Abstract
The concept of genetic canalization has had an abiding influence on views of complex-trait evolution. A genetically canalized system has evolved to become less sensitive to the effects of mutation. When a gene product that supports canalization is compromised, the phenotypic impacts of a mutation should be more pronounced. This expected increase in mutational effects not only has important consequences for evolution, but has also motivated strategies to treat disease. However, recent studies demonstrate that, when putative agents of genetic canalization are impaired, systems do not behave as expected. Here, we review the evidence that is used to infer whether particular gene products are agents of genetic canalization. Then we explain how such inferences often succumb to a converse error. We go on to show that several candidate agents of genetic canalization increase the phenotypic impacts of some mutations while decreasing the phenotypic impacts of others. These observations suggest that whether a gene product acts as a ‘buffer’ (lessening mutational effects) or a ‘potentiator’ (increasing mutational effects) is not a fixed property of the gene product but instead differs for the different mutations with which it interacts. To investigate features of genetic interactions that might predispose them toward buffering versus potentiation, we explore simulated gene-regulatory networks. Similarly to putative agents of genetic canalization, the gene products in simulated networks also modify the phenotypic effects of mutations in other genes without a strong overall tendency towards lessening or increasing these effects. In sum, these observations call into question whether complex traits have evolved to become less sensitive (i.e., are canalized) to genetic change, and the degree to which trends exist that predict how one genetic change might alter another’s impact. We conclude by discussing approaches to address these and other open questions that are brought into focus by re-thinking genetic canalization.
Keywords: Canalization, buffering, potentiation, epistasis, gene-regulatory networks, evolution of complex traits
1. Introduction
Identical genetic changes do not always have identical effects on phenotype. Because organisms are composed of interacting parts, the effect of perturbing any one part can offset or magnify perturbations to others, resulting in mutations with context-dependent effects on function [1,2]. A major goal of the genomic age is to predict traits, such as disease risk or behavior, from genetic data alone. Context dependence makes achieving this goal much more challenging. In the effort to map genotype to phenotype, one type of context dependence – called genetic buffering – has received special attention [3–5]. Genetic buffering, in which some gene products render changes in other genes less consequential [6–14], appears to be common [3–5] and reversible. A mutation, environmental shift or pharmacological agent can reveal phenotypic effects of previously inconsequential genetic differences [15–17]. Understanding the mechanisms that support buffering, or any trends that predict which phenotypes are revealed when a buffer is compromised, might inform questions of high significance, such as how human diseases result from common genetic variants [18], or how a species will respond to climate change [19–22]. The ability to modulate buffering also lies at the core of Waddington’s concept of genetic canalization, whereby a system evolves to become less sensitive to the effects of mutations [16,23]. Although the concept is compelling, the evidence for genetic canalization has been challenged [3–5,24].
The challenge relates to a key consequence of genetic canalization, that mutations with buffered effects — often called cryptic genetic variation — will accumulate in populations because their effects on phenotype are shielded from natural selection. If the system is decanalized (i.e., the buffering mechanism is impaired), then cryptic variation will be revealed in the form of greater phenotypic diversity across the population. Cryptic genetic variation is common, having been observed in a variety of systems for a variety of phenotypes [25,26]. Perhaps the most well-known example is the cryptic genetic variation that is revealed upon impairment of the molecular chaperone Hsp90. In diverse eukaryotes, reducing Hsp90 function increases phenotypic diversity among genetic backgrounds [6,15,27–30]. A common line of argument holds that the abundance of cryptic genetic variation implies that biological systems tend to be genetically canalized, and indeed Hsp90 has been described as an agent of genetic canalization [6–14]. However, this argument suffers from the fallacy of the converse: although genetic canalization implies that cryptic genetic variation should accumulate, the accumulation of cryptic genetic variation does not necessarily imply that genetic canalization exists [3–5].
1.1. Observations of buffering are not evidence of canalization
The existence of cryptic genetic variation requires only that some mutations’ effects are buffered, not that the system has evolved to have greater capacity to buffer mutations’ effects. That is, to avoid the converse-fallacy trap, it is critical not to confuse observations of genetic buffering (e.g., particular cryptic genetic variants with effects that are revealed by impairment of Hsp90) with evidence of genetic canalization or mutational robustness (i.e., an overall tendency for mutations to have less effect in the presence of Hsp90 or another putative buffer). Doing so can have negative consequences, such as inaccurate predictions about how systems might respond when a putative agent of canalization is impaired by pharmaceutical agents. Nonetheless, despite a lack of evidence (or in some cases evidence to the contrary), the view has persisted that genetic canalization is widespread [8,14]. In other words, the literature on genetic canalization has itself become canalized, difficult to perturb from the view that biological systems have evolved to buffer the effects of mutations (or that particular gene products such as Hsp90 do the buffering). Even works that pointed out flaws and uncertainties in canalization research, such as Scharloo’s influential 1991 review [31], have not doubted the existence of genetic canalization. After an incisive critique of some of Waddington’s inferences, Scharloo nonetheless answered his own question, “Does this mean that we have to abandon the canalization concept?” with “Of course not. … We cannot escape the conclusion that development in wild-type individuals is geared to produce constant phenotypes notwithstanding the presence of genetic variability …” [31].
In this paper, we aim to decanalize the way context-dependent genetic effects are conceptualized. Starting with Hsp90 and another chaperone, GroEL, as case studies, we examine the observations commonly used to identify robust systems, highlighting hypotheses other than genetic canalization that can explain these observations. Next we briefly summarize our recent work demonstrating that Hsp90 does not canalize cell-morphological features of the budding yeast Saccharomyces cerevisiae against the effects of genetic change [5]. Indeed we find the opposite: although Hsp90 buffers the phenotypic effects of some mutations, it enhances or ‘potentiates’ the phenotypic effects of others, and its overall (albeit weak) tendency is to potentiate [5]. We go on to illustrate the generality of this phenomenon — that highly interactive gene products both buffer and potentiate — by discussing how key molecular players in common examples of ‘robust’ biological systems, such as the circadian clock, can often potentiate, rather than buffer, the effects of genetic change.
Waddington supported the idea that the mechanism underlying canalization involves the interaction of many gene products [32,33]. Indeed, molecular networks are discussed throughout this special issue on canalization [32,34–38]. In this article, using simulated gene-regulatory networks, we develop null expectations for the relative prevalences of buffering and potentiating interactions in nature, and we explore network features that might tip the balance toward buffering or potentiation. Consistent with intuition laid out in other articles in this special issue [34,36,37], we find that network hubs (akin to Hsp90) stand out in their ability to modify the phenotypic effects of mutation. We find that simulated networks mimic trends we observe in natural systems. For example, they demonstrate that most gene products modify the phenotypic effects of mutations in other genes without a strong overall tendency towards buffering or potentiation.
The experimental results we review and the in silico results we present both support a model [5,39,40] in which buffering and potentiation are not seen as distinct phenomena, but as context dependencies that result from the interaction networks that underlie biological systems. These results call into question whether any single gene product can be classified as a genetic canalizer. It is important to note that the doubts we cast on canalization in this review apply to genetic canalization (i.e. robustness to genetic perturbations), and not to environmental canalization (i.e. robustness to environmental perturbations); for evidence supporting the latter see, for example, Hallgrimsson et al in this special issue [38]. We conclude our review by offering some thoughts on how to experimentally address outstanding questions about genetic canalization, buffering and potentiation. More generally, we also discuss whether there exist any trends amidst pervasive context dependence that can improve prediction of phenotype from genotype.
