Abstract
The eukaryotic cell is organized as a complex grid system where membrane bound cellular compartments, organelles, must be localized to the right place at the right time. One way to facilitate correct organelle localization and organelle cooperation is through membrane contact sites, areas of close proximity between two organelles that are bridged by protein/lipid complexes. It is now clear that all organelles physically contact each other. The main focus of this review is contact sites of peroxisomes, central metabolic hubs whose defects lead to a variety of diseases. New peroxisome contacts, their tethering complexes and functions have been recently discovered. However, if and how peroxisome contacts contribute to the development of peroxisome-related diseases is still a mystery.
Keywords: peroxisomes, organelles, contact sites, peroxisomal disorders, tethering, mitochondria, endoplasmic reticulum
The cellular grid – peroxisomes stay on the line
Peroxisomes are hubs of cellular metabolism, performing essential cellular functions such as α- and β-oxidation of fatty acids, amino acid synthesis and metabolism of reactive oxygen species (ROS) [1,2]. As key players in several cellular functions, peroxisomes have established a network of interactions with other organelles to cooperatively perform these roles. To facilitate these interactions, peroxisomes form membrane contact sites, areas of close proximity between two organelle membranes which are tethered by protein and/or lipid components [3–6]. These cellular microenvironments have defined membrane compositions and are formed between most, if not all, cellular membranes [7]. Contact sites are implicated in a growing number of processes from organelle morphology and inheritance, to lipid metabolism and intracellular signaling [3,4,8,9], but identifying their molecular components has proven to be a challenging task.
In recent years, a growing number of peroxisome contacts and their tethers have been identified (Figure 1) [5] and the functional importance of these contacts is becoming clearer. As the components and functions of these contacts are deciphered, it begs the question: what is the role of peroxisome contact sites in disease?
Figure 1. The cellular peroxisome contact site grid.
In the past five years several new peroxisome contact sites have been identified, and their molecular components and functions are now being explored. In mammalian cells, ACBD5 and ACBD4 have been shown to interact with the ER proteins VAPA and VAPB, in a contact site suggested to regulate peroxisome biogenesis and metabolism. Peroxisomes have also been shown to interact with lysosomes for cholesterol trafficking (SYT7-PI(4,5)P2). In S. cerevisiae, peroxisomes have been shown to interact with the ER for inheritance during cell division (Pex3-Inp1-Pex3) and with mitochondria to perform metabolic functions (Pex34-?; Fzo1-Fzo1/?; Pex11-Mdm34). In the filamentous fungus U. maydis, peroxisomes form a contact site with endosomes that allows then to hitchhike and move through the cell (?-PxdA-?).
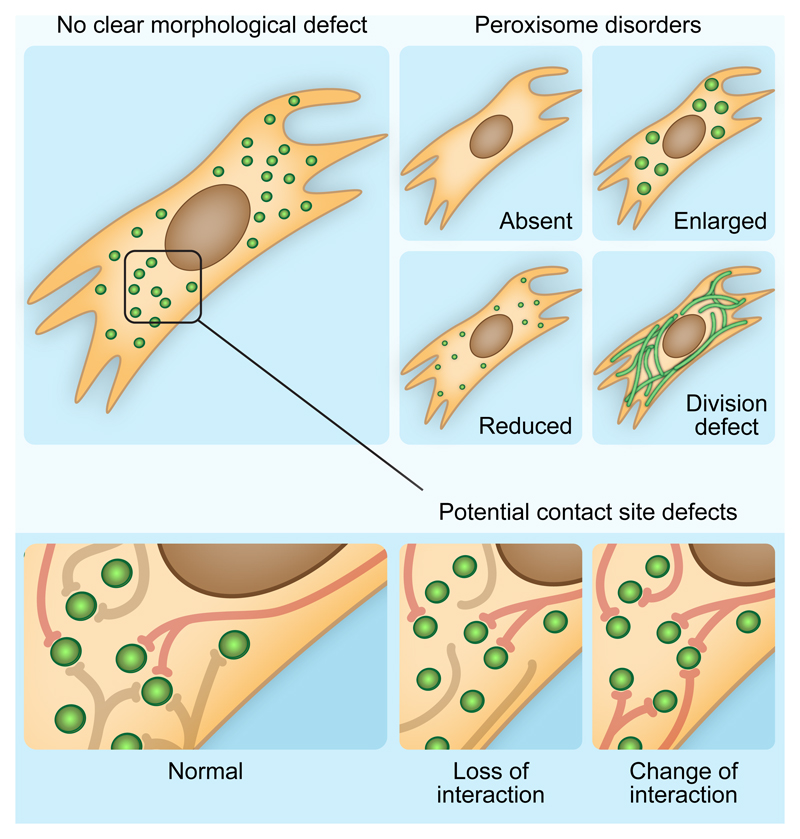
Conventional peroxisomal disorders are characterized by mild to severe neurodevelopmental defects, sight and hearing impairments, and organ specific deficiencies, particularly affecting the liver and kidneys (reviewed in [10,11]). These diseases are typically caused by the absence of, or mutations in, essential peroxisomal proteins, known as peroxins (peroxisomal biogenesis factor, PEX), or in enzymes involved in peroxisomal metabolic functions. Many of these disorders and their associated proteins can be readily identified using standardized biochemical techniques [12,13]. Since peroxisomes play an essential role in the catabolism and anabolism of many lipid species, these tests include measuring the levels of several metabolites in the patient’s plasma, such as increases in long- and very-long-chain fatty acids (LCFA and VLCFA, respectively), lower levels of the ether phospholipid plasmalogen and of docosahexaenoic acid (omega 3), as well as testing for α- and β-oxidation of fatty acids in patient skin fibroblasts. Additionally, since many patients with these disorders present altered peroxisome number, size and distribution, microscopic analysis of peroxisomes in patient skin fibroblasts can facilitate the diagnosis. However, none of these tests are tailored to identify alterations in the interaction of peroxisomes with other organelles, and since in such cases the metabolic functions of peroxisomes themselves may not be affected and peroxisome shape, number and size may remain unaltered (Figure 2), this implies that peroxisome contact site disorders might have, to date, fallen under the radar.
