Abstract
Hepatitis B with precore stop codon mutation is related with severe liver damage in HBeAg negative patients. It is of utmost importance to screen the G1896A precore mutation. The study was designed to assess the impact of G1986A mutations in patients with different clinical spectra of the liver disease by PCR–LCR. 210 HBV positive patients with HBeAg negative serology of different kind of liver diseases (AVH = 72, FH = 21, CH = 79, Cirrhosis = 20 and HCC = 18) were screened. Patients were screened for the presence or absence of precore G1896A mutation by PCR–LCR. Direct nucleotide sequencing was done to confirm the results of LCR. Precore mutant in HCC was 94.4% (17/18), 85.7% (18/21) in FH, 60% (12/20) in liver cirrhosis, 48.1% (38/79) in chronic hepatitis and 27.7% (20/72) in AVH cases. The serum ALT level was statistically significant between HBeAg negative WT and G1896A mutants in chronic hepatitis cases. ALT level and HBV DNA level was slightly raised in the pre core mutant but and was not significant. Genotype D had a higher prevalence (79.5%) as compared to genotype A (20.5%). The mutations detected by PCR–LCR were in 100% concordance with direct sequencing. The exceptionally high prevalence of G1896A in FH and HCC demonstrates that the precore mutants are strongly associated with the progression of liver diseases in patients with HBeAg negative serology. The findings are also suggestive of screening HBV precore G1896A mutation particularly in HBeAg negative cases. The precore G1896A mutation increases proportionately in severe form of liver diseases. LCR can be a suitable tool for screening of G1896A mutations.
Keywords: Hepatitis B, G1896A, Mutation, Ligase chain reaction, Direct sequencing, Serology
1. Introduction
Hepatitis B virus (HBV) infection leads to a wide-variety of clinical manifestations ranging from acute self-limited illness to different forms of chronic infection progressing to liver failure in some patients. India is known to be the largest pool of HBV carriers in the world next to China. There are 43 million estimated HBV carriers in India (Datta, 2008, Gupta et al., 2008, Konstantinou et al., 2015). One-third of the patients with acute hepatitis and two thirds of cases with chronic liver disease and hepatocellular carcinoma in India are due to HBV infection. HBV infection is a public health problem in India and its importance for morbidity and mortality has not been substantiated (Acharya, 2014).
The prevalence of precore mutants worldwide has been expressed as 7–30% albeit with wide differences (Schalm et al., 1990). In Asia, as many as 55% of chronic hepatitis B patients have a disease, which is sustained by mutant viruses. A study by (Guptan et al., 1996) has demonstrated that one fourth of hepatitis B related chronic liver disease in Asian Indians is due to mutant hepatitis B virus (precore (15.5%) or surface mutant (10.8%) forms). In another study from North India, prevalence of precore mutant in HBV related chronic liver diseases is as high as 25.8% and this mutation have found to be associated with increased severity of liver diseases (Guptan et al., 1996). Preliminary studies conducted in South India suggest that G to A switch at nucleotide 1896 is the most common mutation of HBV genome in patients with chronic liver disease from the Indian subcontinent. There are also no data on association of precore mutants with fulminant hepatic failure and acute failure from this part of the world.
The point mutation involving the pre-core or other regions can be detected by direct sequencing of the genome, but this methodology is too costly and time consuming for routine clinical use. An adequately rapid, specific and sensitive method for detection of this precore G1896A mutant is needed (Minamitani et al., 1997). Landegreen et al. (1988) reported an easier method to detect point mutation with ligase enzyme. Barany (1991) presented a method that uses thermostable ligase for ligase chain reaction (LCR) based on the theory of Landegreen. This method of LCR has been successfully used for the detection of pre-core mutants of HBV (Minamitani et al., 1997). They have also shown that sensitivity of this method can be increased by performing LCR on PCR amplified DNA and the results obtained are comparable with that of direct sequencing.
