Abstract
Numerous studies indicated that microRNAs are critical in the regulation of cellular differentiation, by controlling the expression of underlying genes. The aim of this study was to investigate the effect of miR-210 upregulation on differentiation of human umbilical cord blood (HUCB)-derived mesenchymal stem cells (MSCs) into osteoblasts. MSCs were isolated from HUCB and confirmed by their adipogenic/osteogenic differentiation and flow cytometric analysis of surface markers. Pre-miR-210 was amplified from human DNA, digested and ligated with plenti-III-mir-green fluorescent protein (GFP) vector, and cloned in STBL4 bacteria. After confirmation with polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), the plenti-III-GFP segment bearing pre-miR-210 was transfected into MSCs by electroporation. Two control vectors, pmaxGFP and Scramble, were transfected separately into MSCs. The expression of miR-210 and genes related to osteoblast differentiation, i.e. runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP) and osteocalcin gene, in the three groups of transfected MSCs was analyzed 0, 7, 14, and 21 days of transfection by quantitative reverse transcription PCR (qRT-PCR). Overexpression of miR-210 was observed in MSCs transfected with miR-210-bearing plasmid, and this was significantly different compared to Scramble group (p < 0.05). Significantly increased expression of Runx2 (at day 7 and 14), ALP and osteocalcin genes (at all time points for both genes) was observed in MSCs with miR-210-bearing plasmid compared to controls. Overall, the overexpression of miR-210 in MSCs led to MSC differentiation into osteoblasts, most probably by upregulating the Runx2, ALP, and osteocalcin genes at different stages of cell differentiation. Our study confirms the potential of miRNAs in developing novel therapeutic strategies that could target regulatory mechanisms of cellular differentiation in various disease states.
Keywords: microRNAs, miR-210, MSC, osteoblast, cord blood, cell differentiation
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs, approximately 22 nucleotides in length, that regulate gene expression at the post-transcriptional level, by cleavage (degradation) of target mRNA or translation repression. Different studies indicated that, in addition to the known essential molecules and mechanisms of cellular differentiation, such as cytokines involved in cell signaling and transcription factors and other proteins associated with (epigenetic) regulation of gene expression, microRNAs are also critical in the processes of transition between different cell types, by regulating the expression of underlying genes [1-4]. For example, a study showed that the expression modulation of specific miRNAs (miR-451 upregulation and miR-150 downregulation) promoted erythroid differentiation of cluster of differentiation (CD)133+ hematopoietic stem cells (HSCs), even without the presence of erythroid-specific cytokines [2]. Furthermore, miR-143 could promote differentiation of pre-adipocytes, miR-206 induced differentiation of myoblasts, while miR-1 and miR-133 were involved in the control of skeletal muscle proliferation and differentiation [5].
miR-210 belongs to hypoxamirs, a group of miRNAs which are highly upregulated by hypoxia. The miR-210 gene is located in the intron of a noncoding RNA on chromosome 11p15.5 [4]. Different studies demonstrated that, under hypoxic conditions, miR-210 regulates gene expression in a number of cellular processes [4], such as inhibition of cell proliferation, promotion of stem cell survival, inhibition of mitochondrial metabolism, silencing of DNA repair system, and induction of angiogenesis in endothelial cells. Moreover, upregulation of miR-210 could promote erythroid [6,7], adipogenic, and osteoblastic cell differentiation [4,5,8].
Stem cells are unspecialized cells able to self-renew for long periods and, under specific conditions, to differentiate into one or more specialized cell types [9,10]. There are two main types of stem cells: embryonic stem cells (ESCs) are pluripotent cells found in the inner cell mass of blastocysts, and adult or somatic stem cells are multipotent cells that reside in different tissues/organs of an adult organism. Bone marrow is the source of two adult stem cell types: HSCs that can differentiate into all types of blood cells and mesenchymal (stromal) stem cells (MSCs) responsible for regeneration of non-hematopoietic cells, such as cartilage, bone and fat cells. MSCs are capable of differentiation into several cells (including bone cells) in vitro, but they reside in a latent state in vivo [11]. In addition to bone marrow, other sources of MSCs among adult tissues include peripheral blood, adipose tissue, the lung, heart and fallopian tubes, while MSCs associated with fetus development and neonatal birth can be isolated from fetal liver and lungs, placenta, umbilical cord and cord blood.
Differentiation of bone marrow MSCs into osteoblasts, after the initial bone resorption by osteoclasts, is one of the critical steps in a highly orchestrated process of bone remodeling, and is crucial for maintaining skeletal homeostasis. This process of osteoblast differentiation from MSCs is tightly regulated by different factors, including the regulation of related genes/proteins by miRNAs [12]. Several properties may be used for the isolation and identification of MSCs, including their plastic adherence under standard culture conditions, surface antigen expression (e.g. they express CD90, CD105, CD73, and CD44 but lack the expression of CD45, CD14, CD11b, CD79a, CD19 and human leukocyte antigen – antigen D related [HLA-DR]), and their multilineage potential in vitro [13].
