Abstract
This study assessed genetic contributions to six cognitive domains, identified by the MATRICS Cognitive Consensus Battery as relevant for schizophrenia, cognition-enhancing, clinical trials. Psychiatric Genomics Consortium Schizophrenia polygenic risk scores showed significant negative correlations with each cognitive domain. Genome-wide association analyses identified loci associated with attention/vigilance (rs830786 within HNF4G), verbal memory (rs67017972 near NDUFS4), and reasoning/problem solving (rs76872642 within HDAC9). Gene set analysis identified unique and shared genes across cognitive domains. These findings suggest involvement of common and unique mechanisms across cognitive domains and may contribute to the discovery of new therapeutic targets to treat cognitive deficits in schizophrenia.
Keywords: schizophrenia, PRS, GWAS, neuropsychology, MCCB
1. Introduction
Schizophrenia is highly heritable (h2=0.8) (McGuffin et al., 1984). Moreover, cognitive impairments are core heritable features of schizophrenia (h2=0.20–0.80) (Blokland et al., 2017), yet contributing genes remain to be determined.
Psychiatric Genomics Consortium (PGC) case-control genome-wide association (GWA) studies have found 108 schizophrenia risk loci, including several in ZNF804A, NRGN, TCF4, MIR137, and major histocompatibility complex regions (Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
PGC schizophrenia polygenic risk scores (PRS) have been associated with lower general cognitive ability (Lencz et al., 2014; McIntosh et al., 2013), speed of emotion identification and verbal reasoning (Germine et al., 2016), as well as verbal-numerical reasoning, reaction time, and memory (Hagenaars et al., 2016). Additionally, several control (Davies et al., 2015; Need et al., 2009) and schizophrenia GWAS using cognitive traits have been reported on (Hashimoto et al., 2013; Ohi et al., 2015; Ren et al., 2015; Sanchez-Roige et al., 2018; Smeland et al., 2017; Trampush et al., 2017). However, no PRS or GWA studies have assessed cognitive domains identified by the MATRICS Cognitive Consensus Battery (MCCB), developed for clinical trials of cognition-enhancing treatments for schizophrenia (Kern et al., 2008; Nuechterlein et al., 2008).
Here we report PRS, GWA, and gene set findings from genetic analyses with six MCCB cognitive domain scores (speed of processing, attention/vigilance, working memory, verbal learning, visual learning, and reasoning/problem solving) previously shown impaired in schizophrenia (Cohen’s d=−0.67 to d=−1.14) (van Erp et al., 2015).
2. Materials and Methods
2.1 Participants
This study includes data from 127 clinically stable individuals with schizophrenia (DSM-IV-TR, no medication changes within the last two months, no tardive dyskinesia) and 136 healthy volunteers (Table 1). Individuals with a history of major medical illness, drug dependence in the last 5 years (except nicotine), or current substance abuse disorder were excluded. Healthy volunteers with a history of major neurological or psychiatric illness or with a first-degree relative with an Axis-I psychotic disorder were also excluded. Participants’ cognitive domain scores, based on the Computerized Multiphasic Interactive Neurocognitive System (CMINDS®) neuropsychological test battery (O’Halloran et al., 2008), were published in a prior report (van Erp et al., 2015). Genotyping of blood samples from unrelated and mixed ethnicity subjects was performed using the Illumina MEGA+Psych chip (Illumina, SD, USA). All subjects signed written informed consent approved by institutional review boards.
Table 1.
Sample Demographics and Clinical Characteristics
| Schizophrenia Patients (n=127) | Healthy Volunteers (n=136) | Statistic | p-value | |
|---|---|---|---|---|
| Mean Age (SD) | 39.1 (11.2) | 38.6 (11.4) | t261=0.35 | 0.73 |
| Sex (Male/Female) | 106/21 | 98/38 | χ21=4.91 | 0.03 |
| Handednessa (bilateral/left/right) | 3/10/114 | 2/6/128 | FET | 0.46 |
| Subject Educationb (SD) | 4.6 (1.0) | 5.8 (0.9) | t261=11.46 | <0.0001 |
| Parental Educationb (SD) | 5.7 (1.8) | 5.8 (1.5) | t261=0.20 | 0.66 |
| Race | FET | 0.31 | ||
| American Indian or Alaskan Native | 2 | 2 | ||
| Asian | 18 | 10 | ||
| Black or African American | 20 | 17 | ||
| Native Hawaiian or Pacific Islander | 1 | 1 | ||
| White | 86 | 106 | ||
| NAART | 29.3 (12.8) | 40.7 (11.4) | t258= −7.55 | <0.0001 |
| Age at Onset | 21.5 (6.6) | |||
| Duration of Illness | 17.7 (11.2) | |||
| PANSS positive | 15.4 (5.0) | |||
| PANSS negative | 14.6 (5.5) | |||
| PANSS general | 28.5 (7.5) | |||
| PANSS composite | 0.8 (6.4) |
FET=Fisher’s Exact Test; NAART=North American Adult Reading Test; PANSS=Positive and Negative Syndrome Scale;
Based on the Edinburgh Handedness Inventory;
Based on the Hollingstead Socioeconomic Status Scale.