With the above background, the current study was designed to look at the prevalence of precore G1896A mutations in HBeAg negative serology patients with different kinds of liver diseases. Also, the effect of mutation in the underlying disease and the sensitivity and specificity of the methods employed for the detection of mutation.
2. Materials and methods
2.1. Study subjects
A total of HBeAg negative 210 patients (acute viral hepatitis: 72), (fulminant hepatitis: 21), (chronic hepatitis: 79), (liver cirrhosis: 20), (hepatocellular carcinoma: 18) who were admitted in the wards of Lok Nayak Hospital, New Delhi, India were enrolled in this study with prior informed consent of the patients. These patients were examined on the basis of history, clinical examination, liver function profile and various serological markers of Hepatitis B (HBsAg, HBeAg, AntiHBc(IgM/ IgG) and AntiHBe) using commercially available ELISA kits (DIA PRO Diagnostic Bioprobes, Srl., Italy). Upper GI endoscopy and ultra sound examination of the patient was done whenever indicated. The patients enrolled in the study were regularly followed up at the gastroenterology clinic of our hospital. Prior information about different risk factors such as intramuscular injections, Intravenous medication, blood transfusions, tattooing and any past history of jaundice in the family was collected on a pre-designed questionnaire. The study was ethically approved by the Institutional Ethics Committee of Maulana Azad Medical College, New Delhi.
2.2. Extraction of HBV DNA
DNA was extracted by phenol–chloroform method. Briefly, to the 100 μl of serum 400 μl of distilled water and 500 μl of TE saturated phenol was added and kept in a water bath for two hours and centrifuged for 20 min. The supernatant was added with 500 μl of TE saturated phenol and 200 μl chloroform: isoamyl alcohol (24:1) and centrifuged for 10 min at 12,000 rpm. To the supernatant 1/3 volume of ammonium acetate, 1.5 V of absolute ethanol were added and left overnight in −40 °C. It was then centrifuged for 20 min. The supernatant was discarded and saturated with 70% ethanol. Pellet was then dried and suspended in 25 μl of distilled water and amplified by polymerase chain reaction (PCR).
2.3. Polymerase chain reaction (PCR)
PCR was done in a 50 μl of reaction mixture with primers HP1: 1771-1790 # 5′-TA(C/T) TAG GAG GCT GTA GGC AT-3′ and HP2: 2079-2060 # 5′-AGA ATA GCT TGC CTG AGT GC-3′ in a thermo cycler and amplification reaction carried out in 40 cycles of 94 °C for 1 min., 55 °C for 1 min. and 72 °C for 1 min., with a 10 min. extension step at 72 °C at the end. 10 μl of the reaction mixture was analysed by electrophoresis in 2% agarose gel that was stained with ethidium bromide and could detect 309 bp PCR product under UV light (Fig. 1).
Figure 1.
Representative photographs showing PCR products of HBV DNA of 309 bp from the precore region. Lane 1: ϕX174 HaeIII digested marker, Lane 2: negative control and Lanes 3, 4,5,6,7, 8 & 9: Showing samples positive for HBV DNA precore region.