In this study, we investigated the effect of miR-210 upregulation on differentiation of human umbilical cord blood (HUCB)-derived MSCs into osteoblasts, to explore the potential use of this miRNA in the treatment of diseases such as hematopoietic malignancies and osteoporosis.
MATERIALS AND METHODS
Isolation, culture, and confirmation of MSCs
The umbilical cord blood of two full-term newborns, not intended for therapeutic purposes, was collected in cord blood bags at the Iranian Blood Transfusion Organization (IBTO) in Tehran, after the approval from the local ethics committee (Number: IR.MUMS.REC.1392.704). First, primary mononuclear cells (MNCs) were isolated from the cord blood using Ficoll-Paque PLUS (Amersham Pharmacia, Piscataway, NJ, USA), and then MSCs were isolated from MNC fraction based on their ability of adhesion to culture flasks. MSCs were cultured at 37 °C under humid conditions with 95% O2 and 5% CO2, in low glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin.
To confirm the presence of MSCs, upon reaching a confluency of 80%, 6×104 cells were transferred to 24-well tissue culture plates and adipogenic or osteogenic differentiation was induced. For fat cell differentiation, MSCs were cultured in a medium containing 100 nM dexamethasone, 50 µm indomethacin and 0.5 mM 3-isobutyl-1-methylxanthine (IBMX) for 14 days, and then were stained with Oil Red O (Sigma, USA). For differentiation of bone cells, MSCs were cultured in a medium containing 50 µM ascorbic acid, 10 mM beta-glycerol-3-phosphate and 100 nM dexamethasone for 21 days, and stained with Alizarin Red (Sigma, USA).
Flow cytometry was performed on primary passages to evaluate the surface expression of CD90, CD73, CD45 and CD103 markers, and data were analyzed with FlowJo software (https://www.flowjo.com/). The antibodies were purchased from Santa Cruz Biotechnology (USA). For this purpose, 105 cells were counted, washed with phosphate-buffered saline (PBS) and suspended in 100 µl bovine serum albumin (BSA, 3%). Then 10 µl of unlabeled primary antibodies were added and the mixture was incubated for 30–45 minutes at 4 °C. Next, cells were washed with 1 ml cold PBS and suspended in 100 µl BSA (3%), 2 µl of secondary fluorescein isothiocyante (FITC)-labeled antibodies was added and the mixture was incubated 30–45 minutes at 4 °C in the dark. Finally, 300 µl of cold PBS was added and the cells were analyzed by flow cytometer.
Pre-miR-210 cloning
Sequence data for human precursor miR-210 (pre-miR-210) was retrieved from miRBase Sequence database (http://www.mirbase.org/) and the corresponding genomic location was analyzed at Ensembl site (https://www.ensembl.org/index.html). After identifying the coding region for pre-miR-210, the primers were designed using Gene Runner software (Version 5.0.9 1 beta) and blasted against the National Center for Biotechnology Information (NCBI) database. DNA was extracted from human blood using a VioGene kit (Taiwan). Pre-miR-210 was amplified using PCR (Ampliqon, Denmark). STBL4 bacteria containing plenti-III-mir-green fluorescent protein (GFP) vector (ABM, Canada) were cultured in lysogeny broth (LB) medium and plasmid extraction was performed using a plasmid extraction kit (Viogene, Taiwan). The plasmid DNA and pre-miR-210 PCR products were digested with XhoI and EcoRI restriction enzymes (Fermentas, USA). After digestion, the desired segment (pre-miR-210) was ligated to plasmid DNA using T4 DNA ligase (Fermentas, USA).
The STBL4 recombinant bacteria were transformed with the plenti-III segment bearing pre-miR-210 and cultured overnight in LB agar medium containing kanamycin (1000 µg/ml). After 24 hours, single colonies were randomly selected and cultured in LB agar medium. The plasmid DNA was extracted and single clones with plenti-III-GFP bearing pre-miR-210 were confirmed by digestion of the PCR products with XhoI and EcoRI restriction enzymes (polymerase chain reaction-restriction fragment length polymorphism [PCR-RFLP]).
Transfection
One day prior to transfection, MSCs from the 4th passage were grown to a confluency of 85% and 1.5×106 cells were counted. The plasmid bearing pre-miR-210 (vector with the target gene) and two control plasmids, one containing pmaxGFP (an empty vector) and the other containing Scramble (vector with a sequence that does not belong to any known miRNA), were transfected separately into MSCs using a Human MSC Nucleofector kit (Lonza, USA) and U23 program.
