotherapy drugs can develop with extended treatment.11 Pharmacogenomic studies point to the evolution of drug resistance being centered on the upregulation of the variety of genes involved in controlling the efflux of anticancer drugs or directing transcriptional activation among others.
Nuclear Factor Erythroid 2-Related Factor (NRF2) is considered the master regulator of 100–200 target genes involved in cellular responses to oxidative and/or electrophilic stress. Targets include GSH mediators, antioxidants, and genes controlling efflux pumps.12 NRF2 is also known to regulate the expression of genes involved in protein degradation and detoxification, and it is negatively regulated by Kelch-like ECH-associated protein 1 (KEAP1), a substrate adaptor for the Cul3-dependent E3 ubiquitin ligase complex. Under normal conditions, Keap1 constantly targets NRF2 for ubiquitin-dependent degradation, maintaining a low expression of NRF2 on downstream target genes. However, chemotherapy has been shown to activate transcriptional activity of the NRF2 target genes, often triggering a cytoprotective response; enhanced expression of NRF2 occurs in response to environmental stress or detrimental growth conditions. Other mechanisms that lead to NRF2 upregulation include mutations in KEAP1 or epigenetic changes of the promoter region. The upregulation of NRF2 expression leads to an enhanced resistance of cancer cells to chemotherapeutic drugs, which by their very action induce an unfavorable environment for cell proliferation. Indeed, Hayden et al.12 have clearly demonstrated that increased NRF2 expression leads to the resistance of cancer cells to chemotherapeutic drugs, including cisplatin. Singh et al.13 also showed that constitutive expression of NRF2 leads to radioresistance and inhibition of NRF2 causes increased endogenous reactive oxygen species (ROS) levels as well as decreased survival. Recently, Torrente et al.14 identified crosstalk between NRF2 and the homeodomain-interacting protein kinase to HIPK2, demonstrating that HIPK2 exhibits a cytoprotective effect through NRF2. This critically important work further establishes our foundational understanding of the NRF2 transcriptional network and circuitry, and it strengthens the argument for promoting NRF2 as a target for genetic manipulation and sensitization of cancer cells to chemotherapeutic drugs.
Furthermore, Tang et al.15 and Ren et al.16 demonstrated the use of NRF2 inhibitors, luteolin and brusatol, to enhance the efficacy of chemotherapeutic drugs in various cancer cell types, as well as xenografts. The use of luteolin causes significant reductions in mRNA and protein levels of NRF2 and downstream target genes, and it sensitizes A549 cells to chemotherapeutic drugs. The use of brusatol was shown to increase sensitivity of A549 cells through enhanced ubiquitination and degradation of NRF2. This work was extended in vivo using an A549 xenograft, and it demonstrated decreased proliferation and growth as well as increased apoptosis when tumors are co-treated with brusatol and cisplatin. Thus, the concept of utilizing the inhibition of NRF2 as a supplemental approach to cancer treatment had been proposed.
With an increased focus on the development of combinatorial strategies and taking into consideration the role of NRF2 in chemo-resistance, we developed a treatment strategy that combines CRISPR-directed gene editing17, 18, 19, 20 with traditional chemotherapy. The overall strategy is to design and utilize a CRISPR/Cas9 gene-editing tool to disable the NRF2 gene in lung cancer cells, rendering it incapable of producing a functional protein. Cells with this gene knockout should then be more sensitive to chemotherapeutic agents, such as cisplatin, carboplatin, and vinorelbine, as the genes responsible for efflux of anticancer drugs would not be activated, even under the most environmentally stressful conditions. Coupling gene editing with chemotherapy is only feasible now with development of the CRISPR/Cas9 gene-editing system, which has advanced the field dramatically over the past 4 years. It is now possible to precisely knock out genes from any eukaryote or prokaryote with unprecedented specificity and efficiency. The CRISPR/Cas9 complex aligns in homologous register with the target gene, which enables it to execute a double-stranded DNA break. This action is followed by an attempt by the cell to reclose scission, most often through a process known as non-homologous end joining (NHEJ). The reclosure is often imperfect and unfaithful as a number of nucleotides are lost during the process, resulting in a genetic frameshift and the subsequent production of nonfunctional transcripts, a gene knockout. Here we report the successful knockout of the NRF2 gene using CRISPR/Cas9 in chemo-resistant A549 lung cancer cells, with the subsequent demonstration of increased effectiveness of the anticancer drugs cisplatin, carboplatin, and vinorelbine in both culture and a xenograft mouse model.
Results
Creation of NRF2-Knockout Clonal A549 Cell Lines Using a CRISPR-Directed Gene-Editing Approach
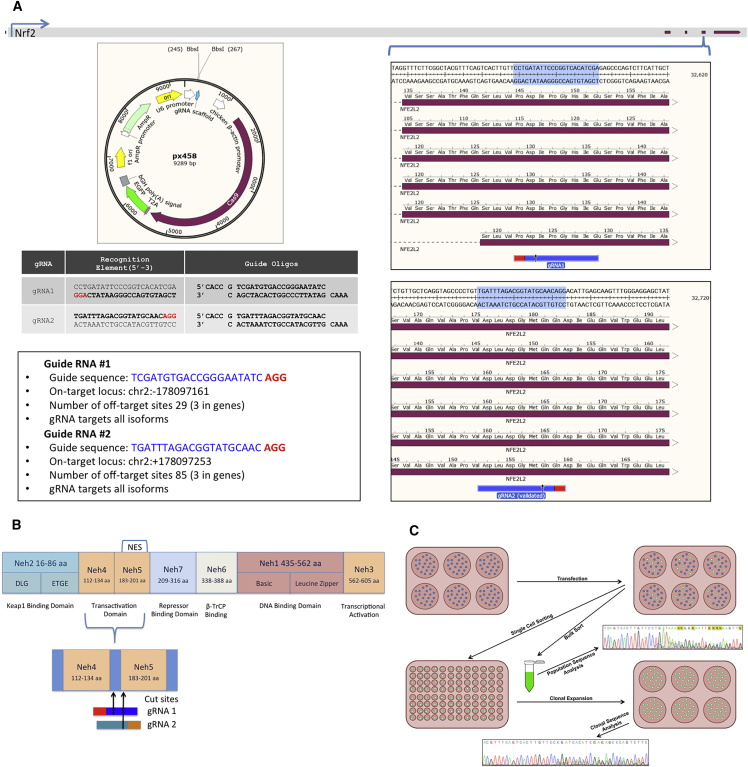
Our strategy was to use CRISPR-directed gene editing to functionally disable NRF2 alleles in A549 cells. It is critical to establish the fact that a gene-editing technology can knock out a target gene. Below we provide the strategy details, which were utilized to generate the genetic tools used to disable NRF2 in A549 cells. Figure 1A illustrates the CRISPR/Cas9 machinery designed to target and knock out NRF2. The gray bar running along the top of the panels represents the genomic sequence of NRF2, with the red blocks indicating coding regions. The blue brackets indicate the relative region where each CRISPR/Cas9 is designed to cleave the DNA. Each guide RNA (gRNA) was designed to target the fourth exon of NRF2 in a region that contains all known isoforms to ensure complete ablation of the gene (https://www.ncbi.nlm.nih.gov/gene/4780). The gRNA with the highest score, according to the Broad Institute’s CRISPR Design software (http://crispr.mit.edu/), was chosen for gRNA1, and a previously validated gRNA21 was chosen for gRNA2. The gRNAs were assembled by annealing the CRISPR RNA (crRNA) oligos and ligating them to complementary restriction site overhangs in plasmid px458 (Addgene 48138) digested with BbsI, as depicted in each panel.
Figure 1.
CRISPR Design and NRF2-Knockout Experimental Workflow
NRF2-coding regions containing the six known genetic isoforms were utilized for targeting by CRISPR/Cas9. (A) The gRNA sequences, along with their chromosomal loci and cloning details, are displayed. (B) The structural domains and location of CRISPR-directed gene editing of the NRF2 protein. (C) The experimental workflow for testing the efficiency of CRISPR/Cas9 knockout of NRF2 in a targeted population and in isolated and expanded clonal cell lines.
