Abstract
Rho-associated protein kinases (ROCK) 1 and 2 are attractive drug targets for a range of neurologic disorders; however a critical barrier to ROCK-based therapeutics is ambiguity over whether there are isoform-specific roles for ROCKs in neuronal structural plasticity. Here, we used a genetics approach to address this long-standing question by analyzing both male and female adult ROCK1+/− and ROCK2+/− mice compared to littermate controls. Individual pyramidal neurons in the medial prefrontal cortex (mPFC) were targeted for iontophoretic microinjection of fluorescent dye, followed by high-resolution confocal microscopy and neuronal 3D reconstructions for morphometry analysis. Increased apical and basal dendritic length and intersections were observed in ROCK1+/− but not ROCK2+/− mice. Although dendritic spine densities were comparable among genotypes, apical spine length was decreased in ROCK1+/− but increased in ROCK2+/− mice. Spine head and neck diameter were reduced similarly in ROCK1+/− and ROCK2+/− mice; however certain spine morphologic subclasses were more affected than others in a genotype-dependent manner. Biochemical analyses of ROCK substrates in synaptic fractions revealed that phosphorylation of LIM kinase and cofilin were reduced in ROCK1+/− and ROCK2+/− mice, while phosphorylation of myosin light chain was decreased exclusively in ROCK1+/− mice. Collectively, these observations implicate ROCK1 as a novel regulatory factor of neuronal dendritic structure and detail distinct and complementary roles of ROCKs in mPFC dendritic spine structure.
Keywords: Rho kinase, LIM kinase, Dendritic Spines, Prefrontal Cortex, Dendritic Arbor, Morphometry, Synaptosomes
Introduction
Originally isolated as GTP-bound RhoA interacting proteins, the Rho-associated coiled-coil containing kinases (ROCK) are members of the AGO family of serine/threonine kinases and are extensively studied regulators of actin–myosin-mediated cytoskeleton contractility (Ishizaki et al. 1996; Leung et al. 1996; Leung et al. 1995; Matsui et al. 1996; Nakagawa et al. 1996). Two mammalian ROCK isoforms exist, ROCK1 and ROCK2, and share 65% similarity in their amino acid sequences and 92% identity in their kinase domains (Nakagawa et al. 1996). ROCK1 and ROCK2 expression patterns are largely similar in humans with higher transcript levels of ROCK1 in thymus and blood and ROCK2 in brain (Julian and Olson 2014). ROCKs phosphorylate a number of substrates predominantly tied to cellular morphology, adhesion, and motility. These actions implicate ROCK1 and ROCK2 as putative therapeutic targets for a variety of human conditions, such as cancer, asthma, insulin resistance, kidney failure, osteoporosis, and erectile dysfunction (Albersen et al. 2010; Komers et al. 2011; Lee et al. 2009; Olson 2008; Rath and Olson 2012; Schaafsma et al. 2008). Moreover, studies have identified pathogenic roles for ROCKs or explored the potential to repurpose ROCK inhibitors in neurologic disorders, including glaucoma, spinal cord injury, stroke, Alzheimer’s disease, Frontotemporal Dementia, Parkinson’s disease, and Amyotrophic Lateral Sclerosis (Challa and Arnold 2014; Duffy et al. 2009; Gentry et al. 2016; Gunther et al. 2017; Henderson et al. 2016; Herskowitz et al. 2013; Koch et al. 2014; Sellers et al. 2018; Shibuya et al. 2005; Tatenhorst et al. 2016).
Pharmacologic studies have driven much of our understanding of ROCKs in the brain, with Fasudil and Y-27632 being the most widely characterized inhibitors. However these and other current ROCK inhibitors are not isoform-specific and likely inhibit other kinases, including PKA and PKC, at higher doses used for in vivo experiments (Davies et al. 2000). Therefore, it is challenging to assign functions to ROCK1 or ROCK2 based on the kinase inhibitors, and this raises important questions about the contribution of ROCK1 or ROCK2 to the observed beneficial effects of pan-ROCK inhibitors. Although ROCKs share protein substrates, including myosin light chain (MLC), myosin light chain phosphatase, and LIM kinases (LIMK), evidence from genetic approaches in cell-based assays suggests distinct functions of ROCK isoforms (Amano et al. 1996; Kimura et al. 1996; Sumi et al. 2001). Older studies from homozygous knockout mice revealed major developmental problems or embryonic lethality in ROCK1−/− or ROCK2−/−, respectively; however, a different genetic background alleviated some of the effects from ROCK1 deletion (Shimizu et al. 2005; Thumkeo et al. 2003; Zhang et al. 2006). Working with ROCK1+/− or ROCK2−/− mouse embryonic fibroblasts, Shi et al. studied differential roles for ROCK1 and ROCK2 in regulating actin cytoskeleton reorganization after doxorubicin exposure. These findings suggested ROCK1 destabilizes actin via MLC phosphorylation whereas ROCK2 stabilizes actin through cofilin phosphorylation (Shi et al. 2013). Genetic exploration of ROCK1 and ROCK2 function in brain has been limited due to the complications of homozygous knockout mice on mixed backgrounds. Yet despite reports indicating that ROCK1+/− and ROCK2+/− mice develop normally, in vivo studies of the heterozygous models are rare (Duffy et al. 2009; Zhang et al. 2006). To this end, we independently generated new ROCK1+/− and ROCK2+/− mice on the C57BL/6N background to define ROCK isoform-specific functions related to dendritic structural plasticity. The findings herein provide novel distinct yet complimentary roles for ROCK1 and ROCK2 in the prefrontal cortex.
Materials and methods
Animals
All experimental procedures were performed under a protocol approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham (UAB). Generation of ROCK1+/− mice are described as follows: C57BL/6N-Rock1<tm1b(NCOM)Mfgc>/Tcp were made as part of the NorCOMM2 project with C57BL/6N-Rock1<tm1a(NCOM)Mfgc>/Tcp mice made from NorCOMM embryonic stem (ES) cells at the Toronto Centre for Phenogenomics (Bradley et al. 2012). C57BL/6N-Rock1<tm1b(NCOM)Mfgc>/Tcp mice were obtained from the Canadian Mouse Mutant Repository. ROCK2+/− mice were generated as follows: C57BL/6N-Rock2tm1a(KOMP)Wtsi mice were made from ES cells purchased from the International Mouse Phenotyping Consortium at the University of California, Davis. ES cell injections were performed by the UAB Transgenic & Genetically Engineered Models Core. The C57BL/6N-Rock2tm1a(KOMP)Wtsi ES cells used for this research project were generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). For more information or to obtain KOMP products go to www.komp.org or email service@komp.org. For all experiments, age-matched and sex-matched controls were used. All mice were 5.5-6 months old when sacrificed. When necessary, additional details on mouse sex are provided in figure legends.
Genotyping.
3-4mm tail clippings were taken from 21 day old mice. Tails were digested at 55°C overnight in Proteinase K Buffer (Fisher Scientific, BP1700-100). Samples were diluted 1:50 in nuclease free water for genotyping. The PCR reaction consisted of 12.5 μl Amplitaq Gold 360 Master Mix (Life Technologies, 4398886), 8.5 μl nuclease free water (Promega, P1199), 1 μl each of the forward and reverse primers (Integrated DNA Technologies), and 2 μl diluted DNA sample. Primers used for ROCK1+/− mice were: 5’- CCATCTACACCAACGTAACC -3’ and 5’ CATAACCACCACGCTCATC -3’. Primers used for ROCK2+/− mice were: 5’- GGGATCTCATGCTGGAGTTCTTCG -3’ and 5’- TAAGGTCCTAGGTTCAATCCCCAGC -3’.
Perfusions and tissue processing
PFA, vibratome, storage.
Animals were anesthetized with Fatal Plus (Vortech Pharmaceuticals, Catalog #0298-9373-68). The abdominal cavity and pericardium were carefully opened to expose the heart. Mice were transcardially perfused with cold 1% paraformaldehyde (PFA) for 1 min, followed by 10 min of 4% paraformaldehyde (Sigma Aldrich, Catalog #P6148) with 0.125% glutaraldehyde (Fisher Scientific, Catalog #BP2547). Peristaltic pump (Cole Parmer) was used for consistent administration of PFA. Immediately following perfusion, the mouse was decapitated, and the whole brain was removed and drop fixed in 4% PFA containing glutaraldehyde for 8-12 hours at 4°C. After post-fixation, the brains were sliced in 250 μm coronal increments using a Leica vibratome (VT1000S) with a speed of 70, and frequency of 7. The platform was filled with cold 0.1 M PB buffer, and the brain was glued (Loctite) perpendicular to the stage, cerebellum side down. All slices were stored one slice per well in a 48-well plate containing 0.1% sodium azide (Fisher, Catalog#BP922l) in 0.1M PB. Plates covered in parafilm are stored long term at 4°C. Notably, these procedures were performed according to (Dumitriu et al. 2011).
PBS, storage.
Animals were anesthetized with Fatal Plus. The abdominal cavity and pericardium were carefully opened to expose the heart. Mice were transcardially perfused with cold 1X PBS for 2 minutes. Immediately following perfusion, the brain was extracted and dissected into two hemispheres. Each hemisphere was immediately flash frozen in 2-methylbutane (Sigma, Catalog#320404), placed on dry ice, and transferred to the −80°C for storage.