2. Only weak or indirect evidence supports chaperones as agents of genetic canalization
Protein folding chaperones, such as Hsp90 in eukaryotes [41], and GroEL in bacteria [12], represent some of the most well-known putative agents of genetic canalization [14]. These chaperones have been proposed to buffer the phenotypic effects of genetic change by helping proteins to fold despite mutation [6,12]. Researchers across diverse fields have hypothesized about the impact of increased mutational robustness provided by chaperone proteins, proposing that it might: 1) protect cancers from deleterious effects of elevated mutation rate [17,42,43], 2) allow organisms evolving under strong genetic drift to avoid death as a result of mutation accumulation [12], 3) promote evolvability by allowing cryptic genetic variation to accumulate in genomes [11,29], and 4) provide a selective advantage that encouraged the maintenance of chaperone systems over evolutionary time [29].
But what is the evidence that protein-folding chaperones increase mutational robustness? One line of evidence comes from studies that show inhibiting Hsp90 reveals the phenotypic effects of cryptic genetic variation in flies [15], fish [6,28], plants [27,30], and yeast [5,29]. However, these studies do not demonstrate that Hsp90 increases robustness to the phenotypic effects of new mutations [3]. An alternative hypothesis to explain these observations is that, just as stabilizing selection favors synonymous over non-synonymous genetic changes [44], selection could favor Hsp90-buffered over Hsp90-potentiated mutations, leaving the false signal that Hsp90 tends to buffer mutational effects [3,5,39]. If this is the case, Hsp90 may more often potentiate, rather than buffer, the effects of new mutations, but selection obscures this bias by enriching for Hsp90-buffered genetic variation. This alternative hypothesis echoes our earlier point that the capacity of biological systems to accumulate cryptic genetic variation should not be taken as evidence that new mutations tend to have cryptic effects.
A second line of evidence that protein-folding chaperones increase mutational robustness is that clients of these chaperones, i.e. proteins known to require chaperones for folding, tend to accumulate more mutations over evolutionary time than non-clients [36,45] and also accumulate more mutations when chaperones are overexpressed [46]. Even if chaperones make clients more robust to mutation, they might not genetically canalize the physiological or developmental traits to which these clients contribute. Studies in budding yeast have found that the effects of new genetic perturbations (either spontaneous mutations or natural variants segregating in a cross of genetically divergent parents) on complex phenotypes such as growth rate or single-cell morphology are potentiated by Hsp90 almost as often as, or even more often than, they are buffered by Hsp90 [5,29]. Laboratory evolution experiments in E. coli demonstrated that, in particular client proteins, GroEL overexpression allows strongly deleterious mutations to accumulate by buffering their deleterious effects [12]. However, across whole genomes, GroEL overexpression decreased the number of mutations that accumulated by twofold [12]. This result might be explained by a tendency for the chaperone to amplify the effects of mutations, most of which are deleterious. In other words, there could be a genome-wide tendency for GroEL to potentiate rather than buffer mutational effects. In general, if GroEL and Hsp90 both allow mutations to accumulate in client proteins, but overall do not make organisms more robust to mutational effects, it does not seem appropriate call either protein an agent of genetic canalization.
Further, the observation that mutations accumulate in client proteins [45,46] is not necessarily evidence that chaperones tend to buffer the phenotypic effects of these mutations. Mutations with phenotypic effects that are potentiated can also accumulate in clients. For example, GroEL can act as a buffer of a client phosphotriesterase (PTE) by preserving native function despite mutation, and GroEL can also act as a potentiator by mitigating the destabilizing effects of mutations that impart novel esterase activity [46]. Mutations that improve the novel esterase activity of the PTE enzyme are more common and have larger effects when GroEL is overexpressed [46]. Therefore, whether GroEL is classified as a buffer or potentiator depends on whether native or novel PTE function is examined, and in both cases the number of mutations that support function is increased upon GroEL overexpression.
The example of GroEL highlights a primary source of confusion in the identification of mechanisms that support genetic canalization. Although GroEL can allow mutations to accumulate without perturbing native PTE function, saying that GroEL is an agent of genetic canalization ignores other observations and in general oversimplifies the complicated effects a chaperone may have. Overall, these observations support a model in which chaperones modify the phenotypic effects of mutations in diverse ways, both direct and indirect, and the types of interactions that persist in genomes depend on what selection has favored [5]. Important questions about how context-dependent effects interact with selection pressures to shape complex-trait variation [2,40] become accessible upon decanalizing the way we think about (and therefore test) canalization.
2.1. Hsp90 is best described as neither a buffer nor a potentiator
As explained above, a rigorous test of whether a particular protein or mechanism contributes to genetic canalization requires overcoming potentially false signals of canalization left by natural selection (i.e. accumulation of buffered genetic variants). This test can be carried out by assaying how a putative canalizer modifies the effects of new mutations, which have not been filtered by natural selection [3]. We performed such a test using a set of 94 yeast strains that had been propagated in a way that allows mutations to accumulate under minimal selection (i.e. through repeated population bottlenecks) [44,47]. We demonstrated that Hsp90 tends to potentiate, rather than buffer, the impacts of these mutations on cell shape and size [5]. In sharp contrast, we found that Hsp90 tends to buffer the effects of polymorphisms present in nature, providing support for the hypothesis that natural selection biases which context-dependent genetic effects persist in populations, leaving a false signal that organisms are canalized against genetic perturbation [5]. Our previous study, applying the same approach of using mutation-accumulation strains to another putative agent of canalization, the histone variant H2A.Z, also rejected this candidate protein as increasing mutational robustness [4]. These studies both call for reconsideration of: 1) the extent to which organisms are canalized against genetic change, and 2) what (if any) mechanisms support genetic canalization.
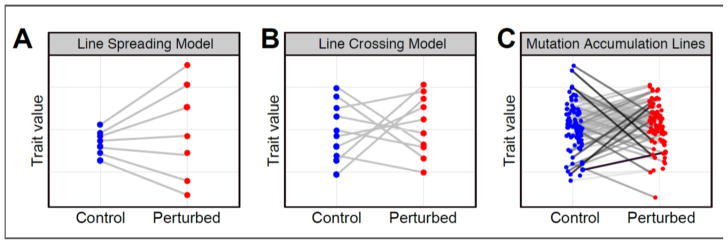
The context-dependent phenotypic impacts of new mutation also suggest that epistasis is common. We found that, for both Hsp90 and H2A.Z, their predominant influence on the mapping from genotype to phenotype is best described as neither potentiation nor buffering [5]. That is, both Hsp90 and H2A.Z affect the phenotypic impacts of many mutations without a strong overall tendency toward reducing or magnifying these impacts. This influence can be described as ‘line-crossing’ epistasis because of the appearance of a particular type of plot demonstrating how a perturbation, such as Hsp90 impairment, changes the rank order of strain means (Fig 1). Line crossing can be contrasted with line spreading, in which rank order does not change (and the lines therefore do not intersect each other) but the amount of phenotypic variation does change (Fig 1). Mechanistically, a predominance of line-crossing epistasis is not consistent with the idea that Hsp90 tends to buffer ancestral phenotypes by helping mutant proteins to fold. Alternative explanations as to how Hsp90 influences the phenotypic outcomes of different mutations in different ways include that it can help mutant proteins fold into states that impart novel phenotypes [41,46,48], and that it can encourage the degradation rather than the refolding of some mutant proteins [49,50].