Figure 1B illustrates the functional domains of the NRF2 protein, including the KEAP1-binding domain, transactivation domain, repressor-binding domain, β-TrCP-binding domain, DNA-binding domain, and the transcriptional activation domain.22, 23, 24 The Neh5 domain spans exons 4 and 5 and contains a redox-sensitive nuclear export signal (NES), which regulates the intracellular localization of NRF2.23 In theory, by disrupting the gene and/or protein within the Neh4 and Neh5 domains, the NES is shifted, rendering it nonfunctional. Figure 1C exhibits the experimental workflow beginning with the transfection of pX458, containing either gRNA1 or gRNA2, into A549 lung adenocarcinoma cells and progressing through to the final step of allelic analyses of an individual clonal population.
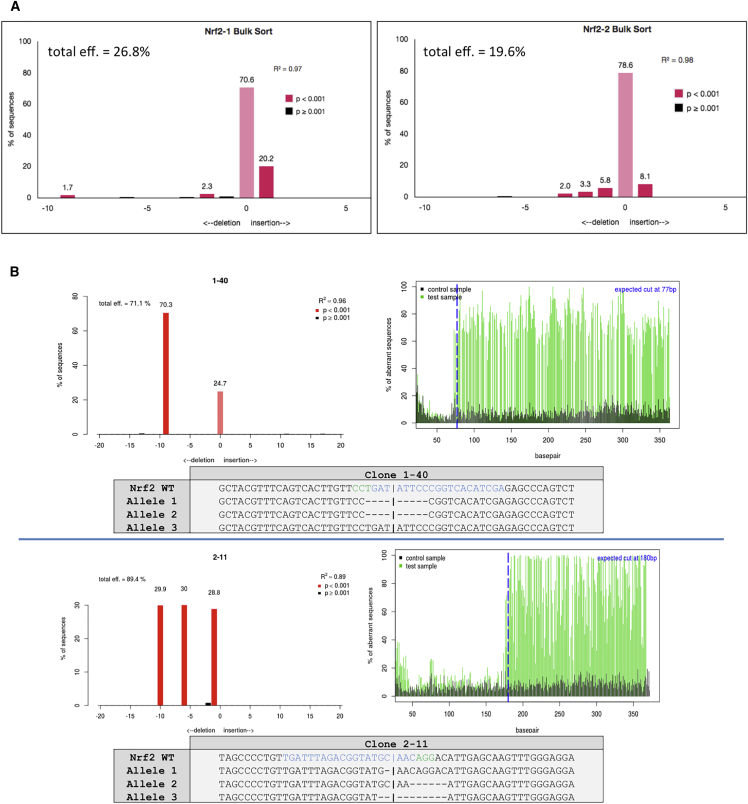
Importantly, plasmid pX458 contains an EGFP reporter, which allows for the isolation of individual transfected cells by fluorescence-activated cell sorting (FACS). To evaluate the efficiency of CRISPR-directed NRF2 knockout in the total targeted population, EGFP+ cells were isolated as a population, and the degree of genetic disruption at the NRF2 locus in cells, transfected with either gRNA1 or gRNA2 pX458, was determined. The sorted populations were Sanger sequenced, and the resulting trace files were analyzed for the presence of insertions or deletions (indels), a marker for gene disruption. These data were obtained using a program known as Tracking of Indels by Decomposition (TIDE).25 As represented in Figure 2A, both CRISPR/Cas9 designs generated a significant amount of indels, evidenced by TIDE results, indicating a high degree of NRF2 disruption. These results validate the approach and indicate that disruption of the NRF2 is possible in A549 cells via CRISPR/Cas9.
Figure 2.
Genomic Analyses of NRF2-Knockout Clones
(A) Bulk-sorted GFP+ A549 cells transfected with either gRNA1 or gRNA2 were Sanger sequenced and analyzed for indel activity by TIDE. (B) Clonally isolated NRF2-targeted cells were genomically analyzed for CRISPR/Cas9-induced NHEJ activity. Genomic DNA was Sanger sequenced and TIDE was used to develop the indel spectrums, sequence decompositions, and allelic patterns of NRF2, as shown for clones 1-40 and 2-11.
Next, the same experiment was carried out, except, in this case, individual cells were isolated by FACS to obtain single-cell clonal expansions. When the single-cell isolates had expanded to sufficient quantity, half of each clonal population was cryo-preserved, and allelic sequence analysis was performed on the other half using the same strategy and method described above. Figure 2B displays the allelic analyses of two clones, 1-40 and 2-11, derived from gRNA1- and gRNA2-transfected cells, respectively, which were chosen from a total of nine (Figure S1) for subsequent experimentation and analyses. It soon became apparent that all isolated clones generated from this parental lot of A549 cells obtained from ATCC harbored three alleles at the NRF2 locus. The red columns indicate the indel sizes present and their respective representative ratios within that clone. Clone 1-40 contained a 9-bp deletion with a 2:1 ratio to 0-bp indels, revealing a heterozygous knockout of NRF2. Clone 2-11 contained a 10-bp deletion, a 6-bp deletion, and a 1-bp deletion at a 1:1:1 ratio, a homozygous knockout of NRF2. The specific indel patterns on each allele of both clones were characterized by manually aligning the sequence trace files to the wild-type sequence, with the TIDE indel data as a guide. For convenience, we refer to clone 1-40 as a heterozygous knockout and clone 2-11 as a homozygous knockout. Figure S1 provides a listing of the large population of clonally isolated and expanded A549 cell lines bearing various allelic combinations of NRF2 disruption.
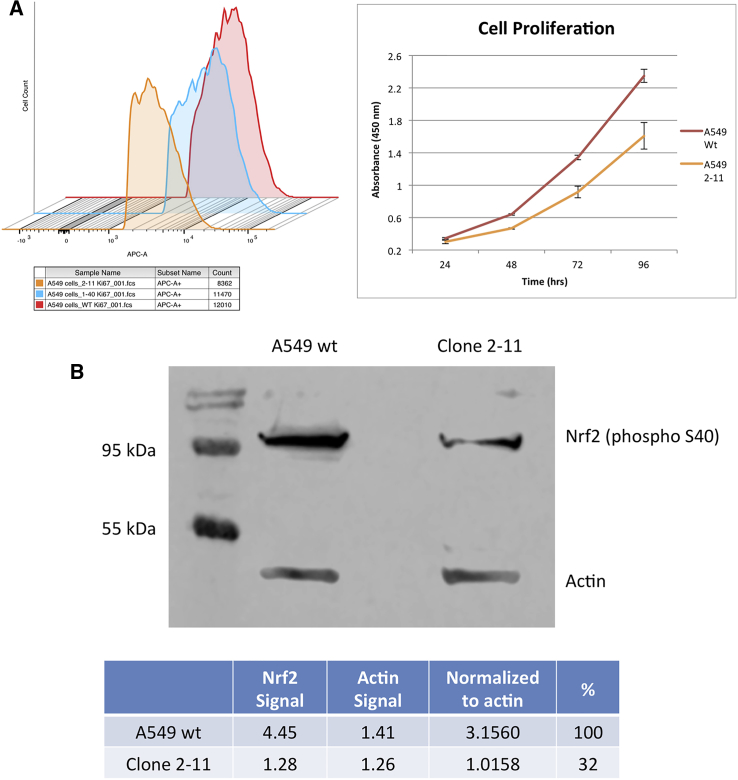
A fundamental cellular phenotype that could be affected by the lack of an NES is the rate at which cells proliferate in culture.26, 27 Wild-type A549 cells typically have a doubling time of 24 hr, however, it was noted that clone 1-40, but more clearly 2-11, grew slower in the clonal expansion process (data not shown). This observation prompted us to further investigate the proliferation profile of clones 1-40 and 2-11 by staining the cells with antibodies against Ki67, followed by FACS analysis. Ki67 is a nuclear antigen expressed in actively proliferating cells. Therefore, one might predict a decrease in Ki67 expression based on the growth characteristics seen in cell culture. Ethanol-fixed cells were stained with Alexa Fluor 647 Anti-Ki67, analyzed by FACS, and plotted as a histogram (Figure 3A, left panel). Nonspecific binding was controlled for using the mouse immunoglobulin G (IgG)1 κ isotype control provided and gated on FlowJo. The x axis represents fluorescence intensity of the allophycocyanin (APC)-conjugated Anti-Ki67, of which a shift to the left can be seen in clone 2-11, indicating a decrease in fluorescence intensity, correlating to a decrease in cellular proliferation.
Figure 3.
Cellular Proliferation Profile of NRF2-Knockout A549 Cells and Western Blot Analysis
Cells were fixed with ethanol for 72 hr and stained with Alexa Fluor 647 Anti-Ki67. (A) Fluorescence-activated cell sorting (FACS) was used to capture the intensity of Ki67-stained cells and plotted as a histogram using FlowJo software (left panel). Cell proliferation was measured via bioreduction of MTS to a formazan product, plotted as mean raw absorbance values (right panel), and error bars represent ± SEM. (B) Western blot analysis of wild-type A549 cells and NRF2-knockout 2-11 cells using an antibody for phosphorylated NRF2.