Iontophoretic microinjection of fluorescent dye
Microinjections were executed using previously described methods (Dumitriu et al. 2011). We used a Nikon Eclipse FN1 upright microscope with a 10X objective and a 40X water objective placed on an air table. We used a tissue chamber assembled in the lab consisting of a 50×75 mm plastic base with a 60×10 mm petri dish epoxied to the base. A platinum wire is attached in a way that the ground wire can be connected to the bath by an alligator clip. The negative terminal of the electric current source was connected to the glass micropipette filled with 2 μl of 8% Lucifer yellow dye (ThermoFisher, Catalog#L453). Micropipettes (A-M Systems, Catalog #603500) with highly tapered tips were pulled fresh the day of use. The manual micromanipulator was secured on the air table with magnets that provided a 45° angle for injection. Using a brush, the brain slice was placed into a small petri dish containing 1X PBS and 4’,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature. After incubation in DAPI, the slice was placed on dental wax, and then a piece of filter paper was used to adhere the tissue. The filter paper was then transferred to the tissue chamber filled with 1X PBS and weighted down for stability. We used the 10X objective to advance the tip of the micropipette in XY and Z until the tip was just a few micrometers above the tissue. The 40X objective was then used to advance to tip into layers II and III of the prefrontal cortex. Once the microelectrode contacted a neuron, 2 nA of negative current was used for 5 min to fill the neuron with lucifer yellow. After the 5 min, the current was turned off and the micropipette was removed from the neuron. Neuron impalement within layers II and III occurs randomly in a blind manner. If the entire neuron does not fill with dye after penetration, the electrode is removed and the neuron is not used for analysis. Multiple neurons were injected in each hemisphere of the medial prefrontal cortex of each animal. After injection the filter paper containing the tissue was moved back into the chamber containing 1X PBS. Using a brush, the tissue was carefully lifted off the paper and placed on a glass slide with two 125 μm spacers (Electron Microscopy Sciences, Catalog #70327-20S). Excess PBS was carefully removed with a Kimwipe, and the tissue was air-dried for 1 min. One drop of Vectashield (Vector Labs, Catalog #H1000) was added directly to the slice; the coverslip (Warner, Catalog #64-0716) was added and sealed with nail polish. Injected tissue was stored at 4°C in the dark.
Confocal Microscopy
Confocal microscopy was used to capture images of pyramidal cells and dendrites from prefrontal cortex layers 2 and 3 and our methods were based on (Dumitriu et al. 2011). All imaging was performed by a blinded experimenter. Images were captured with a Leica LAS AF TCS MP5, Leica Microsystems 2.6.3, and either 20X or 63X oil immersion objective (Leica HCX PL Apo CS, N.A. 1.40). The following parameters were used for all images: Argon laser: 20% power; 458 Laser: 100% power; smart offset: 1.5%; smart gain: 800 V; Pinhole: 1 airy unit; image size: 1024 × 1024px. The experimenter focused on each dye-impregnated neuron and captured three-dimensional z-stacks of those meeting criteria. Criteria for imaging of neuronal arbors included: (1) located within 80 μm working distance of microscope, (2) fully impregnated with dye, (3) unobstructed by any artifact. For each neuron, z-stacks were captured with the following parameters: z-step: 0.503 μm; image size: 1024 × 1024 px (0.223 μm × 0.223 μm × 0.503 μm); zoom: 3×; line averaging: 2; acquisition rate: 700 Hz. Image stacks were collected in.lif format. The experimenter identified secondary dendrites from dye-impregnated neurons and captured three-dimensional z-stacks of those meeting criteria. Criteria for imaging neuronal dendrites included: (1) within 80 μm working distance of microscope; (2) relatively parallel with the surface of the coronal section; (3) no overlap with other branches; (4) minimum of 50 μm from the soma; (5) maximum of 110 μm from the soma. Secondary dendrites of pyramidal arbors were viewed and selected at low magnification. This segment was then viewed and imaged at 63X magnification. For each neuronal dendrite, z-stacks were captured with the following parameters: z-step: 0.1259 μm; image size: 1024 × 1024 px (0.0501 μm × 0.0501 μm × 0.0501 μm); zoom: 4.8x; line averaging: 4; acquisition rate: 400 Hz. Images were registered in ImageJ using linear stack alignment with Scale Invariant Feature Transform (SIFT) and recommended settings. Registered images were saved in .tif format. Captured images were deconvolved using Huygens Deconvolution System (16.05, Scientific Volume Imaging, the Netherlands) and the following settings: GMLE; maximum iterations: 10; signal to noise ratio: 15; quality: 0.003. Prior to deconvolution, imaging meta-data from original .lif files were extracted to Huygens template files in .hgsm format and attributed to each corresponding registered image. Deconvolved images were saved in .tif format.
Neuronal reconstructions
Neuronal arbor and dendritic spine reconstruction and analysis were performed with Neurolucida 360 (2.70.1, MBF Biosciences, Williston, Vermont). Dendritic spine analysis was performed as previously described with the noted adjustments (Boros et al. 2017). Briefly, image stacks of neuronal dendrites were imported to Neurolucida 360, and the full dendrite length was traced with semi-automatic directional kernel algorithm. The experimenter manually confirmed that all assigned points matched dendrite diameter and position in x, y, and z planes and adjusted each reconstruction if necessary. Dendritic spine reconstruction was performed automatically using a voxel-clustering algorithm and the following parameters: outer range: 5.0 μm; minimum height: 0.3 μm; detector sensitivity: 80%; minimum count: 8 voxels; backbone length. Next, the experimenter manually verified that the classifier correctly identified all protrusions. When necessary, the experimenter added any semi-automatically by increasing detector sensitivity. The morphology and backbone points of each spine were verified to ensure a representative spine shape, and merge and slice tools corrected inconsistencies. Each dendritic protrusion was automatically classified as a dendritic filopodium, thin spine, stubby spine, or mushroom spine based on morphological measurements using previously described parameters (Boros et al. 2017). Reconstructions were collected in Neurolucida Explorer (2.70.1, MBF Biosciences, Williston, Vermont) for branched structure analysis, and then exported to Microsoft Excel (Redmond, WA). Spine density was calculated as the number of spines per μm of dendrite length. To characterize neuronal arbors, image stacks were imported to Neurolucida 360. First, the soma was detected in three-dimensions semi-automatically using sensitivity at 40-50%. Neuronal arbors were automatically detected using the following parameters: seeding: medium density; smallest segment: 30 μm; sensitivity: 75-100%. Then, the experimenter closely examined the accuracy of each arbor reconstruction in each x, y, and z dimension. When necessary, the experimenter semi-automatically joined, separated, or extended segments using detach, connect, and remove tools. The experimenter ensured that the base of each arbor was as near to the soma as possible. All extensions from the soma were manually classified as apical dendrites, basal dendrites, or axons. Apical and basal arbor reconstructions were imported to Neurolucida Explorer and separately analyzed. Sholl analysis characterized the number of intersections or length using 10 μm concentric shells (radii), and these results were exported and collected in Microsoft Excel. The number of intersections and length between Sholl radii were calculated.
Biochemistry
Brain region subdissection.
Hemibrains were bathed in a petri dish of ice-cold PBS with protease (Sigma S8820) and phosphatase inhibitors (Thermo Scientific 1861277). Using a dissecting microscope, brain anatomical atlas, and surgical instruments, the medial prefrontal cortex was isolated from each hemibrain. All brain regions were stored at −80°C.
Synaptosomal preparations.
Synaptosomes were prepared using the following biochemical fractionation protocol as previously described (Hallett et al. 2008; Warmus et al. 2014). Sub-dissected tissue samples were bathed and homogenized for 30 seconds in TEVP buffer (10 mM Tris base, 5 mM NaF, 1 mM Na3VO4, 1 mM EDTA, 1 mM EDTA) with 320 mM sucrose and protease and phosphatase inhibitors. A small volume was saved as whole homogenate (WH). Remaining sample was centrifuged at 800 × g for 10 min at 4°C. The supernatant (S1) was removed, and the pellet (P1) was stored in TEVP + inhibitors. S1 was centrifuged at 9200 × g for 10 minutes at 4°C. The supernatant (S2) was removed and stored. The pellet (P2) was resuspended in TEVP + 32 mM sucrose + inhibitors and centrifuged at 25000 × g for 20 minutes at 4°C. The supernatant (LS1) was removed and stored. The pellet (synaptosome fraction) was resuspended in TEVP + inhibitors and stored. Protein concentration was determined for all samples by bicinchoninic acid method (Pierce), and Western blots were performed to quantify protein content according to our previously described methods (Herskowitz et al. 2011).
Western blotting.
Twenty-five micrograms of protein per sample were loaded into each well. Primary antibodies were incubated overnight at 4°C. Primary antibodies include: ROCK1 (Abcam, 45171), ROCK2 (Santa Cruz 5561), LIMK (Cell Signaling 3842S), Phospho-LIMK (Cell Signaling 3841), PAK1 (Cell Signaling 2602), Phospho-Myosin Light Chain (Cell Signaling 3671), Myosin Light Chain (Cell Signaling 3672), Phospho-Cofilin (Cell Signaling 3313), and Cofilin (Cell Signaling 3318). Secondary antibodies include: AlexaFluor 680 goat anti-rabbit (Life Technologies A21109) and Goat anti-mouse (Li-Cor 926-32210). Odyssey Image Station (Li-Cor) captured images, and Odyssey Application Software (3.0, Li-Cor) quantified band intensities.