Figure 2. Potential involvement of peroxisome contact sites in peroxisomal disorders.
Healthy human cells have a large population of round, well distributed peroxisomes. In classical peroxisome disorders, these organelles can be completely absent (defects in PEX3, PEX16 and PEX19), reduced and/or enlarged (defects in DBP, ACOX1), or can have altered shape and distribution (defects in MFF, DLP1 and PEX11β). Notably, alterations in peroxisome number or morphology could also be a secondary effect of a defect in contact sites. In these cases, the defect can still be diagnosed microscopically but identifying its genetic cause will require further investigation. We hypothesize that in some cases of peroxisomal disorders peroxisome shape, size and distribution will appear normal microscopically, however alterations in the number of interactions with specific organelles, or changes in the balance of interaction with several organelles will be the cause of the disease. New tools should be developed to diagnose these cases at the cellular level.
To shine a light on the possibility that defects in peroxisome contacts might lead to diseases and stimulate research in the area, here we explore the known components and roles of peroxisome contact sites, how they affect overall peroxisome function and morphology and suggest their potential role in peroxisome diseases.
1. How can defects in peroxisome contact sites lead to disease?
In recent years, several contact sites have been identified for peroxisomes and their function is now starting to be explored. In this section we will discuss these emerging discoveries and explore how defects in such contacts may affect peroxisome function and lead to diseases. Although in recent years peroxisome contact sites have been studied in various organisms such as plants [14,15], we will be focusing on studies that explore peroxisome contact sites in mammalian cells, for their relevance in disease, and in fungi, as good model systems to study the cell biology of contact sites.
Growth and division
To grow in size peroxisomes must take up membrane lipids, most of which are synthesized in the ER. Both vesicular [16,17] and non-vesicular pathways [18] have been demonstrated to underlie lipid transport from the ER to peroxisomes. Non vesicular pathways rely on contact sites for direct transfer through lipid transfer proteins. Indeed, Electron Microscopy (EM) evidence [19–21] demonstrates contact sites between peroxisomes and the ER in various organisms, including humans.
In addition to being a lipid source for membrane expansion, the ER might also play a major role in peroxisome division, both by facilitating the assembly of the division machinery or by physically enabling the constriction and fission of the organelle. This ER-contact site function, which has been previously described for both mitochondrion and endosome fission (reviewed in [9]), could shed some light on the mechanical forces that enable peroxisome membranes to constrict prior to fission. The absence of such an interaction could have pathological effects similar to those observed in peroxisome division disorders (Figure 2) [10], with peroxisomes being unable to or having delays in division.
The role of mammalian peroxisome-ER tethering proteins in lipid transfer
Two recent publications have identified the first peroxisome-ER tethering complex in mammalian cells [21,22]. The tether is formed by the peroxisomal membrane protein acyl-CoA binding domain containing 5 (ACBD5) that interacts with the ER proteins vesicle-associated membrane protein-associated proteins A and B/C (VAPA and VAPB) (Figure 1). This interaction relies on the recognition of a two phenylalanines in an acidic tract (FFAT) motif in ACBD5 by the major sperm protein (MSP) domain in the VAP proteins. FFAT motifs are present in other VAP interacting proteins [23] that are essential for formation of contact sites with mitochondria [24] or late endosomes [25]. Disruption of the ACBD5/VAP tethering complex significantly alters peroxisome-ER interaction and prevents the growth of peroxisomal membranes, suggesting that this tethering complex could facilitate and/or regulate lipid transport from the ER to peroxisomes. Furthermore, a second ACBD family protein, ACBD4, has also been shown to interact with VAPB [26].