2.4. Ligase chain reaction (LCR)
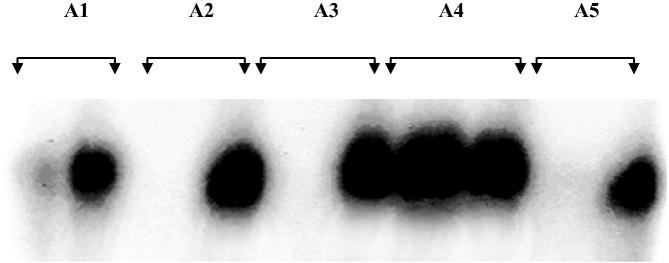
LCR was performed in the samples, which tested positive for HBV DNA for the detection of wild virus and mutant virus (precore mutant with a point mutation from G–A switch at nucleotide 83 from the precore region) (Fig. 2). The LCR was carried out to detect the HBV precore G1896A mutants with the strategy as described by Minamitani et al. (1997). Briefly, common primers CP1 & CP II each at concentration of 10 mmol/L were 5′ end labelled in 25 μl of 50 mmol/L Tris HCl (ph 8.0), 10 mmol/L MgCl2, T4PNK (NEB), 10 mmol/L (g-32p) adenosine triphosphate (Board of radiation and isotope technology, Department of Atomic Energy, Government of India). The reaction mixture was incubated at 37 °C for 1 h and at 95 °C for 10 min. The LCR reaction then took place in 30 μl reaction mixture with 5 μl of target DNA in 20 mmol Tris–HCI (pH 7.5) containing 20 mmol KCI, 10 mmol MgCl2, 1 mmol Dithiothreitol, 0.1 mmol ATP, 0.1% NP-40, 4 μg of herring sperm DNA, 6 U of Pfu ligase (Stratagene, La Jolla, CA), 20 nmol with each discriminating primers (WS & WA or MS & MA) and 20 nmol each common primers (CS and CA). The samples were incubated at 94 °C for 1 min. and then at 60 °C for 1 min. and this cycle was repeated a total of 30 times. 10 μl of LCR products were run on 8% polyacrylamide gel at 175 volts for 2 h. The gel was dried and exposed to X-ray film for 40 min. at room temperature. The signals generated by LCR were measured densitometrically (Fig. 2). The sequences used are as follows: CPI: 5′GGG CAT GGA CAT (T/C) GAC CC(G/T) TAT AAA GAA TTT GGA GC(T/A)(T/A)C(T/C) G-3′; CPII: 5′ AAA GCCA(C/T) CCA AGG CAC AGC TT(G/A) GAG GCT TGA ACA GT(A/G) G-3′, WPI: 5′-C(T/C) ACT GTT CAA GCC TC (C/T) AAG CTG TGC CTT GG(G/A) TGG CTT TG-3′, WPII: 5′-C(A/G) G(A/T) (A/T) GCT CCA AAT TCT TTA TA(C/A) GGG TC(A/G) ATG TCC ATG CCCC-3’, MP1: 5′-C(T/C) ACT GTT CAA GCC TC(C/T) AAG CTG TGC CTT GG(A/G) TGG CTT TA-3′, MPII: 5′-C(A/G) G(A/T) (A/T) GCT CCA AAT TCT TTA TA(C/A)GGG TC(A/G) ATG TCC ATG CCCT-3′.
Figure 2.
Representative photograph showing detection of HBV by PCR coupled LCR. Wild type HBV is shown in the left lane and mutant HBV is shown in the right lane for each patient.
2.5. Nucleotide sequencing
The target PCR products within the agarose gel were purified for sequencing using the Perfectprep Gel cleanup Kit (Eppendorf, Wetsbuty, NY), according to the manufacturer’s instructions and then sequenced using 8 μ1 of dye terminator from a DNA sequencing kit (Big Dye Terminator V.3.0 Cycle Sequencing Ready Reaction, Foster City, CA) on commercial basis.
2.6. Statistical analysis
Statistics was performed using a Chi square test, Fisher’s exact test and Mann–Whitney’s U test as appropriate. Data were expressed as mean ± standard deviation (SD). Difference was considered significant for p values less than 0.05. The statistical analysis software used was SPSS software, version 12.0.
3. Results
72 HBV (HBeAg −ve) related acute viral hepatitis B were enrolled in the study (Table. 1). Males constituted 47.2% (34/72) of the study while females constituted 52.8% (38/72) of the study. The mean age ± S.D of the patients with acute viral hepatitis B inducted in the study were 33.06 ± 16.36 years. These 72 HBV DNA positive cases were serologically suspected as precore mutants as they had HBsAg positive, IgMAnti-HBc positive, HBeAg negative, and anti-HBe positive serology. The precore G1896A mutation was confirmed by PCR coupled with LCR in 20 out of 72 (27.7%) serologically suspicious precore mutant cases (Table 1) which was further confirmed by direct sequencing. The ALT level was comparable and statistically non significant between HBeAg negative WT (272.52 ± 487.128 IU/L) and precore G1896A (289.57 ± 585.43 IU/L) mutants in AVH cases. The HBV DNA level (log copies/mL) was also found comparable and statistically non significant between HBeAg negative WT (3.4 ± 1.6) and precore G1896A (3.7 ± 1.4) mutants in AVH cases. Genotype D & A accounted for 79.5% and 20.5% of the cases respectively (Table 2).