Statistical analysis
The effect of ROCK1 or ROCK2 heterozygosity was determined by comparing neuronal structure or biochemistry with ROCK1 or ROCK2 homozygotes, respectively. Experimental power and sample sizes per condition were determined by referencing similar studies (Anderson et al. 2014; Radley et al. 2015). Pair-wise comparisons were separately performed for ROCK1+/+ and ROCK1+/− and for ROCK2+/+ and ROCK2+/−. All analyses were conducted with Prism 6.0 (GraphPad Software, La Jolla, CA). Significance level was set at P < 0.05, and data were reported as mean ± standard error of the mean. For spine density or biochemical comparisons, animal means were averaged to generate a genotype mean. Comparison of genotype means was conducted by two-tailed student’s t-test, unless non-normally distributed or otherwise indicated. Comparisons of spine densities and morphology distributions were performed as previously described (Boros et al. 2017). Spines of apical and basal dendrites were analyzed separately. Two-way analysis of variance (ANOVA) was used to compare densities of morphological classes between genotypes (factors: spine class and genotype). Morphological spine characteristics were compared using cumulative frequency distributions of individual spines from each genotype. Normality of distribution was assessed by D’Agostino & Pearson omnibus normality test. If non-normally distributed, a non-parametric two-sample Kolmogorov-Smirnov test compared whether genotypes segregated in each spine morphology distribution. Sholl analysis was performed in Neurolucida Explorer by drawing concentric shells at 10 μm intervals from the soma. Apical and basal arbors were analyzed separately. The arbor lengths and number of intersections for each individual neuron and radius were averaged to generate means for each genotype-radius pair. Two-way ANOVA (factors: genotype and radius from soma) compared arbor length or number of intersections between genotypes.
Results
ROCK1 reduction alters prefrontal dendritic morphology
To begin the investigation, we focused on the prefrontal cortex because it is a region tightly linked to cognitive performance, including working memory, and is highly vulnerable in several neurologic disorders that implicate ROCKs (Boxer et al. 2006; Gentry et al. 2016; Henderson et al. 2016; Herskowitz et al. 2013; Sellers et al. 2018; van Veluw et al. 2012; Wong et al. 2014). Individual layer 2 or 3 pyramidal neurons in the prelimbic medial prefrontal cortex (mPFC) were targeted for iontophoretic microinjection of the fluorescent dye Lucifer yellow, followed by high-resolution confocal laser scanning microscopy and neuronal 3D reconstructions for morphometry analysis (Fig. 1A, B). Examination of dendritic length in ROCK1+/+ and ROCK1+/− mice by sholl analysis revealed a main effect of genotype by two-way ANOVA on apical and basal dendrites (F(1, 426) = 10.89, P = 0.0010 and F(1, 370) = 7.083, P = 0.0081, respectively) (Fig. 1C, E). Furthermore, apical and basal dendrite intersections were increased in ROCK1+/− mice (F(1, 426) = 10.86, P = 0.0011 and F(1, 370) = 13.08, P = 0.0003, respectively) (Fig. 1D, F). Examination of apical and basal dendritic length or intersections in ROCK2+/+ and ROCK2+/− mice revealed no statistically significant main effects of genotype (Fig. 1, G-J). These data suggest that ROCK1 heterozygosity induces increased dendritic length and complexity at the apical and basal poles of pyramidal neurons in the mPFC. However, mPFC pyramidal neuron architecture at the dendrite level remains relatively normal with ROCK2 heterozygosity.
Figure 1.
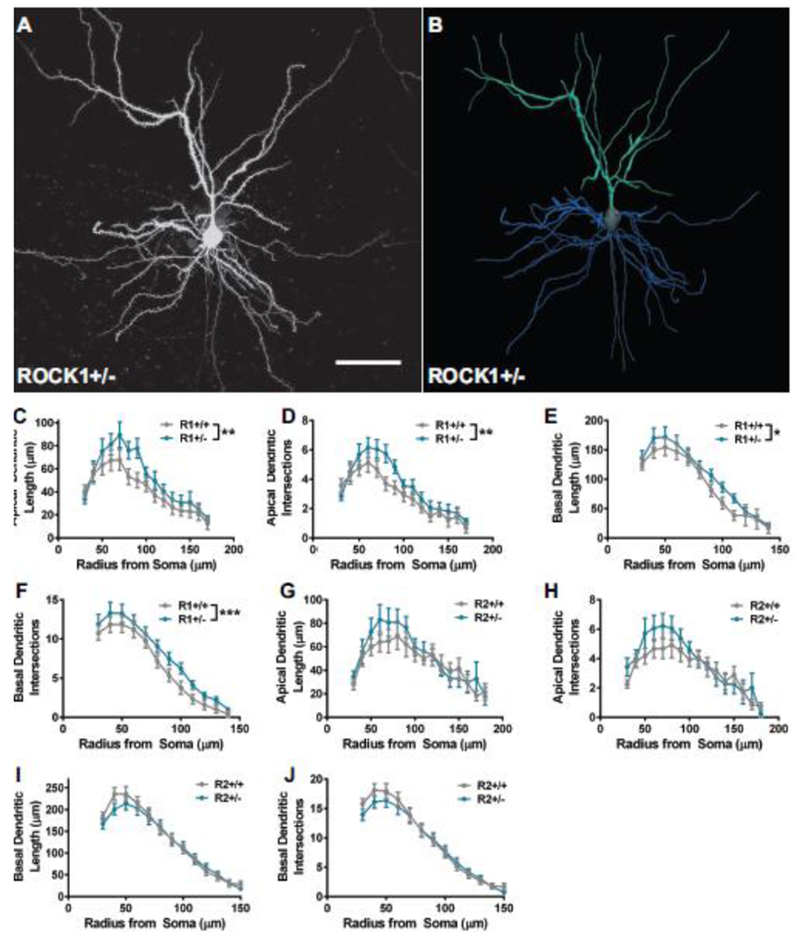
ROCK1 heterozygosity induces increased dendritic length and complexity. A. Representative maximum-intensity image of iontophoretically-filled layer 3 pyramidal neuron in the medial prefrontal cortex. Scale bar represents 50 μm. B. Corresponding three-dimensional digital reconstruction generated in Neurolucida360. Apical arbors are depicted in teal; basal arbors are shown in blue; and soma and axon are in grey. Sholl analysis with 10 μm shells was performed for apical and basal arbors from (C-F) ROCK1+/+ (N=17 neurons from 4 mice) and ROCK1+/− (N=19 neurons from 6 mice) or (G-J) ROCK2+/+ (N=23 neurons from 5 mice) and ROCK2+/− (N=25 neurons from 6 mice). Sholl curves for ROCK1+/+ and ROCK1+/− indicate that genotypes segregate based on C apical dendrite length (Two-way analysis of variance: **P=0.0010, F1,426 = 10.89), D apical dendrite intersections (Two-way ANOVA: **P=0.0011, F1, 426 = 10.86), E basal dendrite length (Two-way ANOVA: **P=0.0081, F1, 370 = 7.083), and F basal dendrite intersections (Two-way ANOVA: **P = 0.0003, F1, 370 = 13.08). Sholl curves for ROCK2+/+ and ROCK2+/− indicate that genotypes do not segregate based on G apical dendrite length, H apical dendrite intersections, I basal dendrite length, or J basal dendrite intersections. Lines represent the mean ± standard error of the mean. R1+/+, ROCK1+/+ ; R1+/−, ROCK1+/− ; R2+/+, ROCK2+/+ ; R2+/−, ROCK2+/−.
Effects of ROCK1 or ROCK2 heterozygosity on prefrontal dendritic spine morphology
In vitro and in vivo studies indicate that small molecule inhibitors of ROCKs can increase or decrease dendritic spine density, depending on the experimental paradigm (Hodges et al. 2011; Kang et al. 2009; Newell-Litwa et al. 2015). Our previous work demonstrated that pan-ROCK inhibition alters spine morphology in cultured hippocampal neurons (Swanger et al. 2015). However due to the lack of isoform-specificity with pan-ROCK inhibitors, the contribution of ROCK1 or ROCK2 to the observed structural phenotypes could not be determined. Thus, we addressed how genetic depletion of ROCK1 or ROCK2 may affect mPFC dendritic spine morphology. Architectural features of spines were delineated from high-resolution optical stacks of dendrites from the Lucifer yellow-filled pyramidal neurons that were analyzed in Figure 1 (Fig. 2).
Figure 2.