GFP expression assay
The medium was changed after 24 hours of transfection. Following 48 hours of transfection, the expression of GFP was detected in the cells under a fluorescent microscope, indicating the percentage of transfected cells. Flow cytometry was also performed to determine the contamination level of MSCs.
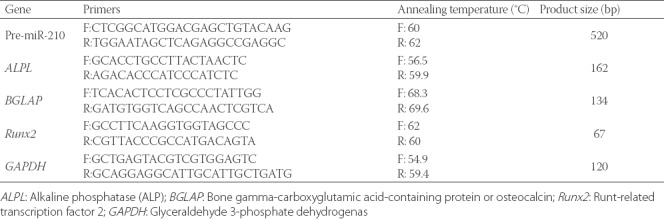
To determine the expression level of miR-210, runt-related transcription factor 2 (Runx2, RUNX2 gene), alkaline phosphatase (ALP, ALPL gene) and osteocalcin or bone gamma-carboxyglutamic acid-containing protein (BGLAP, BGLAP gene) in transfected MSCs, RNA was isolated 0, 7, 14, and 21 days of transfection from MSC cells at a confluency of 80% and analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR, ABI 7500 QRT-PCR System Perkin-Elmer, Applied Biosystems, USA). Total RNA was extracted from transfected cells, using bioZOL™-G RNA Isolation Reagent. Briefly, 500 µl biozol was added to the cells, followed by phenol/chloroform extraction according to the manufacturer’s instructions. A miRNA isolation kit (Stratagene, USA) was used for purification of miRNAs. The quality of RNA samples was determined spectrophotometrically at 260/280nm and using gel electrophoresis. Complementary DNA (cDNA) was synthesized from 6.5 µl of total RNA using a Takara kit (Japan). Due to the short length, miRNAs were first elongated in a polyadenylation reaction using a 1st-strand cDNA synthesis kit (Stratagene, USA) and then reverse transcribed into QPCR-ready cDNA (initial denaturation for one cycle at 95 °C for 10 minutes, denaturation, amplification and quantification for 40 cycles at 95 °C for 15 seconds, 60 °C for 15 seconds and 72 °C for 20 seconds, respectively). The difference in gene expression was calculated using the 2-ΔΔCT method (these steps were repeated for pmaxGFP and Scramble as controls and pre-miR-210-bearing plasmid as target). The U6 small nuclear (snRNA) gene (GGGCAGGAAGAGGGCCTAT) and human GAPDH were used as the reference genes to normalize the expression levels of target genes. The respective primers and their specifications are presented in Table 1. The expression of miR-210, Runx2, ALP and osteocalcin genes in MSC cells transfected with pre-miR-210 was compared with the control groups (pmaxGFP and Scramble) on days 0, 7, 14 and 21 of transfection.
TABLE 1.
Primers used for quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Statistical analysis
Data were analyzed in SPSS for Windows, Version 11.5. (SPSS Inc., Chicago) first using descriptive statistics and then by comparative analysis. We compared the mean of continuous variables within groups using paired-sample t-test and between groups using one-way analysis of variance (ANOVA). A value of p ≤ 0.05 was considered significant.
RESULTS
Isolation, culture, and differentiation of MSCs
The human MSCs isolated from umbilical cord blood were observed in the form of spindle-shaped cells under the microscope. MSCs were confirmed by their adherence to plastic surface, flattened, spindle shaped morphology, and the ability to differentiate into fat and bone cells (Figure 1). In mature fat cells, intracellular lipid vesicles were stained bright red with Oil-Red-O, and in bone cells extracellular calcium deposition was stained bright red-orange with Alizarin-Red-S (Figure 1). Flow cytometric analysis showed that the cells were negative for CD45 but positive for CD90, CD103, and CD73 markers (Figure 2).
FIGURE 1.