Table 1.
Prevalence of precore G1896A mutation in HBeAg negative cases of HBV related patients with the different spectra of liver diseases as detected by LCR.
| Spectrum of liver disease | HBV precore G1896G (wild) | HBV precore G1896A (mutant) |
|---|---|---|
| Acute viral hepatitis (72) | 52 (72.2%) | 20 (27.7%) |
| Fulminant hepatitis (21) | 3 (14.3%) | 18 (85.7%) |
| Chronic hepatitis (79) | 41 (51.9%) | 38 (48.1%) |
| Liver cirrhosis (20) | 8 (40%) | 12 (60%) |
| Hepatocellular carcinoma (18) | 1 (5.6%) | 17 (94.4%) |
Table 2.
Distribution of HBV genotype among the patients having different spectra of liver diseases.
| Spectrum of liver disease | HBV Genotype A | HBV Genotype D |
|---|---|---|
| Acute viral hepatitis (72) | 19.4% (14/72) | 80.6% (58/72) |
| Fulminant hepatitis (21) | 19.05% (4/21) | 80.95% (17/21) |
| Chronic hepatitis (79) | 21.5% (17/79) | 78.5% (62/79) |
| Liver cirrhosis (20) | 20% (4/20) | 80% (16/20) |
| Hepatocellular carcinoma (18) | 22.2% (4/18) | 77.8% (14/18) |
| Total cases (210) | 20.5% (43/210) | 79.5% (167/210) |
21 patients with fulminant hepatitis B (HBeAg −ve) were enrolled in the study (Table 1). Males comprised 10 (47.6%) of the study while females constituted 11 (52.4%) of the study. The mean age ± S.D of the patients with fulminant hepatitis B inducted in the study were 31.08 ± 11.3 years. These 21 cases were serologically suspected as precore mutants as they had HBsAg positive, HBcIgM positive, HBeAg negative, and anti-HBe positive serology. The precore G1896A mutation was confirmed by PCR coupled with LCR in 18 out of 21 (85.7%) serologically suspicious precore mutant cases (Table 1) which was further confirmed by direct sequencing. The ALT level was significantly raised in all the FH cases than the normal limit but was comparable and statistically non significant between HBeAg negative WT (1749.66 ± 648.27 IU/L) and precore G1896A mutant (1272.52 ± 887.128 IU/L) cases. The HBV DNA level (log copies/mL) was also found comparable and statistically non significant between HBeAg negative WT (3.76 ± 0.4) and precore G1896A mutants (3.9 ± 1.1).
Totally 79 cases with histologically proven chronic hepatitis B (HBeAg −ve) were enrolled in the study (Table 1). Males constituted 51 (64.5%) of the study while females constituted 28 (35.5%) of the study. The mean age ± S.D of the patients with chronic hepatitis B inducted in the study were 34.47 ± 13.52 years. These 79 cases were serologically suspected as precore mutants as they had HBsAg positive, HBcIgG positive, HBeAg negative, and anti-HBe positive serology. The precore G1896A mutation was confirmed by PCR coupled with LCR in 38 out of 79 (48.1%) serologically suspicious precore mutant cases (Table 1) which was further confirmed by direct sequencing. Significant difference (p < 0.005) was observed in ALT level of HBeAg negative WT (67.6 ± 32.7 IU/L) and precore G1896A (109.82 ± 38.27 IU/L) mutant chronic hepatitis B cases. The HBV DNA level (log copies/mL) was found comparable and statistically non significant between HBeAg negative WT (4.36 ± 1.4) and precore G1896A mutants (4.9 ± 1.27).