Interestingly, both ACBD5 and VAPB have been implicated in human disorders. ACBD5 mutations were recently identified in four patients, all of whom display retinal dystrophy. Additionally, three of the patients (from the same family) have severe white matter disease and spastic paraparesis [27,28], and the fourth patient presents progressive leukodystrophy, syndromic cleft palate and ataxia [29]. All patients have a complete absence of ACBD5 and show a significant accumulation of VLCFA, which are exclusive substrates of peroxisomal β-oxidation [28,29]. As little is known about the function of ACBD5, this suggests that in the absence of ACBD5 there is a reduction of peroxisomal function – either because the growth of their membranes is hampered or more directly because ACBD5 recognizes ER-derived and cytosolic VLCFA, and recruits them to peroxisomes for degradation. However, it is not clear if these defects result from the loss of tethering between the ER and peroxisomes and whether ACBD5’s tethering function contributes to the patients’ pathology. It would be interesting to analyze patient cells in light of this newly discovered function and see the effects of mutations in the acyl-CoA binding domain and FFAT motif on VLCFA levels. Furthermore, if this proves to be a peroxisome contact site disorder, it is interesting to observe that there are no changes in peroxisome number and distribution in the studied cells.
Mutations in VAPB are associated with the development of rare inherited motor neuron diseases, in particular late-onset spinal muscular atrophy and amyotrophic lateral sclerosis (ALS) type 8 [30,31]. These disorders are characterized by degeneration of specific groups of motor neurons, associated with a progressive loss of muscle function. In these patients there is a strong reduction in VAPB levels, as the mutated protein tends to aggregate and be targeted for degradation. However, due to the extensive number of interactions that VAPB establishes with other proteins, pinpointing the molecular processes that trigger these disorders will be more complicated. Conversely, the late onset and progressive nature of these diseases demonstrates the ability of the cell to partially compensate for the absence of an important contact site protein, likely by finding alternative pathways to perform VAPB-related functions. To date, no disease variants of VAPA have been identified.
Membrane growth in yeast
In comparison to humans, the role of peroxisome-ER contact sites in the transfer of lipids to peroxisomes for membrane growth in the yeast Saccharomyces cerevisiae (simply termed yeast from here on) is still poorly understood. This is made more difficult by the fact that membrane contact sites between these two organelles are hard to experimentally distinguish from subdomains of pre-peroxisomal vesicle biogenesis. These subdomains do not constitute contact sites since the process of vesicle formation predicates the existence of a shared membrane and contact sites require opposing membranes. These vesicles however, might be capable of forming contact sites with the ER after budding.
In yeast it has not yet been shown if and how ER-Peroxisome contact sites enable lipid transfer. A group of proteins which might play a role in both budding and contact site formation are Pex28, Pex29, Pex30, Pex31 and Pex32. These proteins affect peroxisome proliferation, as shown by deletion strains for each protein [32,33]. In these cells, peroxisomes are either reduced in number but enlarged, or smaller but in a higher number. These proteins have homology to ER reticulons, pointing to a membrane-bending activity that might either facilitate lipid transport between the ER and peroxisomes, or the bending of the ER membrane, in which several of these family members reside [34], to form pre-peroxisomal vesicles [34–36].
Inheritance and motility
Peroxisome inheritance and motility are two interconnected processes that regulate peroxisome distribution during cell division.
Moving Yeast peroxisomes
In yeast, peroxisomes form a contact site with the ER network during cell division to maintain part of the peroxisome population in the mother cell [37]. Tethering to the ER is mediated by inheritance of peroxisomes 1 (Inp1) protein, which forms a complex with two Pex3 molecules, one at the peroxisomal membrane and one at the cortical ER membrane (Figure 1) [38]. A second protein, Inp2, links peroxisomes to myosin 2 motors and enables the transport of the remaining peroxisomes to the budding cell [39]. In the absence of Inp1, and by proxy of this specific peroxisome-ER contact site, all peroxisomes are transported to the bud, whereas overexpression of Inp1 leads to the sequestration of all peroxisomes at the mother cell [37].
In the filamentous fungus Ustilago maydis, peroxisomes rely on contact sites and motility for inheritance and distribution in hyphae. In this organism, peroxisomes move bi-directionally via the microtubule network, a process which facilitates their distribution during hyphae growth. However, instead of binding directly to motor complexes, peroxisomes form a contact site with early endosomes, hitchhiking on this organelle to move in the cell (Figure 1) [40,41]. This contact is mediated by peroxisome distribution mutant A (PxdA), an endosome-associated protein. Other elements of this contact are yet to be discovered.
Mammals – peroxisomes on the move
Similar mechanisms for peroxisome inheritance in mammalian cells are yet to be described. Unlike yeast, mammalian cells frequently have hundreds of peroxisomes evenly distributed within the cytosol. Early models of peroxisome distribution during cell division reasoned that this is a stochastic process due to symmetric cell division and the very low probability of any of the daughter cells to be completely depleted of peroxisomes. However, it is recently starting to be appreciated that inheritance may be a more actively regulated process [42]. Specifically, when the peroxisomal protein PEX11β, which is involved in membrane dynamics and division, was depleted from mouse epidermal cells, peroxisome distribution and inheritance was altered. Importantly, these alterations led to delays in cell cycle progression and defects in the orientation of cellular spindles, resulting in differentiation defects of the epidermis in a mouse model. Even though the role of PEX11β in this process is not yet understood, this demonstrates for the first time the existence of peroxisome-specific regulation during the cell cycle.