2.3 Polygenic Risk Score, Genome-wide Association, and Gene Set Analysis
Genotyping data were filtered to remove single-nucleotide polymorphisms (SNPs) with low minor allele frequency (MAF<0.01), deviations from Hardy-Weinberg Equilibrium (p<1×10−6), or poor genotyping call rate (<95%) using PLINK (Purcell et al., 2007). Filtered data were imputed to the 1000 Genomes Project reference panel (1000 Genomes Project Consortium et al., 2015) (phase 1, version 3) using the Michigan Imputation Server (Das et al., 2016). For each individual, a PRS was generated using the GWAS summary of Psychiatric Genomics Consortium (PGC)-schizophrenia meta-analysis (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), with linkage disequilibrium pruning parameters of R2=0.5 over 250 kb windows using 1000 Genomes Project reference panel (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502). The current sample was not part of the PGC analysis on which the PRS was based. PRS was used to test for association with cognitive domain scores using Pearson’s correlations (two-tailed), statistically controlling for age, sex, and 4 multidimensional-scaling components (MDS). Genomewide linear regression analyses predicted each neuropsychological domain with each SNP, statistically controlling for diagnosis, age, sex, site, and four MDS. Fast and flexible gene- or set-based association tests, using GWAS summary data from the neuropsychological domain scores, were performed using Genome-wide Complex Trait Analysis (GCTA) (Bakshi et al., 2016; Yang et al., 2011). This method overcomes the limitations of the resampling-based methods by calculating the p-value for a set of SNPs (±50 Kb of a gene) from an approximated distribution of the sum of χ2-statistics over the SNPs using GWAS summary data and linkage disequilibrium correlations between SNPs from 1000 Genomes Project samples as a reference. We listed the top 40 identified genes and their p-values.
3. Results
The positive study results are that: 1) PGC-schizophrenia PRS showed significant negative correlations with each cognitive domain; 2) GWA identified significant associations for 3 out of 6 cognitive domains; and 3) gene-set analyses found unique and common contributing genes across the cognitive domains.
3.1 Polygenic Risk Score Analyses
PRS showed significant negative correlations with speed of processing (r260=−0.20, p=0.001), attention/vigilance (r258=−0.15, p=0.015), working memory (r261=−0.19, p=0.0018), verbal learning (r261=−0.19, p=0.0018), visual learning (r258=−0.28, p=2.8×10−6), reasoning/problem solving (r260=−0.21, p=0.0005), and the CMINDS composite (r256=−0.29, p=1.6×10−6).
3.2 Genome-wide Association Analyses
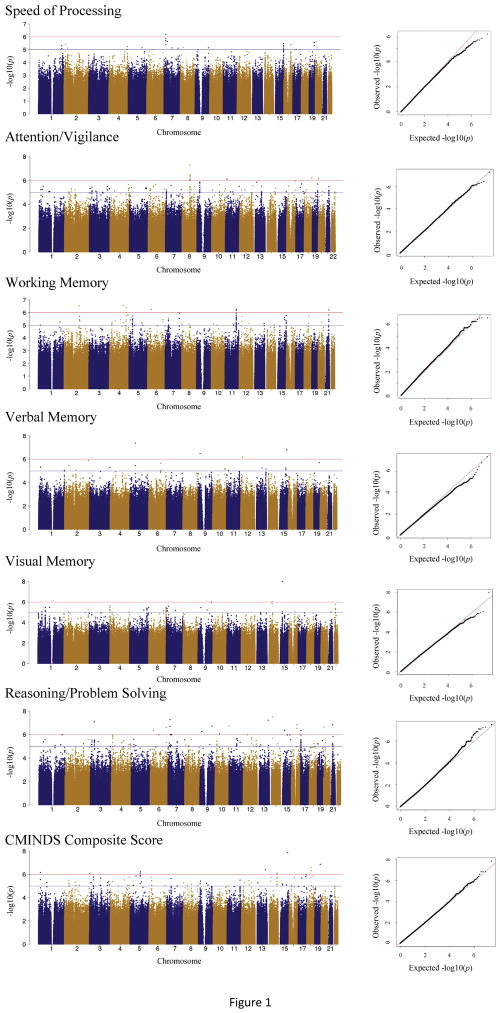
GWA analyses identified significant associations for attention/vigilance (rs830786 within HNF4G), verbal memory (rs67017972 100bp upstream of NDUFS4), and reasoning/problem solving (rs114499642, rs74412765 within LOC102724945, and rs76872642 within HDAC9) domain scores (p<5×10−8; Figure 1; Table 2).
Figure 1.
GWAS Manhattan and QQ Plots for each Cognitive Domain. The Manhattan plots display the association p-value for each SNP in the genome (displayed as −log10 of the p-value). Red and blue lines display p=1×10−6 line and p=1×10−5 respectively. Quantile-quantile plot for the empirical and theoretical distributions are shown as black and red lines, respectively.
Table 2.
Genetic variants associated with cognitive domains.