20 HBV related liver cirrhosis (HBeAg −ve) were enrolled in the study (Table 1). Males constituted 16 (80%) while females constituted 4 (20%) of the study. The mean age ± S.D of the patients with liver cirrhosis inducted in the study were 47.36 ± 16.13 years. These 20 cases were serologically suspected as precore mutants as they had HBsAg positive, HBcIgG positive, HBeAg negative, and anti-HBe positive serology. The precore G1896A mutation was confirmed by PCR coupled with LCR in 12 out of 20 (60%) serologically suspicious precore mutant cases (Table 1) which was further confirmed by direct sequencing. The ALT level of precore G1896A mutant (103.23 ± 98.71 IU/L) cases was slightly higher than HBeAg negative WT (89.87 ± 92.43 IU/L) but statistically non significant in liver cirrhosis cases. The HBV DNA level (log copies/mL) was also found comparable and statistically non significant between HBeAg negative WT (5.15 ± 1.36) and precore G1896A mutants (5.36 ± 1.47).
18 hepatocellular carcinoma (HBeAg −ve) were enrolled in the study (Table 1). Males constituted 16 (88.8%) of the study while females constituted 2 (11.2%) of the study. The mean age ± S.D of the patients with hepatocellular carcinoma inducted in the study was 49.23 ± 14.39 years. These 18 cases were serologically suspected as precore mutants as they had HBsAg positive, HBcIgG positive, HBeAg negative, and anti-HBe positive serology. The precore G1896A mutation was confirmed by PCR coupled with LCR in 17 out of 18 (94.4%) serologically suspicious precore mutant cases (Table 1) which was further confirmed by direct sequencing. The ALT level was comparable between HBeAg negative WT (92 IU/L) and precore G1896A (89.23 ± 78.71 IU/L) mutants in HCC cases. The HBV DNA level (log copies/mL) was also found comparable between HBeAg negative WT (5.1) and precore G1896A (5.36 ± 1.47).
G1896A mutation detected by LCR was also confirmed by direct sequencing. The study suggests that the LCR is 100% sensitive for the detection of HBV precore G1896A mutation (Fig. 3).
Figure 3.
Sequencing chromatogram showing wild type in upper panel and G1896A mutation in lower.
4. Discussion
HBV continues to be the most important aetiological agent of acute viral hepatitis, fulminant hepatitis, chronic hepatitis, liver cirrhosis and hepatocellular carcinoma in the developing and underdeveloped world. HBV is prone to mutations with nucleotide substitutions estimated at a rate of 1 × 10−5 to 3 × 10−5 per site per year (Eloy et al., 2013). Mutations have been detected in all the regions of the HBV genome but the significance of many of these mutations is unclear.
The presence of precore mutations was significantly high (p < 0.005) in AVH (27.7% 20/72) in comparison to FH (85.7% 18/21). This suggests that precore mutation may be one of the important factors for FHF. There is a very limited data from India on the prevalence and epidemiology of precore mutants. Various studies from the western world have reported that the precore mutants are responsible for 5–10% of HBV related liver diseases (Zuccaro et al., 2015), whereas studies from Japan have reported a much higher frequency (Imamura et al., 2003). There have been very few studies, which have looked into the frequency of precore mutants in patients with acute viral hepatitis and fulminant hepatic failure. Triki et al. from Tunisia have reported that 40% of patients with acute viral hepatitis had infection by a mutant virus (A1896) (Ayed et al., 2007). Minamitani et al. (1997) from Japan, using ligase chain reaction assay reported that mutant HBV was present in 100% (4/4) of patients with acute viral hepatitis and 100% (7/7) in patients with fulminant hepatic failure. Similarly Singh et al. (2005) from India using DNA sequencing technique reported the presence of precore/core mutants in 100% (5/5) of patients with fulminant hepatic failure due to HBV which is in contrast with our study where the precore mutation accounted for 27.7% acute viral hepatitis cases and 85.7% in fulminant hepatitis cases which were serologically suspected for precore mutants (Khare et al., 2012, Bhattacharya et al., 2014).