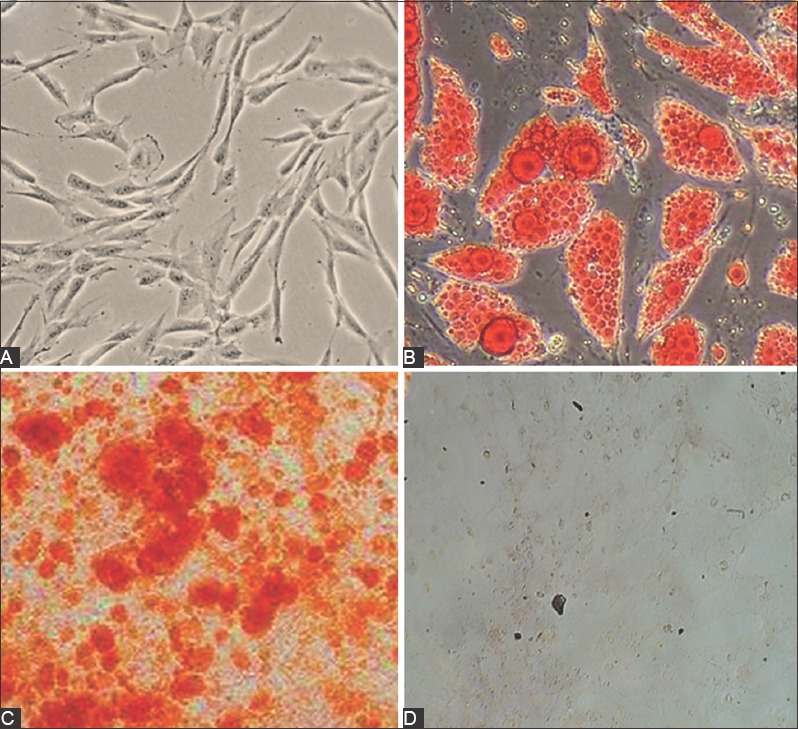
Confirmation of mesenchymal stem cells (MSCs) isolated from human umbilical cord blood by morphological analysis and their ability to differentiate into fat cells (Oil-Red-O) and bone cells (Alizarin-Red-S). (A) MSCs at passage 3 were observed in the form of spindle-shaped cells under the microscope. (B) In adipocytes, intracellular lipid vesicles were stained bright red with Oil-Red-O, and (C) in osteocytes extracellular calcium deposition was stained bright red-orange with Alizarin-Red-S. (D) Control group: MSCs without staining.
FIGURE 2.
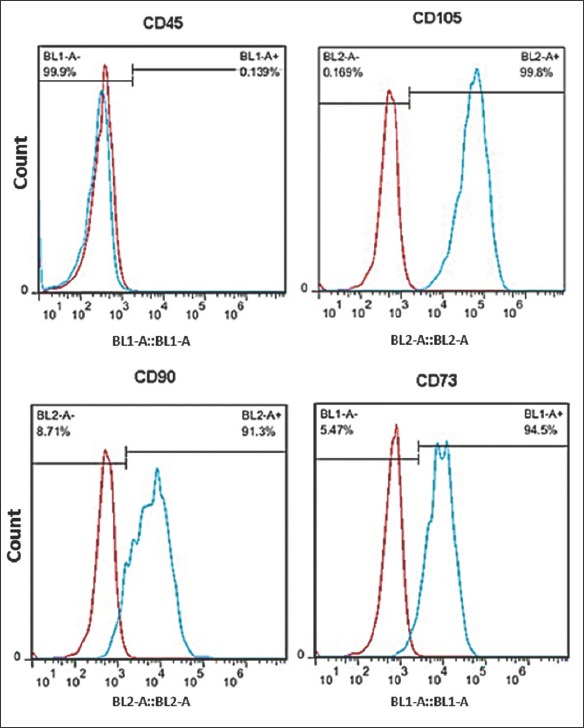
Confirmation of mesenchymal stem cells (MSCs) isolated from human umbilical cord blood by flow cytometry. Isolated MSCs were positive for cluster of differentiation (CD)73, CD105 and CD90 but negative for CD45 (a marker of hematopoietic cells).
Cloning and transfection
Pre-miR-210 was successfully amplified from human DNA and a PCR product of 520 bp was generated (Figure 3). Transformation of STBL4 recombinant bacterium with plenti-III-GFP plasmid bearing pre-miR-210 was confirmed by PCR-RFLP analysis of individual bacterial clones. MSCs were transfected with plenti-III-GFP plasmid bearing pre-miR-210 by electroporation. Approximately 63% of transfected MSCs emitted green fluorescence under the inverted fluorescence microscope, 48 hours after the transfection. Flow cytometric analysis also confirmed successful transfection of MSCs (Figure 4).
FIGURE 3.
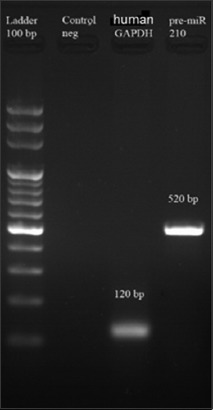
Verification of amplification of pre-miR-210 from human DNA by gel electrophoresis (agarose gel, 2%). The figure shows that a 520-bp PCR product was generated for pre-miR-210 versus 120-bp PCR product for human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene and 100 pb DNA ladder.
FIGURE 4.
