Abstract
The White Collar complex is responsible for sensing light and transmitting that signal in many fungal species. In Cryptococcus neoformans and C. deneoformans the complex is involved in protection against damage from ultraviolet (UV) light, repression of mating in response to light, and is also required for virulence. The mechanism by which the Bwc1 photoreceptor contributes to virulence is unknown. In this study, a bwc1 deletion mutant of C. neoformans was transformed with three versions of the BWC1 gene, the wild type, BWC1C605A or BWC1C605S, in which the latter two have the conserved cysteine residue replaced with either alanine or serine within the light-oxygen-voltage (LOV) domain that interacts with the flavin chromophore. The bwc1+BWC1 strain complemented the UV sensitivity and the repression of mating in the light. The bwc1+BWC1C605A and bwc1+BWC1C605S strains were not fully complemented for either of the phenotypes, indicating that these BWC1 alleles impair the light responses for strains with them. Transcript analysis showed that neither of the mutated strains (bwc1+BWC1C605A and bwc1+BWC1C605S) showed the light-inducible expression pattern of the HEM15 and UVE1 genes as occurs in the wild type strain. These results indicate that the conserved flavin-binding site in the LOV domain of Bwc1 is required for sensing and responding to light in C. neoformans. In contrast to defects in light responses, the wild type, bwc1+BWC1, bwc1+BWC1C605A and bwc1+BWC1C605S strains were equally virulent, whereas the bwc1 knock out mutant was less virulent. Furthermore, pre-exposure of the strains to light prior to inoculation had no influence on the outcome of infection. These findings define a division in function of the White Collar complex in fungi, in that in C. neoformans the role of Bwc1 in virulence is independent of light sensing.
Keywords: Cryptococcus, Galleria mellonella, Medical mycology, Photobiology
Graphical abstract
1. Introduction
The effects of light on fungi impact aspects of their biology as diverse as sporulation, circadian clock function, primary metabolism, and the biosynthesis of secondary metabolites such as toxins or pigments (Fuller et al., 2016; Tisch and Schmoll, 2010). Those responses are controlled by a small number of photosensory proteins (Fischer et al., 2016; Idnurm et al., 2010; Rodriguez-Romero et al., 2010). Among them the blue light receptor White Collar-1 (WC-1), first characterized in the model ascomycete Neurospora crassa, appears to have originated early and then been well-conserved during the evolution of the fungi.
In N. crassa the light-regulated processes are controlled by WC-1 and WC-2, which form a GATA-type zinc finger transcription factor complex. WC-1 contains a number of domains: for DNA binding, two PAS domains (named after the Per, Arnt and Sim proteins, in which they were first observed) involved in protein-protein interactions, two putative transcriptional activation domains, a nuclear localization signal, and a LOV domain which is a specialized type of PAS domain involved in environmental sensing of light, oxygen and voltage (Froehlich et al., 2002; He et al., 2002; Wang et al., 2015). The chromophore flavin adenine dinucleotide (FAD), accommodated in the LOV domain, is essential for the light sensing activity of WC-1. A motif of NCRFLQ amino acids that contributed to the α’A helix is a highly conserved region among LOV domains, as the cysteine (C) residue is essential for the covalent attachment of the flavin chromophore to the sensor protein in the presence of light (Cheng et al., 2003). WC-1 interacts with a partner protein WC-2, another GATA-type transcription factor, to form the White Collar complex (WCC) that activates the expression of light-responsive genes upon light stimulation (Collett et al., 2002; Froehlich et al.,2002; Liu et al., 2003).
The availability of fungal genome sequences has allowed the identification of photoreceptors, especially wc-1 and wc-2 orthologs, in varied fungal species. Mutation of these white collar genes in most cases abolishes the effects of blue light on the species. However, analysis of such mutants has also revealed phenotypes in the absence of light, such as changes in sporulation or secondary metabolite production (Fuller et al., 2016; Fuller et al., 2015), leading to questions about how this complex functions above and beyond its perception of light.
One property that may be unrelated to light status is virulence in pathogenic species. In the human pathogenic fungi Cryptococcus neoformans and C. deneoformans [their previous names are C. neoformans var. grubii/serotype A and C. neoformans var. neoformans/serotype D, respectively (Hagen et al., 2015)], light inhibits mating and induces a protective response against ultraviolet light. Mutation of the orthologs of N. crassa wc-1 and wc-2 impairs these effects (Idnurm and Heitman, 2005; Lu et al., 2005; Verma and Idnurm, 2013). An additional phenotype associated with mutating the BWC1 or BWC2 genes in C. neoformans and the related species C. deuterogattii is a reduction in virulence in animal models of disease (Idnurm and Heitman, 2005; Zhu et al., 2013). Cryptococcosis refers to a set of diseases caused by the C. neoformans species complex, currently made up of seven species that are most problematic when growing in the lungs and central nervous systems of people (Heitman et al., 2011). While a number of factors and genes have been identified that control virulence, mutation of the WCC does not impact on any of these that are known to date (Alspaugh, 2015; Brown et al., 2014).
The role of light or the WCC in fungal virulence is more widespread than the Cryptococcus species. For instance, mutation of the WC homologs also alters the pathogenicity in plant pathogens. In the maize leaf pathogen Cercospora zeae-maydis, the WC-1 ortholog is required for tropism toward the host stomata and lesion formation (Kim et al., 2011a). In the rice blast pathogen Pyricularia (Magnaporthe) oryzae, light represses asexual spore releases and a dark-phase immediately after pathogen–host contact plays a critical role in successful disease development, with spore release and the light-dependent repression accomplished by the photoreceptor MGWC-1 (Kim et al., 2011b; Lee et al., 2006). A T-DNA insertion within the WC-2 ortholog also causes a decrease in pathogenicity of P. oryzae (Jeon et al., 2007). Mutation of wc1 in Fusarium oxysporum, which is normally a plant pathogen, causes a decrease in virulence in a mouse model of disease (Ruiz-Roldán et al., 2008). In the gray mold pathogen Botrytis cinerea, loss of BcWCL1 results in attenuated virulence in the host plant when light is present, and this is due to BcWCL1 being required for coping with excessive reactive oxygen species generated by the host’s oxidative burst and photooxidative stress (Canessa et al., 2013). A light-responsive transcription factor, BcLTF1, downstream of BcWCL1 is responsible for anti-oxidative stress activities and is thus required for advanced host infection by B. cinerea (Schumacher et al., 2014). In addition, the influence of the circadian clock, which is controlled by light, on the disease outcome in the Botrytis cinerea-Arabidopsis thaliana interaction is dependent on the effects of the clock and light from the fungal pathogen, rather than changes in plant defense systems (Hevia et al., 2015). In addition to the genetic contributions of the WCC to virulence, pre-exposing fungi to light can impact subsequent virulence outcomes (Campbell and Berliner, 1973; Oliveira et al., 2018).
It is not yet clear in the pathogenic fungi how light or the light-sensing proteins contribute to virulence. This study used C. neoformans as the model organism to test if light sensing is responsible for the function of the conserved White Collar complex in virulence.