| rsID | CHR | BP | A1 | BETA | STAT | p-value | Gene | |
|---|---|---|---|---|---|---|---|---|
| Speed of Processing | rs1149530 | 7 | 16848375 | T | 0.77 | 5.1 | 6.4×10−7 | |
| rs818800 | 7 | 16846521 | G | 0.73 | 4.9 | 1.4×10−6 | ||
| rs11763030 | 7 | 16887616 | A | 0.62 | 4.9 | 1.9×10−6 | ||
| Attention/Vigilance | rs830786 | 8 | 76355875 | T | −1.5 | −5.6 | 4.9×10−8 | HNF4G |
| rs75131442 | 8 | 76831979 | T | −1.8 | −5.3 | 3.3×10−7 | ||
| rs79963003 | 8 | 76874205 | A | −1.6 | −5.2 | 4.0×10−7 | ||
| Working Memory | rs17511050 | 4 | 133842651 | T | −1.4 | −5.3 | 2.8×10−7 | |
| rs148396385 | 2 | 151173721 | G | −1.0 | −5.3 | 2.9×10−7 | ||
| rs17396139 | 4 | 162285366 | C | −0.42 | −5.2 | 4.0×10−7 | ||
| Verbal Memory | rs67017972 | 5 | 53071288 | A | 0.60 | 5.7 | 4.3×10−8 | NDUFS4 |
| rs7164861 | 15 | 89125815 | T | −1.3 | −5.4 | 1.6×10−7 | ||
| rs78096325 | 9 | 20207963 | A | −1.2 | −5.2 | 3.3×10−7 | ||
| Visual Memory | rs776010265 | 9 | 126073408 | G | −0.49 | −5.0 | 1.0×10−6 | |
| rs2900031 | 14 | 48879362 | T | 0.59 | 5.0 | 1.4×10−6 | ||
| rs7156750 | 14 | 48885146 | T | 0.48 | 4.9 | 1.7×10−6 | ||
| Reasoning/Problem Solving | rs74412765 | 14 | 34522904 | T | −1.7 | −5.7 | 3.2×10−8 | LOC102724945 |
| rs76872642 | 7 | 18669403 | A | −3.4 | −5.6 | 5.3×10−8 | HDAC9 | |
| rs114499642 | 13 | 104628385 | G | −2.6 | −5.6 | 6.0×10−8 | ||
| CMINDS Composite Score | rs150466277 | 19 | 50957600 | C | −2.8 | −5.4 | 1.4×10−7 | |
| rs80284955 | 18 | 41042547 | G | −1.1 | −5.3 | 2.5×10−7 | ||
| rs185342442 | 13 | 98503979 | G | −3.1 | −5.2 | 3.9×10−7 |
rsID=SNP ID; CHR=chromosome; BP=base pair position; A1=minor allele; BETA=regression coefficient; STAT=t-statistic; Gene=lists a known gene near SNPs that are significant at about p<5.0×10−8.
3.3 Gene Set Analyses
Gene set analyses identified unique and shared genes associated with cognitive domain scores (Table 3).
Table 3.
Gene set analysis based on GWAS results identified unique (non-shaded) and shared genes (shaded) associated with cognitive domain scores
| Speed of Processing | p-value | Attention/Vigilance | p-value | Working Memory | p-value | Verbal Memory | p-value | Visual Memory | p-value | Reasoning/Problem Solving | p-value | CMINDS Composite Score | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGR2 | 0.0002 | ACD | 0.00077 | ACD | 0.00017 | C10orf25 | 0.00095 | ADH1A | 0.00115 | APIP | 0.00044 | ACD | 0.00041 |
| AGR3 | 0.0004 | ABCB1 | 0.00076 | BRF2 | 0.00010 | DTYMK | 0.00057 | AGAP11 | 0.00109 | BAG4 | 0.00068 | ABCA11P | 0.00069 |
| ANGEL1 | 0.0009 | ACTR3 | 0.00092 | C5orf22 | 2.31E-005 | FAM200B | 0.00092 | ALOX12 | 0.00011 | CATSPER2 | 0.00052 | ALOX12 | 0.00032 |
| ANGEL2 | 0.0010 | C16orf86 | 0.00077 | C16orf86 | 0.00017 | GCSAML | 0.00097 | ARL6 | 0.00064 | CHN1 | 0.00048 | C16orf86 | 0.00041 |
| ATF1 | 0.0016 | C19orf48 | 0.00014 | CCDC42B | 0.00029 | GTF2H1 | 0.00011 | C17orf49 | 0.00058 | CKMT1B | 0.00048 | C17orf49 | 0.00078 |
| BTD | 0.0010 | CBWD6 | 0.00074 | CTCF | 0.00033 | HOXD-AS1 | 0.00035 | CAMK2N2 | 0.00064 | CLTA | 0.00082 | BLVRA | 0.00105 |
| CDH1 | 0.0002 | CEBPA | 0.00084 | DDX54 | 0.00045 | HOXD-AS2 | 0.00048 | COL28A1 | 0.00039 | CYP2A13 | 0.00033 | BRINP2 | 0.00061 |
| CDH3 | 0.0002 | CEBPA-AS1 | 0.00065 | E2F2 | 0.00029 | HOXD1 | 0.00080 | DACT2 | 5.89E-005 | CYP2F1 | 0.00058 | ATP6V1B1 | 0.00110 |
| DIO3 | 0.0017 | ENKD1 | 0.00077 | ENKD1 | 0.00017 | HOXD3 | 0.00017 | DDOST | 0.00100 | DDHD2 | 0.00031 | ENKD1 | 0.00041 |
| DIO3OS | 0.0007 | EVPLL | 0.00063 | FAM102A | 0.00048 | HOXD4 | 0.00010 | DNALI1 | 9.88E-005 | FGFR1 | 0.00011 | CCDC116 | 0.00033 |
| FLJ43681 | 0.0017 | FKBP9 | 0.00086 | GPR124 | 0.00016 | HOXD8 | 0.00094 | ECE2 | 0.00061 | FKBP9 | 0.00015 | DCLK3 | 0.00034 |
| HIF1AN | 0.0011 | FOXD4L6 | 0.00075 | GFOD2 | 0.00025 | HSPA13 | 0.00044 | FAM25A | 0.00044 | HIC1 | 0.00065 | GFOD2 | 0.00083 |