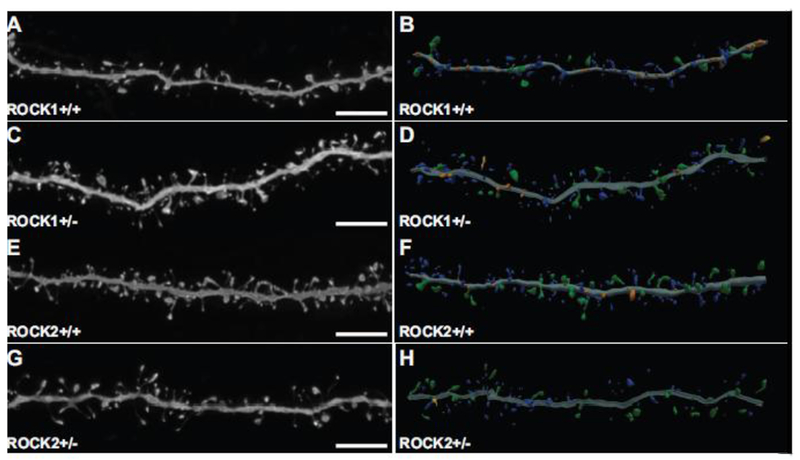
Representative high-resolution confocal microscope images and corresponding three-dimensional digital reconstruction models of dendrites. (A, C, E, G) Representative maximum-intensity confocal images of dye-filled dendrites after deconvolution. Scale bars represent 5 μm. (B, D, F, H) Three-dimensional digital reconstructions of dendrites depicted on the left. Colors correspond to dendritic protrusion classes: blue represents thin spines; orange, stubby spines; green, mushroom spines; and yellow, dendritic filopodia.
Spine morphology influences excitatory neurotransmission and synaptic plasticity, and spines can be classified on the basis of their three-dimensional structure as stubby, mushroom, or thin (Harris et al. 1992; Hayashi and Majewska 2005; Hering and Sheng 2001). Comparison of dendritic spine density on apical and basal dendrites among ROCK1+/+ and ROCK1+/− mice revealed no main effect of genotype by two-way ANOVA. This analysis included thin, stubby, and mushroom spine populations (Fig. 3A). To further analyze spine structure, the cumulative distributions of apical and basal spine length, head diameters, and neck diameters for ROCK1+/+ and ROCK1+/− mice were plotted. The cumulative frequency plots of apical spines indicated that the distribution of lengths was decreased in ROCK1+/− mice (Kolmogorov-Smirnov: D=0.06705, P<0.0001), whereas distributions of head diameter and neck diameter were similar to ROCK1+/+ (Fig. 3, B-D). The cumulative frequency plots of basal spines indicated that distribution of lengths was slightly decreased in ROCK1+/− mice (Kolmogorov-Smirnov: D=0.0465, P=0.0372) (Fig. 3E). Distribution of head diameter was reduced significantly in ROCK1+/− mice (Kolmogorov-Smirnov: D=0.1382, P<0.0001) (Fig. 3F), and this effect was largely driven by robust decreases in thin spine head diameters (Kolmogorov-Smirnov: D=0.1304, P<0.0001) (Fig. 3G). Neck diameter distribution was also decreased significantly in ROCK1+/− mice (Kolmogorov-Smirnov: D=0.0879, P<0.0001) (Fig. 3H), and this effect was driven by mushroom spine populations (Kolmogorov-Smirnov: D=0.2416, P<0.0001) (Fig. 3I).
Figure 3.
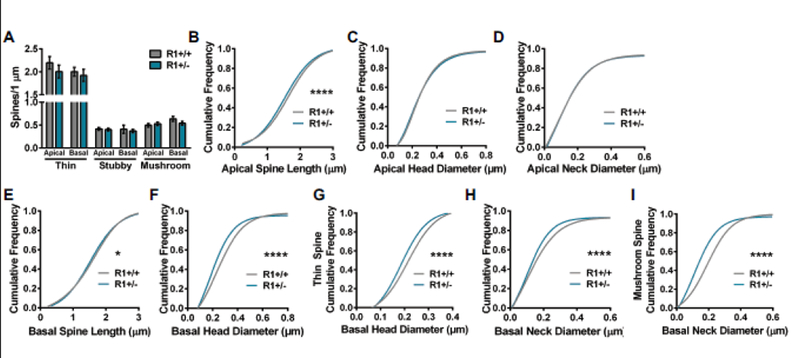
Comparison of apical and basal spine densities and morphology in ROCK1+/+ (N=36 dendrites from 5 mice) and ROCK1+/− (N=40 dendrites from 6 mice). A. Mean number of thin, stubby, or mushroom spines per 1 μm of apical or basal dendrite was similar between ROCK1+/+ and ROCK1+/−. Bar graphs plot the mean with standard error of the mean. (B-I) The cumulative frequency distributions for individual spines were plotted for each genotype and morphological parameter. Comparison of apical spine distribution plots indicate that ROCK1+/+ and ROCK1+/− segregate based on B apical spine length (Kolmogorov-Smirnov [KS]: D=0.0671, ****p<0.0001); but not C apical head diameter or D apical neck diameter. Comparison of basal spine distribution plots show that genotypes segregate based on E basal spine length (KS: D=0.0465, *P=0.0372); F basal head diameter (KS: D=0.1382, ****P<0.0001); G basal thin spine head diameter (KS: D=0.1304, ****p<0.0001); H basal neck diameter (KS: D=0.0879, ****P<0.0001); and I basal mushroom spine neck diameter (KS: D=0.2416, ****P<0.0001). R1+/+, ROCK1+/+ ; R1+/−, ROCK1+/−.
Examination of dendritic spine density, including thin, stubby, and mushroom spine populations, on apical and basal dendrites among ROCK2+/+ and ROCK2+/− mice revealed no main effect of genotype by two-way ANOVA (Fig. 4A). To analyze spine structure, the cumulative distributions of apical and basal spine lengths, head diameters, and neck diameters were plotted. The cumulative frequency plots of apical spines indicated that the distribution of lengths was increased in ROCK2+/− mice (Kolmogorov-Smirnov: D=0.0702, P<0.0001), predominantly driven by increased length of mushroom spines (Kolmogorov-Smirnov: D=0.1195, P=0.0063) (Fig. 4 B, C). Distributions of apical spine head and neck diameters were reduced significantly in ROCK2+/− mice (Kolmogorov-Smirnov: D=0.0610, P=0.0002; and Kolmogorov-Smirnov: D=0.0778, P<0.0001, respectively) (Fig. 4 D, E). Thin, stubby, and mushroom spine populations contributed approximately equivalently to the effects on head and neck diameter in ROCK2+/− mice. The cumulative frequency plots of basal spines indicated that the distribution of lengths was similar in ROCK2+/+ and ROCK2+/− mice, whereas head and neck diameters were reduced in ROCK2+/− mice (Kolmogorov-Smirnov: D=0.0567, P<0.0001; and Kolmogorov-Smirnov: D=0.0541, P=0.0002, respectively) (Fig. 4, F-H). Notably, thin spines drove the reduction in neck diameter (Kolmogorov-Smirnov: D=0.0653, P=0.0003), while all spine classes contributed to decreased head diameter in ROCK2+/− mice (Fig. 4I).
Figure 4.
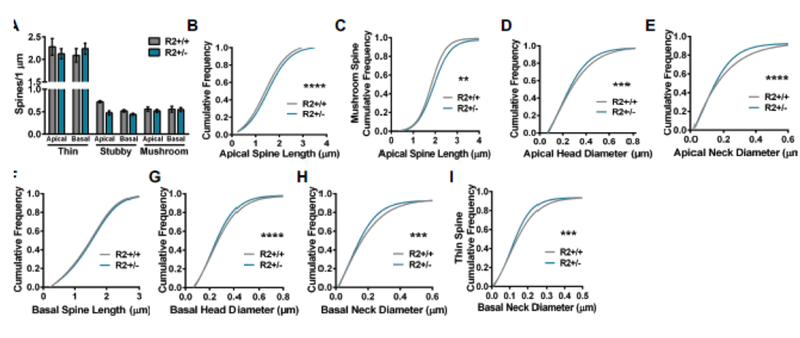
Comparison of apical and basal spine densities and morphology in ROCK2+/+ (N=32 dendrites from 4 mice) and ROCK2+/− (N=58 dendrites from 5 mice). A. Mean number of thin, stubby, or mushroom spines per 1 μm of apical or basal dendrite was similar between ROCK2+/+ and ROCK2+/−. Bar graphs plot the mean with standard error of the mean. (B-I) The cumulative frequency distributions for individual spines were plotted for each genotype and morphological parameter. Comparison of apical spine distribution plots indicate that ROCK2+/+ and ROCK2+/− segregate based on B apical spine length (Kolmogorov-Smirnov [KS]: D=0.0702, ****p<0.0001); C apical mushroom spine length (KS: D=0.1195, **P=0.0063); D apical head diameter (KS: D=0.0610, ***P=0.0002); and E apical neck diameter (KS: D=0.0778, ****P<0.0001). (F-I) Comparison of basal spine distribution plots show that genotypes segregate based on G basal head diameter (KS: D=0.0567, ****p<0.0001); H basal neck diameter (KS: D=0.0541, ***P=0.0002); and I basal thin spine neck diameter (KS: D=0.0653, ***P=0.0003); but not F basal spine length. R2+/+, ROCK2+/+; R2+/−, ROCK2+/−.