2. Materials and Methods
2.1. Generation of plasmids
The LITMUS 28i vector (New England Biolabs, Ispwich, MA) was double digested with the restriction enzymes KpnI and BamHI. The BWC1 open reading frame and adjacent 5′ and 3′ regulatory regions were amplified from the genomic DNA of strain KN99α with the primer pair PK003/PK004, and then cloned into linearized LITMUS 28i using T4 DNA ligase, yielding the LITMUS 28i-BWC1 vector. Primer sequences are given in Table 1. A plasmid without errors in BWC1 was identified by sequencing the inserts. The URA5 gene was also amplified from KN99α DNA with the primer pair PK027/PK028, and the product was cloned into the pCR®2.1-TOPO vector (Invitrogen, Grand Island, NY), resulting in plasmid pCR®2.1-TOPO-URA5. Subsequently, both the LITMUS 28i-BWC1 and pCR®2.1-TOPO-URA5 plasmids were digested by EcoRI, and the URA5 fragment was ligated with the linearized LITMUS 28i-BWC1, resulting in the plasmid LITMUS 28i-BWC1-URA5. Specific mutations were introduced into this plasmid at the site of the conserved codon for cysteine within the LOV domain, by means of the QuikChange® Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). The cysteine residue is amino acid number 605 of GenBank accession XP_012048765.1. The primers used for the mutations are listed in Table 1. After mutagenesis, the resultant plasmids were confirmed to be otherwise error-free by DNA sequencing.
Table 1.
Oligonucleotide primers used in this study.
| Name | Sequence (5′−3′) | Function and location |
|---|---|---|
| PK003 | GGTACCATAAGTTTGCTGCGAAGC |
Forward primer to amplify BWC1, with KpnI cut site |
| PK004 | GGATCCGTGATGTGGGCAGTTACG |
Reverse primer to amplify BWC1, with BamHI cut site |
| PK010 | CGGTATCGAAGACAATCC |
Forward primer to amplify URA5 (outside the URA5 used in construction) |
| PK011 | TGGGAGGAAGAGGATGAG |
Reverse primer to amplify URA5 (outside the URA5 used in construction) |
| PK018 |
GGGACTTTGAAGAAAACGAGCATTA CGACCGAGAAGTTG |
Reverse primer to amplify the left part used to replace Cys site with Ala in BWCI gene |
| PK019 |
CAACTTCTCGGTCGTAATGCTCGTT TTCTTCAAAGTCCC |
Forward primer to amplify the rigt part used to replace Cys site with Ala in BWCI gene |
| PK020 | GGCTGACGGAATCTCTAG |
Forward primer to sequence the Cys site mutation |
| PK021 | TATAGGCACAACAGTGAC |
Reverse primer to sequence the Cys site mutation |
| PK027 |
CAACTTCTCGGTCGTAATTGCAAAG AGCGAAGTTGC |
Forward primer to amplify URA5 |
| PK028 |
GGGACTTTGAAGAAAACGGATTGTT GGCTGTCAATC |
Reverse primer to amplify URA5 |
| PK035 |
GGGACTTTGAAGAAAACGTCTATTA CGACCGAGAAGTTG |
Reverse primer to amplify the left part used to replace Cys site wirh Ser in BWCI gene |
| PK036 |
CAACTTCTCGGTCGTAATAGACGTT TTCTTCAAAGTCCC |
Forward primer to amplify the right part used to replace Cys site wirh Ser in BWCI gene |
| HEM15F | TGGTAGAGGCTTTTGCAC |
Real time PCR of HEM15 |
| HEM15R | TCACCACGGTTGACAATG |
Real time PCR of HEM15 |
| UVE1F | TGCATCCTGGTCAGTTTACGC |
Real time PCR of UVE1 |
| UVE1R | CATATGGATAATCATGACAC |
Real time PCR of UVE1 |
| ACTINF | CAATGGTATTGCCGACCG |
Real time PCR of ACTIN |
| ACTINR | CTTCGCGATCCACATCTG |
Real time PCR of ACTIN |
2.2. Fungal strains used in the experiments
C. neoformans strains used in this study are listed in Table 2. They were maintained on yeast peptone dextrose (YPD) medium. Crosses, between strains of MATa and MATα mating types, were conducted on V8 medium (5% Campbell’s V8 juice, 4% agar, pH 5). The bwc1 mutant strain AI81 (MATα) and ura5 base pair substitution mutant strain JF99 (MATa) were crossed to generate progenies of the bwc1 ura5 double mutant, via micromanipulation of the basidiospores. A bwc1 ura5 strain of MATα plated on minimal medium (yeast nitrogen base; YNB) plus 1 M sorbitol was transformed with the plasmids LITMUS 28i-BWC1-URA5, LITMUS 28i-BWC1C605A-URA5, or LITMUS 28i-BWC1C605S-URA5 via the biolistic system (Bio-Rad, Hercules, CA), with standard parameters (Toffaletti et al., 1993). Transformants were screened by PCR to select those in which the vectors were correctly incorporated into the genome at the URA5 locus by homologous recombination. The bwc1 ura5 MATα strains transformed with either wild type or site-mutated BWC1 alleles were crossed with the bwc1 ura5 MATa strain obtained earlier, to isolate progeny of the MATa mating type with the BWC1 alleles integrated into the URA5 locus.
Table 2.
Strains of Cryptococcus used in this study.
| Name | Species* | MAT | Genotype | Origin and notes |
|---|---|---|---|---|
| JEC30 | D | a | lys1 | (Moore and Edman, 1993) |
| AI42 | D | a | lys1 bwc1::NAT | (Idnurm and Heitman, 2005) |
| AI81 | A | alpha | bwc1::NAT | (Idnurm and Heitman, 2005) |
| KN99α | A | alpha | Wild type | (Nielsen et al., 2003) |
| AI202 | A | a | ade2 | (Idnurm, 2010) |
| JF99 | A | a | ura5 | (Nichols et al., 2004) |
| AIZCN1 | A | alpha | bwc1::NAT ura5 | Progeny of JF99 × AI81 |
| AIZCN2 | A | a | bwc1::NAT ura5 | Progeny of JF99 × AI81 |
| AIZCN4 | A | alpha |
bwc1::NAT ura5::URA5+BWC1 |
Created by transforming P-Litmus28i-Bwc1(serot ype A)-URA5(serotype A) into strain AIZCN1 |
| AIZCN12 | A | a |
bwc1::NAT ura5::URA5+BWC1 |
Progeny of AIZCN4 × AIZCN2 |
| AIZCN30 | A | alpha |
bwc1::NAT ura5::URA5+BWC1 |
Progeny of AIZCN4 × AIZCN2 |
| AIZCN34 | A | alpha |
bwc1::NAT ura5::URA5+BWC1C605S |
Created by site-directed mutation |
| AIZCN36 | A | alpha |
bwc1::NAT ura5::URA5+BWC1C605A |
Created by site-directed mutation |
| AIZCN66 | A | alpha | ade2 | Progeny of AI81 × AI202 |
| AIZCN68 | A | alpha | bwc1::NAT ade2 | Progeny of AI81 × AI202 |
| AIZCN70 | A | alpha |
bwc1::NAT ura5::URA5+BWC1 ade2 |
Progeny of AIZCN30 × AI202 |
| AIZCN72 | A | alpha |
bwc1::NAT ura5::URA5+BWC1C605A ade2 |
Progeny of AIZCN36 × AI202 |
| AIZCN74 | A | alpha |
bwc1::NAT ura5::URA5+BWC1C605S ade2 |
Progeny of AIZCN34 × AI202 |
species: A = C. neoformans; D = C. deneoformans
2.3. Assessment of inhibition of mating by light in crosses between strains
Dikaryotic filaments form during the sexual cycle of C. neoformans. Crosses between strains were compared to each other for their filamentous growth ability on V8 medium at pH 7 as influenced with continuous white fluorescent light (1600 lux) in contrast to culturing in darkness. To quantify the effect of light on mating, a cell fusion assay was conducted (Idnurm and Heitman, 2005). An auxotrophic C. neoformans strain AI202 (MATa, ade2) was crossed with the MATα strains of bwc1, bwc1+BWC1, bwc1+BWC1C605A, and bwc1+BWC1C605S. Among the progenies from each cross, the MATα isolates with genotypes of ade2, bwc1 ade2, bwc1+BWC1 ade2, bwc1+BWC1C605A ade2, and bwc1+BWC1C605S ade2 were obtained. These strains were subsequently used to make inter-species hybrids by crosses with the bwc1 mutant or wild type strains of C. deneoformans with the MATa lys1 genotype. After 24 h incubation in dark or continuous white light at room temperature, each set of mixed yeast cells was suspended in distilled water to the concentration of 107/ml. Aliquots (200 μl) of each cell suspension were pipetted to YNB medium, and the cells were subsequently spread evenly on each plate. Colony growth on the minimum medium indicates a prototrophic dikaryotic or diploid strain resulting from successful cell fusion between two auxotrophic parents (C. neoformans ade2 and C. deneoformans lys1). Colony numbers were recorded to quantify the cell fusion events for each set of crosses.
