Abstract
The discovery of induced pluripotent stem cells (iPSCs) has solidified the concept of transcription factors as major players in controlling cell identity and provided a tractable tool to study how somatic cell identity can be dismantled and pluripotency established. A number of landmark studies have established hallmarks and roadmaps of iPSC formation by describing relative kinetics of transcriptional, protein and epigenetic changes, including alterations in DNA methylation and histone modifications. Recently, technological advancements such as single-cell analyses, high-resolution genome-wide chromatin assays and more efficient reprogramming systems have been used to challenge and refine our understanding of the reprogramming process. Here, we will outline novel insights into the molecular mechanisms underlying iPSC formation, focusing on how the core reprogramming factors OCT4, KLF4, SOX2 and MYC (OKSM) drive changes in gene expression, chromatin state and 3D genome topology. In addition, we will discuss unexpected consequences of reprogramming factor expression in in vitro and in vivo systems that may point towards new applications of iPSC technology.
Keywords: iPS cells, reprogramming, OKSM, pluripotency, chromatin organization, epigenetic remodeling, transcriptional activation, transcriptional silencing
Introduction
An identical set of transcription factors (TFs) – the now famous “Yamanaka factors” OCT4, KLF4, SOX2 and MYC (OKSM) – can convert mouse and human fibroblasts into embryonic stem cell (ESC)-like induced pluripotent stem cells (iPSCs) with species-specific properties [1,2]. This breakthrough discovery suggested the existence of shared molecular mechanisms that can establish pluripotency in somatic cells. Indeed, the universal applicability of OKSM to induce pluripotency has been demonstrated by successful reprogramming of fibroblastic cells from multiple species and of diverse adult cell types [3]. iPSC derivation has also been achieved by alternative TF combinations, containing some or none of the original Yamanaka factors [4,5] and, strikingly, by prolonged exposure to a mixture of chemical compounds that alter the activity of signaling pathways and chromatin modifying enzymes [6]. Nevertheless, OKSM reprogramming is currently more efficient and applicable to a wider range of cell types than alternative approaches to induce pluripotency and is consequently most widely used to derive patient-specific cell lines and study the mechanisms of iPSC formation.
A number of recent technological advances have facilitated monitoring iPSC formation driven by OKSM. For example, novel panels of surface antigens have been identified that dynamically change early or late during mouse [7–9] and human [10,11] cell reprogramming. This has allowed for the isolation and subsequent molecular dissection of better defined intermediate cell populations. In parallel, different strategies that reduce the inherent stochasticity of OKSM-mediated reprogramming have been described, including expression of additional TFs [12], interference with repressive chromatin-modifying complexes in specific media environments [13] or combined modulation of cellular signaling pathways [14–16], thereby enabling high-resolution, genome-wide approaches for the dissection of reprogramming mechanisms. In the following, we will describe recent advances in our understanding of general reprogramming trajectories, before discussing the molecular roles of OKSM factors in inducing pluripotency.
Reprogramming trajectories and alternative consequences of OKSM expression
Early work in mouse fibroblasts suggested that iPSC formation proceeds via an ordered sequence of events, each of which represents a bottleneck that aspiring iPSCs have to pass [17,18]. This provided the empirical basis for the current understanding of reprogramming as a – with respect to its outcome at the single cell level – stochastic process [19] that can be facilitated experimentally [20]. In addition to refining the “classic” reprogramming hallmarks of early down-regulation of somatic genes, epithelialization and ultimate reactivation of pluripotency-associated genes, research in recent years has also identified surprising intermediate stages of iPSC formation. For example, before embarking on the essential mesenchymal-to-epithelial transition (MET) [21,22], fibroblasts undergoing reprogramming transiently become more mesenchymal [23]. Accordingly, transcription factors promoting an epithelial-to-mesenchymal transition (EMT) such as SNAI1 are paradoxically enhancing iPSC formation [24,25]. The timing of EMT during fibroblast reprogramming correlates with transient metabolic changes, including hyperactivation of oxidative phosphorylation [26,27] and increase in reactive oxygen species signaling [28], that occur shortly after OKSM induction. This highlights the need for a better understanding of the links between changes in metabolism and other aspects of reprogramming [29].