Chronic hepatitis B continues to be the paramount cause of chronic liver disease in India. The chance of developing progressive liver disease is higher in subjects with prolonged replicative phase. Infection of infants and children frequently leads to persistent HBV carriage with a significant risk for the development of chronic active hepatitis, cirrhosis and hepatocellular carcinoma. Host factors such as age, gender and immunocompetence, are the main reasons which determine the outcome of HBV infection. Precore stop codon has also been associated with severe liver damage in HBeAg negative individuals in chronic hepatitis B (Sharma et al., 2010, Ozgenc et al., 2007).
The presence of precore core mutant in 94.4% of HCC cases demonstrates that precore mutant showed high association with HCC. The serum ALT level between HBeAg negative and wild type was statistically significant but was insignificant in the other spectrum of liver diseases.
Therefore, it is of immense importance to screen precore G1896A mutations in patients with HBeAg negative serology and we have tried to establish LCR as a mutation screening tool. LCR can act as a useful tool for the management of hepatitis B virus.
Mutant HBV with precore stop codon at position G1896A has been reported to be associated with severe liver damage in chronic liver disease patients (Rezende et al., 2005, Arankalle et al., 2011). A study from North India, showed the prevalence of precore mutant in chronic liver diseases to be as high as 25.8% and is also associated with increased severity of liver diseases. Preliminary studies conducted in South India suggests that G to A switch at nucleotide G1896A is a most common mutation of HBV genome in patients with chronic liver disease from the Indian subcontinent (Vivekanandan et al., 2004). Tunisia has reported that precore mutants could be detected in 65% of HBsAg positive patients with chronic infection (Ayed et al., 2007). Our results are consistent with those of studies from Japan that mutations in the core region can be frequently detected in patients with chronic HBV infections and that these mutations were more often found in HBeAg negative patients (Tu et al., 2015). Our study is also in accordance with the study from South India suggesting that the precore and core variants of HBV which are defective in HBeAg synthesis are relatively more common in Indian patients with chronic HBV infection (Agarwal et al., 2015).
The survival rate among the patients diagnosed as chronic hepatitis B was 100% after 5 years. Cirrhosis and hepatocellular carcinoma are the two major complications of chronic hepatitis B infection that incomparably increase the death rate. The death rate at 5 years was 16% for those with compensated cirrhosis and 65–86% for decompensated cirrhosis. HBeAg-negative patients had a higher rate of cirrhosis when compared with HBeAg-positive patients. Older age and persistent viral replication are predictors for the development of cirrhosis as well as mortality.
Various factors involving the host and the virus contribute to the development of HCC. The rate of mortality was high in HBeAg negative patients than in HBeAg positive patients in HCC. Precore mutation accounted for 66.6% (12/20) in Cirrhosis and 94.4% (17/18) in HCC. This is also in accordance with the other studies by Hori et al. (2015) and Barreto et al. (2015).
Genotypes D and A were the most common genotype detected HBV associated liver disease patients in India, and genotype D was the most predominant accounting for 79.5% of the cases.
Ligase chain reaction was used as a tool for screening G1896A mutation in all the clinical categories of HBV patients. The detection of the G1896A mutation or Wild type samples by LCR was in 100% agreement with the results of direct sequencing. The usage of ligase chain reaction is comparatively more economical, less time consuming and equally reliable as direct DNA sequencing. However, the use of radioisotope remains a matter of concern due to its biohazardous nature. Thus, ligase chain reaction is 100% specific and sensitive technique when compared with the gold standard direct sequencing and it can be used very efficiently where a large population of precore 1896 mutation needs to be screened.