2.4. Analysis of UV sensitivity
Strains were first cultured to logarithmic phase in 5 ml of liquid YPD medium contained in light-proof 50 ml volume black tubes (Argos Technologies, Elgin, IL), and then the cell suspensions were serially diluted in a dark room with illumination by a dim red light (GBX LED safelight, Kodak, Rochester, NY). The serial dilutions were dotted onto two YPD agar plates: one was irradiated with a UV dose of 120 J/m2 using a XL-1500 UV Crosslinker (Spectronics Corporation, Long Island, NY), and the other was not irradiated. The plates were then incubated at 30°C for two days.
2.5. Northern blot and qPCR analyses to measure transcript abundances of genes
Strains were first grown to logarithmic phase in liquid YPD medium, and equal quantities of yeast cells for each strain were inoculated onto YPD agar plates (15 cm diameter) and grown in complete darkness for 23 h, after which one set was exposed to light for 15 min or 1 h (1600 lux, cool-white fluorescent lights). After growth and light induction, the cells of each strain were collected from the agar surface, washed with distilled water, and pelleted in a centrifuge. This procedure was conducted in a dark room with illumination by dim red light. RNA was extracted from lyophilized yeast cells using TRIzol RNA reagent (Invitrogen). Total RNA (20 μg) was resolved on denaturing formaldehyde agarose gels, blotted to Zeta-Probe membranes (Bio-Rad, Hercules, CA), and probed with [α−32P] dCTP radiolabeled DNA fragments of genes amplified with the primers listed in Table 1. Levels of transcripts on northern blots were quantified in ImageJ (Schneider et al., 2012).
For qPCR, total RNA was reverse transcribed with Superscript III (Invitrogen). SYBR Green (Fermentas, Vilnius, Lithuania) was used to detect the amplification of DNA from the cDNAs by quantitative PCR in an ABI7300 machine (Bio-Rad), with the amplification primers for PCR listed in Table 2. The 2−ΔΔCT method was used for relative quantification of genes (Livak and Schmittgen, 2001) whereby the actin (ACT1) gene was the constitutively expressed gene.
2.6. Yeast two-hybrid assays to test for protein interactions between Bwc2 and the Bwc1 isoforms
The plasmids pGBD.c1 and pGAD.c1 containing the cDNA of BWC1 or BWC2 had been created previously (Idnurm and Heitman, 2005; James et al., 1996). Both pGBD.c1-BWC1 and pGAD.c1-BWC1 were used as templates to introduce a point mutation to the flavin-binding site of the LOV domain in BWC1 via the QuikChange Site-Directed Mutagenesis kit. The correct mutations in the resultant plasmids were confirmed by sequencing. Subsequently, the Saccharomyces cerevisiae reporter strain PJ69–4A (James et al., 1996) was co-transformed with the following plasmid combinations: pGBD.c1 + pGAD.c1, pGAD.c1-BWC1 + pGBD.c1-BWC2, pGBD.c1-BWC1 + pGAD.c1-BWC2, pGAD.c1-BWC1C605A + pGBD.c1-BWC2, pGBD.c1-BWC1C605A + pGAD.c1-BWC2, pGAD.c1-BWC1C605S + pGBD.c1-BWC2, pGBD.c1-BWC1C605S + pGAD.c1-BWC2. Double transformants were selected on yeast nitrogen base medium lacking leucine and tryptophan. The interactions between the Bwc1 isoforms and Bwc2 were measured by growth in the absence of adenine and β-galactosidase activity.
2.7. Virulence assays for C. neoformans strains in the insect model Galleria mellonella
Larvae of wax moth (Galleria mellonella) purchased from Vanderhorst Wholesale (Saint Marys, OH) were infected as previously described (Mylonakis et al., 2005). Aliquots of 200 μl C. neoformans cells (2×107) were spread evenly on solid YPD medium and placed in light or dark (wrapped with foil). After overnight culture, the yeast cells were harvested by flooding with phosphate buffered saline (PBS) and scraping with cell scrapers. The cultures in dark were harvested in the dark room with the aid of red safe light. The concentration of resulted cell suspensions was adjusted to 2×107 per ml. For inoculation, 5 μl of the cells were injected into the rear proleg of each larva via a 10 μl Hamilton syringe. For each strain, 12 larvae were inoculated and subsequently incubated at 37°C in darkness, and examined daily for survival, with survival between groups compared using log-rank tests. The virulence experiments were performed on two separate occasions.
2.8. RNA-seq analysis
Five strains of C. neoformans were inoculated in tissue culture medium (10% horse serum, 90% RPMI) at a cell concentration of 105/ml. They were cultured in complete darkness at 5% CO2 and 37°C for 24 hours. The yeast cells were subsequently harvested under dim red light. Total RNA of these samples was extracted using the TRIzol reagent. 15 μg of each RNA sample was treated with DNase I, and the DNase I-treated samples were then diluted to 100 μg/μl. Each sample was processed and sequenced at the DNA Core Facility, University of Missouri, on an Illumina HiSeq2000 with 100-bp paired reads. All reads were mapped to the C. neoformans reference genome (EnsemblFungi, Genome Assembly: CNA3) by using TopHat v1.3.0. The aligned reads were then analyzed for the number of fragments per kilobase of transcript per million mapped reads using Cufflinks v.1.0.3. Statistically-significant differences between strains were determined by CuffDiff. The differentially expressed genes among strains were clustered by R Heatmap2 package. RNA-sequence data are available at GenBank under accession PRJNA491293.