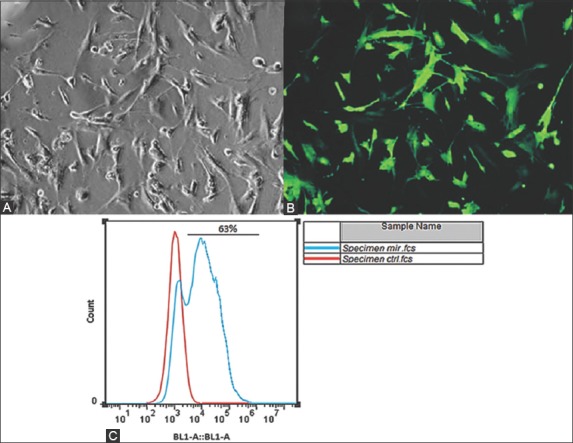
Mesenchymal stem cells (MSCs) were successfully transfected with plenti-III-mir-green fluorescent protein (GFP) plasmid bearing pre-miR-210 by electroporation, (A) as observed under optical microscope. (B) Approximately 63% of transfected MSCs emitted green fluorescence under the inverted fluorescence microscope, 48 hours after the transfection. (C) Flow cytometric analysis also confirmed successful transfection of MSCs, i.e., infection rate of 63% was observed.
miR-210 upregulation
The expression of miR-210 in MSCs transfected with miR-210 and Scramble was analyzed by qRT-PCR on day 7, 14 and 21 following transfection, and a statistically significant difference between the two groups was observed as follows: in MSCs with miR-210-bearing plasmid there was 4.8-fold overexpression of miR-210 on day 7, 2.9-fold on day 14, and 2.2-fold on day 21; in Scramble group, there was 0.9-fold overexpression of miR-210 on day 7, 1.5-fold on day 14, and 1.5-fold on day 21 (p < 0.05). The expression of miR-210 in the subsequent period (until day 21), gradually decreased in both groups and this was expected because electroporation is a transient method for transfection (Figure 5).
FIGURE 5.
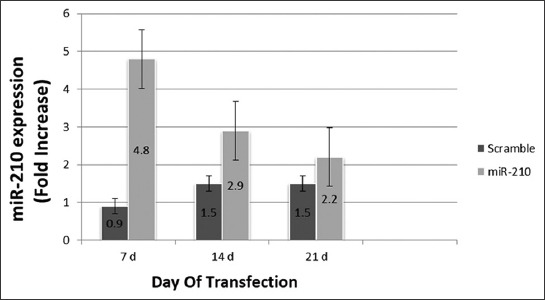
Overexpression of miR-210 in mesenchymal stem cells (MSCs) on different days of transfection. Fold increase ± standard error of the mean (SEM) of miR-210 expression was compared between MSCs transfected with plenti-III-mir-green fluorescent protein (GFP) plasmid bearing pre-miR-210 and MSCs transfected with Scramble on days 7, 14 and 21. The highest expression level of miR-210 was detected on day 7. The difference in gene expression was calculated using the 2-ΔΔCT method (p < 0.05). Expression of miR-210 in MSCs transfected with miR-210-bearing plasmid was significantly different compared to Scramble group (p < 0.05).
Expression of osteocalcin, ALP, and Runx2 genes
The expression of Runx2, ALP and osteocalcin genes in MSCs transfected with miR-210, pmaxGFP and Scramble was analyzed by qRT-PCR on days 0, 7, 14 and 21 following transfection.
The expression of Runx2 gene in cells transfected with miR-210-bearing plasmid, Scramble and pmaxGFP was increased 1.2, 1.3 and 0.9 fold, respectively on day 0 (pre-transfection); 14.6, 0.9 and 1 fold, respectively on day 7; 5.4, 0.9 and 0.9 fold, respectively on day 14; and 1.9, 0.8 and 1 fold, respectively on day 21 (Figure 6). The difference in Runx2 gene expression was statistically significant between miR-210-bearing plasmid and two control groups on day 7 (p < 0.01) and 14 (p < 0.027), but not on day 21.
FIGURE 6.
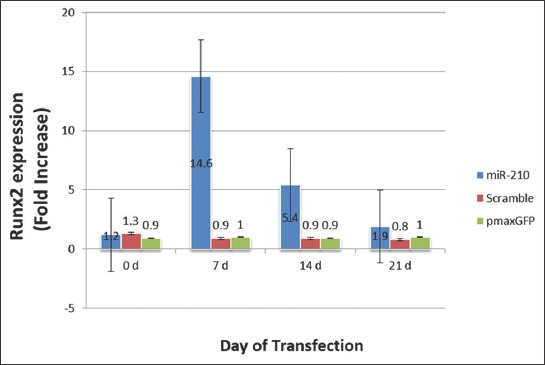
The expression level of runt-related transcription factor 2 (Runx2) in mesenchymal stem cells (MSCs) on different days of transfection. Fold increase ± standard error of the mean (SEM) of Runx2 expression was compared between MSCs transfected with plenti-III-mir-green fluorescent protein (GFP) plasmid bearing pre-miR-210, MSCs transfected with Scramble and MSCs transfected with pmaxGFP vector on days 0, 7, 14 and 21. The highest expression of Runx2 was detected on day 7. The difference in gene expression was calculated using the 2-ΔΔCT method (p < 0.05). The difference in Runx2 gene expression was statistically significant between miR-210-bearing plasmid and two control groups on day 7 (p < 0.01) and 14 (p < 0.027), but not on day 21.