Figure 1. Hsp90 is best described as neither a buffer nor a potentiator.
These plots display how the phenotypic effects of new mutations can change when the system is perturbed. Each circle represents a distinct genotype. The placement of the circles on the vertical axis represents the phenotypic variation among these genotypes, for a trait of interest. (A) A model of a perturbation that reveals previously buffered effects of mutations and therefore increases phenotypic diversity. This effect can be described as line spreading. A perturbation that reduces phenotypic diversity, causing line spreading in the opposite direction, is characteristic of potentiation. (B): A model of a perturbation that modifies the effects of new mutations in diverse, genotype-specific ways but shows no tendency toward increasing or reducing phenotypic diversity. This effect can be described as line crossing. (C): Inhibiting Hsp90 in yeast mutation accumulation lines changes cell morphology in line-specific ways. For most morphological features, including the one shown, this effect is more consistent with line crossing than line spreading. Lines are shaded darker the more different a line’s response to Hsp90 inhibition is relative to the response of the common ancestor of all the lines. The data in the rightmost panel, as well as the line crossing and spreading models, are adapted from Geiler-Samerotte et al 2016 [5]; original figures copyright the authors.
We propose that the most likely explanation for why Hsp90’s influence on new mutations leads predominantly to line-crossing epistasis follows directly from thinking about biological systems as being composed of networks of interacting parts. Many proteins that depend on Hsp90 for folding are regulators of signaling [36,41]. Although Hsp90 directly influences the impacts of mutations in these clients, it can also indirectly influence the impacts of mutations in non-clients that interact in some way with the signaling pathways in which clients participate. As we discuss in the next section, we perhaps can best understand Hsp90’s influence on the phenotypic impacts of mutations by expanding our view to look at full networks of interacting proteins [14,41].
3. Simple regulatory relationships create buffering or potentiating interactions
The impact of perturbing one protein in a regulatory network can often be modified by perturbations to other proteins. Depending on the network structure, particular proteins might have a tendency to suppress or enhance the phenotypic impacts of mutations in other proteins. Buffering and potentiation can result from very simple molecular interactions (Fig 2) and are ubiquitous across gene regulatory networks, although they often go by other names [51] (e.g. positive or negative interactions [52], genetic suppression [53], synthetic lethality [54]). Many mechanisms by which a molecule interacts with other molecules (e.g., helping other proteins to fold, binding an enhancer motif, blocking a protein’s active site) may result in genetic variation having context-dependent effects. In addition, interactions that do not involve direct molecular interactions may also produce context-dependent effects (e.g., epistasis between genes that independently promote cell proliferation and cell expansion [55]). Using Hsp90 as a case study, we next illustrate that whether a protein acts as a buffer or a potentiator does not necessarily depend on the mechanism by which it interacts with other molecules, but instead may depend on the network structure underlying those interactions and on the phenotype being surveyed.
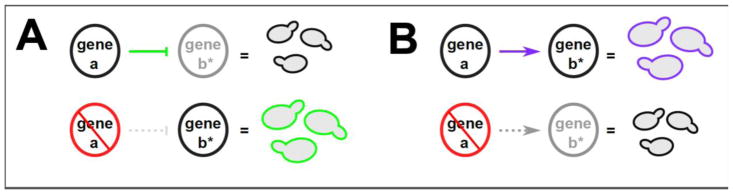
Figure 2. Buffering and potentiating interactions are prevalent within gene-regulatory networks.
Very simple molecular interactions result in buffering and potentiation. (A) A repressor can mask the phenotypic effects of mutations in its target. Shown here, a mutation in gene b, marked with an asterisk, would act to increase cell size, but this effect is buffered by gene a, which encodes a repressor of gene b’s expression. Loss of function of gene a (red) abrogates buffering. (B) An activator can enhance the phenotypic effects of mutations in its target. Shown here again is a mutation in gene b that increases cell size, but this time the effect is potentiated by gene a, which here encodes an activator of gene b’s expression. Loss of function of gene a (red) abrogates potentiation. Throughout the figure, green color indicates regulatory interactions that underlie buffering or formerly buffered traits; purple color indicates regulatory interactions that underlie potentiation or potentiated traits; and gray color indicates regulatory interactions that do not exist or genes that are not expressed in the particular scenario.
3.1. Regulatory role might explain when Hsp90 appears to be a buffer or potentiator
Because the effects of some Hsp90-interacting variants are revealed by Hsp90 inhibition whereas the effects of others are suppressed, there has been a confusing rift in the way previous literature describes the impact of Hsp90 on phenotypic variation. Hsp90 has been described as a genetic canalizer that promotes the stability of phenotypes over long periods [14,29]. But in other literature, Hsp90 has been described as a potentiator that promotes the emergence of new phenotypes [56–58]. Thinking of buffering and potentiation as separate phenomena confounds efforts to understand the mechanism that underlies Hsp90’s pervasive influence on mutant phenotypes. We propose that Hsp90’s buffering and potentiating abilities do not result from disparate mechanistic effects (e.g. whether Hsp90 folds mutant proteins into native or novel conformations [48,59]), but instead result from differences in the network architecture underlying the phenotype of interest. Indeed, there are some gene-regulatory networks in which Hsp90 predominantly acts as a buffer, and others in which it tends to act as a potentiator.
3.1.1. Hsp90 represses filamentation in yeast and buffers filamentation-inducing mutations
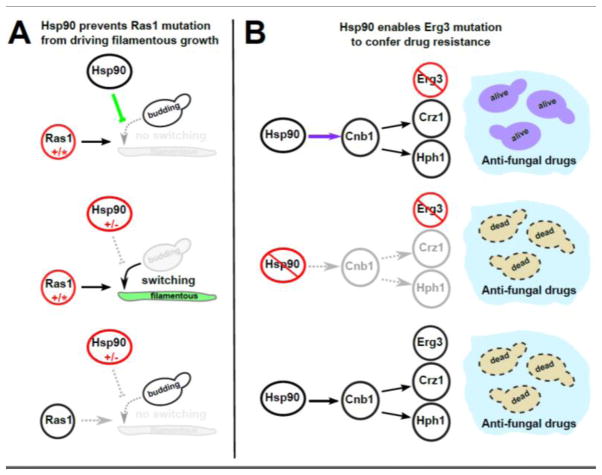
Yeasts such as Candida albicans and S. cerevisae undergo a switch from a budding cell morphology to a filamentous morphology that increases their virulence [60]. This switch is activated by Ras1-PKA signaling [61], and is repressed downstream of PKA function by Hsp90 [60]. Mutations that activate Ras1-PKA signaling, encouraging the switch from budding to filamentous morphology, are buffered by Hsp90 [60] (Fig 3A). Hsp90’s repressing and buffering roles go hand in hand; by independently repressing the switch to a filamentous morphology, Hsp90 buffers mutations that would activate this switch.