3. Results
3.1. The flavin-binding site in the LOV domain of Bwc1 is required for full light responses in C. neoformans
Plasmid vectors were created with three versions of the BWC1 gene: a wild type copy, and those that cause a cysteine to alanine or serine substitution in the flavin-binding part of the encoded protein. These substitutes were based on previous work on the Neurospora crassa WC-1 and oat phototropin proteins (Cheng et al., 2003; Salomon et al., 2000), with the rationale that the new isoforms would retain their other functions apart from a loss in the ability to transmit the light signal. These constructs were transformed into a bwc1 knock-out mutant strain and integrated into the URA5 locus in the genome by single cross-overs through homologous recombination to ensure no genome positional variation in gene expression, with strains with integration of the plasmids into the correct site identified by PCR analysis (data not shown). A strain with the bwc1 deletion allele and carrying the empty URA5 plasmid integrated at the URA5 locus was created for comparison.
The UV sensitivity was compared among these strains (Figure 1). After UV exposure, the complemented strain (bwc1+BWC1) recovered as well as the wild type strain. However, the bwc1 strains with point mutations at the conserved cysteine site, BWC1C605A or BWC1C605S (this allelic information is abbreviated to BWC1C-A or BWC1C-S, respectively, for simplicity on all the figures), were considerably more sensitive to UV irradiation.
Fig. 1.
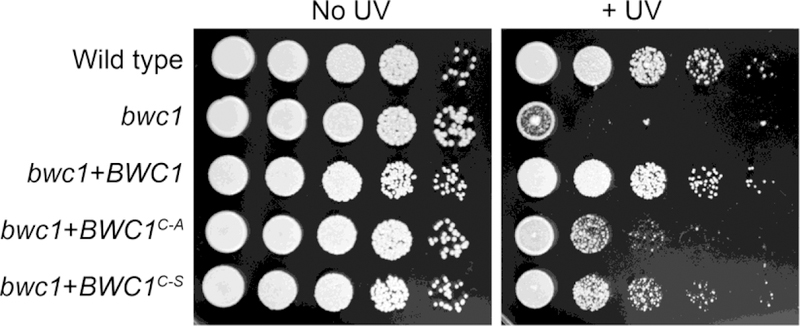
The UV sensitivity is higher in strains of C. neoformans with either the bwc1 deletion mutation or the two amino acid substitutions. Ten-fold serial dilutions were plated onto YPD medium, and one set irradiated with UV light (120 J/m2). Photographs show colonies 2 days later.
A second response to light in C. neoformans and C. deneoformans is the inhibition of mating. Compared to in darkness, the production of the dikaryotic mating filaments was prominently repressed by light in the wild type strain and the complemented strain (Figure 2). However, light treatment did not effectively block filamentation in the bwc1 knock out strain or the bwc1+BWC1C605A and bwc1+BWC1C605S site mutation strains. The effect of light on mating of these strains was then quantified via a cell fusion assay in which the parental strains have auxotrophic mutations, that are cross-complemented when the strains fuse to form a diploid or intra-species dikaryon (Idnurm and Heitman, 2005). The ratios of cell fusion events in the light versus those in darkness were lower in both wild type and complemented strains than in the knock out and site mutation strains (Figure 3). These results confirmed that a functional LOV domain is required for sensing light by Bwc1 to inhibit mating in Cryptococcus species.
Fig. 2.
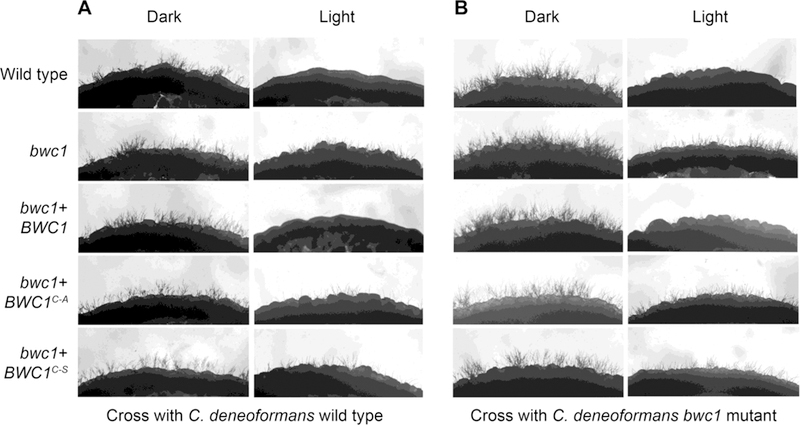
Production of the sexual filaments is inhibited in response to light in wild type strains, but not in crosses with the bwc1 mutant or the two strains carrying the mutant allele of BWC1. Strains of C. neoformans was crossed to either the C. deneoformans strain (A) JEC30 with the wild type copy of BWC1 or (B) strain AI42 with the bwc1 gene deleted.
Fig. 3.
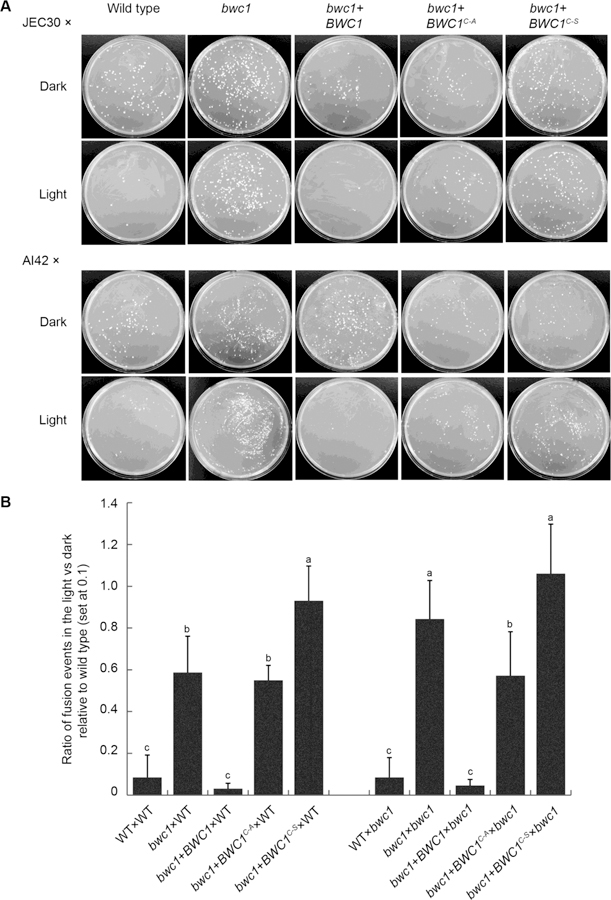
Cell fusion inhibition by light is lost in the two LOV domain mutant alleles of Bwc1. Strains carrying the ade2 mutation with the genotypes listed were mixed with C. deneoformans strains JEC30 (lys1) or AI42 (lys1 bwc1) and cultured in the light or darkness for 24 h. Cultures were scraped from the plates and plated onto minimal medium. Colony forming units are a measure of cell fusion events. (A) Colonies indicate diploid or dikaryotic strains after cell fusion. (B) Quantification of cell fusion events; significant differences between data values are indicated by different letters above the histograms.