The expression of ALP gene in cells transfected with miR-210-bearing plasmid, Scramble and pmaxGFP was increased 0.9, 1.1 and 1.2 fold, respectively on day 0 (pre-transfection); 5.5, 1.4 and 1.2 fold, respectively on day 7 (p < 0.026); 7, 1.3 and 1.8 fold, respectively on day 14 (p < 0.02); and 8.5, 1.1 and 1.8 fold, respectively on day 21 [p < 0.017] (Figure 7). The difference in ALP gene expression was statistically significant between miR-210-bearing plasmid and two control groups at all time points.
FIGURE 7.
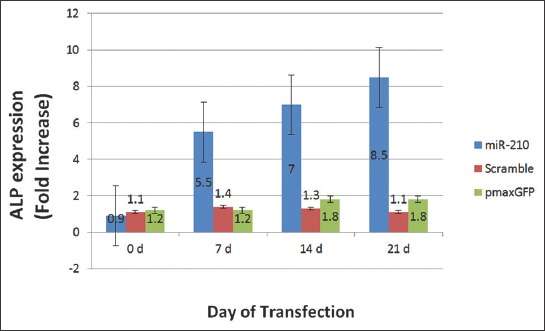
The expression level of alkaline phosphatase (ALP, ALPL) gene in mesenchymal stem cells (MSCs) on different days of transfection. Fold increase ± standard error of the mean (SEM) of ALP expression was compared between MSCs transfected with plenti-III-mir-green fluorescent protein (GFP) plasmid bearing pre-miR-210, MSCs transfected with Scramble and MSCs transfected with pmaxGFP vector on days 0, 7, 14 and 21. The highest ALP gene expression was detected on day 21. The difference in gene expression was calculated using the 2-ΔΔCT method (p < 0.05). The difference in ALP gene expression was statistically significant between miR-210-bearing plasmid and two control groups at all time points (p < 0.026 at day 7, p < 0.02 at day 14, p < 0.017 at day 21).
The expression of osteocalcin gene in cells transfected with miR-210-bearing plasmid, Scramble and pmaxGFP was increased 0.3, 0.9 and 1 fold, respectively on day 0 (pre-transfection); 8.6, 1.1 and 1.3 fold, respectively on day 7 (p < 0.017); 9.4, 1.2 and 0.5 fold, respectively on day 14 (p < 0.015); and 7.6, 1.9 and 2.4 fold, respectively on day 21 [p < 0.019] (Figure 8). The difference in osteocalcin gene expression was statistically significant between between miR-210-bearing plasmid and two control groups at all time points.
FIGURE 8.
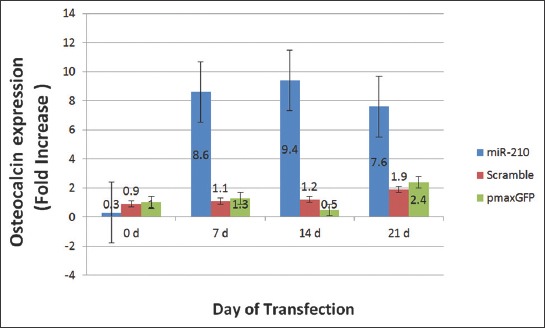
The expression level of osteocalcin or bone gamma-carboxyglutamic acid-containing protein (BGLAP) gene in mesenchymal stem cells (MSCs) on different days of transfection. Fold increase ± standard error of the mean (SEM) of osteocalcin gene expression was compared between MSCs transfected with plenti-III-mir-green fluorescent protein (GFP) plasmid bearing pre-miR-210, MSCs transfected with Scramble and MSCs transfected with pmaxGFP vector on days 0, 7, 14 and 21. The highest expression of osteocalcin gene was detected on day 14. The difference in gene expression was calculated using the 2-ΔΔCT method (p < 0.05). The difference in osteocalcin gene expression was statistically significant between miR-210-bearing plasmid and two control groups at all time points (p < 0.017 at day 7, p < 0.015 at day 14, p < 0.019 at day 21).
DISCUSSION
miR-210 belongs to a group of miRNAs that are highly upregulated under hypoxic conditions, in different cellular processes. Moreover, the upregulation of miR-210 promotes differentiation of blood, fat and bone cells, although the regulation of these processes is still not completely clear. In this study, we investigated the effect of miR-210 upregulation on MSC differentiation into osteoblasts, using gene transfection in vitro.