In conclusion, G1896A precore mutation is highly prevalent in patients with HBeAg negative serology in particular with the clinical categories like FH, CH, Cirrhosis and HCC. The precore G1896A mutation increases proportionately in severe form of liver diseases. The exceptionally high prevalence of G1896A in FH and HCC demonstrates that the precore mutants are strongly associated with the progression of liver diseases in patients with HBeAg negative serology. This observation suggests the need of screening HBV precore G1896A mutation in this region particularly in HBeAg negative cases. The LCR technology can be a suitable tool for screening G1896A mutations.
Acknowledgements
The authors extend their appreciation to the Research Center at the College of Applied Medical Sciences, and Deanship of Scientific Research, King Saud University for funding this work.
Footnotes
Peer review under responsibility of King Saud University.
References
- Acharya S.K. Epidemiology of hepatocellular carcinoma in India. J. Clin. Exp. Hepatol. 2014;4:27–33. doi: 10.1016/j.jceh.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A.K., Sen S., Banerjee D., Srivastava R., Praharaj A.K. Distribution of hepatitis B virus genotype and cancer predicting precore and basal core promoter mutations. Med. J. Armed Forces India. 2015;71:225–232. doi: 10.1016/j.mjafi.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V.A., Gandhi S., Lole K.S., Chadha M.S., Gupte G.M., Lokhande M.U. An outbreak of hepatitis B with high mortality in India: association with precore, basal core promoter mutants and improperly sterilized syringes. J. Viral Hepatitis. 2011;18:20–28. doi: 10.1111/j.1365-2893.2010.01391.x. [DOI] [PubMed] [Google Scholar]
- Ayed K., Gorgi Y., Ayed-Jendoubi S., Aouadi H., Sfar I., Najjar T., Ben Abdallah T. Hepatitis B virus genotypes and precore/core-promoter mutations in Tunisian patients with chronic hepatitis B virus infection. J. Infect. 2007;54:291–297. doi: 10.1016/j.jinf.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Barany F. Proceedings of National Academy of Science (USA) Vol. 80. 1991. Genetic disease detection and DNA amplification using thermostable ligase; pp. 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto S.G., Barreto M., Chaubal R., Dutt A. The fight against cancer: Is it worthwhile? Indian J. Med. Paediatr. Oncol. 2015;36:85–86. doi: 10.4103/0971-5851.158833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya H., Bhattacharya D., Nagarajan M., Reesu R., Roy S., Attayur P.S. Prevalence of mutations in basal core promoter and precore region of hepatitis B virus in vaccinated and nonvaccinated individuals of the aboriginal nicobarese tribe of car nicobar island, India. Intervirology. 2014;57:357–364. doi: 10.1159/000365756. [DOI] [PubMed] [Google Scholar]
- Datta S. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. J. Virol. 2008:155–156. doi: 10.1186/1743-422X-5-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy A.M., Moreira R.C., Lemos M.F., Silva J.L., Coêlho M.R., Eloy A.M., Moreira R.C., Lemos M.F., Silva J.L., Coêlho M.R. Hepatitis B virus in the State of Alagoas, Brazil: genotypes characterization and mutations of the precore and basal core promoter regions. Braz. J. Infect. Dis. 2013;17:704–706. doi: 10.1016/j.bjid.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Joshi Y.K., Singh S. Role of horizontal transmission in hepatitis B virus spread among household contacts in north India. Intervirology. 2008;51:7–13. doi: 10.1159/000118790. [DOI] [PubMed] [Google Scholar]
- Guptan R.C., Thakur V., Sarin S.K. Frequency and clinical profile of precore and surface hepatitis B mutants in Asian – Indian patients with chronic liver disease. Am. J. Gastroenterol. 1996;14:1312–1317. [PubMed] [Google Scholar]