3.2. The mutation of the cysteine in the flavin-binding site of LOV domain in Bwc1 has no effect on the physical interaction between Bwc1 and Bwc2 proteins
In contrast to N. crassa WC-1 and homologs of this protein in the ascomycetes, the predicted Bwc1 protein in C. neoformans lacks a DNA binding domain. Thus, Bwc1 must interact with Bwc2 such that the two proteins can together transcriptionally regulate expression of downstream genes. Yeast two-hybrid analysis was conducted to test whether the mutation at the conserved cysteine site of the Bwc1 protein rendered any negative effect on the interaction between Bwc1 and Bwc2. In S. cerevisiae strains expressing the fusions with the Gal4 activation domain (AD) or DNA binding domain (BD), i.e. AD-Bwc1 and BD-Bwc2, AD-Bwc2 and BD-Bwc1, AD-Bwc1C605A and BD-Bwc2, AD-Bwc2 and BD-Bwc1C605A, AD-Bwc1C605S and BD-Bwc2, AD-Bwc2 and BD-Bwc1C605S, the reporter genes ADE2 and lacZ were induced, as shown by cell growth in the absence of adenine and increased levels of β-galactosidase activity (Figure 4). In contrast, the S. cerevisiae strain expressing the AD and BD domains alone showed no growth on the adenine-deficient medium and expressed a minimal level of β-galactosidase activity. These findings demonstrate that Bwc1 and Bwc2 can physically interact, irrespective of the mutation at the conserved cysteine site in the LOV domain of the Bwc1. This indicates that changing the cysteine residue to alanine or serine does not abolish protein folding or stability of Bwc1, at least as measurable through a yeast two hybrid assay.
Fig 4.
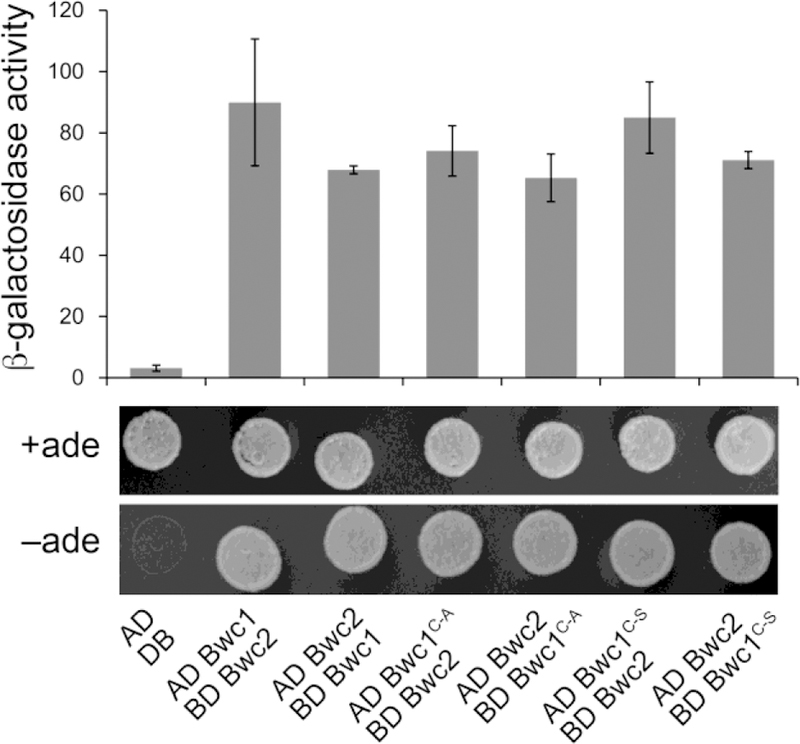
The Bwc1C605A and Bwc1C605S isoforms physically interact with Bwc2 in yeast two hybrid assays. The cDNAs of either gene were cloned adjacent to the activation (AD) or DNA binding (BD) domains of S. cerevisiae Gal4. Constructs were transformed into an S. cerevisiae reporter strain. The β-galactosidase activity and growth on medium without adenine (ade) indicate interactions between Bwc2 and the Bwc1 alleles to reconstitute the Gal4 protein. There was no statistical difference between the β-galactosidase activities of the wild type vs. mutant isoforms.
3.3. The flavin-binding site in the LOV domain of Bwc1 is required for the increase in transcript abundance of genes in C. neoformans in response to light
The ferrochelatase-encoding gene HEM15 is light-regulated by White Collar in diverse fungi (Canessa et al., 2013; Idnurm and Heitman, 2010). The transcript level of this gene, in response to light exposure, was measured in wild type, the bwc1 knock out mutant, BWC1 complemented, and the BWC1C605A, BWC1C605S site mutation strains. Northern blot analysis showed that a 15 min or 1 h exposure to light could slightly induce the expression of HEM15 in the wild type, and BWC1, BWC1C605A and BWC1C605S site mutation strains (Supplemental Figure 1). In contrast, the expression of HEM15 remained low in the bwc1 knock out mutant in response to light. HEM15 is expressed at a low level, making its detection and then quantitative comparisons by northern blots challenging. We therefore switched to a PCR-based method to detect this transcript.
Quantitative real time PCR was used to quantify the expression of HEM15 and of UVE1, encoding a DNA repair endonuclease and a downstream target of the light signal via the White Collar complex in fungi (Verma and Idnurm, 2013). Like HEM15, the expression level of UVE1 is low and therefore difficult to detect by northern blotting. Light treatment induced the expression of HEM15 and UVE1 in wild type and the BWC1 complementation strains, while there was only a basal level of expression in the bwc1 knock out mutant, BWC1C605A and BWC1C605S site mutation strains irrespective of the light/dark conditions (Figure 5). These data further indicate that the conserved cysteine site in the LOV domain is required for the ability to respond fully to light signals via the Bwc1-Bwc2 complex in Cryptococcus species.
Fig. 5.
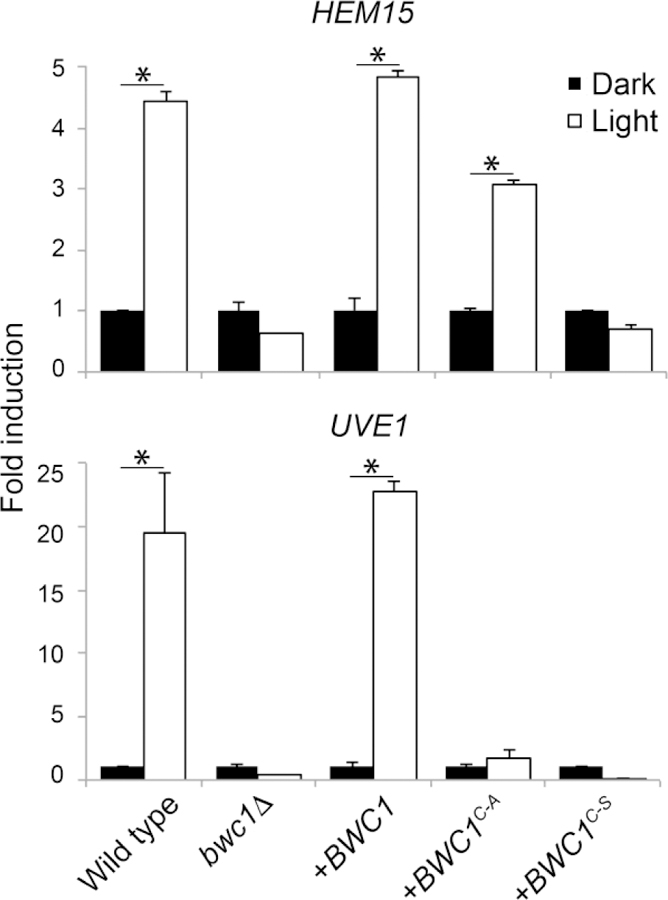
The induction of two conserved light-regulated genes is impaired in the bwc1 mutant and the two strains carrying the modified forms of BWC1. Quantitative real time PCR of the expression of two light-regulated genes in five strains of C. neoformans. Fold change in transcript levels in response to 15 min exposure to light relative to the constant darkness for each strain, normalized against the actin gene. The histograms marked with asterisks are significantly different from each other using Student’s t test (P < 0.05).