It is becoming increasing clear that reprogramming trajectories can be influenced by species as well as, to a surprisingly strong degree, by the somatic cell of origin. For example, human fibroblasts seem to undergo MET not after but coinciding with reactivation of pluripotency loci such as LIN28A, NANOG and TET1 [10]. In the mouse, reactivation of endogenous loci encoding core pluripotency regulators is a comparatively late event during iPSC derivation from fibroblasts and indicative of successful reprogramming [8,18]. However, blood cells reactivate Pou5F1 (the locus encoding OCT4) earlier than the low-stringency marker SSEA1 and before becoming independent of transgenic OKSM expression [30], while neural stem cells and astrocytes activate Nanog before expressing E-CADHERIN and undergoing MET [31]. Interestingly, astrocytes but not fibroblasts expressing OKSM transiently upregulate the neural progenitor marker Sox1, which functionally appears to be redundant with exogenous Sox2 expression [32]. These observations illustrate the complex interactions between OKSM and cell intrinsic factors and point towards using reprogramming technology to probe the molecular basis of cell-type specific pathogenesis, with possible implications for human disease [33]. For example, the possibility to greatly facilitate iPSC derivation from mouse blood progenitor cells by Wnt activation [14,15] is reminiscent of the preeminent role of this pathway in blood cancers and might suggest shared molecular mechanisms between blood cell reprogramming and leukemic transformation.
Aside from successfully inducing pluripotency, expression of Yamanaka factors can result in apoptosis triggered by MYC overexpression [34], senescence as a consequence of activation of the p53 and INK4/ARF pathways [35,36] (Figure 1) or a partial reprogrammed state due to unresolved chromatin barriers such as H3K9 [37] or DNA methylation [38] domains. In addition, several recent studies suggest the activation of alternative developmental programs during or in parallel to iPSC formation. Thus, both mouse [30] and human [10,39] fibroblasts undergoing reprogramming transiently express regulators of later developmental stages including the primitive streak. While the functional relevance of this observation is debated [40], recent observations in a highly efficient mouse reprogramming system suggest that transient upregulation of transcription factors associated with the post-implantation epiblast coincides with acquisition of functional pluripotency as measured by the ability to give rise to entirely iPSC-derived animals [41]. The observation that intermediates of iPSC derivation express pluripotency regulators yet are prone to differentiation might reconcile studies claiming OKSM factors can trigger direct lineage conversions [42–44] with subsequent work demonstrating that such conversions – at least in mouse – entail transient acquisition of pluripotency features such as Nanog expression and X chromosome reactivation [45,46]. As transient reactivation of a primitive streak program does not appear to occur during granulocyte and keratinocyte reprogramming [30], it would be interesting to determine if this affects molecular or functional properties of resultant iPSCs. It should also be noted that iPSC reprogramming, even when initiated from the same somatic cell of origin, is significantly impacted by experimental conditions and may not always proceed via identical molecular checkpoints. This is demonstrated by the role of TET1, which turns from a facilitator to a repressor of iPSC formation in presence of ascorbic acid [47] and the observation that two different KLF4 variants in a context-dependent manner can affect factor stoichiometry and consequently OKSM-driven MET [48,49].
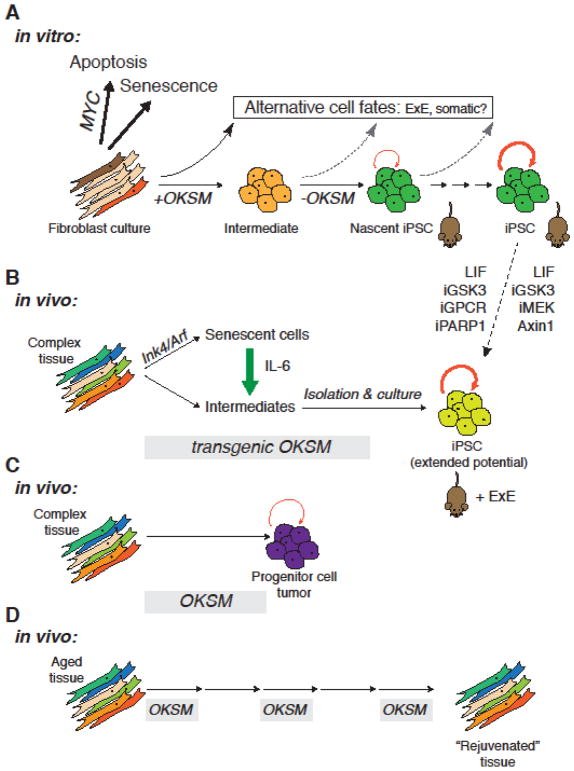
Figure 1. Developmental fates triggered by reprogramming factors.