Our results demonstrated that the overexpression of miR-210 in MSCs led to MSC differentiation into osteoblasts through the upregulation of several osteoblastic genes. The expression of RUNX2 gene was the highest on day 7 and it gradually decreased in the subsequent period (until day 21), indicating that Runx2 is an early marker of osteoblastic differentiation. On the other hand, the expression of ALP gene gradually increased during the 21-day period showing the highest expression on 21 day, suggesting its role as a late marker of osteoblast differentiation. Finally, the expression of osteocalcin gene (BGLAP) was the highest on the 14th day implying its role in the intermediate stages of differentiation. Mizuno et al. [5] demonstrated the positive role of miR-210 in bone morphogenetic protein 4 (BMP-4)-induced differentiation of murine bone marrow-derived ST2 MSC cells into osteoblasts. They showed that miR-210 promotes osteoblastic differentiation by targeting and suppressing the activin A receptor type 1B (AcvR1b) gene and consequently inhibiting the transforming growth factor beta (TGF-β)/activin signaling pathway. Namely AcvR1b, together with the activin type 2 receptor, transmits signals from activin. The activated AcvR1b binds and phosphorylates Smad2 and Smad3 proteins which results in transcription of genes that have an inhibitory effect on cell proliferation [5]. Consistently, our results showed that the increase in Runx2 expression over the 21-day period following transfection led to osteoblastic differentiation. Qiu et al. reported that polyphenol(-)-epigallocatechin-3-gallate (EGCG) had a protective effect on hypoxia-induced apoptosis and osteogenic differentiation in human bone marrow-derived MSCs, via upregulating miR-210 and downregulating the expression of miR-210-targeted receptor tyrosine kinase ligand ephrin-A3 [14]. Furthermore, the study of Liu et al. suggested that miR-210 plays an important role in the regulation of postmenopausal osteoporosis due to estrogen deficiency, by promoting the vascular endothelial growth factor (VEGF) expression and osteoblast differentiation [15].
Comparably, a Runx2/miR-3960/miR-2861 regulatory feedback loop was reported to mediate the differentiation of mouse ST2 stromal cells; specifically, it was suggested that the upregulation of Runx2 induces miR-3960/miR-2861 transcription and that, in turn, the level of Runx2 mRNA is maintained by miR-3960 and miR-2861 via repressing Homeobox A2 and histone deacetylase 5, and thus stabilizing the osteoblast differentiation [16]. This regulatory model of osteoblastic differentiation is largely in agreement with our results, as we indicated the association between high expression of Runx2 at the early stages of osteoblastic differentiation and miR-210 upregulation.
Gong et al. showed that 10 miRNAs, including miR-27a, miR-125a-5p and miR-466f-3p, were downregulated and 18 miRNAs, including miR-17, miR-20a and miR-210, were upregulated during osteogenic differentiation of Satb2-induced murine bone marrow stromal cells [17]. This result is in line with our study as we also demonstrated the upregulation of miR-210 during osteoblastic differentiation. In another study [18] miR-181a promoted osteoblastic differentiation of BMP2-stimulated mouse MC3T3 preosteoblasts by targeting negative regulators Tgf-beta induced and TGF-β type I receptor and thus inhibiting the TGF-β signaling. The study also showed that regulator of G protein signaling 4 (RGS4) and GATA binding protein 6 (GATA6) were direct targets of miR-181a, i.e. decreased expression of RGS4 and GATA6 during osteoblastic differentiation of mouse MC3T3 cells was associated with increased levels of miR-181a and the subsequent increase in osteoblastic differentiation [18]. The studies of Bhushan et al. [18] and Mizuno et al. [5] emphasize the important role of TGF-β signaling and its downregulation by miRNAs in osteoblastic differentiation.
CONCLUSION
Taken together, the upregulation of miR-210 led to differentiation of human umbilical cord blood-derived MSCs into osteoblasts. The regulation of osteogenic differentiation by specific miRNAs may be a promising strategy to improve the healing process of a fractured bone. However, at this stage, additional studies are required to better understand the regulatory mechanisms of osteogenic differentiation and the most efficient use of these mechanisms in tissue engineering and as therapeutic targets in bone regeneration process.
ACKNOWLEDGMENTS
This study was the result of MSc student thesis (Number: 920773) supported financially by the vice president of Research at Mashhad University of Medical Sciences and Allied Medical Sciences of Tehran University of Medical Sciences (grant number: 920773 31-05-92).