One interesting possibility is that the ER/peroxisome contact might have a role in the distribution of peroxisomes during the cell cycle. Since a high percentage of peroxisomes are associated with the ER in mammalian cells and since peroxisome motility significantly increases in the absence of ACBD5 and VAP proteins [21,22], this contact site might restrict peroxisome movement. Hence, by stably interacting with the ER, peroxisomes might “hitchhike” on this organelle during cell division, therefore distributing evenly between daughter cells. Further studies on peroxisome inheritance and motility in mammalian cells, the contribution of peroxisome contact sites to these processes and how they might lead to peroxisomal disorders are forthcoming.
Metabolic pathways and signaling
Peroxisomes play essential roles in numerous metabolic pathways, many of which are performed in close cooperation with other organelles such as the ER and mitochondria [5,43]. This cooperation relies on the ability of organelles to efficiently exchange several metabolites, a process that is likely to be facilitated by the formation of membrane contact sites. This cooperation might also contribute to the heterogeneity of the peroxisome population, as peroxisomes might specialize in specific functions, depending on which organelle they interact with [44].
As previously described, defects in peroxisomal metabolism are at the basis of many peroxisomal disorders. However, it has not been well explored how cells would respond to having metabolically functioning peroxisomes and partner organelles, but defects in the transport of metabolites between them? In theory, the expectation is that this would lead to the accumulation of toxic substrates or the lack of essential metabolic products therefore manifesting itself as a bona fide metabolic or peroxisomal disorder. For example, inhibiting cholesterol movement through peroxisomes leads to its accumulation at lysosomes, and associated depletion from the plasma membrane [45]. Interestingly, accumulation of cholesterol at lysosomes leads to a rare inherited lysosomal neurodegenerative disorder – Niemann Pick type C [46]. Hence, such accumulation should have fatal results. A second scenario might involve the compensation of the absence of one contact by the expansion of a contact with a different organelle (Figure 2). As was previously shown [47,48], this is a readily available mechanism for the cells which allows the fine tuning of different pairs of contact sites, likely regulated by the cell’s needs in different metabolic conditions. In this case, the absence of contact sites between two specific organelles might not only slow down the metabolic processes shared by them, but induce an exacerbated increase of interaction with another.
Working together – mammalian peroxisomes at play
ER
In mammals, peroxisomes collaborate with the ER for the synthesis of plasmalogens, cholesterol, bile acids and other lipid species [1]. For these processes to occur, several metabolites need to be transported between these two organelles. For example, one of the first steps in the synthesis of plasmalogens (and other etherphospholipids) is the formation of an ether-bond, a process that occurs exclusively in peroxisomes. The resulting metabolite must then be shuttled to the ER for further enzymatic steps leading to the formation of plasmalogens. Although it is not currently known how these metabolites are transported between both organelles, it is tempting to suggest that such a process would preferentially occur at contact sites, where the distance between both organelles is reduced and the function of specific metabolite transporters could be more strictly regulated [46]. Interestingly, silencing of either ACBD5 or the VAP proteins has been shown to reduce the levels of plasmalogens in human HeLa cells [22], suggesting that the contact site itself plays an integral role for the efficient production of plasmalogens (despite the presence of all the enzymatic players for its anabolism). How this process is regulated and the molecular players involved are still to be uncovered.
Mitochondria
Another essential peroxisome interactor in the cell are the mitochondria. These organelles cooperate in β-oxidation of fatty acids and ROS metabolism [49–51]. In mammalian cells both peroxisomes and mitochondria carry out the β-oxidation of fatty acids. However, long chain acyl chains such as those found in LCFAs and VLCFAs, are initially degraded in peroxisomes and must then be transported to mitochondria for further rounds of oxidation. Hence it is not surprising that some patients with peroxisomal disorders also present altered mitochondrial morphology and function [52]. Although not much is known about the peroxisome-mitochondrion contact site in mammalian cells, two proteins that are involved in fatty acid transport and metabolism, ABCD1 and ACBD2, have been suggested to play a role in this connection [53,54].
Lysosomes
A more recently identified peroxisome interactor is the lysosome. In addition to their essential role in peroxisome degradation, lysosomes also form contact sites with peroxisomes held by the lysosomal protein synaptotagmin 7 (SYT7) which interacts with Phosphatidylinositol 4,5-bisphosphate, PI(4,5)P2, at the peroxisomal membrane in mammals (Figure 1). This interaction was shown to play a role in cholesterol trafficking [45]. Interestingly, it was suggested that defects in cholesterol trafficking might play a role in the pathology of peroxisomal disorders, as cholesterol accumulations can be readily identified in a mouse model of X-linked adrenoleukodystrophy. Concomitantly, defects in peroxisome function in peripheral nerves have been shown to alter the turnover of gangliosides, leading to their accumulation at lysosomes and impairment of lysosomal functions, further substantiating a close interaction between both organelles [55]. Additional studies are necessary to further elucidate the interplay between these two organelles in patients and to understand if the peroxisome-lysosome interaction plays a major role in the pathology of peroxisomal disorders.