The decrease in proliferation in 2-11 cells was striking while the reduction in proliferation in 1-40 cells was modest at best. Thus, we decided to continue our studies on the effect of NRF2 knockout using 2-11 cells, only because CRISPR-directed gene editing succeeded in disrupting function more completely in those cells. Based on growth characteristics in cell culture and the FACS analysis, the MTS assay was utilized to assess the proliferation of 2-11 cells compared to wild-type cells (Figure 3A, right panel). Allelic analysis of clone 2-11 indicated that NRF2 is genetically disabled, and, when normalized to beta-actin and compared to wild-type A549 cells, clone 2-11 showed a knockdown of ∼68% (Figure 3B). Since one of the three alleles in clone 2-11 maintained a functional reading frame, this result was not unexpected. Genetic analysis indicated that the Neh5 domain, which contained the NES, was disrupted.28, 29 Thus, we moved forward to characterize this clone, identifying it as a functional knockout.
Chemosensitivity Is Increased in NRF2-Knockout A549 Cell Lines
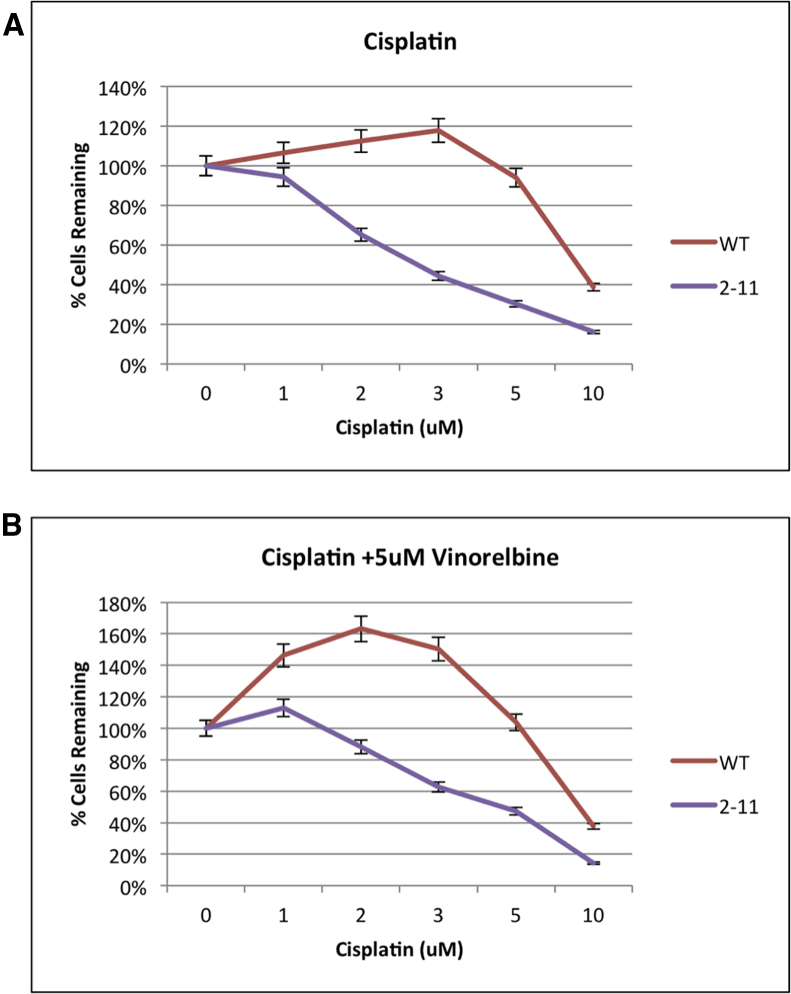
To examine the chemosensitivity of the genetically engineered NRF2-deficient A549 cell lines, the MTS assay, depicted in Figure 4, was utilized. In Figure 4A, wild-type and 2-11 A549 cells were exposed to increasing dosages of cisplatin. After 72 hr, cisplatin was removed, and the MTS reagent was added for 3 hr after which time the population was measured for the absorbance of formazan. The data showed that, as predicted, wild-type A549 cells were resistant to high dosages of cisplatin.30 In fact, wild-type A549 cells displayed a slight increase in cell proliferation up to 3 μM cisplatin before proliferation was adversely affected at the final concentrations of 5 and 10 μM, respectively. In the genetically engineered knockout cell lines, we clearly observed an increase in chemosensitivity in a dose-dependent fashion. The 2-11 homozygous knockout cells displayed a heightened sensitivity, evident even at the lowest dose with a loss of proliferation at concentrations at and above 1 μM. Thus, it is possible that we were observing a gene dosage effect of sorts, in that the heterozygous cell line exhibited more resistance to cisplatin than homozygous knockout cells because it contained at least one viable gene copy.
Figure 4.
Proliferation Capacity of Wild-Type and NRF2-Modified A549 Cells (2-11) in Response to Chemotherapeutic Drugs
Proliferation was measured via bioreduction of MTS to a formazan product. (A and B) Cells were treated with increasing dosages of cisplatin (A) and increasing dosages of cisplatin with 5 μM vinorelbine (B) for 72 hr, then evaluated for cell proliferation. Error bars represent ± SEM.
In Figure 4B, we display a result from cells being exposed to the same increasing amount of cisplatin as described for Figure 4A, except vinorelbine was added to a final concentration of 5 μM. Vinorelbine is an established companion to cisplatin and combinatorial chemotherapeutic regimens for NSCLC.31 The wild-type A549 cells again displayed a dramatic increase in proliferation even at lower doses, and they did not show elevated sensitivity until the dosage surpassed 5 μM; but, the knockout cell line (2-11) displayed increased sensitivity to the combinatorial drug therapy. Carboplatin, a related anticancer drug and commonly used chemotherapy for NSCLC,31 was also evaluated for enhanced chemosensitivity in these genetically engineered A549 cells, and the cell-killing response reflected what was observed in the experiments using cisplatin (data not shown).
Genetically Reengineered A549 Cells Showed a Slower Growth Rate and Increased Chemosensitivity in a Xenograft Mouse Model
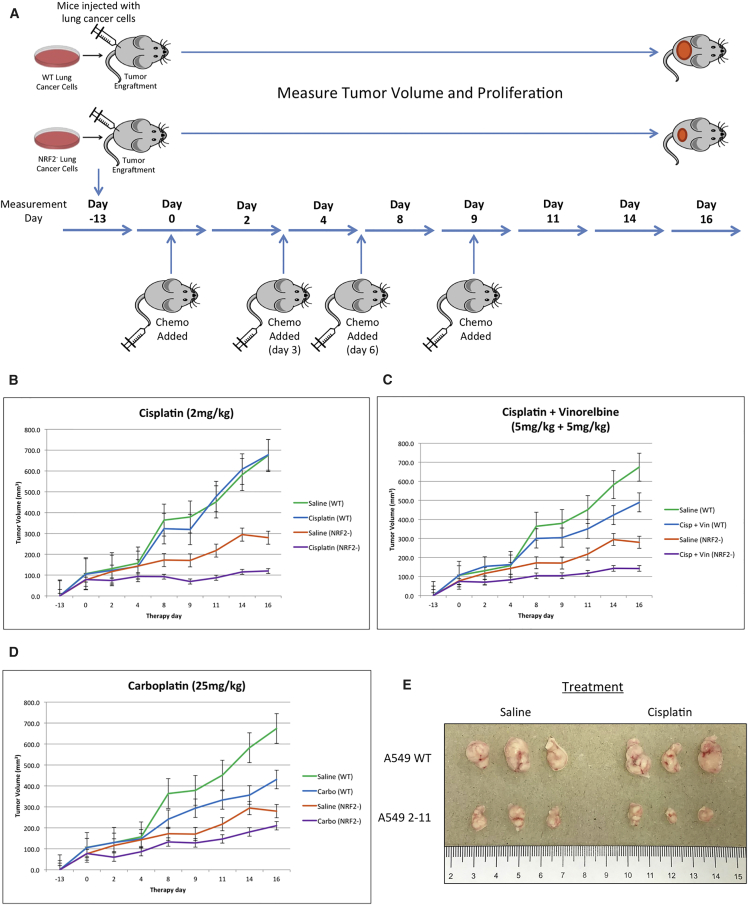
Since CRISPR/Cas9-mediated NRF2 knockdown increased chemosensitivity in A549 cells in vitro, we examined enhanced chemosensitivity driven by gene editing in a xenograft mouse model. The homozygous knockout A549 cells (clone 2-11) and wild-type A549 cells (control group) were implanted into the back of a nude mouse, and the cells (5 × 106 per cell line) were allowed to proliferate into a tumor with a diameter of approximately 100 mm3. The workflow is depicted in Figure 5A. As part of the strategy, the chemotherapeutic agent was added at days 0, 3, 6, and 9, respectively, through tail vein injection, as indicated in the diagram. Tumor growth through volume and proliferation were measured over the course of 16 days, starting at the time of the first injection of the chemotherapeutic agent, day 0, and the results are presented in Figures 5B–5D.