- Hori M., Katanoda K. Morphological distribution of thyroid cancer from cancer incidence in five continents vol. X. Jpn. J. Clin. Oncol. 2015;45:1182. doi: 10.1093/jjco/hyv176. [DOI] [PubMed] [Google Scholar]
- Imamura T., Yokosuka O., Kurihara T., Kanda T., Fukai K., Imazeki F., Saisho H. Distribution of hepatitis B viral genotypes and mutations in the core promoter and precore regions in acuteforms of liver disease in patients from Chiba. Jpn. Gut. 2003;52:1630–1637. doi: 10.1136/gut.52.11.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Negi S.S., Singh S., Singhal M., Kumar S., Prakash C., Venugopal R., Rawat D.S., Chauhan L.S., Rai A. Genetic analysis of precore/core and partial pol genes in an unprecedented outbreak of fulminant hepatitis B in India. Epidemiol. Infect. 2012;140:1823–1829. doi: 10.1017/S0950268812000349. [DOI] [PubMed] [Google Scholar]
- Konstantinou D., Deutsch M. The spectrum of HBV/HCV coinfection: epidemiology, clinical characteristics, viral interactions and management. Ann. Gastroenterol. 2015;28:221–228. [PMC free article] [PubMed] [Google Scholar]
- Landegreen V., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Minamitani S., Nishiguchi S., Kuroki T. Detection by ligase chain reaction of precore mutant of hepatitis B virus. Hepatology. 1997;25:216–222. doi: 10.1053/jhep.1997.v25.pm0008985293. [DOI] [PubMed] [Google Scholar]
- Ozgenc O., Ozacar T., Erensoy S., Inan N., Ari A., Kuruuzum Z., Bilgic A. Clinical significance of basal core promoter and precore mutations in chronic hepatitis B. Hepatogastroenterology. 2007;54:2319–2323. [PubMed] [Google Scholar]
- Rezende R.E., Fonseca B.A., Ramalho L.N., Zucoloto S., Pinho J.R., Bertolini D.A., Martinelli A.L. The precore mutation is associated with severity of liver damage in Brazilian patients with chronic hepatitis B. J. Clin. Virol. 2005;32:53–59. doi: 10.1016/j.jcv.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Schalm S.W., Thomas B., Hadziyannis S.J. Chronic hepatitis B. Progression of liver disease. 1990;9:443–462. [PubMed] [Google Scholar]
- Sharma S., Sharma B., Singla B., Chawla Y.K., Chakraborti A., Saini N., Duseja A., Das A., Dhiman R.K. Clinical significance of genotypes and precore/basal core promoter mutations in HBV related chronic liver disease patients in North India. Dig. Dis. Sci. 2010;55:794–802. doi: 10.1007/s10620-009-1083-y. [DOI] [PubMed] [Google Scholar]
- Singh T.N., Kananbala S., Thongam W., Devi K.H.S., Singh N.B. Increasing trend of HIV seropositivity among commercial sex workers attending the Voluntary and Confidential Counseling and Testing Centre in Manipur, India. Int. J. STD AIDS. 2005;16:166–169. doi: 10.1258/0956462053057684. [DOI] [PubMed] [Google Scholar]
- Tu W.H., Lv Y., Zhang Y.M., Hou W., Wang J.Y., Zhang Y.J., Liu H.Y., Zhu H.X., Qin Y.L., Mao R.C., Zhang J.M. Precore/basal core promoter mutants quantification throughout phases of hepatitis B virus infection by simple probe. World J. Gastroenterol. 2015;21:6639–6648. doi: 10.3748/wjg.v21.i21.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivekanandan P., Abraham P., Sridharan G., Chandy G., Shaji R.V., Daniel D., Raghuraman S., Daniel H.D., Subramaniam T. Precore mutation in patients and blood donors with hepatitis B virus infection from the Indian subcontinent. J. Mol. Diagn. 1896;8(2004):51–56. doi: 10.1007/BF03260047. [DOI] [PubMed] [Google Scholar]
- Zuccaro O., Romanò L., Mele A., Mariano A., Clementi M., Tosti M.E., Taliani G., Galli C., Zanetti A.R., Spada E. Study group, clinical, epidemiological and virological features of acute hepatitis B in Italy. Infection. 2015 doi: 10.1007/s15010-015-0747-0. [DOI] [PubMed] [Google Scholar]