Figure 3. Hsp90 acts as a buffer of some phenotypes and a potentiator of others.
(A) Hsp90 buffers the filamentous growth phenotype that would otherwise result from a Ras1 mutation. In the top panel Ras1 is mutated in a manner that drives filamentous growth in yeast, but Hsp90 represses filamentous growth (green arrow) and therefore buffers the effect of the Ras1 mutation. In the middle panel, Hsp90 inhibition relieves repression of filamentous growth so the cryptic Ras1 mutation now causes filamentous growth. The bottom panel shows that filamentous growth is not a result of HSP90 inhibition alone, but instead is a result of the interaction between Hsp90 inhibition and the Ras1 mutation. (B) Hsp90 potentiates the drug resistance conferred by Erg3 loss of function. In the top panel Hsp90 activates Cnb1 (purple arrow), which in turn activates stress-response proteins that enable Erg3-mutant cells to survive, and therefore potentiates the effect of the Erg3 mutation. In the middle panel, Hsp90 inhibition prevents the Cnb1-dependent stress response so Erg3 loss of function is fatal. The bottom panel shows that wild type yeast are sensitive to the drug; resistance results from the interaction between Hsp90 presence and Erg3 loss of function. Similarly to Fig 2, red color indicates mutations or altered function; green color indicates regulatory interactions that underlie buffering or formerly buffered traits; purple color indicates regulatory interactions that underlie potentiation or potentiated traits; and gray color indicates regulatory interactions or phenotypic states that do not exist or gene products that are not active in the particular scenario.
3.1.2. Hsp90 enables stress responses in yeast and potentiates anti-fungal drug resistance
Whereas Hsp90 represses filamentation, it activates a cascade of proteins involved in the cell’s response to stress by helping another activator of this pathway, Calcineurin (Cnb1), to fold. This stress response allows yeast populations to evolve resistance to anti-fungal drugs via loss-of-function mutations in erg3. When Hsp90 is inhibited, erg3 mutants do not contribute resistance against anti-fungal drugs (Fig 3B); therefore, Hsp90 has been described as potentiating the evolution of drug resistance [58]. Loss-of-function mutations in genes encoding proteins downstream of Hsp90 in the stress response network, such as Crz1 and Hph1/Hph2, impair the anti-fungal resistance of erg3 mutants to varying degrees in different yeast strains [62]. When Hsp90 is inhibited, there are no longer phenotypic differences between these mutants in response to anti-fungal drugs; resistance is abrogated [62]. By activating this cascade of stress response proteins, Hsp90 potentiates the effects of the mutations they possess.
3.1.3. Hsp90 has opposite influences on the same mutation depending on the phenotype of interest
A screen for Hsp90-dependent genetic effects in yeast revealed that the same allele of the MEC1 gene has Hsp90-buffered effects on one phenotype (resistance to hydroxyurea), but Hsp90-potentiated effects on a different phenotype (resistance to ultraviolet light) [29]. Perhaps Hsp90 has a different effect on the folding of the same mutant protein when cells are exposed to hydroxyurea than when they are exposed to ultraviolet light, but a different mechanism to explain these results might be more likely. We propose that this MEC1 allele is buffered in one condition and potentiated in the other because Hsp90 plays different regulatory roles in the networks underlying resistance to hydroxyurea versus ultraviolet light.
4. No absolute distinction exists between buffers and potentiators
Using Hsp90 as a case study, we have shown that whether a protein acts as a buffer or a potentiator depends on context, including the phenotype being examined. Given that the impact of Hsp90 inhibition on a variety of phenotypes is best described as line crossing (rather than line spreading) [5] (Fig 1), using terms such as ‘buffering’ or ‘potentiation’ to summarize how a gene product generally interacts with genetic variation (Fig 1A) may be misleading. It might be safer to use the words ‘buffer’ and ‘potentiate’ to characterize specific interactions (i.e., Hsp90 buffers the effects of one mutation, and potentiates the effects of another). This view shifts the focus of canalization research toward understanding interactions, which are also discussed by other articles in this special issue on canalization (e.g. interactions that arise from gene regulation [35–37]). This view highlights that gene products with diverse mechanisms of action, in addition to protein folding, may buffer the phenotypic effects of some mutations and potentiate the effects of others. Indeed, we showed this previously for the histone variant H2A.Z [4]. Next, we will describe two examples of robust biological systems that drive home the point that buffering and potentiation are properties of interactions, not characteristics of a specific mechanism or gene product. Even in these examples of robust networks, we can find particular genetic changes that are potentiated by gene products that also act as buffers.
4.1. Potentiation in a robust sensory-organ specification network in Drosophila
A Drosophila melanogaster fly typically develops four scutellar bristles (sensory organs) on its posterior thorax, and the variance around this number in a wild-type sample of flies is very low. Loss of the microRNA miR-9a increases variance among isogenic flies, implying that miR-9a contributes to the robustness of sensory-organ specification against random fluctuations in the underlying developmental process [63]. Loss of miR-9a also increases variance among genetic backgrounds; that is, loss of miR-9a reveals cryptic genetic variation affecting the number of scutellar bristles [63]. miR-9a acts by repressing production of Senseless, a transcription factor required for sensory-organ specification. It is proposed that this regulation creates a threshold level of senseless mRNA concentration below which Senseless protein abundance remains low and above which protein abundance increases linearly with mRNA concentration [63]. More generally, this kind of threshold can be viewed as a buffering mechanism that creates a plateau in the relationship between gene activity and phenotypic outcome. Indeed, the number of scutellar bristles was one of the early experimental systems in which genetic canalization was studied, and the system motivated a model for canalization based on thresholds and the phenotypic plateaus they create [64,65]. Therefore, miR-9a serves as a fitting example of a molecule that is tempting to describe as a ‘buffer’.
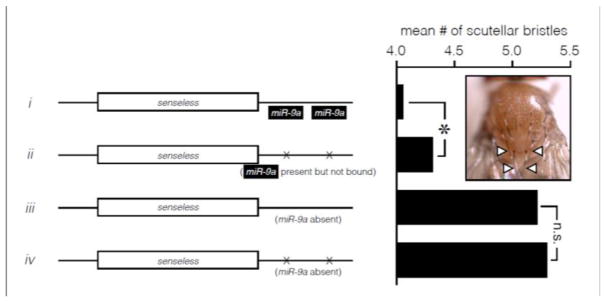
Although scutellar bristle development is a powerful and intuitive system for investigating buffering, it is important to note that miR-9a can act as a potentiator in this system as well. It is easy to envision mutations that would be buffered by miR-9a in their effects on scutellar bristle number. For example, any mutation that caused a slightly higher baseline level of senseless transcription would not cause a shift upwards in the average number of scutellar bristles, as long as the threshold mRNA concentration was not surpassed. Nonetheless, it is also possible to envision mutations that would be potentiated by miR-9a. Indeed, such mutations are known to exist: mutations in the miR-9a binding sites in the 3′ UTR of senseless mRNA are only relevant in the presence of miR-9a [63] (Fig 4). Thus, even if miR-9a is found to have a tendency toward more buffering interactions than potentiating interactions, it would be advisable to describe it in terms of this tendency rather than to call it a buffer.
Figure 4. miR-9a potentiates a mutation in the miR-9a-binding sites in the 3′ UTR of the senseless mRNA.