Sharing is caring – yeast peroxisomes in metabolism
In yeast, where β-oxidation of fatty acids is exclusively performed by peroxisomes, this organelle is found in close proximity to a specific area of the mitochondrial matrix where the pyruvate dehydrogenase (PDH) complex is found [56]. As both peroxisomal β-oxidation of fatty acids and PDH-mediated dehydrogenation of pyruvate lead to the production of acetyl-CoA, the proximity between these two organelles might facilitate the regulation of this compound’s entry into the TCA cycle. This hypothesis was recently further substantiated by showing that expansion of the peroxisome-mitochondria contact site facilitates the transport of β-oxidation products to mitochondria (Schuldiner lab, Personal communications). Interestingly, although the contact site could be expanded by either expressing a synthetic tether or by overexpression of two newly found tethers, Pex34 and Fzo1 (Figure 1), only overexpression of Pex34 increased the transfer of β-oxidation products to mitochondria, suggesting that increasing the area of interaction between two organelles per se is not sufficient to facilitate all levels of metabolic interplay (Schuldiner lab, Personal communications). The role of the Pex34 and Fzo1 tethering proteins, as well as of the previously proposed tethering complex, Pex11-Mdm34 [57], on the peroxisome-mitochondria contact site are yet to be revealed. Interestingly, peroxisomes appear to interact with mitochondria in close proximity to the mitochondria-ER contact site [56], suggesting that a tri-junction between these organelles might help coordinate their metabolic functions.
Degradation
As membrane contact sites in peroxisomes are places of high metabolic transport, peroxisomes that have more prominent contact sites might be more at risk of accumulating damaged proteins and to suffer the effect of ROS [58]. Peroxisomes that are dysfunctional or burdened with damaged proteins are degraded by a selective form of autophagy called pexophagy [59,60]. Organelles tagged for degradation are recognised by specific autophagy adaptors that form a bridge between the organelle and the growing autophagosomal membrane [61].
One interesting role for membrane contact sites might be on the signalling and regulation of peroxisome degradation. Interestingly, ACBD5 has also been suggested to play a role in pexophagy, since in the absence of this protein there is a significant reduction of peroxisome degradation [62].
Recognition and degradation of damaged peroxisomes play an important role in the maintenance of cellular health. Whereas the accumulation of damaged peroxisomes is a common process associated with aging [63], and oxidative damage of these organelles can lead to organ injury [64], excessive pexophagy was suggested to contribute to up to 65% of peroxisomal disorders [65]. This is due to defects in a set of peroxins (PEX1, PEX6 and PEX26) which might protect the organelle from degradation [66]. By playing a role in sensing peroxisome damage, membrane contact sites might help maintain healthy levels of peroxisome turnover, and in their absence a dysregulation of pexophagy.
2. What are peroxisome contact site disorders and how can we characterize them?
Since contact sites play an essential role in the formation, function and degradation of peroxisomes, alterations to contacts should have a serious effect on peroxisome homeostasis and human health. Considering the growing wealth of information now available on peroxisome contact sites, it is surprising that peroxisome contact site-related diseases have not yet been categorically identified (see Outstanding Questions). We will discuss below why identifying these diseases is no easy task:
Outstanding Questions Box.
How can we identify the full list of peroxisome contact sites?
How can we identify the tethers and regulators of peroxisome contacts?
How to separate tethering versus functional activity of proteins, and the potential role of each in disease?
How can we identify the range of functions of each contact site?
How is it possible to diagnose contact site related diseases?
What is the contribution of defects in peroxisome contacts to the development of peroxisomal disorders?
Clinical identification of contact site alterations is complex
Diagnosing a disease in the absence of a molecular mechanism requires the identification and correlation between symptoms to a previous description of the same or similar disease. Hence the first cases of any disease are often extremely hard to categorize, whereas following the first description in the literature many cases rapidly accumulate.
In the last years, identification of diseases is also possible following the discovery of mutations in causal genes that have been characterized on a molecular level as underlying the mechanisms that are inducing the disease.
Unfortunately, for peroxisome contact site-related disorders neither of these aspects exist to date. First, no peroxisome contact site-related disorder has been categorically identified and published, and therefore, no particular set of symptoms has been specifically correlated with this kind of pathology. To complicate matters even more it is probable that changes in one contact site will display very different physiological consequences to changes in a different contact site, making it a necessity to identify each contact site disorder independently. Furthermore, only a few molecular components have been identified to date making it harder to find causal mutations.
Tethering proteins can be multifunctional
A common feature to several contact site proteins is their ability to perform more than one function, such as membrane tethering and metabolic/enzymatic activity. Hence, while some molecular components of peroxisome contact sites have been described, merely associating mutations in these proteins with diseases does not necessarily mean that they are contact site disorders as all of the contact site proteins described to date have at least one additional function. Moreover, some of these proteins are also localized to several contact sites, making it difficult to know what mutations affect which functions, and at which contacts.