Collectively, these findings reveal that apical spine length is decreased in ROCK1+/− but increased in ROCK2+/− mice. While apical spine head and neck diameters were decreased in ROCK2+/− mice compared to ROCK2+/+, these phenotypes were equivalent in ROCK1+/+ and ROCK1+/− mice. Similar reductions in the distributions of head and neck diameter were observed in basal spine populations from ROCK1+/− or ROCK2+/− mice compared to ROCK1+/+ or ROCK2+/+, respectively. These data support the hypotheses that 1) ROCK1 and ROCK2 form a yin and yang to regulate spine length, and 2) both ROCKs influence spine head and neck diameter but ROCK2 may have a more important role in apical arbors.
Biochemical analyses of ROCK substrates in synaptic fractions from ROCK1+/− and ROCK2+/− mice
Because ROCK1+/− and ROCK2+/− mice displayed substantial alterations in mPFC dendritic spine morphology, we explored whether there were biochemical changes in key ROCK substrates at synapses in the prelimbic mPFC. Expectedly, levels of ROCK1 or ROCK2 were decreased approximately 50% in whole homogenates (WH) and synaptosome fractions in heterozygous mice compared to ROCK1+/+ and ROCK2+/+ (Fig. 5). Moreover, ROCK1 and ROCK2 protein levels were unchanged in ROCK2+/− and ROCK1+/−, respectively. Previous reports strongly suggest that ROCKs can regulate actin filaments in dendritic spines by phosphorylating LIMKs at threonine 505 and/or threonine 508 (pLIMK) (Bosch et al. 2014; Sumi et al. 2001). Levels of pLIMK were decreased significantly in synaptosome fractions in ROCK1+/− and ROCK2+/− mice compared to ROCK1+/+ and ROCK2+/+, respectively (P=0.0265, for ROCK1+/− and P=0.0253, for ROCK2+/−) (Fig. 5 B, C, E, F). In contrast, pLIMK levels were increased significantly in WH from ROCK1+/− and ROCK2+/− mice compared to ROCK1+/+ and ROCK2+/+, respectively (P=0.0355, for ROCK1+/− and P=0.0004, for ROCK2+/−) (Fig. 5 A, C, D, F). LIMK protein levels were similar among all genotypes in WH and synaptic fractions, indicating changes in pLIMK were not the result of LIMK fluctuations (Fig. 5 A, B, D, E). Currently, the only known substrate of LIMK is the actin depolymerizing factor cofilin (Arber et al. 1998; Yang et al. 1998). LIMK regulates actin polymerization by phosphorylating cofilin at serine 3 (pcofilin), thereby inactivating cofilin. Levels of pcofilin were decreased significantly in synaptosome fractions in ROCK1+/− and ROCK2+/− mice compared to ROCK1+/+ and ROCK2+/+, respectively (P=0.0008, for ROCK1+/− and P=0.0067, for ROCK2+/−) (Fig. 5 B, C, E, F). Whereas, pcofilin levels were increased significantly in WH from ROCK1+/− and ROCK2+/− mice compared to ROCK1+/+ and ROCK2+/+, respectively (P=0.0315, for ROCK1+/− and P=0.0136, for ROCK2+/−) (Fig. 5 A, C, D, F). Cofilin protein levels were similar among all genotypes in WH and synaptic fractions, indicating that changes in pcofilin were not the result of alterations in total cofilin (Fig. 5 A, B, D, E). Not surprisingly, these findings revealed that pcofilin levels followed upstream changes in pLIMK within WH and synaptosome fractions. This suggests that ROCK1 or ROCK2 heterozygosity leads to similar downstream alterations in LIMK signaling that likely contribute to the overlapping spine morphology phenotypes among ROCK1+/− and ROCK2+/− mice.
Figure 5.
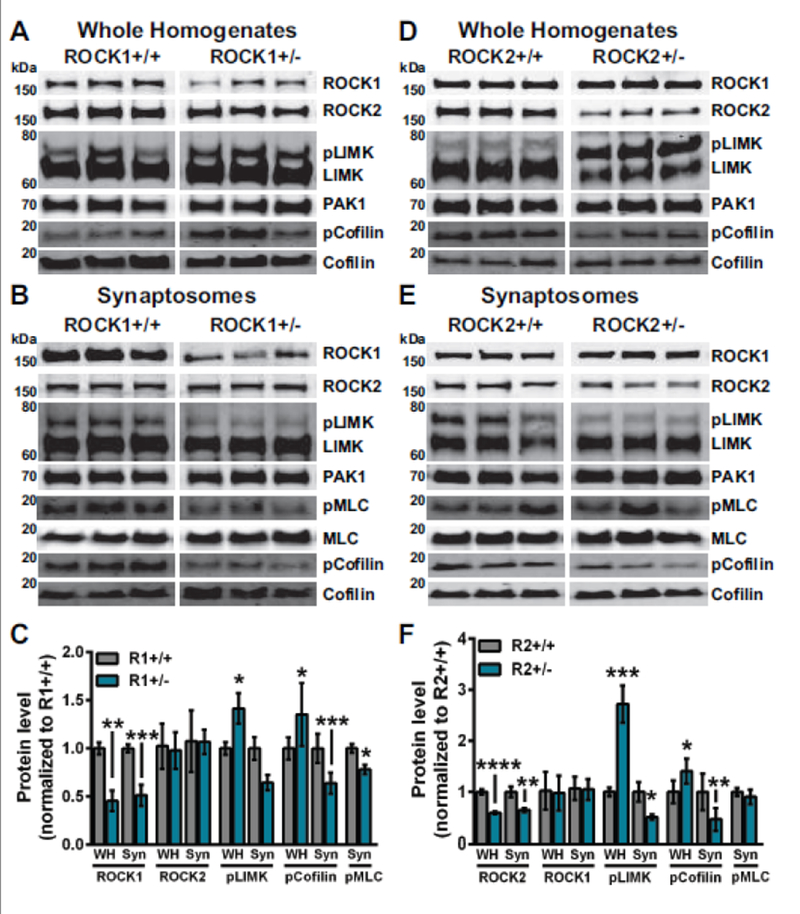
ROCK1 or ROCK2 heterozygosity reduces pLIMK in synaptosome fractions. N=5-10 mice per genotype. Representative Western blots from whole homogenate (WH) and synaptosome (Syn) fractions of (A-B) ROCK1+/+ and ROCK1+/− or (D-E) ROCK2+/+ and ROCK2+/− mice. For densitometry analysis protein levels of WH or Syn homogenates from ROCK1+/− or ROCK2+/− were normalized to ROCK1+/+ or ROCK2+/+, respectively. WH and Syn protein levels measured by densitometry were averaged to generate mean per genotype. C ROCK1+/− WH showed decreased ROCK1 (t-test: **P=0.0024, t=4.369, df=8), elevated pLIMK levels (t-test: *P=0.0355, t=2.429, df=10), and increased pCofilin levels (t-test: *P=0.0315, t=2.499, df=10). ROCK1+/− Syn fractions showed decreased ROCK1 (t-test: ***P=0.0003, t=5.099, df=11), decreased pLIMK (t-test: *P=0.0265, t=2.600, df=10), reduced pCofilin levels (t-test: ***P=0.0008, t=4.580, df= 11), and decreased pMLC (t-test: *P=0.0130, t=3.089, df=9). F ROCK2+/− WH showed decreased ROCK2 (t-test: ****P<0.0001, t=7.273, df=11), elevated pLIMK (t-test: ***P=0.0004, t=5.000, df=11), and increased pCofilin levels (t-test: *P=0.0136, t=2.988, df=10). ROCK2+/− Syn fractions showed decreased ROCK2 (t-test: **P=0.0068, t=3.261, df=12), decreased pLIMK (t-test: *P=0.0253, t=2.554, df=12), and reduced pCofilin levels (t-test: ***P=0.0067, t=3.334 df=11). Bar graphs plot the mean with standard error of the mean. WH, whole homogenate; Syn, synaptosome fractions; R1+/+, ROCK1+/+ ; R1+/−, ROCK1+/− ; R2+/+, ROCK2+/+ ; R2+/−, ROCK2+/−.
ROCKs can regulate actin-myosin contractility by phosphorylating MLC at serine 19 (pMLC), and MLC can play a critical role in dendritic spine morphology and synaptic function (Amano et al. 1996; Ryu et al. 2006). Levels of pMLC were decreased significantly in synaptosome fractions of ROCK1+/− mice compared to ROCK1+/+ (P=0.0130, for ROCK1+/−) but similar in ROCK2+/+ and ROCK2+/− mice (Fig. 5 B, C, E, F). MLC protein levels were similar among all genotypes in synaptic fractions, indicating changes in pMLC among ROCK1+/− mice were not the result of MLC reduction (Fig. 5 B, E). These results indicate that heterozygosity of either ROCK isoform decreases pLIMK in synapses, correlating to reductions in dendritic spine head and neck diameter in the mPFC (Figs. 3–4). Notably, changes in pMLC were selective to ROCK1+/− mice, suggesting a putative mechanism to explain spine length reduction in these animals.