3.4. The cysteine in the LOV domain in Bwc1 is dispensable for virulence of C. neoformans
The larvae of the wax moth (Galleria mellonella) were used as the animal model to determine the virulence of C. neoformans strains. The wild type, bwc1 knock out, bwc1+BWC1 complementation, and bwc1+BWC1C605A and bwc1+BWC1C605S site mutation strains were used to inoculate 12 larvae of the wax moth G. mellonella per strain. The effect of exposure to light on the virulence of each strain was tested by comparing larvae infected with C. neoformans cultured in the light or under constant darkness.
Survival rates were the same for the strains of wild type, bwc1+BWC1, bwc1+BWC1C605A and bwc1+BWC1C605S (Figure 6). However, the larvae infected with the bwc1 knock-out mutant survived longer than the other strains (P = 0.00261 and P = 0.00208 between wild type and bwc1 deletion mutant inoculated with light-exposed or dark cultured strains, respectively). Pre-treatment of strains with light did not alter the virulence of the C. neoformans strains when compared to when the strains were cultured in darkness and all manipulations including the inoculation into the larvae was performed under safe red light (P = 0.66 between larvae infected with the wild type strain cultured in the light vs. dark).
Fig. 6.
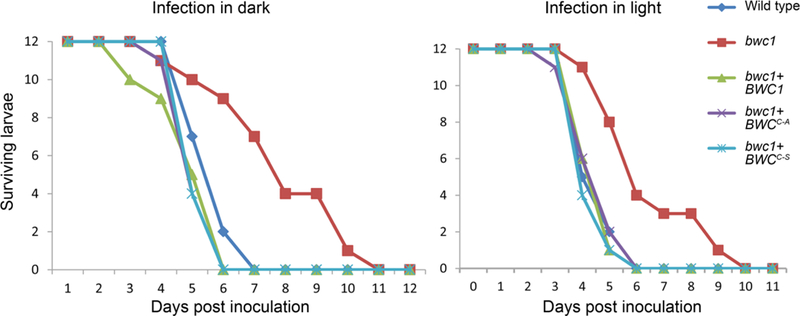
C. neoformans strains with the BWC1C605A and BWC1C605S alleles are as virulent as wild type, and pre-exposure to light does not impact virulence. Wax moth larvae were inoculated with strains grown in complete darkness and manipulated under safe red light, or exposed to light. Survival was monitored each day. The survival difference for larvae inoculated with the bwc1 mutant compared to the other strains is statistically significant (e.g. in light compared to wild type P = 0.00261, log-rank test).
These results indicate that in contrast to its impact on the UV sensitivity and inhibition of mating, that the mutation of the conserved cysteine site for flavin binding in the LOV domain does not attenuate the virulence of C. neoformans. Hence, the Bwc1-Bwc2 transcription factor complex regulates the virulence of C. neoformans in a light-independent manner.
3.5. Analysis of gene expression in C. neoformans bwc1 mutant strains
Towards understanding what genes may be misregulated when the BWC1 gene is mutated, five strains were grown under conditions in constant darkness and aiming to mimic conditions within a vertebrate host. RNA was extracted from the samples, and gene expression estimated by RNA-sequencing and analysis of transcript abundance by comparing the bwc1 deletion strain to the wild type, bwc1+BWC1, bwc1+BWC1C605A and bwc1+BWC1C605S strains (supplemental table 1). genes were up-regulated and 192 down-regulated more than 2-fold in these four strains, as illustrated in the heat map representation of the data (supplemental figure 2). Comparison of the genes to those known to be involved in virulence, e.g. as defined in large scale studies of genes required for virulence (Liu et al., 2008), revealed no candidate that might explain the virulence differences. One exception was one of the most differentially expressed genes, MPR1, which has recently been shown to encode a metalloprotease required for crossing the blood brain barrier in mouse models of cryptococcosis (Na Pombejra et al., 2018; Vu et al., 2014).
4. Discussion
The LOV domains are essential for the light sensing capabilities of photosensory proteins in plants, fungi and bacteria (Briggs and Spudich, 2005), and LOV-domain containing proteins are implicated as virulence factors in pathogenic fungi and bacteria. In fungi, the most common and best-characterized LOV domain protein is White Collar 1 (Fischer et al., 2016). This gene, and a second gene wc-2, were first identified and cloned based on mutant phenotypes in the ascomycete N. crassa (Ballario et al., 1996; Linden and Macino, 1997). Both genes are required for sensing blue light. With the advent of genome sequencing projects for many fungi, it became clear that this gene pair is found throughout many fungal lineages. However, these homologs have roles beyond sensing light, such as during growth under conditions of no light, although such “dark” functions have received little attention to date. Some of the best appreciation of these functions comes from the role the WC complex plays in the circadian clock machinery of N. crassa. In this species the complex allows light to serve as an indicator of time-of-day, and then is part of the feedback loop during the rest of the day that is required for circadian timing (Baker et al., 2012).
The LOV domains have a conserved structure and amino acid residues for interacting with chromophores such as flavin adenine dinucleotide or flavin mononucleotide. Of these residues, an invariant cysteine in the α’A helix is critical for the formation of a covalent adduct with the C (4a) position of the flavin in response to light exposure (Crosson and Moffat, 2001; Crosson and Moffat, 2002). Mutations to one of the nucleotides encoding the cysteine residue are predicted to block the formation of the light-induced adduct between the LOV domain and the flavin chromophore, thus impairing the capability of the protein to transmit the light signal.
We therefore tested the role of light in the virulence-related function of the Bwc1 protein of the human pathogen C. neoformans by making and then testing strains with amino acid substitutions in the LOV domain. The work reported here is on C. neoformans: we also used the same approach to create an equivalent strain set of strains in C. deneoformans using constructs specific to C. deneoformans (data not shown). Because the C. deneoformans background used has a low level of virulence, the results of the C. neoformans strains are reported here. Two specific alleles of BWC1 were generated that substitute either an alanine or serine for the cysteine residue required for adduct formation to the flavin molecule in response to light. The equivalent residue in LOV domains inactivates light sensing in phototropin of Arabidopsis thaliana or WC-1 of N. crassa (Cheng et al., 2003; Christie et al., 2002). These C. neoformans BWC1 alleles have loss-of-function phenotypes for two light-dependent effects, i.e. the inhibition of mating and protection from UV light. However, the two alleles have different effects on the protein properties, with the BWC1C605A allele allowing some light-dependent induction of HEM15, which is consistent with the phenotypes of the equivalent mutations introduced into the N. crassa wc-1 gene. That is, Cheng et al. (2003) found that the substitution of a serine for cysteine blocked the increase in transcript levels of both the frq and al-3 genes in response to light, but the alanine substitution strain had nearly normal induction of frq yet abolished induction of al-3. It is worth noting that while this adduct formation is considered a requirement for transmission of the photon signal, flavin may still interact with these modified LOV domains or with other amino acid residues. The BWC1C605S allele renders the encoded protein even less sensitive to light, and the strains carrying the two altered BWC1 alleles are equally pathogenic as wild type. Taken together, it can be concluded that the cysteine residue within the conserved NCRFLQ motif of the LOV domain in Bwc1 is essential for the capabilities of C. neoformans to fully response to light yet it is dispensable for regulating virulence of this fungal pathogen.