Figure 5.
Restored Chemosensitivity in Mice with NRF2 Knockout in Tumors
(A) Experimental workflow of mouse xenograft. Athymic nude mice were subcutaneously injected with either wild-type A549 cells or NRF2-knockout A549 cells, and, once established tumors reached 100 mm3, they were treated with the first dose of chemotherapy on day 0. Mice were subsequently treated with chemo on days 3, 6, and 9. Tumor volumes were measured daily for 16 days until tumors reached 2,000 mm3. (B–D) Wild-type A549 or NRF2-knockout A549 tumors were treated with 2 mg/kg cisplatin (B), 5 mg/kg cisplatin with 5 mg/kg vinorelbine (C), 25 mg/kg carboplatin (D), or saline, and tumor size was measured for 16 days. Error bars represent ± SEM. Tumors (treated with 2 mg/kg cisplatin or saline) were extracted from both the wild-type A549 and NRF2-knockout A549 (2-11) implanted mice. (E) Representative tumors from each group are shown (n = 3).
Figure 5B depicts the results of tumor growth over the course of 16 days. As expected, proliferation of wild-type A549 cells, treated with either saline or 2 mg/kg cisplatin, was not inhibited by the drug, confirming the well-established resistance of A549 cells to cisplatin. The NRF2-knockout xenograft proliferated in the mouse but at a reduced rate, even without the addition of cisplatin. These data support published observations and results from our own cell culture work; an elevated level of NRF2 in cancer cells has been shown to promote cancer cell proliferation,27 and small interfering RNA (siRNA)-directed inhibition30 of the NRF2 transcript slows proliferation of A549 cells. The most dramatic effect was seen when a combinatorial approach was taken, wherein NRF2-knockout cells were treated with cisplatin over a period of 16 days. In this case, proliferation of the implanted cells was arrested, and the tumor size was maintained at the same level throughout the course of the experiment, which confirms our previous results generated from experiments conducted in cell culture (Figure 4).
Figure 5C depicts similar results when fixed concentrations of 5 mg/kg cisplatin and 5 mg/kg vinorelbine were used in combination following the same xenograft mouse experimental protocol. Interestingly, the wild-type A549 cells appeared to be more sensitive to this combination of drugs. This observation may reflect the synergistic effect that vinorelbine has on cisplatin killing of A549 cells, providing an important internal control that our experimental system recapitulates previously known outcomes. Once again, the homozygous knockout 2-11 cell line proliferated at a slower rate than the wild-type cells in the absence of drug treatment, but the combination of NRF2 knockout and drug treatment led to a cessation of tumor growth and maintenance of tumor size over the course of 16 days. The same response was seen once again in the data presented in Figure 5D, wherein 25 mg/kg carboplatin was injected into the tail vein; the same reduced proliferation and growth trend described above was reproduced. These results suggest that the combination of gene editing in chemotherapy produces an enhanced chemosensitivity in A549 cells, both in cell culture and in a xenograft mouse model.
Analyses of A549 Tumor Proliferation
Representative tumor samples were harvested from four groups (wild-type A549 with saline, wild-type A549 with 2 mg/kg cisplatin, knockout 2-11 with saline, and knockout 2-11 with 2 mg/kg cisplatin) (n = 3 for each group). As shown in Figure 5E, a distinct difference among the four extracted tumor groups was apparent. As described above, tumors generated from wild-type cells proliferated aggressively within the xenograft mouse model in the absence or presence of cisplatin. The NRF2-knockout cell line proliferated more slowly than the wild-type, even in the absence of the drug. But, the smallest tumors were observed in all the samples from mice bearing NRF2-knockout cells treated with cisplatin.
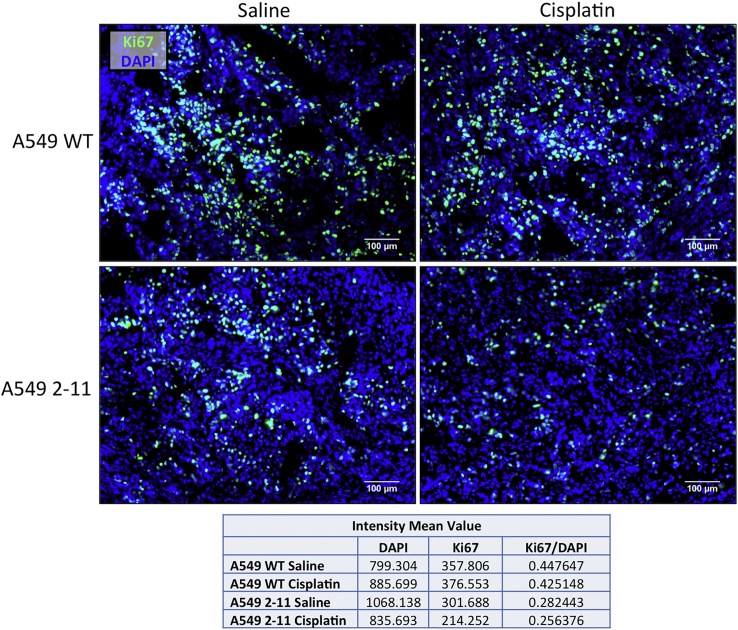
Since A549 2-11 knockout xenograft tumors exhibited smaller tumor volume compared to their wild-type counterparts, we wanted to examine the proliferative activity within the tumors using Ki67, a well-known marker for proliferation, which presents during all active phases of cell cycle (G1, S, G2, and mitosis).32 As shown in Figure 6, within A549 cells treated with only saline, abundant Ki67-positive cells were observed; tumors extracted from mice treated with cisplatin produced similar levels of Ki67-positive cells. In the case of tumors generated from 2-11 cells, Ki67 staining was noticeably decreased, and treatment with cisplatin resulted in even lower levels of Ki67, suggesting cisplatin enhances the response of the knockout 2-11 cells by slowing down proliferation even further.
Figure 6.
Proliferation of Xenograft Tumors
Representative images of xenograft tumors extracted from mice, implanted with either wild-type A549 or NRF2-knockout A549 cells (2-11), 16 days after the initial treatment with either 2 mg/kg cisplatin or saline, were sectioned and stained with Ki67 (green) and DAPI (blue). Fluorescence intensity mean values for DAPI and Ki67 were obtained for the images using the Zeiss Zen software, and relative values were obtained for fluorescence intensity of Ki67. Scale bars represent 100 μm.