Data from Cassidy et al 2013 [63] show that miR-9a can act as a potentiator. Transgenic flies carrying wild-type or mutated copies of senseless were assayed in a wild-type (i, ii) or miR-9a-mutant (iii, iv) genetic background. Left: Schematic representations of the senseless mRNAs in the transgenic flies, with coding sequence as white box and UTRs as lines. X symbols denote mutations to two miR-9a binding sites in the senseless 3′ UTR. Black boxes in (i) show miR-9a bound to two binding sites in the senseless mRNA 3′ UTR. Right: Bar plot of mean scutellar bristle numbers in the four transgenic lines depicted at left. When miR-9a is present, a statistically significant difference in mean scutellar bristle number between flies carrying a wild-type vs. mutant senseless 3′ UTR was seen (i vs. ii, asterisk). But when miR-9a function is eliminated, mutations to the senseless 3′ UTR have no significant effect on mean bristle number (iii vs. iv, n.s.). In other words, miR-9a potentiates the phenotypic effect of mutating the senseless 3′ UTR. The inset image of the dorsal side of a fly thorax shows the stereotypical four scutellar bristles (arrowheads).
Elsewhere in the same robust sensory-organ specification network lies another example of potentiation. The cell fate decision to form a scutellar bristle is controlled by the Notch signaling pathway; lateral inhibition interactions, as well as cell-autonomous interactions, involving Notch and its ligands Delta and Serrate ensure that only four cells attain this fate [66]. Wild-type flies and flies that are heterozygous for a null Serrate allele have very low rates of duplicating a scutellar bristle (approximately 1%). Whereas flies heterozygous for a Notch-null allele have a 28% rate of duplicating a scutellar bristle, flies doubly heterozygous for a Notch-null allele and a Serrate-null allele have less extreme phenotypes (only 13% duplication rate) [66]. This observation demonstrates that, when present at wild-type levels, the Serrate ligand potentiates (i.e., enhances) the impact of the Notch null mutation. This example is a classic case of genetic suppression, and in general any cases of genetic suppression can be reframed as cases of potentiation. Thus, to find additional examples of potentiation, one could simply compile the results of genetic suppressor screens [67,68].
4.2. Potentiation in a robust circadian clock
The circadian clock is a prime example of a biochemical network whose reliability is important for organisms spanning bacteria, fungi, plants and animals. Generally, the clock is composed of proteins undergoing a full production and depletion cycle over a period of 24 hours to match the light/dark cycles of the day. The clock’s period is highly robust to differences in temperature [69], as well as to stochastic fluctuations [70] and mutations [71] that affect the abundances of its key protein components. Disruption of the circadian clock is linked to a number of negative consequences, including higher cancer incidence [72–74] and early aging [75]. For these reasons, the circadian clock is another fitting example of a robust system.
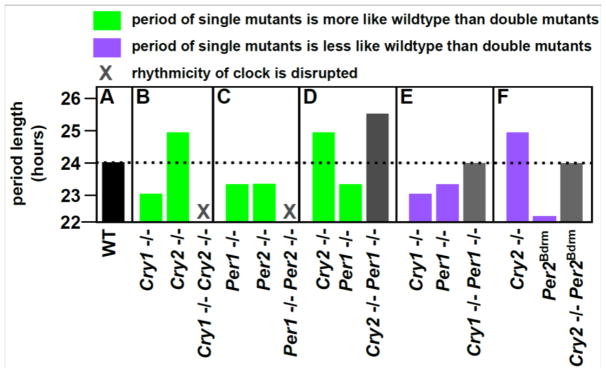
Within the clock system, however, are both buffering and potentiating interactions (Fig 5). The abundances of the Per1, Per2, Cry1, and Cry2 proteins cycle over the clock’s 24-hour period. The oscillation is mediated by negative feedback: the proteins form Per/Cry heterodimers that translocate to the nucleus and inhibit transcription of the Per and Cry genes [76]. There are several buffering interactions among these clock proteins. Loss of function of any one of the four is insufficient to completely disrupt circadian rhythmicity [77]. Per1 and Per2 each buffer to some extent the impact of loss of the other, as do Cry1 and Cry2 [77]. Per1 also acts as a buffer with respect to Cry2 mutations, as Cry2; Per1 double knockouts display more severe period lengthening than Cry2 knockouts alone [76]. However, even in this robust network there are points of sensitivity that permit change [76,78,79]. Per1 potentiates the effects of null mutations in Cry1. Whereas the circadian period of Cry1 mutants is shortened by 1 hour relative to wild-type mice [80,81], normal period length is restored in Cry1; Per1 double mutants [79]. Similarly, Cry2 potentiates the effects of Per2 knockouts on circadian rhythmicity and normal clock gene expression patterns [76]. As a result, none of these four proteins can be classified as a buffer or a potentiator. As with the Hsp90 examples, and with the miR-9a example, buffering and potentiation are properties of interactions not gene products.
Figure 5. Circadian clock proteins Per and Cry exhibit both buffering and potentiating interactions.
The figure depicts the effects of clock protein mutations (horizontal axis) on the period length of the circadian clock (vertical axis). Panel (A) displays the period length of wild-type mice, which is approximately 24 hours. Each subsequent panel shows the approximate period lengths of two individual mutants and of the corresponding double mutant. Panels (B–D) depict buffering: the effects of single gene knockouts are less severe than the effects of the corresponding double knockouts. Panels (E) and (F) depict potentiation: single mutations have a more severe impact on period length than do the corresponding double mutants. Data are from Yu et al 2011, van der Horst et al 1999, and Oster et al 2002 and 2003 [76,77,79,80].
5. Simulated gene-regulatory networks show buffering and potentiation
The presence of potentiation among famous examples of putative ‘buffers’ (e.g. Hsp90, H2A.Z, GroEL, miR-9a, and clock proteins) raises the possibility that many gene products might buffer the effects of some new mutations and potentiate the effects of others, making line-crossing epistasis common. To investigate how typical gene products might interact with new mutations, we used an established modeling framework for simulating complex gene-regulatory networks [82–87]. Specifically, we asked: 1) whether the relative prevalence of line crossing vs. line spreading in nature is an expected feature of interaction networks that underlie biological systems; and 2) whether the regulatory relationship between a gene product and a mutated gene can predict whether buffering or potentiation would be observed.
Our goal here was to use this modeling framework to investigate the extent to which random regulatory relationships produce genetic interactions that resemble buffering versus potentiation. The following analysis is by no means intended to be an exhaustive investigation of different regulatory architectures and complexities. Instead, it is meant as a first step toward understanding empirical observations through modeling of regulatory networks, which will be important in addressing the major open questions in the field (see Section 6 below and the paper by A. Badyaev in this special issue [37]).