A) In cultured fibroblasts, OKSM leads to initiation of senescence or controlled cell death in the majority of cells. A small subset of cells gives rise to unstable reprogramming intermediates that still require exogenous reprogramming factors before giving rise to nascent iPSCs upon factor withdrawal and to established iPSCs upon passaging. Fibroblasts expressing OKSM can also give rise to alternative cell fates, which was most convincingly shown for extraembryonic endoderm, but evidence suggest that intermediates and nascent iPSCs might also be more prone to differentiate into other cell lineages than established iPSCs. Nascent murine iPSCs have less pronounced self-renewal capacity than established iPSCs, but both can efficiently give rise to all somatic cell types upon blastocyst injection. Established iPSC can be endowed with the ability to give rise to extraembryonic (ExE) tissues by culture in two alternative approaches targeting indicated molecules. B) Prolonged OKSM expression in adult transgenic mice yields intermediates as well as senescent cells that support reprogramming by secretion of factors such as IL-6. Culture of circulation-derived intermediates and ex vivo culture yields iPSCs that not only can give rise to extraembryonic and embryonic tissues. C) An intermediate interval of OKSM expression in vivo triggers partial epigenetic remodeling and in tissue such as kidney the emergence of transplantable tumors with molecular features of tissue-specific embryonic progenitor cells. D) Repeated short intervals of OKSM expression in vivo lead to reversal of molecular features of aging and functional restoration of tissue function in aged animals.
OKSM expression in fibroblasts has also been shown to yield extraembryonic endoderm stem (XEN) cells in parallel with iPSCs [50]. This observation is in accordance with high-resolution analysis of fibroblast reprogramming by Mass Cytometry, which revealed appearance of a PDGFRA+NANOG- population late in reprogramming [51]. On the other hand, single cell RNA-sequencing analysis suggest that cells with placenta-like or neuronal-like signatures represent an alternative reprogramming outcome (Schiebinger et al., bioRxiv, doi:10.1101/191056) (Figure 1A). It remains to be determined, for example by analysis of clonal reprogramming cultures, whether these observations reflect increased developmental plasticity due to reprogramming factor expression or are due to the intrinsic heterogeneity of starting fibroblast cultures.
Of note, in vivo expression of OKSM in context of some transgenic mouse models [52] but not others [53,54] has been reported to yield iPSCs with the capacity to give rise to trophectodermal cells. The specific aspect of in vivo reprogramming responsible for endowing iPSCs with these developmental features remains to determined, but it is worth noting that recently described in vitro culture conditions could overcome the inability of mouse and human pluripotent stem cells to differentiate into extraembryonic lineages [55–57] (Figure 1). Intriguingly, in vivo reprogramming is in part driven by secretion of the cytokine IL-6 as part of an OKSM-induced senescence program in a subset of cells [58]. This suggests a dynamic interplay between cells on two different trajectories following OKSM induction (initiation of senescence vs initiation of reprogramming) (Figure 1), which also appears to be relevant for in vitro reprogramming [58,59].
Reprogramming factor induction in vivo – in part depending on the length of expression intervals – can also have developmental outcomes distinct from pluripotency and subsequent teratoma formation. These include dysplasia and generation of partially reprogrammed, transplantable progenitor cell tumors [54] and tissue rejuvenation/regeneration in aged and injured animals [60] (Figure 1C–D). These observations suggest that in vivo OKSM expression has the potential to uncover unexpected aspects of iPSC formation and might represent a versatile tool to study physiological and pathological processes not directly related to pluripotency.
OKSM binding and activities
iPSC generation requires global and dramatic OKSM-driven molecular changes that will successfully erase the somatic identity and establish a stable pluripotent program. Recently, extensive genome-wide chromatin assays combined with transcriptomics and epigenomics datasets from early or later stages of iPSC generation offered new mechanistic insights into the distinct or synergistic properties and effects of OKSM binding. Here, we focus on the direct and indirect effects of OKSM binding on (i) the transcriptional erasure of the somatic identity, (ii) the activation of pluripotency program and (iii) the reorganization of chromatin architecture during reprogramming (Figure 2), highlighting novel findings and critical rate-limiting steps and cofactors.
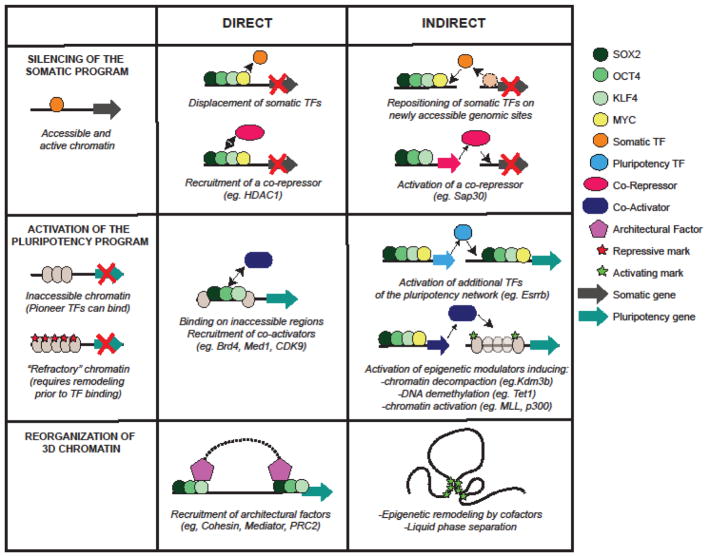
Figure 2. Mechanisms and molecular consequences of OKSM binding.