Discussion
Pharmacologic inhibitor studies have provided much of what we know about ROCKs in the brain, and two pan-ROCK inhibitors, Fasudil and Ripasudil, have been used to treat human disease (Feng et al. 2015). Despite the enormous potential these compounds exhibit to modify human disease-progression, target-selectivity caveats have fogged our view of several fundamental, basic questions. Current ROCK-based therapeutics is moving rapidly toward the development of isoform-specific inhibitors, yet this promising avenue is hampered due to ambiguity over ROCK1- or ROCK2-specific functions in the brain (Feng et al. 2015; Julian and Olson 2014). Moreover, previous genetic attempts to explore ROCK1 and ROCK2 function were severely limited due to complications of knockout mice homozygosity on mixed genetic backgrounds. To address this, we generated new ROCK1+/− and ROCK2+/− mice on the C57BL/6N background. Through a combination of highly optimized three-dimensional modeling of dendritic structure and biochemistry, our study revealed distinct and complementary roles for ROCK1 and ROCK2 in prelimbic mPFC structural plasticity.
The findings herein are informative given the rising need for ROCK-isoform selective inhibitors based on new research indicating isoform-specific roles of ROCKs in several human diseases (Feng et al. 2015). For instance, ROCK2-selective inhibitors would be applicable to disorders that predominantly associate with ROCK2, rather than ROCK1, including neuroblastoma, multiple sclerosis, spinal cord injury, AD, and stroke (Chan et al. 2005; Chiba et al. 2010; Herskowitz et al. 2013; Hirose et al. 1998; Lapchak and Han 2010; Rikitake et al. 2005; Street et al. 2010; Sun et al. 2006). Moreover, blood pressure reduction is an effect of systemic pan-ROCK inhibitors and is hypothesized to be caused predominantly via ROCK1 inhibition, therefore ROCK2-selective drugs may have a better safety profile (Feng and LoGrasso 2014). Consideration for ROCK-isoform selectivity may also be important for therapeutic avenues that target upstream regulators of ROCKs. For example, ROCK1 can be cleaved by caspases which removes the auto-inhibitory domain on ROCK1, while ROCK2 is similarly activated by Granzyme B-mediated cleavage when cells undergo apoptosis (Coleman et al. 2001; Sebbagh et al. 2005; Sebbagh et al. 2001). Although different protease enzymes are involved, both mechanisms result in constitutively active kinases with isoform-specific consequences in neurons.
ROCK1, but not ROCK2, heterozygosity increased dendritic length and complexity of layer 2/3 pyramidal neurons in the mPFC. This implicates ROCK1 as a novel regulatory factor of neuronal dendritic structure. Mirroring our findings in ROCK1 heterozygotes, intracerebroventricular infusion of the pan-ROCK inhibitor Fasudil increased dendrite length in CA1 hippocampal neurons (Couch et al. 2010). Moreover, overexpression of a constitutively active version of ROCK (the isoform was not defined) reduced dendritic complexity of hippocampal neurons (Nakayama et al. 2000). Together, these results suggest that Fasudil inhibition of ROCK1 contributed to the arbor effects. However, other studies using Golgi-impregnation and sholl analysis indicated that homozygous deletion of ROCK2 on the CD-1 background increased dendrite length and intersections in hippocampal neurons (Duffy et al. 2009). We observed no statistically significant changes in dendritic arbors of ROCK2 heterozygous mice, suggesting that more robust depletion of ROCK2 may be necessary or that ROCK1 can compensate under experimental conditions of ROCK2 heterozygosity. From a translational standpoint, our findings highlight that certain beneficial effects of ROCK1 inhibition may be lost with a ROCK2-specific inhibitor.
Past studies in cultured hippocampal neurons provided a starting point for assessing ROCK1 and ROCK2 specific functions. RNAi depletion of ROCK1 increased spine length and decreased spine head diameter, whereas ROCK2 knockdown increased head diameter (Newell-Litwa et al. 2015). The findings herein indicated that ROCK1 heterozygosity reduced spine length but similarly decreased head diameter, while ROCK2 heterozygosity decreased spine head diameter. Moreover, our previous in vitro studies revealed that treatment with Y-27632 increased spine density as well as spine length in hippocampal neurons (Swanger et al. 2015). These contrasting effects could represent in vitro versus in vivo conditions or differences in cell-type (hippocampal versus mPFC pyramidal neurons). The similar global reductions in spine head and neck diameter that were observed in ROCK1+/− and ROCK2+/− mice suggest that decreased pLIMK in synaptosome fractions may explain the changes in spine head and neck morphology. Similarly, older studies demonstrated that genetic deletion of LIMK reduced spine head area in hippocampal neurons (Meng et al. 2002). The observed increase in WH pLIMK is more challenging to explain. It could reflect a dysregulation of alternative Rho-GTPase signaling, such as PAK1-mediated phosphorylation of LIMK (Edwards et al. 1999), under conditions of ROCK1 or ROCK2 heterozygosity, but we did not observe changes in PAK1 among genotypes or biochemical fractions (Fig. 5 A, B, D, E). Alternatively, increased WH pLIMK may be explained by compensatory phosphorylation by ROCK1 or ROCK2 when the other isoform is depleted. That ROCK1 and ROCK2 appear as a yin and yang to regulate spine length is a fascinating observation and correlates with previous cultured neuron experiments and pan-ROCK inhibitors, demonstrating either increased or decreased spine length (Kang et al. 2009; Newell-Litwa et al. 2015; Swanger et al. 2015) (Figs. 3–4). It is possible that pan-ROCK inhibition could reflect ROCK-isoform specific effects depending on the cell-type or balance of ROCK1 and ROCK2 expression in the cells.
Concomitant alternations in dendritic spine length and head diameter among mPFC layer 2 and 3 apical and basal spine populations in ROCK1+/− and ROCK2+/− mice may reflect more rapid plasticity to regulate information processing (Grutzendler et al. 2002). Layer 2 and 3 neurons mediate granular cortico-cortico connections in the rodent mPFC; however interpreting the differential effects of ROCKs on spines among apical and basal dendrites is challenging (Douglas and Martin 2004; Douglas and Martin 2007). Functional distinctions between apical and basal dendrites in this region are poorly understood. Optogenetic experiments revealed that layer 2 prelimbic pyramidal neurons received functional inputs from the ventral hippocampus, basolateral amygdala (BLA), midline thalamic nucleus, and contralateral mPFC. Synapses from these inputs occurred at various dendritic locations but displayed bias toward spines with larger volumes (Little and Carter 2012). Moreover, photostimulation of BLA or contralateral mPFC neurons projecting to layer 2 or 3 pyramidal neurons indicated that projections from the BLA exhibited the strongest synaptic connections and correlated with increased spine density. Inputs from the BLA targeted spines near the soma which elicited stronger excitatory postsynaptic potentials than spines on distal dendrites (Little and Carter 2013). Notably, this interconnectivity of BLA and mPFC dendrites in layers II and III may be critical for behavioral response to emotional stimuli (Euston et al. 2012). These findings provide some circuit level context and implications for the role of ROCKs in the mPFC.
Past live imaging studies revealed that dendritic spines can change shape and size over timescales of seconds to minutes and hours to days (Parnass et al. 2000). Spine morphology is inextricably linked to spine function, and spines can be generally classified on the basis of their three-dimensional morphology as stubby, mushroom, or thin (Chang and Greenough 1984; Harris et al. 1992; Hering and Sheng 2001; Peters and Kaiserman-Abramof 1970). Stubby spines are predicted to be transitional that may enlarge, possibly to mushroom spines, which are more stable structures with a wide head and thin neck. Thin spines are more dynamic, and lack the wide, stable head indicative of mushroom spines. Spine structure strongly affects molecular diffusion with spine neck as the predominant mediator of compartmentalization, facilitating efficient regulation of synaptic biochemical and electrical components (Tonnesen et al. 2014). Enlargement of the spine head follows induction of long-term potentiation (LTP) (Harvey and Svoboda 2007; Lang et al. 2004; Matsuzaki et al. 2004; Tanaka et al. 2008), and time-lapse STED imaging experiments revealed that spine necks are highly plastic in models of LTP (Tonnesen et al. 2014). This indicated that morphological alterations in spine necks and heads were coordinated. While spine head enlargement increased biochemical compartmentalization, neck changes preserved diffusion recovery after LTP. Additionally, structural changes in the neck may facilitate movement of molecules or secondary chemical messengers from the dendrite into the spine (Fonseca et al. 2004). Head diameter reduction in all spine types among ROCK2+/− mice suggests that ROCK2 is a critical mediator of spine head diameter regardless of the structure/function state of the spine (Fig. 4), whereas ROCK1’s influence on head diameter may be more important when dendritic protrusions adopted a thin spine morphology (Fig. 3G). Reductions in basal mushroom or thin spine neck diameter among ROCK1+/− or ROCK2+/− mice, respectively, may reflect unique roles of individual ROCK isoforms during different phases of LTP in the prefrontal cortex. By modulating both head and neck diameter, it is clear that the ROCKs constitute a tightly regulated system with critical and related cell biological activities in dendritic spines.