For example, both ACBD5 and VAPB which have clear diseases associated with them, play several roles each. ACBD5 likely plays two connected roles on peroxisomes, one in the binding of VLCFAs and the other in the tethering of peroxisomes to the ER. Hence in ACBD5 pathologies it is complicated to differentiate which symptoms correlate with loss of which functions. Similarly, it is hard to untangle which contact site underlies the various phenotypes of pathologies caused by loss of VAP proteins, which have been shown to form contacts with several organelles [23].
Contact site tethering is redundant
Another evident issue with identifying the physiological effects of mutations in tethers is redundancy. All contact sites studied to date have multiple tethers ensuring their formation [6]. Moreover, the presence of co-regulation between different contact sites enables expansion of one contact to compensate for the loss of another [47,48]. Redundancy poses two main problems. First, the absence of any one tethering complex or even the loss of an entire contact site, can be hard to detect. Second, most probably each tethering component will have a different physiological outcome making disease characteristics very variable and often subtle. For example, there may not be a single disease associated with loss of the peroxisome/ER contact site, but rather different pathologies depending on which tethering pair or functional molecule is mutated.
3. Concluding remarks: What next? Identifying peroxisome contact site-related diseases
As we start exploring the intricacy of peroxisome contact sites, what steps can we take to identify and characterize peroxisome contact site disorders? First and foremost, recognizing that these disorders might exist stands as a crucial point to develop techniques and projects aimed at characterizing them. This can be particularly relevant for understanding the pathology of patients who currently present poorly understood phenotypes and who would benefit most from a clear diagnosis. The existence of such patients is made clear by the growing number of diseases that have been associated with non-peroxisomal contact sites such as cancer, diabetes, Alzheimer’s and Parkinson’s disease [67–71]. Second, the complete repertoire of peroxisome contacts, tethers and regulators should be unraveled. For this, systematic approaches employing both yeast and mammalian cells, conjugated with techniques to visualize contact sites should significantly increase the number of identified proteins. Once the proteins are identified, mutations in their encoding genes can be specifically examined during the diagnosis process of peroxisomal disorders. These genes can also be retrospectively analyzed in cells from diseased patients with unknown pathologies, effectively increasing our chances of characterizing the symptomology of these disorders. Additionally, nowadays, when exome sequencing is commonly used, mutations in genes that were not previously connected to peroxisomal disorders will be identified. The potential function of these proteins in peroxisome biogenesis, division, metabolic functions etc. might be studied to better understand how the specific genetic alteration is leading to a disease. During such studies it would be important to remember that a possible cause could also be a defect in peroxisome contacts. Another key aspect will be to develop and adapt techniques to analyze patient cells to identify contact site defects. As no apparent symptoms have yet been associated with peroxisome contact site disorders, several techniques will likely have to be used, including microscopy (both fluorescent and electron), organelle motility analysis and induction of peroxisome proliferation. Additionally, metabolic profiling of patient cells might enable the identification of small changes in specific metabolic pathways, such as the accumulation or absence of certain metabolites, potentially leading to the investigation of specific processes. This will require a closer collaboration between clinical and basic research driven laboratories, enabling a better understanding of disease as a whole.
As we have only just scratched the surface of contact site research, it is exciting to imagine what lies ahead and to work on uncovering peroxisome contact sites and their roles in disease. We are only at the beginning of this exciting journey that will require a close dance between basic cell biology and the clinic.
Trends Box.
-
-
Membrane contact sites are areas of close proximity between two organelles that enable close range interactions and rapid transfer of lipids, ions and metabolites.
-
-
Tethering proteins have a primary role in holding the membranes of two organelles together and can be regulated to control the dynamics of the contact. Tethers often have additional functions in contact sites such as metabolite transfer.
-
-
Contact sites play a role in organelle inheritance, division, trafficking, metabolite transport and signaling.
-
-
Peroxisome disorders are a group of genetically inherited disorders caused by defects in peroxisome biogenesis and metabolism.
-
-
Peroxisomes cannot function in isolation since all of their metabolic functions require transfer of metabolites to/from other organelles.
-
-
Loss of contact sites should dramatically affect the capacity of peroxisomes to function optimally.
Acknowledgements
This work was supported by the European Research Council (ERC) (Consolidator grants Peroxisystem 64660), an SFB grant from the DFG (1190) and a VW foundation grant (93092). Maya Schuldiner is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics. We wish to thank Eden Yifrach and Michal Eisenberg-Bord for constructive feedback on this manuscript, and Noa David Geller for fantastic graphic design.