OKSM impact on the silencing of the somatic program, activation of pluripotency program and topological reorganization of the chromatin are illustrated. Direct mechanisms assume that OSK(M) binding is sufficient to induce transcriptional or topological changes in cis by recruiting the necessary cofactors that are already available in the nuclear milieu. Indirect mechanisms rely on epigenetic and transcriptional changes that occur at different genomic sites in an OKSM-dependent or independent manner and result in the activation of critical co-factors (co-activators or co-repressors) that mediate the silencing or activation of OKSM target genes.
Silencing of the somatic program
The necessity of early and robust silencing of the somatic program has been described in many reprogramming systems and is strongly supported by recent chromatin studies that demonstrate an extensive loss of chromatin accessibility around somatic loci and a strong decommissioning of somatic enhancers during the first phase of reprogramming [61–63]. Earlier papers studying the transcriptional effects of individual TF expression proposed an early repressive role of c-MYC and KLF4 [8,64], although evidence for a direct involvement of these factors was missing. Recent studies characterizing OKSM binding at early stages of reprogramming provide to some degree conflicting evidence. Chronis et al describe extensive OKSM co-binding on previously accessible and active fibroblast enhancers in mouse cells [65]. This suggests for the first time an active role of OKSM in somatic gene repression, for example by recruitment of co-repressors such as HDAC1 that directly interact with OCT4 or by displacement of somatic TFs through antagonism with OKSM (Figure 2) [65]. In support of these findings, another paper reported an early OCT4 binding on permissive/active chromatin that correlated with subsequent somatic gene silencing of bound regions in murine fibroblasts [63]. In contrast, a number of other studies using different human or mouse reprogramming systems and/or different time points, reported that Yamanaka factors bind mostly on previously inaccessible regions and do not directly associate with repressed somatic enhancers [61,62,66,67] (Zviran et al., bioRxiv, doi:10.1101/184135), arguing for an indirect role of OKSM in the erasure of somatic identity. A recently proposed indirect mechanism entails redistribution of somatic TFs from somatic enhancers to newly accessible sites generated by OKSM binding (Figure 2) [65]. It is argued that this would lead to decreased expression of critical somatic TFs, thereby triggering a negative feedback loop and culminating in silencing of the somatic program. In agreement, depletion of somatic TFs, such as c-Jun and Fra1 increases iPSC generation, while their overexpression inhibits reprogramming [61,62,65].
In addition to the role of TFs in the maintenance or repression of the somatic program during iPSC formation, recent studies highlighted novel roles of epigenetic modulators (Figure 2). OKSM-induced early activation of Sap30, encoding a SIN3A corepressor, was critical for efficient closing of the chromatin around somatic genes. In agreement, early depletion of SAP30 compromised iPSC generation, while this protein was dispensable at later stages [61]. Similarly, LSD1/HDAC1 were shown to be critical for the silencing of the B lymphocyte program prior to a very efficient and synchronized activation of the pluripotency network [68]. In this system, LSD1 was post-transcriptionally induced not by OSKM, but by CEBP/A, a specifier of the granulocyte-macrophage lineage, which was ectopically expressed prior to OKSM activation. On the other hand, the coactivator BRD4 has been shown to act as a barrier in early stages of reprogramming by bookmarking and preserving somatic gene activity [69], while also being essential for establishment and maintenance of pluripotency [68, 69]. Of note, erasure of the somatic identity during iPSC reprogramming is not always complete, usually due to inefficient resetting of the epigenetic landscape, an observation made initially in transgenic mouse models and recently confirmed for brain-derived human iPSCs using DNA methylation analysis of isogenic cell lines [70].
Activation of the stem-cell program
In contrast with the early and relatively efficient overwriting of the somatic signature, the establishment of the pluripotency program is considered a stochastic and rate-limiting process, involving step-wise selection and activation of pluripotency-related enhancers. Major roadblocks in this process include the inability of OKSM to access “refractory” genomic regions and the insufficiency of OKSM binding to induce gene activation in the absence of critical co-factors (Figure 2). As an example of the first type of barrier, repressive heterochromatin regions enriched in H3K9 methylation have been shown to be overall refractory to OKSM binding in early stages of reprogramming [66]. Critical stem cell regulators reside within these refractory regions, highlighting the necessity for chromatin remodeling and relaxation. Indeed, depletion of H3K9 methyltransferases (G9A, GLP, SETDB1, SUV39H1 and SUV39H2) promotes reprogramming of human and mouse fibroblasts [37,66,71]. Similarly, knock-down of CAF-1 complex, which normally deposits nucleosome in a replication-dependent manner, enables early relaxation of the somatic chromatin and increased accessibility and SOX2 binding at critical pluripotency-related enhancers and superenhancers [72]. High levels of repressive DNA methylation have been also associated with delayed and inefficient reprogramming, and in concordance, proteins involved in DNA demethylation, such as TET proteins, are strong albeit context-dependent [47] (Zviran et al., bioRxiv doi:10.1101/184135) facilitators of reprogramming.