Our investigation represents an important starting point in tackling the question of whether ROCK isoforms have specific neuroanatomical functions; however a caveat to this approach is that developmental compensation for the loss of one ROCK isoform may have contributed to observed phenotypes. Future studies of conditional ROCK1 and ROCK2 knockout mice will determine whether ROCK isoforms exhibit cell-type specific roles in brain or time-dependent functions, such as in aging or development. The results herein provide clues to the translational potential of future ROCK-isoform specific inhibitors. For instance, recent studies on patients that are resilient to AD indicate that cumulative elongation of dendritic spines in the prefrontal cortex may be critical to maintain cognitive function in the presence of AD pathology (Boros et al. 2017). ROCK2 reduction increased whereas ROCK1 depletion decreased apical spine length in the mPFC (Figs. 3–4), highlighting ROCK2, but not ROCK1, as a therapeutic target for AD. Studies using chronic restraint stress paradigms in rats revealed that aging and stress can substantially reduce dendritic arbor length and complexity in the mPFC, implying adverse experience over the lifespan may interact with cognitive aging (Bloss et al. 2011; Radley et al. 2005; Radley et al. 2004). Under these circumstances, it may be more beneficial to target ROCK1 with pan-ROCK inhibitors to curb simplification and shortening of dendritic arbors (Fig. 1). Collectively, our findings highlight distinct and complementary roles for ROCK1 and ROCK2 in prefrontal cortex dendritic structure and provide an initial study of new ROCK models on the C57BL/6N background.
Acknowledgements:
This work was supported by the National Institutes of Health through the National Institute on Aging AG054719 to J.H.H. and NIA AG043552-05 to J.H.H. Additional support stemmed from a New Investigator Research Grant 2015-NIRG-339422 to J.H.H. from the Alzheimer’s Association.
Funding: This study was funded by the National Institute on Aging (grant numbers AG054719 and AG043552-05). This study was funded by the Alzheimer’s Association (grant number NIRG-339422)
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Compliance with ethical standards
Ethical approval: All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Informed consent: Not applicable.
References
- Albersen M, Shindel AW, Mwamukonda KB, Lue TF (2010) The future is today: emerging drugs for the treatment of erectile dysfunction Expert Opin Emerg Drugs 15:467–480 doi:10.1517/14728214.2010.480973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M et al. (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem 271:20246–20249 [DOI] [PubMed] [Google Scholar]
- Anderson RM, Birnie AK, Koblesky NK, Romig-Martin SA, Radley JJ (2014) Adrenocortical status predicts the degree of age-related deficits in prefrontal structural plasticity and working memory J Neurosci 34:8387–8397 doi:10.1523/JNEUROSCI.1385-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase Nature 393:805–809 doi:10.1038/31729 [DOI] [PubMed] [Google Scholar]
- Bloss EB et al. (2011) Evidence for reduced experience-dependent dendritic spine plasticity in the aging prefrontal cortex J Neurosci 31:7831–7839 doi:10.1523/JNEUROSCI.0839-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros BD, Greathouse KM, Gentry EG, Curtis KA, Birchall EL, Gearing M, Herskowitz JH (2017) Dendritic spines provide cognitive resilience against Alzheimer’s disease Ann Neurol 82:602–614 doi:10.1002/ana.25049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y (2014) Structural and molecular remodeling of dendritic spine substructures during long-term potentiation Neuron 82:444–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer AL et al. (2006) Patterns of brain atrophy that differentiate corticobasal degeneration syndrome from progressive supranuclear palsy Archives of neurology 63:81–86 doi:10.1001/archneur.63.1.81 [DOI] [PubMed] [Google Scholar]
- Bradley A et al. (2012) The mammalian gene function resource: the International Knockout Mouse Consortium Mamm Genome 23:580–586 doi:10.1007/s00335-012-9422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa P, Arnold JJ (2014) Rho-kinase inhibitors offer a new approach in the treatment of glaucoma Expert Opin Investig Drugs 23:81–95 doi:10.1517/13543784.2013.840288 [DOI] [PubMed] [Google Scholar]
- Chan CC, Khodarahmi K, Liu J, Sutherland D, Oschipok LW, Steeves JD, Tetzlaff W (2005) Dose-dependent beneficial and detrimental effects of ROCK inhibitor Y27632 on axonal sprouting and functional recovery after rat spinal cord injury Experimental neurology 196:352–364 doi:10.1016/j.expneurol.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Chang FL, Greenough WT (1984) Transient and enduring morphological correlates of synaptic activity and efficacy change in the rat hippocampal slice Brain Res 309:35–46 [DOI] [PubMed] [Google Scholar]
- Chiba Y et al. (2010) Synergistic effects of bone marrow stromal cells and a Rho kinase (ROCK) inhibitor, fasudil on axon regeneration in rat spinal cord injury Neuropathology : official journal of the Japanese Society of Neuropathology 30:241–250 doi:10.1111/j.1440-1789.2009.01077.x [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF (2001) Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I Nat Cell Biol 3:339–345 doi:10.1038/35070009 [DOI] [PubMed] [Google Scholar]
- Couch BA, DeMarco GJ, Gourley SL, Koleske AJ (2010) Increased dendrite branching in AbetaPP/PS1 mice and elongation of dendrite arbors by fasudil administration J Alzheimers Dis 20:1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors Biochem J 351:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA (2004) Neuronal circuits of the neocortex Annual review of neuroscience 27:419–451 doi:10.1146/annurev.neuro.27.070203.144152 [DOI] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA (2007) Recurrent neuronal circuits in the neocortex Current biology : CB 17:R496–500 doi:10.1016/j.cub.2007.04.024 [DOI] [PubMed] [Google Scholar]
- Duffy P, Schmandke A, Sigworth J, Narumiya S, Cafferty WB, Strittmatter SM (2009) Rho-associated kinase II (ROCKII) limits axonal growth after trauma within the adult mouse spinal cord J Neurosci 29:15266–15276 doi:29/48/15266 [pii] 10.1523/JNEUROSCI.4650-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu D, Rodriguez A, Morrison JH (2011) High-throughput, detailed, cell-specific neuroanatomy of dendritic spines using microinjection and confocal microscopy Nat Protoc 6:1391–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN (1999) Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics Nat Cell Biol 1:253–259 doi:10.1038/12963 [DOI] [PubMed] [Google Scholar]
- Euston DR, Gruber AJ, McNaughton BL (2012) The role of medial prefrontal cortex in memory and decision making Neuron 76:1057–1070 doi:10.1016/j.neuron.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, LoGrasso PV (2014) Rho kinase inhibitors: a patent review (2012 - 2013) Expert opinion on therapeutic patents 24:295–307 doi:10.1517/13543776.2014.863279 [DOI] [PubMed] [Google Scholar]
- Feng Y, LoGrasso PV, Defert O, Li R (2015) Rho Kinase (ROCK) Inhibitors and Their Therapeutic Potential J Med Chem [DOI] [PubMed] [Google Scholar]
- Fonseca R, Nagerl UV, Morris RG, Bonhoeffer T (2004) Competing for memory: hippocampal LTP under regimes of reduced protein synthesis Neuron 44:1011–1020 doi:10.1016/j.neuron.2004.10.033 [DOI] [PubMed] [Google Scholar]
- Gentry EG, Henderson BW, Arrant AE, Gearing M, Feng Y, Riddle NC, Herskowitz JH (2016) Rho Kinase Inhibition as a Therapeutic for Progressive Supranuclear Palsy and Corticobasal Degeneration J Neurosci 36:1316–1323 doi:10.1523/JNEUROSCI.2336-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB (2002) Long-term dendritic spine stability in the adult cortex Nature 420:812–816 [DOI] [PubMed] [Google Scholar]
- Gunther R et al. (2017) Rho Kinase Inhibition with Fasudil in the SOD1(G93A) Mouse Model of Amyotrophic Lateral Sclerosis-Symptomatic Treatment Potential after Disease Onset Front Pharmacol 8:17 doi:10.3389/fphar.2017.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett PJ, Collins TL, Standaert DG, Dunah AW (2008) Biochemical fractionation of brain tissue for studies of receptor distribution and trafficking Curr Protoc Neurosci Chapter 1:Unit 1 16 doi:10.1002/0471142301.ns0116s42 [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B (1992) Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation J Neurosci 12:2685–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Svoboda K (2007) Locally dynamic synaptic learning rules in pyramidal neuron dendrites Nature 450:1195–1200 doi:10.1038/nature06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Majewska AK (2005) Dendritic spine geometry: functional implication and regulation Neuron 46:529–532 [DOI] [PubMed] [Google Scholar]
- Henderson BW, Gentry EG, Rush T, Troncoso JC, Thambisetty M, Montine TJ, Herskowitz JH (2016) Rho-associated protein kinase 1 (ROCK1) is increased in Alzheimer’s disease and ROCK1 depletion reduces amyloid-beta levels in brain J Neurochem 138:525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M (2001) Dendritic spines: structure, dynamics and regulation Nat Rev Neurosci 2:880–888 doi:10.1038/35104061 35104061 [pii] [DOI] [PubMed] [Google Scholar]
- Herskowitz JH et al. (2013) Pharmacologic inhibition of ROCK2 suppresses amyloid-beta production in an Alzheimer’s disease mouse model J Neurosci 33:19086–19098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz JH, Seyfried NT, Gearing M, Kahn RA, Peng J, Levey AI, Lah JJ (2011) Rho kinase II phosphorylation of the lipoprotein receptor LR11/SORLA alters amyloid-beta production J Biol Chem 286:6117–6127 doi:M110.167239 [pii] 10.1074/jbc.M110.167239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose M et al. (1998) Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells J Cell Biol 141:1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges JL, Newell-Litwa K, Asmussen H, Vicente-Manzanares M, Horwitz AR (2011) Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology PloS one 6:e24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T et al. (1996) The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase EMBO J 15:1885–1893 [PMC free article] [PubMed] [Google Scholar]