In brief, the model considers N genes, each of which encodes a transcription factor that, in principle, can regulate expression of any of the N genes. The regulatory relationships are captured in an N X N matrix, W. Each element of W, wij, represents the strength and direction of the regulatory influence on gene i of the transcription factor encoded by gene j. To simulate a gene-regulatory network, a W matrix is sampled with a specified probability that wij = 0. Here, this probability was set such that the mean numbers of regulators and targets per gene are 3. The nonzero wij are sampled from the standard Normal distribution. An initial state vector of the network (of length N) is also sampled such that each gene i starts out either not expressed (expressed at a level of 0) or fully expressed (expressed at a level of 1), with equal probability. An individual’s gene-expression phenotype is the vector of steady-state expression levels of its genes. This gene-expression phenotype is obtained by iterated multiplication of W by the current expression-level vector, with each value x in the resulting vector at each iteration passed through a sigmoidal function (here 1/[1+e−x]), to make the response to gene regulation nonlinear in a way that resembles actual transcriptional-regulatory networks. In other words, regulatory inputs to a gene combine by first summing the influences of each transcription factor (the transcription factor’s current expression level multiplied by the relevant wij element), then applying the sigmoidal transformation so that net positive influences of sufficient intensity effectively turn the gene completely on, net negative influences of sufficient intensity effectively turn the gene completely off, and there is a smooth transition between the two extremes. When a mutation is simulated, a single nonzero wij is chosen at random and its value is changed to a new draw from the standard Normal distribution. When a gene is knocked out, all values in the corresponding row and column of W are set to 0, and its steady-state expression level is set to 0. An individual’s gene-expression phenotype, a vector of length N, is converted to a single scalar value, which we refer to simply as the individual’s ‘phenotype’, by taking the Euclidean distance of the gene-expression phenotype of the individual to that of a reference individual and dividing this distance by N. By definition therefore the phenotype of the reference individual is 0, and the maximum phenotype is 1. Not all networks reach steady state through the iterative process described above; an individual is deemed to be inviable (and is assigned a phenotype of 1) if, within 100 iterations, its network does not converge to a steady state [83].
We ran 5000 simulations of 25-gene networks in each of which a randomly sampled, viable ancestral genotype was allowed to accumulate mutations independently in 100 derivative lines. These 100 lines simulate a single collection of mutation accumulation (MA) lines, and each of the 5000 simulations represents an independent MA line collection derived from a different ancestor. For each collection, we measured the phenotype for each of the 100 lines (distance of steady-state gene-expression levels from those of the ancestor), as well as for the 25 single-gene knockout derivatives of each line (2600 phenotypes total, per collection). For each collection of MA lines, the difference in phenotypic variation (i.e., between-line standard deviation) between the lines without any knockout and the lines with each knockout is a measure of how much buffering versus potentiation exists. If more phenotypic variation is seen among the lines with a particular knockout than among the same lines without the knockout, then the knocked-out gene tends to buffer the effects of mutations (as in Fig 1A). If, in contrast, less variation is seen among the lines with a particular knockout than among the same lines without the knockout, then the knocked-out gene tends to potentiate the effects of mutations.
In a result that very closely resembles the experimental result for Hsp90’s effects on single-cell morphological variation in budding yeast [5], we find that gene knockouts tend to reduce phenotypic variation (Fig 6A), indicating that potentiation is more common than buffering in simulated regulatory networks. We simulated knockouts of each gene in turn for each of the 5000 simulations, yielding 125,000 comparisons of wild-type vs. knockout between-line standard deviations. The mean difference obtained by subtracting the wild-type between-line standard deviation from the knockout between-line standard deviation is negative (−1.36 × 10−5), a highly significant departure from the null expectation of zero difference (two-sided one-sample Wilcoxon signed-rank test, P < 2.2 × 10−16). Within each simulation, some genes might tend to potentiate the effects of mutations (Fig 6A; points below line), while other genes might tend to buffer the effects of mutations (Fig 6A; points above line). Nonetheless, for a majority (2693) of the 5000 simulated networks, the median standard-deviation difference was negative, and for 186 of these, a 95% confidence interval around the median standard-deviation difference did not overlap zero (Fig 6A; purple boxplots). For comparison, for 2283 of the 5000 simulations, the median standard-deviation difference was positive, and for 94 of these, a 95% confidence interval around the median standard-deviation difference did not overlap zero. These results therefore suggest a small but significant bias toward potentiation.
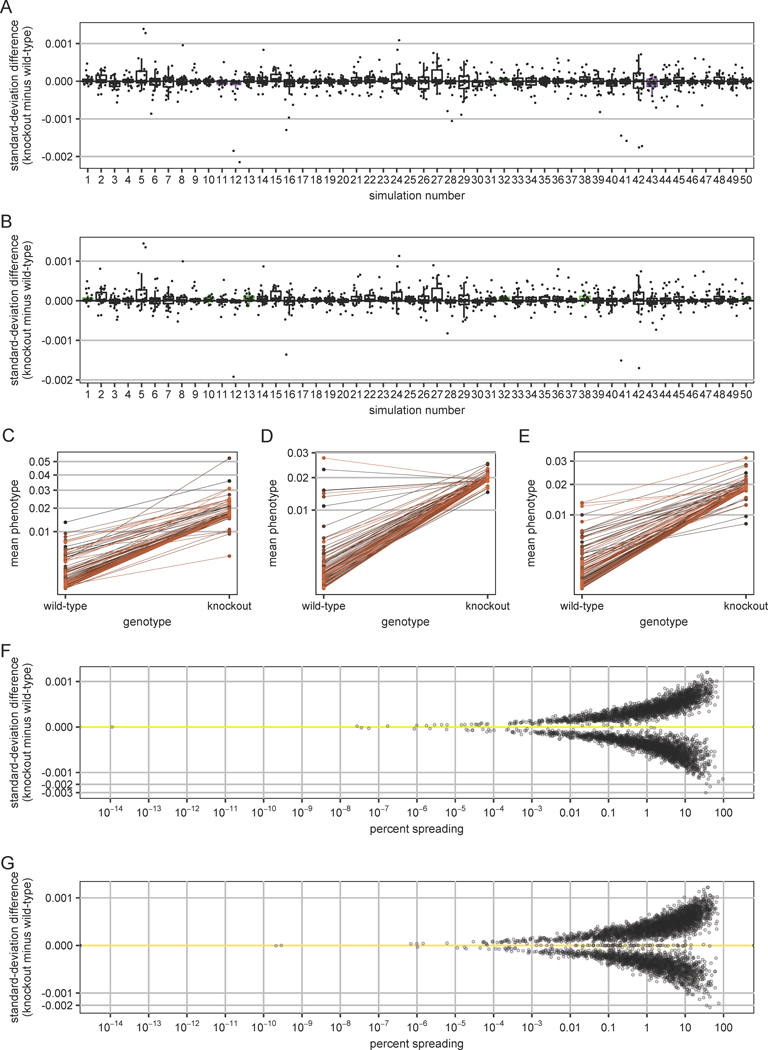
Figure 6. Simulated gene networks recapitulate tendencies toward potentiation and line-crossing epistasis.