Despite the exclusion of reprogramming TFs from selective refractory regions, a number of recent studies strongly support that early binding of OKS –and to a lesser degree of MYC- occurs predominantly on “inaccessible”, nucleosomal and even DNA methylated regulatory elements and induces accessibility for other TFs and cofactors (Figure 2)[62,66] (Zviran et al., bioRxiv doi:10.1101/184135). This early binding depends on (i) the ability of OKS to act as pioneer factors capable of partial motif recognition and binding on nucleosomal chromatin [66,73], (ii) their high affinity for DNA methylated targets [74] and (iii) their strong synergy among each other [65,66]. Importantly, although reprogramming TFs have been well-documented to function as trans-activators in established ESCs, their co-binding early during reprogramming does not guarantee immediate activation of target gene loci, suggesting context-dependent confinements of their activity. In fact, factors involved in chromatin remodeling (eg SWI/SNF subunits), epigenetic modulation (eg. BRD4 and MLL) (reviewed in Ref.31) and release of paused polymerase (eg. CDK9, P-TEFb) [68,75] have been described as critical cofactors for OKSM activity. Synergy between OKSM and other pluripotency-related transcription factors, such as ESRRB and NANOG, which are only induced later during reprogramming is another rate-limiting step that can be eliminated by enforced expression of these TFs during early reprogramming [20,65].
Re-organization of chromatin architecture
Increasing evidence supports that 3D chromatin architecture is cell-type specific and needs to be reorganized during cell fate transitions. Accordingly, chromatin topology differs between somatic and pluripotent cells and is largely reset during iPSC reprogramming [76–81]. However, evidence for the extent of this reorganization, the underlying mechanisms and the relationship to transcriptional and epigenetic changes has only begun to emerge. Previous studies have provided insights in to the chromatin rewiring around select pluripotency-associated loci during reprogramming, reporting that the establishment of chromatin loops usually precedes or coincides with transcriptional change [76,77]. A recent study that characterized global topological changes at various stages of B cell reprogramming [82] further supported and refined that chromatin reorganization often occurs concomitantly or prior to gene expression changes, supporting a potential causal role. In addition, the degree of architectural rewiring around critical pluripotency genes appears to correlate with the timing and probability of their activation. For example, the major topological changes observed at the Nanog and Sox2 loci correlate with their slower reactivation compared to the early-activated Oct4 locus, which does not undergo major conformation changes in this system. These data suggest that architectural constraints in addition to other epigenetic barriers may act as bottlenecks for efficient establishment of the pluripotent program.
Although all the aforementioned topological changes are triggered by ectopic expression of OKSM, the direct involvement of these factors remains unexplored. Evidence for the potential architectural roles of OKSM include (i) bioinformatics analysis showing that OKS binding is enriched around topologically reorganized domains in the course of reprogramming [76–79,82], (ii) knock-down experiments that disrupted predicted KLF4-mediated loops in established ESCs [77] and (iii) protein-protein interactions of OKS with known architectural factors such as Cohesin and Mediator [76,77]. On the other hand, the recent evidence that changes in histone modifications, such as H3K4me2 [82], precede conformational alterations, may support an indirect architectural role of OKSM due to co-factor recruitment and/or epigenetic remodeling around their target sites. In that case, topological alterations may be directly induced and maintained by epigenetic modulators, which may also function as architectural factors -as has been reported for PRC2 [83]. Alternatively, deposition or removal of defined epigenetic marks may change the biophysical properties of the local chromatin contributing to new self-organized and phase-separated multimolecular assemblies due to high affinity with other genomic regions with similar properties [84]. Future experiments will be critical to determine the OKSM- dependent and -independent mechanisms of local and global topological reorganization during iPSC formation, potential rate-limiting steps and interconnections with epigenetic and transcriptional changes.