| HSPA8 | 0.0016 | HSPA8 | 0.00029 | IQCD | 0.00039 | ING5 | 0.00034 | GLUD1 | 0.00123 | KLRG2 | 0.00069 | EIF4E3 | 0.00093 |
| HYAL1 | 0.0015 | HAX1 | 0.00059 | IRF4 | 0.00048 | LDHA | 0.00039 | GNL2 | 0.00043 | LETM2 | 0.00012 | C9orf92 | 8.59E-005 |
| LINC00687 | 0.0012 | KLK1 | 4.38E-006 | KLK1 | 0.00040 | LDHC | 0.00075 | LOC101927780 | 0.00060 | MEPCE | 0.00057 | FARP2 | 0.00053 |
| LOC101927881 | 0.0014 | KLK15 | 3.01E-006 | KLK15 | 0.00030 | LOC101928869 | 0.00052 | LINC00656 | 0.00061 | MIR132 | 0.00074 | GSTM2P1 | 0.00071 |
| NDUFB8 | 0.0007 | KLK3 | 1.66E-006 | KLK3 | 0.00014 | LOC102800310 | 0.00033 | LOC101928035 | 0.00105 | MIR1343 | 0.00067 | LOC100289361 | 0.00068 |
| LOC442497 | 0.0013 | KLK2 | 3.07E-006 | LOC100506178 | 0.00045 | MATR3 | 0.00025 | LINC00458 | 4.16E-005 | MIR212 | 0.00075 | LINC00458 | 0.00068 |
| LRRC74 | 0.0014 | KLKP1 | 6.88E-006 | LOC401320 | 0.00039 | MIR10B | 0.00017 | LOC100506713 | 0.00011 | MIR6840 | 0.00057 | LOC100506713 | 0.00039 |
| LOC341056 | 0.0012 | LOC341056 | 0.00082 | MIR6762 | 0.00048 | MIR1228 | 0.00093 | LOC101929420 | 4.04E-006 | OR5A1 | 0.00083 | LOC101927750 | 0.00068 |
| MIR4311 | 0.0011 | LARS2-AS1 | 0.00100 | MIR7106 | 0.00039 | MIR4781 | 0.00062 | MIR1470 | 0.00024 | OR5A2 | 0.00036 | MIR130B | 0.00067 |
| MIR6872 | 0.0012 | LINC01210 | 0.00066 | MPZ | 0.00045 | MZB1 | 0.00088 | MIR195 | 0.00071 | OR5AN1 | 0.00069 | MIR195 | 0.00094 |
| MYF6 | 0.0012 | LOC100499194 | 0.00031 | NEUROD6 | 3.34E-005 | NDUFA4L2 | 0.00044 | MIR497 | 0.00071 | PILRB | 0.00050 | MIR497 | 0.00094 |
| MCEMP1 | 0.0012 | LAMA4 | 0.00056 | R3HCC1L | 0.00035 | NOMO3 | 0.00049 | MIR497HG | 0.00071 | PMS2P1 | 0.00035 | MIR497HG | 0.00094 |
| NHEG1 | 0.0004 | LOC440896 | 0.00075 | PPM1B | 0.00011 | NXPH4 | 0.00073 | PPM1B | 0.00024 | PPAPDC1B | 0.00029 | PPM1B | 0.00012 |
| NPBWR2 | 0.0010 | MGC45922 | 8.75E-006 | PTN | 0.00013 | OR10S1 | 0.00030 | PAQR9 | 0.00073 | PPP1R35 | 0.00081 | PAQR9 | 0.00074 |
| OPRL1 | 0.0017 | PARD6A | 0.00077 | PARD6A | 0.00017 | OR2G2 | 0.00030 | MN1 | 9.86E-005 | PVRIG2P | 0.00069 | PARD6A | 0.00041 |
| PCP2 | 0.0017 | OR5AR1 | 0.00052 | RASAL1 | 0.00026 | OR4D5 | 0.00014 | PPIL2 | 0.00013 | RP9P | 0.00028 | PPIL2 | 0.00018 |
| RETN | 0.0010 | OR5M11 | 0.00080 | RNU6-83P | 0.00018 | OR6T1 | 9.57E-005 | PINK1-AS | 0.00114 | RNU6-83P | 0.00012 | MIR301B | 0.00067 |
| RPS6KC1 | 0.0006 | OR5M3 | 0.00061 | RLTPR | 0.00044 | OR8D4 | 0.00026 | MIR5581 | 0.00116 | SERF2 | 0.00078 | RLTPR | 0.00069 |
| SEC31B | 0.0009 | RUNDC3B | 0.00057 | RUNDC3B | 0.00011 | PAIP2 | 0.00095 | PROK2 | 0.00047 | SLC4A4 | 0.00063 | RNASE10 | 0.00071 |
| SEMA3B-AS1 | 0.0015 | OR5M9 | 0.00082 | SDHC | 0.00038 | RPF2 | 0.00021 | RNASEK | 0.00046 | SMG6 | 0.00018 | RNASEK | 0.00066 |
| SLC35D3 | 0.0011 | OR9G1 | 0.00084 | SLC25A40 | 0.00033 | SHMT2 | 0.00062 | RNASEK-C17orf49 | 0.00056 | SPDYE3 | 0.00021 | RNASEK-C17orf49 | 0.00074 |
| STXBP2 | 0.0014 | OR9G9 | 0.00084 | SPRR2B | 0.00047 | SLC23A1 | 0.00077 | SNIP1 | 0.00023 | SRSF10 | 0.00030 | SDF2L1 | 0.00038 |
| TFDP2 | 0.0007 | MIR6778 | 0.00097 | SPRR2E | 0.00036 | SNHG4 | 0.00032 | TFAMP1 | 0.00105 | STAG3L5P | 0.00043 | STK25 | 0.00089 |
| TMEM225 | 0.0015 | PGM5P2 | 0.00075 | SPRR2F | 0.00032 | TMEM225 | 0.00055 | TFAP2B | 0.00071 | STAG3L5P-PVRIG2P-PILRB | 0.00040 | TMEM225 | 0.00044 |
| WNT8B | 0.0011 | OR5M8 | 0.00035 | THAP11 | 0.00049 | SPRN | 0.00088 | UBE2L3 | 0.00087 | STRC | 0.00072 | UBE2L3 | 0.00099 |
| TUSC2 | 0.0016 | SNORD88C | 0.00030 | TPCN1 | 0.00048 | STAC3 | 0.00036 | YPEL1 | 0.00033 | TBX21 | 0.00090 | YPEL1 | 0.00033 |
| TSNAXIP1 | 0.0012 | SOX14 | 0.00086 | TSNAXIP1 | 0.00045 | SNORA74A | 0.00036 | WIZ | 0.00039 | WHSC1L1 | 0.00011 | YDJC | 0.00058 |
| ZNF596 | 0.0017 | TOP3A | 0.00084 | ZNF596 | 0.00032 | TMEM59 | 0.00082 | ZNF121 | 0.00105 | ZCWPW1 | 0.00069 | VAX2 | 0.00012 |