- Julian L, Olson MF (2014) Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions Small GTPases 5:e29846 doi:10.4161/sgtp.29846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MG, Guo Y, Huganir RL (2009) AMPA receptor and GEF-H1/Lfc complex regulates dendritic spine development through RhoA signaling cascade Proceedings of the National Academy of Sciences of the United States of America 106:3549–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K et al. (1996) Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science 273:245–248 [DOI] [PubMed] [Google Scholar]
- Koch JC, Tonges L, Barski E, Michel U, Bahr M, Lingor P (2014) ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS Cell Death Dis 5:e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komers R, Oyama TT, Beard DR, Tikellis C, Xu B, Lotspeich DF, Anderson S (2011) Rho kinase inhibition protects kidneys from diabetic nephropathy without reducing blood pressure Kidney Int 79:432–442 doi:10.1038/ki.2010.428 [DOI] [PubMed] [Google Scholar]
- Lang C, Barco A, Zablow L, Kandel ER, Siegelbaum SA, Zakharenko SS (2004) Transient expansion of synaptically connected dendritic spines upon induction of hippocampal long-term potentiation Proceedings of the National Academy of Sciences of the United States of America 101:16665–16670 doi:10.1073/pnas.0407581101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapchak PA, Han MK (2010) Simvastatin improves clinical scores in a rabbit multiple infarct ischemic stroke model: synergism with a ROCK inhibitor but not the thrombolytic tissue plasminogen activator Brain Res 1344:217–225 doi:10.1016/j.brainres.2010.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH et al. (2009) Targeted disruption of ROCK1 causes insulin resistance in vivo J Biol Chem 284:11776–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Chen XQ, Manser E, Lim L (1996) The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton Mol Cell Biol 16:5313–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Manser E, Tan L, Lim L (1995) A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes J Biol Chem 270:29051–29054 [DOI] [PubMed] [Google Scholar]
- Little JP, Carter AG (2012) Subcellular synaptic connectivity of layer 2 pyramidal neurons in the medial prefrontal cortex J Neurosci 32:12808–12819 doi:10.1523/JNEUROSCI.1616-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JP, Carter AG (2013) Synaptic mechanisms underlying strong reciprocal connectivity between the medial prefrontal cortex and basolateral amygdala J Neurosci 33:15333–15342 doi:10.1523/JNEUROSCI.2385-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T et al. (1996) Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho EMBO J 15:2208–2216 [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H (2004) Structural basis of long-term potentiation in single dendritic spines Nature 429:761–766 doi:10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y et al. (2002) Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice Neuron 35:121–133 [DOI] [PubMed] [Google Scholar]
- Nakagawa O, Fujisawa K, Ishizaki T, Saito Y, Nakao K, Narumiya S (1996) ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice FEBS Lett 392:189–193 [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L (2000) Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons J Neurosci 20:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa KA, Badoual M, Asmussen H, Patel H, Whitmore L, Horwitz AR (2015) ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity J Cell Biol 210:225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MF (2008) Applications for ROCK kinase inhibition Curr Opin Cell Biol 20:242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnass Z, Tashiro A, Yuste R (2000) Analysis of spine morphological plasticity in developing hippocampal pyramidal neurons Hippocampus 10:561–568 [DOI] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abramof IR (1970) The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines Am J Anat 127:321–355 [DOI] [PubMed] [Google Scholar]
- Radley JJ, Anderson RM, Cosme CV, Glanz RM, Miller MC, Romig-Martin SA, LaLumiere RT (2015) The Contingency of Cocaine Administration Accounts for Structural and Functional Medial Prefrontal Deficits and Increased Adrenocortical Activation J Neurosci 35:11897–11910 doi:10.1523/JNEUROSCI.4961-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Rocher AB, Janssen WG, Hof PR, McEwen BS, Morrison JH (2005) Reversibility of apical dendritic retraction in the rat medial prefrontal cortex following repeated stress Experimental neurology 196:199–203 doi:10.1016/j.expneurol.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Radley JJ et al. (2004) Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex Neuroscience 125:1–6 doi:10.1016/j.neuroscience.2004.01.006 [DOI] [PubMed] [Google Scholar]
- Rath N, Olson MF (2012) Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy EMBO Rep 13:900–908 doi:10.1038/embor.2012.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikitake Y et al. (2005) Inhibition of Rho kinase (ROCK) leads to increased cerebral blood flow and stroke protection Stroke 36:2251–2257 doi:10.1161/01.STR.0000181077.84981.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J et al. (2006) A critical role for myosin IIb in dendritic spine morphology and synaptic function Neuron 49:175–182 doi:10.1016/j.neuron.2005.12.017 [DOI] [PubMed] [Google Scholar]
- Schaafsma D, Gosens R, Zaagsma J, Halayko AJ, Meurs H (2008) Rho kinase inhibitors: a novel therapeutical intervention in asthma? Eur J Pharmacol 585:398–406 doi:10.1016/j.ejphar.2008.01.056 [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Hamelin J, Bertoglio J, Solary E, Breard J (2005) Direct cleavage of ROCK II by granzyme B induces target cell membrane blebbing in a caspase-independent manner The Journal of experimental medicine 201:465–471 doi:10.1084/jem.20031877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J (2001) Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing Nat Cell Biol 3:346–352 doi:10.1038/35070019 [DOI] [PubMed] [Google Scholar]
- Sellers KJ et al. (2018) Amyloid beta synaptotoxicity is Wnt-PCP dependent and blocked by fasudil Alzheimer’s & dementia : the journal of the Alzheimer’s Association 14:306–317 doi:10.1016/j.jalz.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J et al. (2013) Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment Cell Death Dis 4:e483 doi:10.1038/cddis.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M, Hirai S, Seto M, Satoh S, Ohtomo E (2005) Effects of fasudil in acute ischemic stroke: results of a prospective placebo-controlled double-blind trial Journal of the neurological sciences 238:31–39 doi:S0022-510X(05)00221-2 [pii] 10.1016/j.jns.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Shimizu Y et al. (2005) ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles J Cell Biol 168:941–953 doi:jcb.200411179 [pii] 10.1083/jcb.200411179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street CA et al. (2010) Pharmacological inhibition of Rho-kinase (ROCK) signaling enhances cisplatin resistance in neuroblastoma cells International journal of oncology 37:1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Matsumoto K, Nakamura T (2001) Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase J Biol Chem 276:670–676 doi:10.1074/jbc.M007074200 [DOI] [PubMed] [Google Scholar]
- Sun X et al. (2006) The selective Rho-kinase inhibitor Fasudil is protective and therapeutic in experimental autoimmune encephalomyelitis Journal of neuroimmunology 180:126–134 doi:10.1016/j.jneuroim.2006.06.027 [DOI] [PubMed] [Google Scholar]
- Swanger SA, Mattheyses AL, Gentry EG, Herskowitz JH (2015) ROCK1 and ROCK2 inhibition alters dendritic spine morphology in hippocampal neurons Cell Logist 5:e1133266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H (2008) Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines Science 319:1683–1687 doi:10.1126/science.1152864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatenhorst L et al. (2016) Fasudil attenuates aggregation of alpha-synuclein in models of Parkinson’s disease Acta Neuropathol Commun 4:39 doi:10.1186/s40478-016-0310-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumkeo D et al. (2003) Targeted disruption of the mouse rho-associated kinase 2 gene results in intrauterine growth retardation and fetal death Mol Cell Biol 23:5043–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnesen J, Katona G, Rozsa B, Nagerl UV (2014) Spine neck plasticity regulates compartmentalization of synapses Nat Neurosci 17:678–685 doi:10.1038/nn.3682 [DOI] [PubMed] [Google Scholar]
- van Veluw SJ, Sawyer EK, Clover L, Cousijn H, De Jager C, Esiri MM, Chance SA (2012) Prefrontal cortex cytoarchitecture in normal aging and Alzheimer’s disease: a relationship with IQ Brain structure & function 217:797–808 [DOI] [PubMed] [Google Scholar]
- Warmus BA, Sekar DR, McCutchen E, Schellenberg GD, Roberts RC, McMahon LL, Roberson ED (2014) Tau-mediated NMDA receptor impairment underlies dysfunction of a selectively vulnerable network in a mouse model of frontotemporal dementia J Neurosci 34:16482–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Flanagan E, Savage G, Hodges JR, Hornberger M (2014) Contrasting prefrontal cortex contributions to episodic memory dysfunction in behavioural variant frontotemporal dementia and Alzheimer’s disease PloS one 9:e87778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N et al. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization Nature 393:809–812 doi:10.1038/31735 [DOI] [PubMed] [Google Scholar]
- Zhang YM et al. (2006) Targeted deletion of ROCK1 protects the heart against pressure overload by inhibiting reactive fibrosis FASEB J 20:916–925 doi:20/7/916 [pii] 10.1096/fj.05-5129com [DOI] [PubMed] [Google Scholar]