Conclusions
The study of reprogramming mechanisms has revealed common molecular themes, but also unexpected complexities. The realization that important aspects of OKSM activity are influenced by variables such as starting somatic cell type, culture conditions and factor stoichiometry demonstrates that there likely is no single trajectory of iPSC formation. Rather than being a shortcoming, the context-dependent nature of reprogramming, if applied correctly, will provide novel opportunities to study complex interactions between molecular regulators during controlled experimental cell fate change. As indicated by the wide range of intra- and intercellular processes operational during iPSC formation, such studies will likely provide insight into physiological and pathological events extending far beyond pluripotency.
Acknowledgments
E.A. was funded by National Institute of Health Director’s New Innovator Award (DP2DA043813), the Edward Jr. Mallinckrodt Foundation, and the Tri-Institutional Stem Cell Initiative by the Starr Foundation (2016-018) M.S. was supported by the National Institute of Health (1R01GM111852-01). We apologize to all colleagues whose work we could not cite due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Hochedlinger K, Jaenisch R. Induced Pluripotency and Epigenetic Reprogramming. Cold Spring Harb Perspect Biol. 2015:7. doi: 10.1101/cshperspect.a019448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell Stem Cell. 2014;14:720–734. doi: 10.1016/j.stem.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 7.O’Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S, Kaji K. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013;499:88–91. doi: 10.1038/nature12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujan E, Zunder ER, Ng YH, Goronzy IN, Nolan GP, Wernig M. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature. 2015;521:352–356. doi: 10.1038/nature14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Cacchiarelli D, Trapnell C, Ziller MJ, Soumillon M, Cesana M, Karnik R, Donaghey J, Smith ZD, Ratanasirintrawoot S, Zhang X, et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell. 2015;162:412–424. doi: 10.1016/j.cell.2015.06.016. This study takes advantage of a synchronized transgenic reprogramming system to dissect global transcriptional and chromatins during human iPSC formation, providing evidence for the existence of intermediate stages that resemble specific stages of post-implantation development and exhibit elevated plasticity for the acquisition of alternative cell fates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 12.Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235–239. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- 13.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 14.Vidal SE, Amlani B, Chen T, Tsirigos A, Stadtfeld M. Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Reports. 2014;3:574–584. doi: 10.1016/j.stemcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Nur O, Brumbaugh J, Verheul C, Apostolou E, Pruteanu-Malinici I, Walsh RM, Ramaswamy S, Hochedlinger K. Small molecules facilitate rapid and synchronous iPSC generation. Nat Methods. 2014;11:1170–1176. doi: 10.1038/nmeth.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Liu J, Chen Y, Yang J, Chen J, Liu H, Zhao X, Mo K, Song H, Guo L, et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 2011;21:884–894. doi: 10.1038/cr.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 20.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol. 2013;15:829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- 24.Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S, Onder T, Shibue T, Weinberg RA, Daley GQ. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Reports. 2014;3:691–698. doi: 10.1016/j.stemcr.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingold JA, Fidalgo M, Guallar D, Lau Z, Sun Z, Zhou H, Faiola F, Huang X, Lee DF, Waghray A, et al. A genome-wide RNAi screen identifies opposing functions of Snai1 and Snai2 on the Nanog dependency in reprogramming. Mol Cell. 2014;56:140–152. doi: 10.1016/j.molcel.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawkins KE, Joy S, Delhove JM, Kotiadis VN, Fernandez E, Fitzpatrick LM, Whiteford JR, King PJ, Bolanos JP, Duchen MR, et al. NRF2 Orchestrates the Metabolic Shift during Induced Pluripotent Stem Cell Reprogramming. Cell Rep. 2016;14:1883–1891. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kida YS, Kawamura T, Wei Z, Sogo T, Jacinto S, Shigeno A, Kushige H, Yoshihara E, Liddle C, Ecker JR, et al. ERRs Mediate a Metabolic Switch Required for Somatic Cell Reprogramming to Pluripotency. Cell Stem Cell. 2015;16:547–555. doi: 10.1016/j.stem.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou G, Meng S, Li Y, Ghebre YT, Cooke JP. Optimal ROS Signaling Is Critical for Nuclear Reprogramming. Cell Rep. 2016;15:919–925. doi: 10.1016/j.celrep.2016.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathieu J, Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–551. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Nefzger CM, Rossello FJ, Chen J, Liu X, Knaupp AS, Firas J, Paynter JM, Pflueger J, Buckberry S, Lim SM, et al. Cell Type of Origin Dictates the Route to Pluripotency. Cell Rep. 2017;21:2649–2660. doi: 10.1016/j.celrep.2017.11.029. This study reveals both common and cell type-specific aspects of iPSC formation by to directly comparing genome-wide molecular features of reprogramming intermediates derived from different isogenic somatic cell types. [DOI] [PubMed] [Google Scholar]