The shaded genes are associated with two or more cognitive domains presented on the same row.
4. Discussion
All correlations between the PGC schizophrenia PRS and the cognitive domains scores were negative, consistent with the interpretation that higher schizophrenia genetic risk is associated with worse cognitive performance, corroborating prior findings (Germine et al., 2016; Hagenaars et al., 2016; Lencz et al., 2014; McIntosh et al., 2013).
With regard to the GWAS locus associated with attention/vigilance, HNF4G is expressed in the brain (http://www.brain-map.org). HNF4G is regulated by miR-194, which is dysregulated in individuals with 22q11.2 deletion syndrome who have a 20–30 fold increased risk for psychosis (Sellier et al., 2014). Additionally, mouse Hnf4g was found to be upregulated after toxoplasma gondii infection (He et al., 2016) which is a putative risk factor for schizophrenia (Webster et al., 2013). With regard to reasoning/problem solving, little is known about the LOC102724945 and rs114499642 loci. However, HDAC9, histone deacetylase 9, is expressed in brain and has previously been associated with schizophrenia (Kebir et al., 2014; Tam et al., 2010). HDAC9 is involved in transcriptional regulation, cell cycle progression, and neuronal development and transmission. Rs67017972, associated with verbal memory, is located 100bp upstream of NDUFS4. Lower prefrontal cortex and hippocampal NDUFS4 expression has been found in schizophrenia (Altar et al., 2005; Arion et al., 2015), and Ndufs4 cKO mice show impaired cognitive function and increased anxiety-like behavior (Choi et al., 2017).
Gene set analyses, based on the GWAS results, found that several genes contribute to multiple cognitive domains. For example, PPM1B is associated with working memory, visual memory, and CMINDS composite. PPM1B is a member of the PP2C family of Ser/Thr protein phosphatases, and is expressed in brain. PP2C family members are known negative regulators of cell stress response pathways, and are involved in neuroprotection and neurodegeneration (Klumpp et al., 2006, 2002). Protein interactions between PPM1B, NRG1, and DTNBP1, putative schizophrenia susceptibility genes, have also been reported (Tsuang, 2000). HSPA8, associated with attention/vigilance and speed of processing, was previously identified as a schizophrenia risk locus (Bozidis et al., 2014). HSPA8, Heat shock 70 kDa protein 8, is known to contribute to many biological processes, including signal transduction, apoptosis, autophagy, protein homeostasis, and cell growth and differentiation. PLCB3-PARD3-PARD6A complex, associated with the CMINDS composite and working memory, was found to be associated with schizophrenia in BA22 RNA-Seq study (Huang et al., 2014). PARD6A, partitioning defective 6 homolog alpha, is involved in asymmetrical cell division and cell polarization processes. ALOX12, associated with CMINDS composite and visual learning, had been identified in a Korean schizophrenia study (Kim et al., 2010). MIR497, related to the CMINDS composite and visual learning, was differentially expressed in the prefrontal cortex exosome in schizophrenia and bipolar disorder (Delgado-Morales, 2017). Finally, MIR195, associated with visual memory and CMINDS composite, was found to be upregulated in the superior temporal gyrus of individuals with schizophrenia (Beveridge et al., 2010, 2008). MIR195 regulates numerous schizophrenia-related genes, such as BDNF, RELN, Visinin-like 1, 5-hydroxytryptamine (serotonin) receptor 2a, and glutamate receptor, ionotropic, N-methyl-d-aspartate 3A. Each of the cognitive domains, especially the CMINDS composite, shares several genes with another cognitive domain. Given that the CMINDS composite is an average across all 6 cognitive domains, overlap in genes with the CMINDS composite is expected.