(A) Simulated networks show a tendency toward potentiation, as gene knockouts tend to decrease the amount of phenotypic variation among mutant lines. Each boxplot represents one of 5000 simulated gene-regulatory networks. For legibility, only the first 50 of the 5000 simulations are shown; these 50 are representative of the rest. For each simulation, we calculate the standard deviation of the phenotypes of 100 mutated networks that each differ from a shared ancestor by a single mutation. The vertical axis represents how the standard deviation across these mutated networks changes upon another mutation — a knockout of one gene in the network — where each point represents a knockout of a different gene. Points are randomly offset from vertical alignment to make them more visible. The hinges of each boxplot show the interquartile range of the change in standard deviation for 25 different gene knockouts. The whiskers of each box extend in each direction to the most extreme values that are no further than 1.5 times the interquartile range from the hinge. Boxes corresponding to simulations for which the estimated 95% confidence interval around the median does not overlap zero are shown in purple (negative median) or green (positive median). Confidence intervals are estimated as the median plus or minus 1.58 times the interquartile range divided by the square root of the number of genes in each network. (B) The same plot as in (A) is shown except that phenotype calculations excluded the knocked-out gene. The random offsets from vertical alignment in (A) are preserved so as to allow some comparison of how changing the phenotype calculation in this way changes the standard-deviation difference. (C) A line-crossing plot is shown for the strongest buffering effect observed in any simulation (i.e., the gene knockout causing the largest increase in between-line standard deviation) except for those simulations in which any of the MA lines is lethal with or without the knockout, as the absolute largest increase in between-line standard deviation corresponds to a case in which the knockout makes a few previously viable MA lines inviable. Each point represents the phenotype (excluding the knocked-out gene) for one of the 100 mutated networks of this simulation (vertical axis) with or without gene knockout (horizontal axis); lines connect values corresponding to the same mutated network. Lines are shaded different colors as a visual aid. (D) The same plot as in (C) except shown is the strongest potentiation effect observed in any simulation (i.e., the gene knockout causing the biggest decrease in between-line standard deviation) except for those simulations in which any of the MA lines is lethal with or without the knockout, as the absolute largest decrease in between-line standard deviation corresponds to a case in which the knockout makes a few previously inviable MA lines viable. (E) The same plot as in (C) is shown except for a case where line crossing dominates. This case is the simulation and knocked-out gene for which the total interaction variance (crossing plus spreading) was in the highest one percent, for which the spreading component was lowest, and for which no MA line is lethal with or without the knockout. (F) Line-crossing epistasis dominates in these simulations. The difference in between-line standard deviation upon gene knockout is plotted against the percent spreading. The difference in between-line standard deviation is shown on a cube-root scale to better show points with differences near zero. The percent spreading, which is often < 1%, is shown on a logarithmic scale. Each point represents 1 of 5000 simulated regulatory networks. For legibility, only the result of knocking out the first gene of each network is plotted; these results are representative of those obtained from each of the other genes. (G) The same plot as in (F) is shown except that phenotype calculations excluded the knocked-out gene.
It is important to note that this bias toward potentiation depends on the way we compute phenotypes after gene knockouts. Above, we included each gene’s contribution in the phenotype calculation, including the knocked-out gene. Thus, if the knocked-out gene varies among the 100 MA lines then it may contribute to phenotypic variation among the 100 wild-type MA lines but not among the 100 MA lines with the knockout. This difference in contribution to total variation therefore tilts in the direction of potentiation. Consistent with this reasoning, the bias shifts towards buffering when we exclude the knocked-out gene from phenotype calculations. The mean difference obtained by subtracting the wild-type between-line standard deviation from the knockout between-line standard deviation is now significantly positive (2.91 × 10−5; two-sided one-sample Wilcoxon signed-rank test, P < 2.2 × 10−16) and for 3927 of the 5000 simulated networks, the median standard-deviation difference is now positive. For 529 of these, a 95% confidence interval around the median standard-deviation difference did not overlap zero (Fig 6B; green boxplots).
It is unclear which represents a more valid assumption: including the knocked-out gene in phenotype calculations or excluding it. Biologically, excluding the knocked-out gene means assuming that the transcription factor it encodes only influences phenotype by regulating the activities of the other transcription factors in the network. It seems likely that the vast majority of transcription factors that regulate other transcription factors also have other direct targets through which they influence phenotype, in which case including the knocked-out gene in phenotype calculations is more realistic. More broadly, knocking out a regulator of any kind means that whatever variation had existed in that regulator before the knockout — either cis variation in the gene encoding the regulator itself or trans variation in targets or clients that changes the strength of interaction with the regulator — can no longer influence phenotype. This simple observation might therefore suggest that a bias toward potentiation (as seen for Hsp90) should be expected rather than a surprise.
To a large extent, it does not matter whether the difference in between-line standard deviation runs in the direction of potentiation or the direction of buffering, because the more important result is that the difference is small, particularly in relation to the total amount of epistasis. As in the previous experimental work, we can partition the line-by-knockout interaction variance into components representing line crossing (changes in rank order upon knockout; Fig 1B) and line spreading (change in overall variance upon knockout; Fig 1A). Again the result very closely resembles the experimental result for Hsp90 (Fig 1C). Some knockouts in some simulations show substantial spreading, either in the direction of the gene tending to act as a buffer (Fig 6C) or in the direction of the gene tending to act as a potentiator (Fig 6D). However, line crossing is far more salient than line spreading (Fig 6E). Across all simulations and knockouts, the median percentage of the interaction variance in the line-spreading component is merely 1.6% (2.1% when the knocked-out gene is excluded from phenotype calculations). In greater than 80% of cases, the percentage of the interaction variance due to spreading is less than 10% (whether including the knocked-out gene in phenotype calculations or not) (Fig 6F–G). In other words, epistasis between knocked-out genes and mutations elsewhere in the network is not usually biased toward buffering or potentiation, but may be described as line-crossing epistasis. When we do observe a bias (i.e. significant line spreading), it tends toward potentiation when the knocked-out gene is included in phenotype calculations (Fig 6F; more points below the yellow line) and toward buffering when the knocked-out gene is excluded from phenotype calculations (Fig 6G; more points above the yellow line).
5.1. Regulatory relationships in simulated gene-regulatory networks predict buffering and potentiation
The biological networks that connect Hsp90 to morphological phenotypes are largely unknown, but simulated networks are of course completely known. The simulated networks therefore afford an opportunity to pose questions that cannot be posed otherwise. In particular, we can ask whether any regulatory features are associated with the tendency to buffer or potentiate. First, we asked whether the number of targets a gene regulates is correlated with the effect of its knockout on between-line standard deviation. Indeed, the more a gene is an outgoing hub the more it potentiates the effects of mutations: there is a significant negative correlation between a gene’s influence on between-line standard deviation and its weighted outgoing node degree, a measure of a gene’s targets that accounts for different strengths of interaction (Spearman’s rho = −0.183, P < 2.2 × 10−16 when the knocked-out gene is included in phenotype calculations and Spearman’s rho = −0.047, P < 2.2 × 10−16 when the gene is excluded).
Our examples above (Fig 2 & 3) hinted toward a relationship between whether a regulator is an activator or a repressor and whether it tends to act as a potentiator or buffer. In the simulated networks, any given transcription factor can be an activator of some targets and a repressor of others. We can use the sum of outward influences of a transcription factor to measure the extent to which it plays an activating versus repressing role. The association between this sum and potentiation is consistent with our interpretation of specific Hsp90 cases above: the more a transcription factor tends to act as an activator, the more it potentiates the effects of mutations (Spearman’s rho = −0.112 for sum of outward influences vs. standard-deviation difference, P < 2.2 × 10−16 when the knocked-out gene is included in phenotype calculations and Spearman’s rho = −0.120, P < 2.2 × 10−16 when the knocked-out gene is excluded from phenotype calculations).