- 31.Jackson SA, Olufs ZP, Tran KA, Zaidan NZ, Sridharan R. Alternative Routes to Induced Pluripotent Stem Cells Revealed by Reprogramming of the Neural Lineage. Stem Cell Reports. 2016;6:302–311. doi: 10.1016/j.stemcr.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima-Koyama M, Lee J, Ohta S, Yamamoto T, Nishida E. Induction of Pluripotency in Astrocytes through a Neural Stem Cell-like State. J Biol Chem. 2015;290:31173–31188. doi: 10.1074/jbc.M115.683466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim EJY, Anko ML, Flensberg C, Majewski IJ, Geng FS, Firas J, Huang DCS, van Delft MF, Heath JK. BAK/BAX-Mediated Apoptosis Is a Myc-Induced Roadblock to Reprogramming. Stem Cell Reports. 2018 doi: 10.1016/j.stemcr.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 38.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi K, Tanabe K, Ohnuki M, Narita M, Sasaki A, Yamamoto M, Nakamura M, Sutou K, Osafune K, Yamanaka S. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat Commun. 2014;5:3678. doi: 10.1038/ncomms4678. [DOI] [PubMed] [Google Scholar]
- 40.Raab S, Klingenstein M, Moller A, Illing A, Tosic J, Breunig M, Kuales G, Linta L, Seufferlein T, Arnold SJ, et al. Reprogramming to pluripotency does not require transition through a primitive streak-like state. Sci Rep. 2017;7:16543. doi: 10.1038/s41598-017-15187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Amlani B, Liu Y, Chen T, Ee LS, Lopez P, Heguy A, Apostolou E, Kim SY, Stadtfeld M. Nascent Induced Pluripotent Stem Cells Efficiently Generate Entirely iPSC-Derived Mice while Expressing Differentiation-Associated Genes. Cell Rep. 2018;22:876–884. doi: 10.1016/j.celrep.2017.12.098. This study shows that hallmark functional properties of pluripotent cells – such as the ability to give rise to all somatic cell types – is acquiring at a defined and surprisingly early time point during iPSC formation. Provides evidence that nascent and established iPSCs exhibit molecular and functional differences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert JM, Pulecio J, et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods. 2013;10:77–83. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maza I, Caspi I, Zviran A, Chomsky E, Rais Y, Viukov S, Geula S, Buenrostro JD, Weinberger L, Krupalnik V, et al. Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat Biotechnol. 2015;33:769–774. doi: 10.1038/nbt.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bar-Nur O, Verheul C, Sommer AG, Brumbaugh J, Schwarz BA, Lipchina I, Huebner AJ, Mostoslavsky G, Hochedlinger K. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat Biotechnol. 2015;33:761–768. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H, Wang X, Hu X, Gu T, Zhou Z, et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- 48.Chantzoura E, Skylaki S, Menendez S, Kim SI, Johnsson A, Linnarsson S, Woltjen K, Chambers I, Kaji K. Reprogramming Roadblocks Are System Dependent. Stem Cell Reports. 2015;5:350–364. doi: 10.1016/j.stemcr.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SI, Oceguera-Yanez F, Hirohata R, Linker S, Okita K, Yamada Y, Yamamoto T, Yamanaka S, Woltjen K. KLF4 N-terminal variance modulates induced reprogramming to pluripotency. Stem Cell Reports. 2015;4:727–743. doi: 10.1016/j.stemcr.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Parenti A, Halbisen MA, Wang K, Latham K, Ralston A. OSKM Induce Extraembryonic Endoderm Stem Cells in Parallel to Induced Pluripotent Stem Cells. Stem Cell Reports. 2016;6:447–455. doi: 10.1016/j.stemcr.2016.02.003. This study demonstrates that OKSM expression in mouse fibroblast cultures can yield a second, fully defined cell type in addition to iPSCs, likely without transition through a pluripotent intermediate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. Uses the simultaneous measurement of dozens of intracellular and surface proteins yields to develop a trajectory of mouse fibroblast reprogramming, which refines previous findings and identifies new aspects of iPSC formation including the late emergence of cells with features of mesoendodermal progenitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, Grana O, Megias D, Dominguez O, Martinez D, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 53.Choi HW, Kim JS, Hong YJ, Song H, Seo HG, Do JT. In vivo reprogrammed pluripotent stem cells from teratomas share analogous properties with their in vitro counterparts. Sci Rep. 2015;5:13559. doi: 10.1038/srep13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W, et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell. 2017;169:243–257. e225. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, Ezashi T, Roberts RM. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci U S A. 2015;112:E2337–2346. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J, Ryan DJ, Wang W, Tsang JC, Lan G, Masaki H, Gao X, Antunes L, Yu Y, Zhu Z, et al. Establishment of mouse expanded potential stem cells. Nature. 2017;550:393–397. doi: 10.1038/nature24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Mosteiro L, Pantoja C, Alcazar N, Marion RM, Chondronasiou D, Rovira M, Fernandez-Marcos PJ, Munoz-Martin M, Blanco-Aparicio C, Pastor J, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016:354. doi: 10.1126/science.aaf4445. Demonstrated the potential of in vivo reprogramming as a discovery tool by identifying the INK4/ARF pathway as necessary for iPSC formation and demonstrated paracrine communication between senescent cells and reprogramming intermediates in situ. [DOI] [PubMed] [Google Scholar]