Study strengths are the use of cognitive domain scores that are considered important targets for cognition-enhancing treatments for schizophrenia. Study limitations include sample size and lack of a replication sample, though the observed negative correlations between the schizophrenia PRS and the cognitive domain scores strengthen the confidence in our GWA and gene set findings. Nevertheless, replication of the findings in larger cohorts is warranted.
In conclusion, we found that the PGC-based schizophrenia PRS was significantly negatively correlated with CMINDS cognitive domain performance. In addition, we identified novel loci associated with cognitive domain performance. Finally, gene-based analysis revealed that cognitive domains share contributing genes. These findings suggest involvement of novel unique and common biological mechanisms in cognitive domain deficits in schizophrenia and may contribute to the discovery of new therapeutic targets to treat cognitive deficits in schizophrenia, which do not respond well to traditional antipsychotic treatments (Kahn and Keefe, 2013).
Acknowledgments
Funding/Support: This work was supported by National Institutes of Health grant numbers: NIH 1 U24 RR021992 (Function Biomedical Informatics Research Network), NIH 1 U24 RR025736-01 (Biomedical Informatics Research Network Coordinating Center), 5R01MH094524, P20GM103472, and UL1 TR000153.
Footnotes
Author Contributions: Drs. Nakahara and Van Erp had full access to all of the data in the study, conducted the statistical analysis, and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs. Nakahara and Van Erp wrote the first draft of the manuscript. All authors critically reviewed the manuscript, provided comments, and approved the manuscript for publication.
Previous Presentation: This study has not been previously presented.
Additional Contributions: We are thankful to Mrs. Liv McMillan, BS for overall study coordination, Harry Mangalam, PhD, Joseph Farran, BS, and Adam Brenner, BS, for administering the University of California, Irvine High-Performance Computing cluster, and to the research subjects for their participation.
Financial Disclosure: Dr. Soichiro Nakahara’s effort was supported by Astellas Pharma Inc. while he was a visiting scholar in University of California, Irvine. Dr. Bustillo consulted with Novartis and Otsuka Pharmaceuticals. Dr. Mathalon consulted for Boerhinger Ingelheim and Takeda. Dr. Preda consulted for Boehringer-Ingelheim. Dr. Potkin has financial interests in Bristol-Myers Squibb, Eisai, Inc., Eli Lilly, Forest Laboratories, Genentech, Janssen Pharmaceutical, Lundbeck, Merck, Novartis, Organon, Pfizer, Roche, Sunovion, Takeda Pharmaceutical, Vanda Pharmaceutical, Novartis, Lundbeck, Merck, Sunovion and has received grant funding from Amgen, Baxter, Bristol-Myers Squibb, Cephalon, Inc., Eli Lilly, Forest Laboratories, Genentech, Janssen Pharmaceutical, Merck, Otsuka, Pfizer, Roche, Sunovion, Takeda Pharmaceutical, Vanda Pharmaceutical, NIAAA, NIBIB, NIH/NCRR, University of Southern California, UCSF, UCSD, Baylor College of Medicine. The remaining authors declare no potential conflict of interest.
Role of the Sponsor: The funding sources had no role in the study design, conduct of the study, data collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, Cacace AM, Zaczek R, Albright CF, Tseng G, Lewis DA. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015;20:1397–1405. doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi A, Zhu Z, Vinkhuyzen AAE, Hill WD, McRae AF, Visscher PM, Yang J. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci Rep. 2016;6:32894. doi: 10.1038/srep32894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Blokland GAM, Mesholam-Gately RI, Toulopoulou T, Del Re EC, Lam M, DeLisi LE, Donohoe G, Walters JTR, Seidman LJ, Petryshen TL GENUS Consortium. Heritability of Neuropsychological Measures in Schizophrenia and Nonpsychiatric Populations: A Systematic Review and Meta-analysis. Schizophr Bull. 2017;43:788–800. doi: 10.1093/schbul/sbw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozidis P, Hyphantis T, Mantas C, Sotiropoulou M, Antypa N, Andreoulakis E, Serretti A, Mavreas V, Antoniou K. HSP70 polymorphisms in first psychotic episode drug-naïve schizophrenic patients. Life Sci. 2014;100:133–137. doi: 10.1016/j.lfs.