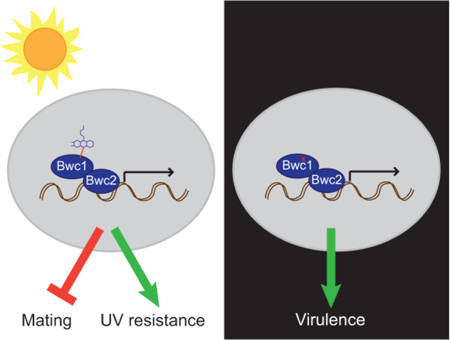
Since the WCC proteins possess domains for both signal input (LOV domain) and output (Zn finger DNA binding domain), it is possible for them to have both light dependent- and independent-regulatory roles. Fungi in which deletion of the genes encoding the WCC results in phenotypes when cultured in darkness include Trichoderma atroviride (Friedl et al., 2008; Sánchez-Arreguín et al., 2012), Trichoderma reesei (Castellanos et al., 2010; Gyalai-Korpos et al., 2010; Tisch and Schmoll, 2013), Aspergillus nidulans (Bayram et al., 2010; Purschwitz et al., 2008), Alternaria alternata (Pruß et al., 2014) and Fusarium fujikuroi (Castrillo and Avalos, 2015). This study therefore provides the genetic evidence to divide the role of Bwc1 into two aspects, one being light sensing to impact UV sensitivity and mating, and the other being light independent and impacting virulence (Figure 7).
Fig. 7.
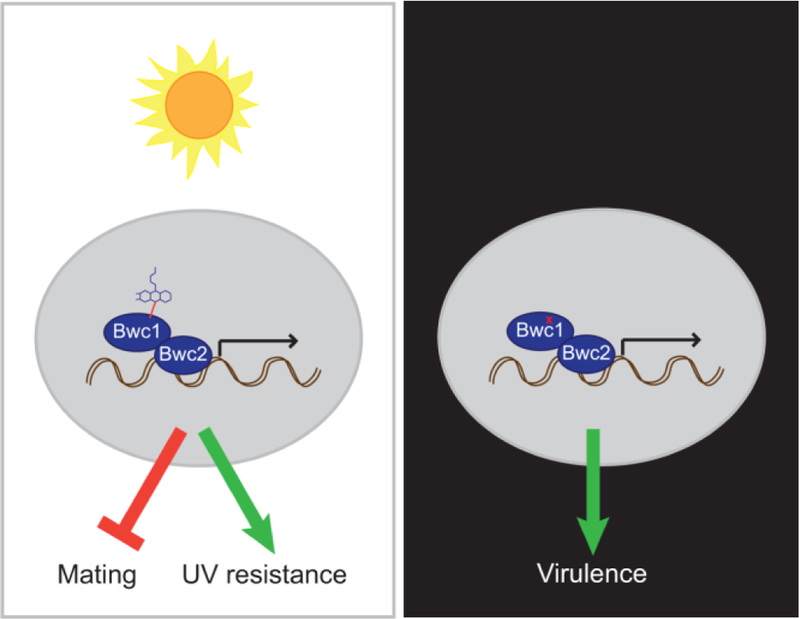
A model for how the Bwc1-Bwc2 complex impacts on C. neoformans biology. In the presence of light, blue wavelengths are absorbed by a flavin molecule, the signal transmitted to alter transcription, and mating inhibited while resistance to UV is increased. In darkness, or when the flavin-binding capabilities of Bwc1 are impaired, transcription still occurs to regulate genes that are required for virulence. Such virulence genes currently remain undiscovered.
Photosensory proteins with LOV domain are also found in the bacteria, although they use different signal transmission domains compared to the zinc finger DNA binding domains in fungal White Collar complexes (Herrou and Crosson, 2011; Losi et al., 2015). Of those bacteria characterized to date, animal pathogens in which light sensing components impact virulence are Brucella abortus (Swartz et al., 2007) and plant pathogens are Pseudomonas syringae and Xanthomonas citri (Kraiselburd et al., 2012; Moriconi et al., 2013). Experiments that generate specific alleles that alter these photosensors may further elucidate how LOV domain proteins contribute to microbial pathogenesis.
One additional consideration, as suggested by the ‘O’ in the abbreviation of the domain name LOV, is that proteins containing this domain may also have the ability to sense changes levels of reactive oxygen species (ROS). The bwc1 mutant has no change in sensitivity to ROS under in vitro conditions (Idnurm and Heitman, 2005); however, this does not preclude a possibility of sensing altered ROS levels within an animal during disease progression.
Future directions for research in C. neoformans would be to define which genes that are regulated by the White Collar complex contribute to virulence. A preliminary experiment used RNA-sequencing on the strains grown in constant darkness to reveal over 300 genes encoding hypothetical proteins that may be regulated by the complex, but because most of those genes have no established role in virulence in C. neoformans or other fungi it is currently unclear why loss of the genes causes the reduction in virulence. Identifying such genes that contribute to virulence could reveal unknown factors that affect the disease outcomes of this major human pathogenic fungus.
Supplementary Material
Highlights.