- 59.Brady JJ, Li M, Suthram S, Jiang H, Wong WH, Blau HM. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-Seq. Nat Cell Biol. 2013;15:1244–1252. doi: 10.1038/ncb2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E, et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell. 2016;167:1719–1733. e1712. doi: 10.1016/j.cell.2016.11.052. Demonstrated unexpected and novel consequences of reprogramming factor activity by showing that careful titration of OKSM expression in transgenic animals can revert age-associated functional decline without the induction of teratomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S, et al. Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell. 2017;21:819–833. e816. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 62.Knaupp AS, Buckberry S, Pflueger J, Lim SM, Ford E, Larcombe MR, Rossello FJ, de Mendoza A, Alaei S, Firas J, et al. Transient and Permanent Reconfiguration of Chromatin and Transcription Factor Occupancy Drive Reprogramming. Cell Stem Cell. 2017;21:834–845. e836. doi: 10.1016/j.stem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Chen J, Chen X, Li M, Liu X, Gao Y, Kou X, Zhao Y, Zheng W, Zhang X, Huo Y, et al. Hierarchical Oct4 Binding in Concert with Primed Epigenetic Rearrangements during Somatic Cell Reprogramming. Cell Rep. 2016;14:1540–1554. doi: 10.1016/j.celrep.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 64.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017;168:442–459. e420. doi: 10.1016/j.cell.2016.12.016. The first comprehensive analysis of OKSM binding during early and later stages of reprogramming and its association to chromatin accessibility, state and transcriptional outcome. This study provides evidence for multiple direct and indirect mechanisms of OKSM-mediated silencing of the somatic program. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narayan S, Bryant G, Shah S, Berrozpe G, Ptashne M. OCT4 and SOX2 Work as Transcriptional Activators in Reprogramming Human Fibroblasts. Cell Rep. 2017;20:1585–1596. doi: 10.1016/j.celrep.2017.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Stefano B, Collombet S, Jakobsen JS, Wierer M, Sardina JL, Lackner A, Stadhouders R, Segura-Morales C, Francesconi M, Limone F, et al. C/EBPalpha creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat Cell Biol. 2016;18:371–381. doi: 10.1038/ncb3326. [DOI] [PubMed] [Google Scholar]
- 69.Shao Z, Yao C, Khodadadi-Jamayran A, Xu W, Townes TM, Crowley MR, Hu K. Reprogramming by De-bookmarking the Somatic Transcriptional Program through Targeting of BET Bromodomains. Cell Rep. 2016;16:3138–3145. doi: 10.1016/j.celrep.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 70.Roost MS, Slieker RC, Bialecka M, van Iperen L, Gomes Fernandes MM, He N, Suchiman HED, Szuhai K, Carlotti F, de Koning EJP, et al. DNA methylation and transcriptional trajectories during human development and reprogramming of isogenic pluripotent stem cells. Nat Commun. 2017;8:908. doi: 10.1038/s41467-017-01077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–224. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017:356. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Xu Y, He M, Zhang M, Cui F, Lu L, Yao M, Tian W, Benda C, Zhuang Q, et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell. 2014;15:574–588. doi: 10.1016/j.stem.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 76.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 80.Krijger PH, Di Stefano B, de Wit E, Limone F, van Oevelen C, de Laat W, Graf T. Cell-of-Origin-Specific 3D Genome Structure Acquired during Somatic Cell Reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, Hu G, Hsu SC, Shields EJ, Lyu X, Apostolou E, et al. Local Genome Topology Can Exhibit an Incompletely Rewired 3D-Folding State during Somatic Cell Reprogramming. Cell Stem Cell. 2016;18:611–624. doi: 10.1016/j.stem.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82••.Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, Gomez A, Collombet S, Berenguer C, Cuartero Y, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018 doi: 10.1038/s41588-017-0030-7. This is the first study that describes the order and the principles of 3D chromatin reorganization during iPSC formation and its correlation to transcriptional changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mas G, Di Croce L. The role of Polycomb in stem cell genome architecture. Curr Opin Cell Biol. 2016;43:87–95. doi: 10.1016/j.ceb.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 84.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]