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
REFERENCES
- [1].Khori V, Shalamzari SA, Isanejad A, Alizadeh AM, Alizadeh S, Khodayari S, et al. Effects of exercise training together with tamoxifen in reducing mammary tumor burden in mice: Possible underlying pathway of miR-21. Eur J Phamacol. 2015;765:179–87. doi: 10.1016/j.ejphar.2015.08.031. https://doi.org/10.1016/j.ejphar.2015.08.031. [DOI] [PubMed] [Google Scholar]
- [2].Kouhkan F, Hafizi M, Mobarra N, Mossahebi-Mohammadi M, Mohammadi S, Behmanesh M, et al. miRNAs: A new method for erythroid differentiation of hematopoietic stem cells without the presence of growth factors. Appl Biochem Biotechnol. 2014;172(4):2055–69. doi: 10.1007/s12010-013-0633-0. https://doi.org/10.1007/s12010-013-0633-0. [DOI] [PubMed] [Google Scholar]
- [3].Minayi N, Alizadeh S, Dargahi H, Soleimani M, Khatib ZK, Tayebi B, et al. The effect of miR-210 up-regulation on proliferation and survival of mouse bone marrow derived mesenchymal stem cell. Int J Hematol Oncol Stem Cell Res. 2014;8(1):15–23. [PMC free article] [PubMed] [Google Scholar]
- [4].Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: The master hypoxamir. Microcirculation. 2012;19(3):215–23. doi: 10.1111/j.1549-8719.2011.00154.x. https://doi.org/10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583(13):2263–8. doi: 10.1016/j.febslet.2009.06.006. https://doi.org/10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- [6].Kosaka N, Sugiura K, Yamamoto Y, Yoshioka Y, Miyazaki H, Komatsu N, et al. Identification of erythropoietin-induced microRNAs in haematopoietic cells during erythroid differentiation. Br J Hematol. 2008;142(2):293–300. doi: 10.1111/j.1365-2141.2008.07151.x. https://doi.org/10.1111/j.1365-2141.2008.07151.x. [DOI] [PubMed] [Google Scholar]
- [7].Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep. 2009;42(8):493–9. doi: 10.5483/bmbrep.2009.42.8.493. https://doi.org/10.5483/BMBRep.2009.42.8.493. [DOI] [PubMed] [Google Scholar]
- [8].Huang X, Le QT, Giaccia AJ. MiR-210 - Micromanager of the hypoxia pathway. Trends Mol Med. 2010;16(5):230–7. doi: 10.1016/j.molmed.2010.03.004. https://doi.org/10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Anderson DJ, Gage FH, Weissman IL. Can stem cells cross lineage boundaries? Nat Med. 2001;7(4):393–5. doi: 10.1038/86439. https://doi.org/10.1038/86439. [DOI] [PubMed] [Google Scholar]
- [10].Mohammadi Z, Afshari JT, Keramati MR, Alamdari DH, Ganjibakhsh M, Zarmehri AM, et al. Differentiation of adipocytes and osteocytes from human adipose and placental mesenchymal stem cells. Iran J Basic Med Sci. 2015;18(3):259–66. [PMC free article] [PubMed] [Google Scholar]
- [11].Bruder SP, Jaiswal N, Haynesworth SE. Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation. J Cell Biochem. 1997;64(2):278–94. doi: 10.1002/(sici)1097-4644(199702)64:2<278::aid-jcb11>3.0.co;2-f. https://doi.org/10.1002/(SICI)1097-4644(199702)64:2<278:AID-JC B11>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [12].Vimalraj S, Selvamurugan N. MicroRNAs: Synthesis, gene regulation and osteoblastdifferentiation. Curr Issues Mol Biol. 2013;15(1):7–18. [PubMed] [Google Scholar]
- [13].Williams AR, Hare JM. Mesenchymal stem cells: Biology, pathophysiology, translational findings, and therapeutic implications for cardiac disease. Circulation research. 2011;109(8):923–40. doi: 10.1161/CIRCRESAHA.111.243147. http://doi.org/10.1161/CIRCRESAHA.111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qiu Y, Chen Y, Zeng T, Guo W, Zhou W, Yang X. EGCG ameliorates the hypoxia-induced apoptosis and osteogenic differentiation reduction of mesenchymal stem cells via upregulating miR-210. Mol Biol Rep. 2016;43(3):183–93. doi: 10.1007/s11033-015-3936-0. https://doi.org/10.1007/s11033-015-3936-0. [DOI] [PubMed] [Google Scholar]
- [15].Liu XD, Cai F, Liu L, Zhang Y, Yang AL. microRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. Biol Chem. 2015;396(4):339–47. doi: 10.1515/hsz-2014-0268. https://doi.org/10.1515/hsz-2014-0268. [DOI] [PubMed] [Google Scholar]
- [16].Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY, et al. A Runx2/miR-3960 /miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011;286(14):12328–39. doi: 10.1074/jbc.M110.176099. https://doi.org/10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, et al. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem. 2014;387(1-2):227–39. doi: 10.1007/s11010-013-1888-z. https://doi.org/10.1007/s11010-013-1888-z. [DOI] [PubMed] [Google Scholar]
- [18].Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott CE, Knaus P. miR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int J Biochem Cell Biol. 2013;45(3):696–705. doi: 10.1016/j.biocel.2012.12.008. https://doi.org/10.1016/j.biocel.2012.12.008. [DOI] [PubMed] [Google Scholar]