The blue light sensing complex is required for virulence of Cryptococcus neoformans
Mutation of a specific cysteine residue for transmission of the light signal blocks light responses
Strains with the cysteine substituted with other amino acid residues have wild type levels of virulence
The contribution of the White Collar 1 homolog to virulence is independent of light signaling
Acknowledgments
We thank Barbara Howlett for suggestions and edits on the manuscript. We are grateful for support from the Chinese Scholarship Council to P.Z. and National Institutes of Health NIAID (grant AI094364) and Australian Research Council (grant FT130100146) to A.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alspaugh JA, 2015. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol 78, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC, 2012. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev 36, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G, 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J 15, 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Bayram Ö, Braus GH, Fischer R, Rodriguez-Romero J, 2010. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol 47, 900–908. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Spudich JL, 2005. Handbook of Photosensory Receptors Wiley-VCH, Weinheim. [Google Scholar]
- Brown JCS, Nelson J, VanderSluis B, Deshpande R, Butts A, Kagan S, Polacheck I, Krysan DJ, Myers CL, Madhani HD, 2014. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell 159, 1168–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CC, Berliner MD, 1973. Virulence differences in mice of type A and B Histoplasma capsulatum yeasts grown in continuous light and total darkness. Infect. Immun 8, 677–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa P, Schumacher J, Hevia MA, Tudzynski P, Larrondo LF, 2013. Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the White Collar Complex. PLoS One 8, e84223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos F, Schmoll M, Martínez P, Tisch D, Kubicek CP, Herrera-Estrella A, Esquivel-Naranjo EU, 2010. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet. Biol 47, 468–476. [DOI] [PubMed] [Google Scholar]
- Castrillo M, Avalos J, 2015. The flavoproteins CryD and VvdA cooperate with the white collar protein WcoA in the control of photocarotenogenesis in Fusarium fujikuroi. PLoS One 10, e0119785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, Yang Y, Wang L, Liu Y, 2003. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA 100, 5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Swartz TE, Bogomolni RA, Briggs WR, 2002. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J 32, 205–219. [DOI] [PubMed] [Google Scholar]
- Collett MA, Garceau N, Dunlap JC, Loros JJ, 2002. Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics 160, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Moffat K, 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98, 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Moffat K, 2002. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Aguirre J, Herrera-Estrella A, Corrochano LM, 2016. The complexity of fungal vision. Microbiol. Spectrum 4, FUNK-0020–2016. [DOI] [PubMed] [Google Scholar]
- Friedl MA, Schmoll M, Kubicek CP, Druzhinina IS, 2008. Photostimulation of Hypocrea atroviridis growth occurs due to a cross-talk of carbon metabolism, blue light receptors and response to oxidative stress. Microbiology 154, 1229–1241. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC, 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297, 815–819. [DOI] [PubMed] [Google Scholar]
- Fuller KK, Dunlap JC, Loros JJ, 2016. Fungal light sensing at the bench and beyond. Adv. Genet 96, 1–51. [DOI] [PubMed] [Google Scholar]
- Fuller KK, Loros JJ, Dunlap JC, 2015. Fungal photobiology: visible light as a signal for stress, space and time. Curr. Genet 61, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyalai-Korpos M, Nagy G, Mareczky Z, Schuster A, Réczey K, Schmoll M, 2010. Relevance of the light signaling machinery for cellulase expression in trichoderma reesei (hypocrea jecorina). BMC Res. Notes 3, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T, 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol 78, 16–48. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y, 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297, 840–843. [DOI] [PubMed] [Google Scholar]
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, Eds., 2011. Cryptococcus: From Human Pathogen to Model Yeast. American Society for Microbiology Press, Washington, D.C. [Google Scholar]
- Herrou J, Crosson S, 2011. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol 9, 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Müller-Esparza H, Larrondo LF, 2015. A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc Natl Acad Sci USA 112, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J, 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3, 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J, 2010. Ferrochelatase is a conserved downstream target of the blue light-sensing white collar complex in fungi. Microbiology 156, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Verma S, Corrochano LM, 2010. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet. Biol 47, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Park S-Y, Chi M-H, Choi J, Park J, Rho H-S, Kim S, Goh J, Yoo S, Choi J, Park J-Y, Yi M, Yang S, Kwon M-J, Han S-S, Kim BR, Khang CH, Park B, Lim S-E, Jung K, Kong S, Karunakaran M, Oh H-S, Kim H, Kim S, Park J, Kang S, Choi W-B, Kang S, Lee Y-H, 2007. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet 39, 561–565. [DOI] [PubMed] [Google Scholar]
- Kim H, Ridenour JB, Dunkle LD, Bluhm BH, 2011a. Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog 7, e1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Singh P, Park J, Park S, Friedman A, Zheng T, Lee Y-H, Lee K, 2011b. Genetic and molecular characterization of a blue light photoreceptor MGWC-1 in Magnaporth oryzae. Fungal Genet. Biol 48, 400–407. [DOI] [PubMed] [Google Scholar]
- Kraiselburd I, Alet AI, Tondo ML, Petrocelli S, Daurelio LD, Monzón J, Ruiz OA, Losi A, Orellano EG, 2012. A LOV protein modulates the physiological attributes of Xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS One 7, e38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Singh P, Chung W-C, Ash J, Kim TS, Hang L, Park S, 2006. Light regulation of asexual development in the rice blast fungus, Magnaporthe grisea. Fungal Genet. Biol 43, 694–706. [DOI] [PubMed] [Google Scholar]
- Linden H, Macino G, 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J 16, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM, 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135, 174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He Q, Cheng P, 2003. Photoreception in Neurospora: a tale of two White Collar proteins. Cell Mol. Life Sci 60, 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Losi A, Mandalari C, Gärtner W, 2015. The evolution and functional role of flavin-based prokaryotic photoreceptors. Photochem. Photobiol 91, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Lu Y-K, Sun K-H, Shen W-C, 2005. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol. Microbiol 56, 280–291. [DOI] [PubMed] [Google Scholar]
- Moore TDE, Edman JC, 1993. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol 13, 1962–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriconi V, Sellaro R, Ayub N, Soto G, Rugnone M, Shah R, Pathak GP, Gärtner W, Casal JJ, 2013. LOV-domain photoreceptor, encoded in a genomic island, attenuates the virulence of Pseudomonas syringae in light-exposed Arabidopsis leaves. Plant J 76, 322–331. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A, 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun 73, 3842–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Pombejra S, Jamklang M, Uhrig JP, Vu K, Gelli A, 2018. The structure-function analysis of the Mpr1 metalloprotease determinants of activity during migration of fungal cells across the blood-brain barrier. PLoS One 13, e0203020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, Fraser JA, Heitman J, 2004. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell 15, 4476–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J, 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun 71, 4831–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AS, Braga GUL, Rangel DEN, 2018. Metarhizium robertsii illuminated during mycelial growth produces conidia with increased germination speed and virulence. Fungal Biol 122, 555–562. [DOI] [PubMed] [Google Scholar]
- Pruß S, Fetzner R, Seither K, Herr A, Pfeiffer E, Metzler M, Lawrence CB, Fischer R, 2014. Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl. Environ. Microbiol 80, 2582–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschwitz J, Müller S, Kastner C, Schöser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R, 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol 18, 255–259. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Romero J, Hedtke M, Kastner C, Müller S, Fischer R, 2010. Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol 64, 585–610. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MIG, 2008. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot. Cell 7, 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR, 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39, 9401–9410. [DOI] [PubMed] [Google Scholar]
- Sánchez-Arreguín A, Pérez-Martínez AS, Herrera-Estrella A, 2012. Proteomic analysis of Trichoderma atroviride reveals independent roles for transcription factors BLR-1 and BLR-2 in light and darkness. Eukaryot. Cell 11, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Simon A, Cohrs KC, Viaud M, Tudzynski P, 2014. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet 10, e1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz TE, Tseng T-S, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA, 2007. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317, 1090–1093. [DOI] [PubMed] [Google Scholar]
- Tisch D, Schmoll M, 2010. Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol 85, 1259–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch D, Schmoll M, 2013. Targets of light signalling in Trichoderma reesei. BMC Genomics 14, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR, 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol 175, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Idnurm A, 2013. Uve1 endonuclease is regulated by White collar to protect Cryptococcus neoformans from UV damage. PLoS Genet 9, e1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu K, Tham R, Uhrig JP, Thompson GR 3rd, Na Pombejra S, Jamklang M, Bautos JM, Gelli A, 2014. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. mBio 5, e01101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhou X, Loros JJ, Dunlap JC, 2015. Alternative use of DNA binding domains by the Neurospora White Collar Complex dictates circadian regulation and light responses. Mol. Cell. Biol 36, 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhai B, Lin X, Idnurm A, 2013. Congenic strains for genetic analysis of virulence traits in Cryptococcus gattii. Infect. Immun 81, 2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