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Choi W-S, Kim H-W, Tronche F, Palmiter RD, Storm DR, Xia Z. Conditional deletion of Ndufs4 in dopaminergic neurons promotes Parkinson’s disease-like non-motor symptoms without loss of dopamine neurons. Sci Rep. 2017;7:44989. doi: 10.1038/srep44989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P-R, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, Hofer E, Ibrahim-Verbaas CA, Kirin M, Lahti J, van der Lee SJ, Le Hellard S, Liu T, Marioni RE, Oldmeadow C, Postmus I, Smith AV, Smith JA, Thalamuthu A, Thomson R, Vitart V, Wang J, Yu L, Zgaga L, Zhao W, Boxall R, Harris SE, Hill WD, Liewald DC, Luciano M, Adams H, Ames D, Amin N, Amouyel P, Assareh AA, Au R, Becker JT, Beiser A, Berr C, Bertram L, Boerwinkle E, Buckley BM, Campbell H, Corley J, De Jager PL, Dufouil C, Eriksson JG, Espeseth T, Faul JD, Ford I, Gottesman RF, Griswold ME, Gudnason V, Harris TB, Heiss G, Hofman A, Holliday EG, Huffman J, Kardia SLR, Kochan N, Knopman DS, Kwok JB, Lambert J-C, Lee T, Li G, Li S-C, Loitfelder M, Lopez OL, Lundervold AJ, Lundqvist A, Mather KA, Mirza SS, Nyberg L, Oostra BA, Palotie A, Papenberg G, Pattie A, Petrovic K, Polasek O, Psaty BM, Redmond P, Reppermund S, Rotter JI, Schmidt H, Schuur M, Schofield PW, Scott RJ, Steen VM, Stott DJ, van Swieten JC, Taylor KD, Trollor J, Trompet S, Uitterlinden AG, Weinstein G, Widen E, Windham BG, Jukema JW, Wright AF, Wright MJ, Yang Q, Amieva H, Attia JR, Bennett DA, Brodaty H, de Craen AJM, Hayward C, Ikram MA, Lindenberger U, Nilsson L-G, Porteous DJ, Räikkönen K, Reinvang I, Rudan I, Sachdev PS, Schmidt R, Schofield PR, Srikanth V, Starr JM, Turner ST, Weir DR, Wilson JF, van Duijn C, Launer L, Fitzpatrick AL, Seshadri S, Mosley TH, Jr, Deary IJ Generation Scotland. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949) Mol Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Morales R. Neuroepigenomics in Aging and Disease. Springer; 2017. [Google Scholar]
- Germine L, Robinson EB, Smoller JW, Calkins ME, Moore TM, Hakonarson H, Daly MJ, Lee PH, Holmes AJ, Buckner RL, Gur RC, Gur RE. Association between polygenic risk for schizophrenia, neurocognition and social cognition across development. Transl Psychiatry. 2016;6:e924. doi: 10.1038/tp.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ, Marioni RE, Fawns-Ritchie C, Cullen B, Malik R, Worrall BB, Sudlow CLM, Wardlaw JM, Gallacher J, Pell J, McIntosh AM, Smith DJ, Gale CR, Deary IJ METASTROKE Consortium International Consortium for Blood Pressure GWAS, SpiroMeta Consortium CHARGE Consortium Pulmonary Group CHARGE Consortium Aging Longevity Group. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry. 2016;21:1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ikeda M, Ohi K, Yasuda Y, Yamamori H, Fukumoto M, Umeda-Yano S, Dickinson D, Aleksic B, Iwase M, Kazui H, Ozaki N, Weinberger DR, Iwata N, Takeda M. Genome-wide association study of cognitive decline in schizophrenia. Am J Psychiatry. 2013;170:683–684. doi: 10.1176/appi.ajp.2013.12091228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-J, Ma J, Elsheikha HM, Song H-Q, Zhou D-H, Zhu X-Q. Proteomic Profiling of Mouse Liver following Acute Toxoplasma gondii Infection. PLoS One. 2016;11:e0152022. doi: 10.1371/journal.pone.0152022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-C, Yang K-C, Lin H, Tsao TT-H, Lee S-A. Transcriptome alterations of mitochondrial and coagulation function in schizophrenia by cortical sequencing analysis. BMC Genomics. 2014;15(Suppl 9):S6. doi: 10.1186/1471-2164-15-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RS, Keefe RSE. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–1112. doi: 10.1001/jamapsychiatry.2013.155. [DOI] [PubMed] [Google Scholar]
- Kebir O, Chaumette B, Fatjó-Vilas M, Ambalavanan A, Ramoz N, Xiong L, Mouaffak F, Millet B, Jaafari N, DeLisi LE, Levinson D, Joober R, Fañanás L, Rouleau G, Dubertret C, Krebs M-O. Family-based association study of common variants, rare mutation study and epistatic interaction detection in HDAC genes in schizophrenia. Schizophr Res. 2014;160:97–103. doi: 10.1016/j.schres.2014.09.029. [DOI] [PubMed] [Google Scholar]
- Kern RS, Nuechterlein KH, Green MF, Baade LE, Fenton WS, Gold JM, Keefe RSE, Mesholam-Gately R, Mintz J, Seidman LJ, Stover E, Marder SR. The MATRICS Consensus Cognitive Battery, part 2: co-norming and standardization. Am J Psychiatry. 2008;165:214–220. doi: 10.1176/appi.ajp.2007.07010043. [DOI] [PubMed] [Google Scholar]
- Kim T, Kim H-J, Park J, Kim J, Chung J-H. Association between polymorphisms of arachidonate 12-lipoxygenase (ALOX12) and schizophrenia in a Korean population. Behav Brain Funct. 2010;6:44. doi: 10.1186/1744-9081-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Selke D, Ahlemeyer B, Schaper C, Krieglstein J. Relationship between protein phosphatase type-2C activity and induction of apoptosis in cultured neuronal cells. Neurochem Int. 2002;41:251–259. doi: 10.1016/s0197-0186(02)00020-7. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Thissen M-C, Krieglstein J. Protein phosphatases types 2Calpha and 2Cbeta in apoptosis. Biochem Soc Trans. 2006;34:1370–1375. doi: 10.1042/BST0341370. [DOI] [PubMed] [Google Scholar]
- Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, Christoforou A, Reinvang I, Mukherjee S, DeRosse P, Lundervold A, Steen VM, John M, Espeseth T, Räikkönen K, Widen E, Palotie A, Eriksson JG, Giegling I, Konte B, Ikeda M, Roussos P, Giakoumaki S, Burdick KE, Payton A, Ollier W, Horan M, Donohoe G, Morris D, Corvin A, Gill M, Pendleton N, Iwata N, Darvasi A, Bitsios P, Rujescu D, Lahti J, Hellard SL, Keller MC, Andreassen OA, Deary IJ, Glahn DC, Malhotra AK. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT) Mol Psychiatry. 2014;19:168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Farmer AE, Gottesman II, Murray RM, Reveley AM. Twin concordance for operationally defined schizophrenia. Confirmation of familiality and heritability. Arch Gen Psychiatry. 1984;41:541–545. doi: 10.1001/archpsyc.1984.01790170015002. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Gow A, Luciano M, Davies G, Liewald DC, Harris SE, Corley J, Hall J, Starr JM, Porteous DJ, Tenesa A, Visscher PM, Deary IJ. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol Psychiatry. 2013;73:938–943. doi: 10.1016/j.biopsych.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuechterlein KH, Green MF, Kern RS, Baade LE, Barch DM, Cohen JD, Essock S, Fenton WS, Frese FJ, 3rd, Gold JM, Goldberg T, Heaton RK, Keefe RSE, Kraemer H, Mesholam-Gately R, Seidman LJ, Stover E, Weinberger DR, Young AS, Zalcman S, Marder SR. The MATRICS Consensus Cognitive Battery, part 1: test selection, reliability, and validity. Am J Psychiatry. 2008;165:203–213. doi: 10.1176/appi.ajp.2007.07010042. [DOI] [PubMed] [Google Scholar]
- O’Halloran JP, Kemp AS, Gooch KN, Harvey PD, Palmer BW, Reist C, Schneider LS. Psychometric comparison of computerized and standard administration of the neurocognitive assessment instruments selected by the CATIE and MATRICS consortia among patients with schizophrenia. Schizophr Res. 2008;106:33–41. doi: 10.1016/j.schres.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Ikeda M, Yamamori H, Yasuda Y, Fujimoto M, Umeda-Yano S, Fukunaga M, Fujino H, Watanabe Y, Iwase M, Kazui H, Iwata N, Weinberger DR, Takeda M. Glutamate Networks Implicate Cognitive Impairments in Schizophrenia: Genome-Wide Association Studies of 52 Cognitive Phenotypes. Schizophr Bull. 2015;41:909–918. doi: 10.1093/schbul/sbu171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhang C, Huang C, Li N, Li M, Li Y, Deng W, Ma X, Xiang B, Wang Q, Li T. Unravelling Genes and Pathways Implicated in Working Memory of Schizophrenia in Han Chinese. Int J Mol Sci. 2015;16:2145–2161. doi: 10.3390/ijms16012145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, Pandit A, Schmidt EM, Foerster JR, Abecasis GR, Gray JC, de Wit H, Davis LK, MacKillop J, Palmer AA 23andMe Research Team. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat Neurosci. 2018;21:16–18. doi: 10.1038/s41593-017-0032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Hwang VJ, Dandekar R, Durbin-Johnson B, Charlet-Berguerand N, Ander BP, Sharp FR, Angkustsiri K, Simon TJ, Tassone F. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS One. 2014;9:e103884. doi: 10.1371/journal.pone.0103884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, Krull F, Bettella F, Eriksen JA, Witoelar A, Davies G, Fan CC, Thompson WK, Lam M, Lencz T, Chen C-H, Ueland T, Jönsson EG, Djurovic S, Deary IJ, Dale AM, Andreassen OA NeuroCHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Cognitive Working Group. Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA Psychiatry. 2017;74:1065–1075. doi: 10.1001/jamapsychiatry.2017.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GWC, van de Lagemaat LN, Redon R, Strathdee KE, Croning MDR, Malloy MP, Muir WJ, Pickard BS, Deary IJ, Blackwood DHR, Carter NP, Grant SGN. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38:445–451. doi: 10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- Trampush JW, Yang MLZ, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, Christoforou A, Reinvang I, DeRosse P, Lundervold AJ, Steen VM, Espeseth T, Räikkönen K, Widen E, Palotie A, Eriksson JG, Giegling I, Konte B, Roussos P, Giakoumaki S, Burdick KE, Payton A, Ollier W, Horan M, Chiba-Falek O, Attix DK, Need AC, Cirulli ET, Voineskos AN, Stefanis NC, Avramopoulos D, Hatzimanolis A, Arking DE, Smyrnis N, Bilder RM, Freimer NA, Cannon TD, London E, Poldrack RA, Sabb FW, Congdon E, Conley ED, Scult MA, Dickinson D, Straub RE, Donohoe G, Morris D, Corvin A, Gill M, Hariri AR, Weinberger DR, Pendleton N, Bitsios P, Rujescu D, Lahti J, Le Hellard S, Keller MC, Andreassen OA, Deary IJ, Glahn DC, Malhotra AK, Lencz T. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry. 2000;47:210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- van Erp TGM, Preda A, Turner JA, Callahan S, Calhoun VD, Bustillo JR, Lim KO, Mueller B, Brown GG, Vaidya JG, McEwen S, Belger A, Voyvodic J, Mathalon DH, Nguyen D, Ford JM, Potkin SG FBIRN. Neuropsychological profile in adult schizophrenia measured with the CMINDS. Psychiatry Res. 2015;230:826–834. doi: 10.1016/j.psychres.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JP, Kaushik M, Bristow GC, McConkey GA. Toxoplasma gondii infection, from predation to schizophrenia: can animal behaviour help us understand human behaviour? J Exp Biol. 2013;216:99–112. doi: 10.1242/jeb.074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]