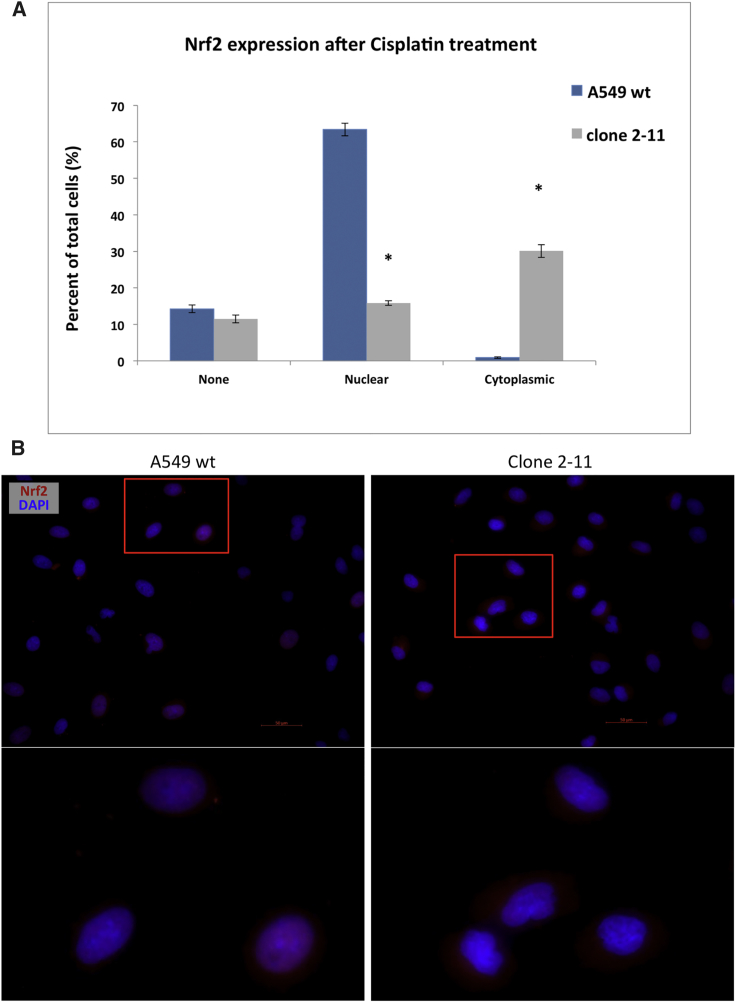
Taken together, the accumulated data build a strong case for clone 2-11 as a functional knockout, since these cells enabled a higher sensitivity to chemotherapy as compared to the wild-type counterpart. We sought to provide some explanation for this phenotype observed in both cell culture and in the mouse. The effect of the disruption of the NES region located in the Neh5 domain of NRF2 was further characterized using immunocytochemistry. Wild-type A549 and clone 2-11 cells were pre-treated with 2 μM cisplatin to stimulate NRF2 expression. Random fields of each cell sample were identified and imaged and total cell counts were determined. Cells were quantified based on the following observed outcomes: no staining of NRF2, nuclear staining only, or cytoplasmic staining only. Figure 7A represents the average quantification of multiple replicates of several experiments, with at least 10 fields of view incorporated into the dataset. We observed a statistically significant difference in the degree of nuclear localization of NRF2 between wild-type A549 and clone 2-11 cells, respectively. In wild-type cells, the majority of NRF2 was located in the nucleus, while in the functional knockout cell line (2-11), NRF2 was predominantly found in the cytoplasm, as seen in Figure 7B. The images of cisplatin-induced wild-type and knockout cells (Figure 7B) reflect the data presented in Figure 7A.
Figure 7.
Cisplatin-Induced Nuclear and Cytoplasmic Localization of NRF2 in Wild-Type A549 Cells and Clone 2-11
Cells were treated with 2 μM cisplatin, fixed, and stained for NRF2. Immunocytochemistry was performed using fluorescence microscopy. Random fields were imaged and the total number of cells per field was counted. (A) The percentage of NRF2-positive stained cells over the total cells analyzed in each category was plotted in this graph. Error bars represent ± SEM; *p < 0.05, Student’s t test. (B) Representative images of nuclear and cytoplasmic localization of NRF2 in wild-type A549 and NRF2-knockout A549 (2-11) cells. Scale bars represent 50 μm.
Discussion
It is becoming increasingly apparent that CRISPR-directed gene editing will have a significant impact on the development of new therapeutic approaches to cancer and inherited diseases. These genetic tools can identify and execute DNA cleavage, at specific sites within the chromosome, at a surprisingly high efficiency and with an improved precision.33 The natural activity of CRISPR/Cas9 is to disable a viral genome infecting a bacterial cell,34, 35 and subsequent genetic reengineering of CRISPR/Cas9 function in human cells presents the possibility of disabling human genes at a significant frequency. Our goal has been to utilize specific gene disruption catalyzed by CRISPR/Cas9 to improve the effectiveness of commonly used anticancer treatments, such as chemotherapy or immunotherapy. In this case, we targeted the NRF2 gene because it is a central regulator of cellular detoxification and response to oxidative and electrophilic stresses.14, 36, 37 NRF2 expression increases when the cell enters a stressful environment, such as encountering a toxic substance. Thus, by disrupting NRF2, we hypothesize that chemotherapeutic agents, such as cisplatin and carboplatin, would work more effectively and at lower dosages. In the broader sense, such an approach would ultimately lead to a reduced level of chemotherapy required to produce the same tumor-killing activity, leading to an improvement in the quality of life of a cancer patient. We built our experimental strategy around the well-established non-small-cell lung adenocarcinoma cell line A549, because it harbors a mutation in the Kelch domain of KEAP1 causing the overexpression of NRF2,30 and it has been used often as a gold standard for the discovery of novel therapeutic agents directed against cancer.
Initialization of the gene disruption process starts with the design of the CRISPR gRNA, which bears complementarity to the target site. It is wise to utilize two separate designs in case chromosomal access to a DNA sequence reduces the capacity of one complex to execute double-strand DNA breakage. We designed and synthesized two gRNAs to target different sites within the NRF2 gene-coding region. Both CRISPRs, when coupled with the expressed Cas9 protein, did in fact lead to gene disruption. The two genetically engineered cell lines, 1-40 and 2-11, used in this work were generated by gRNA1 and gRNA2, respectively. It is also important to identify multiple gRNAs, as human genes consisting of multiple exons can produce several protein isoforms upon translation. We illustrate the importance of identifying gRNA1 and gRNA2 because both will initiate gene disruption of all known NRF2 isoforms. Our genetic analysis of the total population of targeted cells led to the conclusion that CRISPR-directed gene editing had taken place, as evidenced by the emergence of specific indel patterns, insertions and deletions, within the targeted region. Subsequent single-cell cloning and expansion produced two cell lines, 1-40 and 2-11, a heterozygous knockout and a homozygous knockout, respectively. It is important to note that, when dealing with transformed cells, multiple copies (>2) of alleles can be present, as was the case in the A549 cells utilized herein. For our purposes, this tri-allelic arrangement did not affect our results, but it is important to identify the true allelic composition of cell lines used in experiments designed to evaluate potential therapeutics. The CRISPR-directed gene-editing system was designed to disable the NES domain, which reduces the capacity of the protein to re-enter the nucleus and activate the transcription factor. Thus, even though we see NRF2 protein being produced, its functional activity has been significantly disabled. Transcriptional activity is central to the control of the expression of the various genes known to enable chemoresistance. Our data suggest that a functional knockout can be as effective as a genetic knockout. Our observation of significant reduction in cell proliferation in a homozygous knockout encouraged us to proceed forward with analyzing chemosensitivity in the disrupted clone.
NRF2 has been a target of interest for other investigators seeking to disrupt its function to maximize the activity of anticancer drugs.30 To our knowledge, however, we are the first to generate NRF2-deficient A549 cells bearing either heterozygous or homozygous knockouts. Cell line 2-11 exhibited a heightened sensitivity to increasing dosages of cisplatin and, to a lesser extent, in response to increasing concentrations of carboplatin. When cisplatin was combined with vinorelbine, a heightened sensitivity was also observed. Cell killing was determined by the standard MTS assay. Chemosensitivity of the homozygous knockout cell line 2-11 was then evaluated in a xenograft mouse model, wherein the cells were implanted in the back of a nude mouse and allowed to proliferate for 16 days. Subsequent tail vein injection of cisplatin, carboplatin, or cisplatin and vinorelbine, at various days after the tumor had grown to approximately 100 mm3, led to a reduction in tumor proliferation over the course of the next 16 days. Interestingly, cell line 2-11 alone exhibited a slower growth phenotype in a xenograft mouse model, even without the addition of chemotherapeutic drugs. This result indicates that the disruption of the NRF2 gene itself reduces proliferative activity to a small degree, although the addition of cisplatin, carboplatin, or cisplatin and vinorelbine leads to a significant reduction in tumor cell proliferation.
Tumors isolated from mice implanted with wild-type A549 cells or the clonal knockout cells treated with either cisplatin or saline were sectioned and stained for Ki67, a commonly used marker for cell proliferation. Ki67 is strictly associated with cell proliferation and is present during all active phases of the cell cycle, but it is absent in resting cells.32 Our results suggest that there is no difference in Ki67 levels in treated or untreated wild-type A549 cells grown in the xenograft model, again reflecting the well-known resistance of A549 cells to cisplatin. In contrast, Ki67 levels in the NRF2-knockout cells treated with cisplatin were found to be substantially lower when compared to the wild-type counterparts. These results provide a plausible explanation for the reduced size of the tumor found in mice implanted with NRF2-knockout cells, a reduction in tumor cell proliferation as a function of CRISPR-directed gene editing. These results reflect those of Velma et al.38 who reported that cisplatin-treated cells are arrested at the G0/G1 border as a function of increasing concentrations. Cisplatin reduces proliferation or retardation of cell cycle progression with an impact at the interface between G0 and G1. These data certainly indicate that disruption of NRF2 in A549 cells leads to a reduced proliferative phenotype, which may preclude the appearance of apoptosis in tumors analyzed at 16 days. It is possible that apoptosis may be evident shortly after the introduction of any of the four treatments of cisplatin taking place in the early part of the experiment.