Abbreviations
- ROS
reactive oxygen species
- LCFA and VLCFA
long- and very-long-chain fatty acids
- ER
endoplasmic reticulum
- EM
electron microscopy
- PEX
peroxin/peroxisomal biogenesis factor
References
- 1.Lodhi IJ, Semenkovich CF. Peroxisomes: A Nexus for Lipid Metabolism and Cellular Signaling. Cell Metab. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prinz WA. Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrader M, et al. The different facets of organelle interplay—an overview of organelle interactions. Front Cell Dev Biol. 2015;3:56. doi: 10.3389/fcell.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shai N, et al. No peroxisome is an island - Peroxisome contact sites. Biochim Biophys Acta - Mol Cell Res. 2016;1863:1061–1069. doi: 10.1016/j.bbamcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberg-Bord M, et al. A Tether Is a Tether Is a Tether: Tethering at Membrane Contact Sites. Dev Cell. 2016;39:395–409. doi: 10.1016/j.devcel.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Valm AM, et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa AD, et al. Lipid droplet-organelle interactions: Emerging roles in lipid metabolism. Curr Opin Cell Biol. 2015;35:91–97. doi: 10.1016/j.ceb.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterham HR, et al. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta - Mol Cell Res. 2016;1863:922–933. doi: 10.1016/j.bbamcr.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Argyriou C, et al. Peroxisome biogenesis disorders. Transl Sci rare Dis. 2016;1:111–144. doi: 10.3233/TRD-160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braverman NE, et al. Peroxisomes biogenesis disorders: biological, clinical and pathophysiological perspectives. Dev Disabil Res Rev. 2013;17:187–196. doi: 10.1002/ddrr.1113. [DOI] [PubMed] [Google Scholar]
- 13.Wanders RJA, et al. Clinical and Laboratory Diagnosis of Peroxisomal Disorders. Humana Press; New York, NY: 2017. pp. 329–342. [DOI] [PubMed] [Google Scholar]
- 14.Gao H, et al. In Vivo Quantification of Peroxisome Tethering to Chloroplasts in Tobacco Epidermal Cells Using Optical Tweezers. Plant Physiol. 2016;170:263–272. doi: 10.1104/pp.15.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oikawa K, et al. Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat Plants. 2015;1 doi: 10.1038/nplants.2015.35. 15035. [DOI] [PubMed] [Google Scholar]
- 16.Lam SK, et al. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2010;107:21523–21528. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal G, et al. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2011;108:9113–9118. doi: 10.1073/pnas.1018749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raychaudhuri S, Prinz Wa. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2008;105:15785–15790. doi: 10.1073/pnas.0808321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novikoff PM, Novikoff AB. Peroxisomes in absorptive cells of mammalian small intestine. J Cell Biol. 1972;53:532–560. doi: 10.1083/jcb.53.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaar K, et al. Association of isolated bovine kidney cortex peroxisomes with endoplasmic reticulum. Biochim Biophys Acta - Biomembr. 1987;897:135–142. doi: 10.1016/0005-2736(87)90321-x. [DOI] [PubMed] [Google Scholar]
- 21.Costello JL, et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J Cell Biol. 2017;216:331–342. doi: 10.1083/jcb.201607055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua R, et al. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J Cell Biol. 2017;216:367–377. doi: 10.1083/jcb.201608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy SE, Levine TP. VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta - Mol Cell Biol Lipids. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Stoica R, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5 doi: 10.1038/ncomms4996. 3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alpy F, et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci. 2013;126:5500–5512. doi: 10.1242/jcs.139295. [DOI] [PubMed] [Google Scholar]
- 26.Costello JL, et al. Peroxisomal ACBD4 interacts with VAPB and promotes ER-peroxisome associations. Cell Cycle. 2017;16:1039–1045. doi: 10.1080/15384101.2017.1314422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Safieh L, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagita Y, et al. Deficiency of a Retinal Dystrophy Protein, Acyl-CoA Binding Domain-containing 5 (ACBD5), Impairs Peroxisomal β-Oxidation of Very-long-chain Fatty Acids. J Biol Chem. 2017;292:691–705. doi: 10.1074/jbc.M116.760090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferdinandusse S, et al. ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J Med Genet. 2017;54:330–337. doi: 10.1136/jmedgenet-2016-104132. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura AL, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay D, Sengupta S. First evidence of pathogenicity of V234I mutation of hVAPB found in Amyotrophic Lateral Sclerosis. Biochem Biophys Res Commun. 2014;448:108–113. doi: 10.1016/j.bbrc.2014.04.102. [DOI] [PubMed] [Google Scholar]
- 32.Vizeacoumar FJ, et al. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J Cell Biol. 2003;161:321–332. doi: 10.1083/jcb.200210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vizeacoumar FJ, et al. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:665–677. doi: 10.1091/mbc.E03-09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi AS, et al. A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J Cell Biol. 2016;215:515–529. doi: 10.1083/jcb.201602064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David C, et al. A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol Cell Proteomics. 2013;12:2408–2425. doi: 10.1074/mcp.M112.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mast FD, et al. Peroxins Pex30 and Pex29 Dynamically Associate with Reticulons to Regulate Peroxisome Biogenesis from the Endoplasmic Reticulum. J Biol Chem. 2016;291:15408–15427. doi: 10.1074/jbc.M116.728154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fagarasanu M, et al. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol. 2005;169:765–775. doi: 10.1083/jcb.200503083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoblach B, et al. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013;32:2439–2453. doi: 10.1038/emboj.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fagarasanu A, et al. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell. 2006;10:587–600. doi: 10.1016/j.devcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Guimaraes SC, et al. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J Cell Biol. 2015;211:945–954. doi: 10.1083/jcb.201505086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salogiannis J, et al. Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J Cell Biol. 2016;212:289–296. doi: 10.1083/jcb.201512020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asare A, et al. Coupling organelle inheritance with mitosis to balance growth and differentiation. Science. 2017;355 doi: 10.1126/science.aah4701. eaah4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanders RJA, et al. Metabolic Interplay between Peroxisomes and Other Subcellular Organelles Including Mitochondria and the Endoplasmic Reticulum. Front Cell Dev Biol. 2016;3:83. doi: 10.3389/fcell.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galiani S, et al. Super-resolution microscopy reveals compartmentalization of peroxisomal membrane proteins. J Biol Chem. 2016;291:16948–16962. doi: 10.1074/jbc.M116.734038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu B-B, et al. Cholesterol Transport through Lysosome-Peroxisome Membrane Contacts. Cell. 2015;161:291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 46.Carstea ED, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 47.Elbaz-Alon Y, et al. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Lismont C, et al. Redox interplay between mitochondria and peroxisomes. Front Cell Dev Biol. 2015;3:35. doi: 10.3389/fcell.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrader M, et al. Peroxisome-mitochondria interplay and disease. J Inherit Metab Dis. 2015;38:681–702. doi: 10.1007/s10545-015-9819-7. [DOI] [PubMed] [Google Scholar]
- 51.Eisenberg-Bord M, Schuldiner M. Mitochatting – If only we could be a fly on the cell wall. Biochim Biophys Acta - Mol Cell Res. 2017;1864:1469–1480. doi: 10.1016/j.bbamcr.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Salpietro V, et al. Zellweger syndrome and secondary mitochondrial myopathy. Eur J Pediatr. 2015;174:557–563. doi: 10.1007/s00431-014-2431-2. [DOI] [PubMed] [Google Scholar]
- 53.Fan J, et al. ACBD2/ECI2-Mediated Peroxisome-Mitochondria Interactions in Leydig Cell Steroid Biosynthesis. Mol Endocrinol. 2016;30:763–782. doi: 10.1210/me.2016-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGuinness MC, et al. Role of ALDP (ABCD1) and mitochondria in X-linked adrenoleukodystrophy. Mol Cell Biol. 2003;23:744–753. doi: 10.1128/MCB.23.2.744-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleinecke S, et al. Peroxisomal dysfunctions cause lysosomal storage and axonal Kv1 channel redistribution in peripheral neuropathy. Elife. 2017;6 doi: 10.7554/eLife.23332. e23332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen Y, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol Biosyst. 2014;10:1742–1748. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- 57.Mattiazzi Ušaj M, et al. Genome-wide localization study of yeast pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J Mol Biol. 2015;427:2072–2087. doi: 10.1016/j.jmb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbaz-Alon Y, et al. The yeast oligopeptide transporter Opt2 is localized to peroxisomes and affects glutathione redox homeostasis. FEMS Yeast Res. 2014;14:1055–1067. doi: 10.1111/1567-1364.12196. [DOI] [PubMed] [Google Scholar]
- 59.Nordgren M, et al. Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives. Front Physiol. 2013;4:145. doi: 10.3389/fphys.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honsho M, et al. Peroxisome homeostasis: Mechanisms of division and selective degradation of peroxisomes in mammals. Biochim Biophys Acta - Mol Cell Res. 2016;1863:984–991. doi: 10.1016/j.bbamcr.2015.09.032. [DOI] [PubMed] [Google Scholar]
- 61.Behrends C, Fulda S. Receptor proteins in selective autophagy. Int J Cell Biol. 2012;2012 doi: 10.1155/2012/673290. 673290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nazarko TY, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Legakis JE, et al. Peroxisome senescence in human fibroblasts. Mol Biol Cell. 2002;13:4243–4255. doi: 10.1091/mbc.E02-06-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasko R, Goligorsky MS. Dysfunctional lysosomal autophagy leads to peroxisomal oxidative burnout and damage during endotoxin-induced stress. 2013 doi: 10.4161/AUTO.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nazarko TY. Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders. Autophagy. 2017;13:991–994. doi: 10.1080/15548627.2017.1291480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law KB, et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017;13:868–884. doi: 10.1080/15548627.2017.1291470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krols M, et al. Mitochondria-associated membranes as hubs for neurodegeneration. Acta Neuropathol. 2016;131:505–523. doi: 10.1007/s00401-015-1528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrera-Cruz MS, Simmen T. Cancer: Untethering Mitochondria from the Endoplasmic Reticulum? Front Oncol. 2017;7:9–13. doi: 10.3389/fonc.2017.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutter GA, Pinton P. Mitochondria-associated endoplasmic reticulum membranes in insulin signaling. Diabetes. 2014;63:3163–3175. doi: 10.2337/db14-0812. [DOI] [PubMed] [Google Scholar]
- 70.Area-Gomez E, Schon EA. Alzheimer Disease. Springer; Singapore: 2017. pp. 149–156. [DOI] [PubMed] [Google Scholar]
- 71.Hattori N, et al. Mitochondrial-Associated Membranes in Parkinson’s Disease. Springer; Singapore: 2017. pp. 157–169. [DOI] [PubMed] [Google Scholar]