When stimulated with stressors, functional NRF2 translocates to the nucleus where it binds to the ARE (antioxidant response element) sequence and activates transcription of the various downstream cytoprotective genes. The translocation of NRF2 to the nucleus (appears as purple) can be seen in the images of wild-type A549 cells (Figure 7B). However, the genetic knockout of NRF2 in clone 2-11 causes the loss of NRF2 function and appears to halt translocation of the protein, instead remaining in the cytoplasm, also seen in Figure 7B. Functional knockouts may have value as CRISPR moves toward clinical application, particularly for cancer therapy. The long-term goal is to direct CRISPR activity in tumor cells and not normal cells, but that goal remains a daunting challenge. Our report may open a new avenue of investigation in which gene-editing strategies can be designed to disable a functional domain that is more active in tumor cell growth. If such surgical knockouts can be obtained, then a selective effect on tumor cells, but not on normal cells, could be seen.
Our results provide some support for the notion that the combination of gene-editing activity and chemotherapy acts synergistically to reduce tumor cell growth. Beyond the current well-established combinatorial drug strategies used to treat NSCLC, several different combinatorial approaches are also being investigated. For example, the use of an oncolytic virus that infects tumor cells has been found to enhance the activity of chemotherapy. Infection with myxoma virus combined with cisplatin or gemcitabine efficiently destroyed ovarian cancer cells at much lower dosages than needed without viral addition.39 The use of oncolytic virus therapy and cytotoxic chemotherapy for improved effectiveness of cancer treatment is an active area of development.40, 41 Infection with a replication-competent virus before treatment with cisplatin markedly enhances the therapeutic benefit of chemotherapy, results that closely align with our own observations. In our case, the treatment of A549 cells with CRISPR/Cas9 to disable NRF2 at the level of the gene also led to effective killing at lower dosages of multiple chemotherapeutic agents.
With the usage of targeted therapy (targeting EGFR mutation, ALK rearrangement, etc.) and immunotherapy (checkpoint inhibitors, anti-PD1, anti-CTLA4, etc.), the clinical management of NSCLC has greatly improved. Patients can have a longer and better quality of life. However, these therapies can’t solve all the problems. For example, agents that target specific molecules typically have a response rate of ∼70%. However, after a median period of 8–16 months, due to the inevitable resistance, relapse happens in almost all patients.42 With regard to immunotherapy, though pembrolizumab (Keytruda) can be used as first treatment in certain lung cancer patients, only a fraction of them will respond.43
On the other hand, chemotherapy is still indispensable in the lung cancer treatment paradigm. In patients with locoregional NSCLC, chemotherapy is the only systemic therapy proven to improve curability when combined with surgery or radiation.44 In patients with metastasis, chemotherapy is still the mainstay of care for those who have developed resistance to targeted therapy agents. Meanwhile, it also has the potential to stimulate the immune system to boost the effectiveness of immunotherapy. Taken together, given chemotherapy’s critical role in lung cancer treatment, new approaches to improve its effectiveness will generate great impact. That is why, in this study, we leveraged CRISPR/Cas9 to knock down NRF2, a key player in chemo-resistance, to explore its overall effect on chemosensitivity, tumor proliferation, angiogenesis, and apoptosis.
The choice of the target gene for disruption is obviously a crucial component of this type of experimental strategy. For our experiments, NRF2 was chosen because of its well-known association with cyto-protection and its capacity to enable chemo-resistance. An increased level of attention has begun to center on NRF2 dictating some degree of chemo-resistance, as it appears to have a broader impact on resistance to platinum-based chemotherapeutic approaches than once envisioned. Chen et al.37 reported that chemo-resistance in ovarian, cervical, and lung cancer is derived by the enhanced elevated expression and activity of NRF2. Stable knockdowns of NRF2 using small hairpin RNA (shRNA) have shown increased chemosensitivity in a variety of tumor cell lines.30 NRF2 is also known to be the central contributor in the resistance to cisplatin in bladder cancer.
While our results are certainly encouraging, it is also important to realize that, if the method of delivery is not more fully developed, then genetic disruption could take place in both normal cells and tumor cells. While the scope of our work in the objective of this work is not intended to evaluate delivery modalities to lung, direct injection of adenoviral vectors into lung tumors has been a routine procedure in clinical trials evaluating gene therapy of lung cancer.45, 46, 47 Using an adenoviral vector for systemic delivery could certainly work in this case, but it must be chosen wisely because it could prove problematic, considering the tendency of adenoviruses to induce a whole-scale immune response regardless of the structure of the vector variant. Thus, it is imperative that the development of CRISPR-directed gene editing of multipurpose human genes for combinatorial therapy of lung cancer be coupled with the evolution of more effective delivery methods of these genetic tools to lung tissue.
Materials and Methods
Cell Culture Conditions
Human lung carcinoma A549 cells were purchased from ATCC (Manassas, VA, USA). Cells were thawed, according to the manufacturer’s protocol, and grown in F-12K medium (ATCC, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS) (ATCC, Manassas, VA, USA) and 1% Penicillin-Streptomycin Solution (ATCC, Manassas, VA, USA). Cells were cultured and maintained at a concentration between 2 × 103 and 1 × 104 viable cells/cm2, and they were incubated at 37°C and 5% CO2. Cell number was determined using a hemacytometer.
gRNA Design and Construction
The NRF2 gene-coding sequence was entered into the Zhang lab’s online generator (http://crispr.mit.edu/), and the gRNA with the highest score was chosen for gRNA1 (5′-TCGATGTGACCGGGAATATC-3′), and a previously validated gRNA targeting NRF221 was also chosen for gRNA2 (5′-TGATTTAGACGGTATGCAAC-3′). The CRISPR plasmid was cloned using standard cloning methods with single-step digestion-ligation. The CRISPR guide sequences with appropriate 5′ overhangs were cloned into the pX458 backbone vector digested with BbsI (plasmid 48138, Addgene), a human codon-optimized pSpCas9 and chimeric gRNA expression plasmid with a 2AEGFP, purchased through Addgene (https://www.addgene.org). Following construction, plasmids were validated by Sanger sequencing (Genewiz, South Plainfield, NJ, USA).
Transfection and Clonal Isolation
A549 cells were transfected at a concentration of 5 × 105 cells/100 μL in a 4-mm gap cuvette (BioExpress, Kaysville, UT, USA). NRF2-targeting pX458 constructs were separately electroporated (250 V, low voltage [LV], 13-ms pulse length, 2 pulses, 1-s interval) into A549 cells using a Bio-Rad Gene Pulser XCell Electroporation System (Bio-Rad, Hercules, CA, USA). Cells were then recovered in 6-well plates with complete growth media at 37°C for 72 hr prior to sorting. A549 cells were sorted into a 96-well plate with a FACS AriaII flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA), with an individual EGFP+ cell sorted to each well. Clones were expanded and transferred to larger plates as the individual clones reached confluence, with DNA isolation occurring when cells reached confluence in a six-well plate (1 × 106 cells/mL).
Sequencing and Sequence Analyses
CRISPR/Cas9-targeted A549 clones were PCR amplified (forward 5′-gtagtggtgccttagagcttactcatcc-3′, reverse 5′-ctagcatgggcagtactcatgactaag-3′) using Amplitaq Gold Fast PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Briefly, template DNA, primers, water, and master mix were combined and cycled: 95°C for 10 min (96°C for 3 s, 60°C for 3 s, and 68°C for 5 s) × 35 cycles and 72°C for 10 s. The 402-bp products were purified (QIAGEN, Hilden, Germany) and Sanger sequenced using the forward PCR primer. Clonal allelic analyses of individual A549 cell clones were analyzed by the software program TIDE to determine the individual sub-sequences within the multi-peaked breakdown product after CRISPR/Cas9 activity.25 The TIDE analyses provide a visual of the sequence decomposition, the indel patterns of the clone, as well as relative ratios of each clonal indel pattern, serving as an intermediate step in determining each allelic profile. By utilizing the indel patterns and their relative ratios provided by TIDE, the control trace sequence and a clonal trace sequence were manually aligned, allowing for the visualization of the indel patterns of each allele of a clone.
Western Blot Analysis
Total cellular protein was collected from A549 cell lines using a standard RIPA lysis buffer containing a protease inhibitor cocktail. Protein concentrations were determined using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). The samples were heated at 95°C for 10 min and then were subjected to SDS-PAGE on a 10% polyacrylamide gel for 90 min at 100 V. The gel was transferred to a nitrocellulose membrane for 1 hr at 100 V. The blot was placed in 3% BSA and blocked overnight on a shaker at 4°C. Primary antibody incubation was performed overnight on a shaker at 4°C for NRF2 (phospho S40) (1:10,000, Abcam ab76026) and 1 hr at room temperature for beta-actin (1:8,000, Abcam ab8226), and secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) incubations were all done 1 hr at room temperature at a 1:10,000 dilution. The protein bands were visualized via chemiluminescence using a SuperSignal West Dura Extended Duration ECL (Pierce) and detected on the LI-COR Odyssey FC. All bands were quantified for densitometry on the Image Studio software system.
Cell Proliferation by FACS Analysis
A549 cell lines were trypsinized and harvested at 50%–70% confluency. Cells were fixed with ice-cold 70% ethanol dropwise while vortexing and incubated at 4°C for a minimum of 72 hr. Fixed cells were pelleted and washed twice with PBS followed by a 30-min incubation on ice. As indicated in the manufacturer’s protocol (BD Biosciences), 20 μL/106 cells Alexa Fluor 647 Mouse anti-Ki67 (561126, BD Biosciences) was added to the cells and incubated for 30 min. Controls included Alexa Fluor 647 Mouse IgG1 k isotype control (557714, BD Biosciences), at the same dilution. After incubation, cells were washed twice and resuspended in stain buffer (5% BSA in 1× PBS). Cells were analyzed with a FACS AriaII flow cytometer and processed using FlowJo software.
MTS Cell Proliferation Assay
Cell viability was evaluated using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI, USA). A549 cell lines were plated at 2 × 103 cells/well and allowed to culture for 24 hr. The cell media were then aspirated, the cells washed with PBS, then exposed to the MTS reagent for 3 hr. After 3 hr of MTS bioreduction by proliferating cells, the formazan product’s absorbance was measured using a 450-nm filter on an Infinite 2000 PRO microplate reader (Tecan, Mannadorf, Switzerland). Cell viability after drug exposure was evaluated using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay. A549 cell lines were plated at 2 × 103 cells/well and allowed to culture for 24 hr. The cells were then treated with cisplatin, carboplatin, or a combination of cisplatin and vinorelbine for 3 days. The cell media were then aspirated, the cells washed with PBS, then exposed to the MTS reagent for 3 hr. After 3 hr of MTS bioreduction by proliferating cells, the formazan product’s absorbance was measured using a 450-nm filter on an Infinite 2000 PRO microplate reader.
Animal Experiments and Statistical Analysis
The animal trials presented in this report were carried out at Washington Biotech, Simpsonville, MD, USA, under animal use and care protocol (SOP 505, SOP 520, SOP 522, SOP 1610, and SOP1650) approved by the animal care and use committee of Washington Biotechnology (AAALAC-accredited Animal Welfare Assurance number A4192-01). The human xenograft model was established using methodology reported previously.48 Female athymic nude mice (Envigo, 5–6 weeks old) were used in this study. Approximately 5 × 106 cells (wild-type A549 or homozygous knockout [clonal expansion 2-11]) suspended in PBS with 20% Matrigel were injected subcutaneously into the right flank of each mouse. Tumor volume was measured three times a week with a digital caliper once palpable, and it was calculated using the following formula: tumor size = ab2/2, where a is the larger and b is the smaller of the two dimensions. When tumors grew up to a mean volume of around 100 mm3, A549 tumor-bearing mice or A549-2-11 tumor-bearing mice were randomly divided into 7 groups (n = 5 for each group), respectively, and subjected to dose and regimen-finding study. They were treated with tail vein injection of (1) cisplatin (2 mg/kg), (2) carboplatin (25 mg/kg), (3) cisplatin (5 mg/kg) and vinorelbine (5 mg/kg), or (4) saline on days 0, 3, 6, and 9 (day 0 is designated as the day of dose start).21 Tumor volume and body weight (Figure S2) were closely monitored over time. After 16 days, the animals were sacrificed, with tumor removed, weighed, and processed for molecular analysis. Mice were euthanized. The data were expressed as mean ± SD. Student’s t test and one-way or two-way ANOVA were used to assess the significance of difference. A p value < 0.05 was considered significant.
Immunofluorescence Staining
A549 xenografts were resected on day 16, snap frozen in liquid nitrogen, and stored at −80°C until usage. All immunofluorescence staining was performed as previously described.44 Briefly, tumors were embedded in Optimum Cutting Temperature (Tissue Tek, Torrance, CA, USA), and 16-μm-thick sections were obtained with a Leica CM3050 cryostat (Leica Microsystems, Buffalo Grove, IL, USA) and mounted on slides. Slides were fixed (4% paraformaldehyde [PFA] for 30 min at room temperature) and incubated with blocking buffer (5% goat serum and 0.3% Triton X-100 in PBS) for 1 hr at room temperature. Sections were then incubated (overnight at 4°C; 1:100 dilution, dilution buffer of 2% BSA and 0.3% Triton X-100 in PBS) with primary antibody (Ki67 [9129, Cell Signaling Technology]), then washed in PBS and incubated with Alexa Fluor 488-labeled secondary antibody (1:200 dilution; Invitrogen, Grand Island, NY, USA) for 1 hr at room temperature (RT). Sections were washed in PBS, then mounted with SlowFade Gold antifade mountant with DAPI (Invitrogen, Carlsbad, CA, USA). Images were obtained with a Zeiss Observer.Z1 microscope (Carl Zeiss, Gottingen, Germany). The terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was conducted with In Situ Cell Death Detection Kit, Fluorescein (Roche, Basel, Switzerland), following the manufacturer’s instructions.
Immunocytochemistry and Image Quantification
A549 cell lines were seeded in 8-well chamber slides (LabTek II) and allowed to grow for 24 hr. After exposure to 2 μM cisplatin for 48 hr, cells were washed with PBS, fixed, and permeabilized with 4% paraformaldehyde +0.1% Triton X-100 for 45 min while shaking at room temperature. Cells were washed three times with PBS and blocked with a blocking buffer solution (5% normal goat serum + 0.3% Triton X-100 made in 1× PBS) for 2 hr at room temperature. Following blocking, cells were incubated with primary antibody (NRF2 1:500, Abcam ab62352) made in an antibody dilution buffer (1% BSA + 0.3% Triton X-100 made in 1× PBS), overnight in a humidified chamber at 4°C. Cells were washed three times with PBS and incubated with a conjugated secondary antibody (goat anti-rabbit Alexafluor 594, Thermo Fisher Scientific A-11037) made in an antibody dilution buffer at a concentration of 1:200. Controls included secondary-only antibody stains at the same dilutions. Cells were incubated for 1 hr at room temperature in the dark. Cells were washed three times with PBS and the chamber was separated from the glass slide. Immediately following this step, 5 μL Slow Fade Gold antifade reagent with DAPI (S36938, Invitrogen) was added to each section of the slide, and a coverslip was added and sealed. Slides were imaged on the Zeiss Axio fluorescent observer.Z1 microscope, and images were processed on the AxioVision software. Random fields were imaged and the total number of cells per field was counted. Each field was quantified for no staining (none), nuclear staining, or cytoplasmic staining. Two individuals independently counted and quantified the images and values were averaged. The percent of NRF2-positive-stained cells over the total cells analyzed in each category was plotted in the graph. Errors bars represent ± SEM and an asterisk denotes a significant p value that is <0.05 (Student’s t test).
Author Contributions
P.B., Y.W., and K.B. carried out the experiments. P.B. did the genetic analyses. P.B. and E.B.K. conceived the project. P.B., Y.W., K.B., and E.B.K. wrote the manuscript.
Conflicts of Interest
There are no conflicts of interest related to this report.
Acknowledgments
We thank the members of the Kmiec laboratory for input and advice. We thank Dr. Lynn Opdenaker at the CTCR Flow Cytometry Core Facility for running, analyzing, and sorting cells. This project was supported by the Delaware INBRE and COBRE programs, with grants from the National Institute of General Medical Sciences – P20 GM103446 and P20 GM109021 from the NIH and the State of Delaware. Additional funding for this project was provided by the State of Delaware through the Delaware Bioscience Center for Advanced Technology. This content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omto.2018.10.002.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch F.R., Suda K., Wiens J., Bunn P.A., Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 3.Pirker R., Pereira J.R., Szczesna A., von Pawel J., Krzakowski M., Ramlau R., Vynnychenko I., Park K., Yu C.T., Ganul V., FLEX Study Team Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–1531. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 4.Reck M., von Pawel J., Zatloukal P., Ramlau R., Gorbounova V., Hirsh V., Leighl N., Mezger J., Archer V., Moore N., Manegold C. Phase III trial of cisplatin plus gemcitabine with either placebo or bevacizumab as first-line therapy for nonsquamous non-small-cell lung cancer: AVAil. J. Clin. Oncol. 2009;27:1227–1234. doi: 10.1200/JCO.2007.14.5466. [DOI] [PubMed] [Google Scholar]
- 5.Sandler A., Gray R., Perry M.C., Brahmer J., Schiller J.H., Dowlati A., Lilenbaum R., Johnson D.H. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 6.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T., West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C., Wu Y.L., Chen G., Feng J., Liu X.Q., Wang C., Zhang S., Wang J., Zhou S., Ren S. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 9.Hegde P.S., Karanikas V., Evers S. The Where, the When, and the How of Immune Monitoring for Cancer Immunotherapies in the Era of Checkpoint Inhibition. Clin. Cancer Res. 2016;22:1865–1874. doi: 10.1158/1078-0432.CCR-15-1507. [DOI] [PubMed] [Google Scholar]
- 10.Lu H.-Y., Wang X.-J., Mao W.-M. Targeted therapies in small cell lung cancer. Oncol. Lett. 2013;5:3–11. doi: 10.3892/ol.2012.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Li M., Rinehart J.J., Zhang R. Pretreatment with dexamethasone increases antitumor activity of carboplatin and gemcitabine in mice bearing human cancer xenografts: in vivo activity, pharmacokinetics, and clinical implications for cancer chemotherapy. Clin. Cancer Res. 2004;10:1633–1644. doi: 10.1158/1078-0432.ccr-0829-3. [DOI] [PubMed] [Google Scholar]
- 12.Hayden A., Douglas J., Sommerlad M., Andrews L., Gould K., Hussain S., Thomas G.J., Packham G., Crabb S.J. The Nrf2 transcription factor contributes to resistance to cisplatin in bladder cancer. Urol. Oncol. 2014;32:806–814. doi: 10.1016/j.urolonc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Singh A., Bodas M., Wakabayashi N., Bunz F., Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid. Redox Signal. 2010;13:1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrente L., Sanchez C., Moreno R., Chowdhry S., Cabello P., Isono K., Koseki H., Honda T., Hayes J.D., Dinkova-Kostova A.T., de la Vega L. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene. 2017;36:6204–6212. doi: 10.1038/onc.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X., Wang H., Fan L., Wu X., Xin A., Ren H., Wang X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011;50:1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera-Torres N., Banas K., Bialk P., Bloh K.M., Kmiec E.B. Insertional Mutagenesis by CRISPR/Cas9 Ribonucleoprotein Gene Editing in Cells Targeted for Point Mutation Repair Directed by Short Single-Stranded DNA Oligonucleotides. PLoS ONE. 2017;12:e0169350. doi: 10.1371/journal.pone.0169350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey P., Singh A.K., Singh M., Tewari M., Shukla H.S., Gambhir I.S. The see-saw of Keap1-Nrf2 pathway in cancer. Crit. Rev. Oncol. Hematol. 2017;116:89–98. doi: 10.1016/j.critrevonc.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Jung B., Yoo H.S., Shin S., Park Y.J., Jeon S.M. Dysregulation of NRF2 in Cancer: from Molecular Mechanisms to Therapeutic Opportunities. Biomol. Ther. (Seoul) 2018;26:57–68. doi: 10.4062/biomolther.2017.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namani A., Li Y., Wang X.J., Tang X. Modulation of NRF2 signaling pathway by nuclear receptors: implications for cancer. Biochim. Biophys. Acta. 2014;1843:1875–1885. doi: 10.1016/j.bbamcr.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami S., Motohashi H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015;88(Pt B):168–178. doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 29.Li W., Yu S.W., Kong A.N.T. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J. Biol. Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 30.Wang X.J., Sun Z., Villeneuve N.F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G.T. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellmann M.D., Li B.T., Chaft J.E., Kris M.G. Chemotherapy remains an essential element of personalized care for persons with lung cancers. Ann. Oncol. 2016;27:1829–1835. doi: 10.1093/annonc/mdw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholzen T., Gerdes J. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W., Marraffini L.A. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu. Rev. Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y., Wu K., Liu Q., Han N., Zhang L., Chu Q., Chen Y. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J. Hematol. Oncol. 2016;9:83. doi: 10.1186/s13045-016-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Solomides C., Simpkins F., Simpkins H. The role of Nrf2 and ATF2 in resistance to platinum-based chemotherapy. Cancer Chemother. Pharmacol. 2017;79:369–380. doi: 10.1007/s00280-016-3225-1. [DOI] [PubMed] [Google Scholar]
- 38.Velma V., Dasari S.R., Tchounwou P.B. Low Doses of Cisplatin Induce Gene Alterations, Cell Cycle Arrest, and Apoptosis in Human Promyelocytic Leukemia Cells. Biomark. Insights. 2016;11:113–121. doi: 10.4137/BMI.S39445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nounamo B., Liem J., Cannon M., Liu J. Myxoma Virus Optimizes Cisplatin for the Treatment of Ovarian Cancer In Vitro and in a Syngeneic Murine Dissemination Model. Mol. Ther. Oncolytics. 2017;6:90–99. doi: 10.1016/j.omto.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wennier S.T., Liu J., McFadden G. Bugs and drugs: oncolytic virotherapy in combination with chemotherapy. Curr. Pharm. Biotechnol. 2012;13:1817–1833. doi: 10.2174/138920112800958850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson G.R., Relph K., Harrington K., Melcher A., Pandha H. Cancer immunotherapy via combining oncolytic virotherapy with chemotherapy: recent advances. Oncolytic Virother. 2016;5:1–13. doi: 10.2147/OV.S66083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anichini A., Tassi E., Grazia G., Mortarini R. The non-small cell lung cancer immune landscape: emerging complexity, prognostic relevance and prospective significance in the context of immunotherapy. Cancer Immunol. Immunother. 2018;67:1011–1022. doi: 10.1007/s00262-018-2147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianco A., Malapelle U., Rocco D., Perrotta F., Mazzarella G. Targeting immune checkpoints in non small cell lung cancer. Curr. Opin. Pharmacol. 2018;40:46–50. doi: 10.1016/j.coph.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Terrell A.M., Riggio B.A., Anand D., Lachke S.A., Duncan M.K. β1-Integrin Deletion From the Lens Activates Cellular Stress Responses Leading to Apoptosis and Fibrosis. Invest. Ophthalmol. Vis. Sci. 2017;58:3896–3922. doi: 10.1167/iovs.17-21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong M., Li X., Hong L.J., Xie R., Zhao H.L., Li K., Wang H.H., Shin W.D., Shen H.J. Advanced malignant pleural or peritoneal effusion in patients treated with recombinant adenovirus p53 injection plus cisplatin. J. Int. Med. Res. 2008;36:1273–1278. doi: 10.1177/147323000803600614. [DOI] [PubMed] [Google Scholar]
- 46.Li D., Zhang Y., Xie Y., Xiang J., Zhu Y., Yang J. Enhanced tumor suppression by adenoviral PTEN gene therapy combined with cisplatin chemotherapy in small-cell lung cancer. Cancer Gene Ther. 2013;20:251–259. doi: 10.1038/cgt.2013.14. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., Shou F., Zhang H., You Q. Adenovirus-delivered wwox inhibited lung cancer growth in vivo in a mouse model. Cancer Gene Ther. 2016;23:1–6. doi: 10.1038/cgt.2015.56. [DOI] [PubMed] [Google Scholar]
- 48.Kellar A., Egan C., Morris D. Preclinical Murine Models for Lung Cancer: Clinical Trial Applications. BioMed Res. Int. 2015;2015:621324. doi: 10.1155/2015/621324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.