The gene-network simulations presented here are admittedly of narrow scope, leaving many potentially important parameters to explore, such as: the overall size of the network; the topology of the network, as reflected in its overall connectedness as well as the degree distribution and hierarchical organization of its members; the potentially nonrandom clustering of activators and repressors in the network; and the potential association between a regulatory interaction’s current magnitude and sign (activating or repressing) and the probability that a mutation will change the interaction to a particular magnitude and sign. In addition, there might be differences in the conclusions reached when studying random viable networks (as we did here) versus networks that are the product of evolution. Nevertheless, the success of the simulations presented here in recapitulating patterns seen in empirical data suggests that further analysis of this model, and more realistic models related to it [87], might be useful in making sense of context-dependent genetic effects as well as their evolutionary impacts and fates. Because modeled networks are mappings from genotype to phenotype that are at once complex and completely transparent, they provide a unique tool for generating new hypotheses that can motivate new experiments. Below, as we highlight the key open questions in the field, we note the role that modeling might play in answering them.
6. Open Questions
6.1. Do genetic canalization or genetic canalizers exist?
We have argued that buffering and potentiation are properties of interactions, not individual gene products. We have reviewed evidence that Hsp90 is not an agent of genetic canalization but is, instead, a highly interactive protein that buffers some mutational effects and potentiates others. We have highlighted potentiating interactions in systems generally framed as robust. Are, then, any biological systems genetically canalized? Are any gene products genetic canalizers? In other words, do any gene products show a consistent pattern of line-spreading epistasis (Fig 1A), rather than predominantly line-crossing epistasis (Fig 1B)? One goal of our attempt to decanalize thinking about genetic canalization is to raise these questions — to move away from the notion that genetic canalization is common and expected.
Nonetheless, we acknowledge the possibility that genetic canalization exists. For example, a particular protein’s structure (or a functional RNA’s structure) may be canalized against the effects of new mutations [88–90]. However, what we discuss throughout this review is the canalization of complex traits. Given that such traits are the product of interactions between many molecules, proteins, and pathways, we argue that canalization of complex traits may be a systems-level property, rather than an effect contributed by any single gene product that acts as a genetic canalizer. Scutellar-bristle development and the circadian clock might yet be examples of canalized systems; despite our ability to find cases of potentiation in these networks, those cases might be far outnumbered by buffering interactions.
Given natural selection can leave a false signal of robustness (the converse error we described earlier), candidate genetically canalized systems must be studied by assessing how perturbations modify the effects of new mutations, rather than genetic variants that have survived natural selection. Regulatory-network modeling might help identify candidate genetically canalized systems by revealing specific network features that generate a bias toward buffering interactions.
In general, our view motivates a shift from screening for single gene products that act as agents of genetic canalization to asking questions about the network features or genetic interactions that cause (or do not cause) canalization. In addition to these questions, detailed below, we hope that decanalizing the discussion of canalization opens up the floor for others to ask: what is the right way to think about and study canalization?
6.2. What types of genetic interactions cause buffering vs. potentiation?
Understanding the molecular features associated with buffering and potentiation could improve the mapping from genotype to phenotype by suggesting genes or pathways in which changes are likely to be buffered or likely to alter phenotypes. Genes likely to accumulate buffered variation could represent higher priority candidates in the search for cryptic genetic variation that contributes to complex human disease [18], whereas genes likely to have variants with potentiated effects could be candidate targets of adaptive evolution [58]. Simulated regulatory networks suggest that features that bias toward buffering include negative (repressive) regulatory interactions, whereas features that bias toward potentiation include positive (activating) regulatory interactions. Further modeling could confirm these trends or suggest additional ones.
Experiments measuring the context-dependent effects of many mutations could validate inferences from modeling or identify cases where models fail to make accurate predictions. For example, experiments could test whether the connection between activation and potentiation holds up across a large collection of mutations with Hsp90-dependent effects. If such experiments are not limited by natural genetic variation but utilize gene-editing technologies to collect large numbers of mutations in particular genes or pathways, they might have more power to detect trends (and they also have the advantage of being unbiased by selection).
6.3. Which gene products are more likely to participate in interactions with natural genetic variation?
Some gene products, for example chaperones that interact with a large portion of the proteome [36], seem primed to act as ‘global genetic modifiers’, altering the effects of genetic variation in many other genes and demonstrating line-crossing epistasis with many new mutations [40]. Many questions remain about these global modifiers [40], including: 1) what is the full range of molecular functions that they represent, 2) what positions do they occupy in regulatory networks, 3) to what extent do they themselves harbor functional variation in nature, and 4) to what extent do laboratory interactions (usually involving large-effect mutations) predict natural interactions (usually involving mutations of more subtle effect). In addition, one might consider whether ‘local’ genetic modifiers exist that influence the phenotypic effects of variation in specific pathways or regulatory subnetworks, and whether the effects of local modifiers are different in kind, or only in scale, from global modifiers [34]. Answering these questions will require screens designed to identify modifiers, characterization of their molecular functions and cellular interactions, and population-genetic and quantitative-genetic experiments to understand their modulation of the effects of natural variation [40].
6.4. What evolutionary forces lead to accumulation of buffered vs. potentiated variants?
Natural selection can bias genetic interactions toward buffering or potentiation. Our current expectation is that genes under stabilizing selection will tend to accumulate buffered genetic variation [5] whereas genes that experienced recent adaptive evolution will be enriched for potentiated variants [56,58]. Evolution is of course more complicated than these two scenarios, so further attention should be paid to the potential impacts of finite population sizes, selection pressures that vary in time, and demographic and biogeographic effects such as population and range expansion. Moreover, selection on one trait might constrain evolution of a correlated trait, so questions of how pleiotropy and modularity shape buffering and potentiation (and vice versa) should be explored as well. To these ends, modeling can clarify expectations and provide testable hypotheses. For example, simulated networks could be used to investigate whether different types of selection enrich for different types of network features with different buffering or potentiation properties, and whether certain features are more difficult to evolve or to maintain than others.
It has long been hypothesized that selection to buffer the effects of environmental variation would cause genetic canalization to evolve as a by product [91,92]; this is called the congruence hypothesis [24]. Hsp90 and H2A.Z results do not support this hypothesis; both buffer microenvironmental variation but neither increases robustness against the effects of mutations [4,5]. In general, the evidence for congruence is mixed [31,39] and there are theoretical reasons why congruence might not apply to particular regulatory systems [93], but it remains an open question why there is no congruence in these specific cases and instead why there appears to be a link between buffering microenvironmental variation and a predominance of line-crossing epistasis.
6.5. Do networks enriched for buffering vs. potentiation have different evolutionary potential?
Previous studies have pointed out that tuning the levels of a genetic modifier can impact evolutionary trajectories [57]. For example, dialing down the level of a potentiator – a gene product that enhances the impact of genetic changes on phenotype – would reduce phenotypic diversity. This might interfere with adaptive evolution by reducing the chances that a mutant phenotype exists that is able to survive a novel condition. In line with this idea, several studies have suggested inhibiting Hsp90 as a strategy to limit the ability of tumors [56] or infectious microbes [58] to evolve resistance to drugs. Problematically, Hsp90 does not always act as a potentiator. For phenotypes that tend to be buffered by Hsp90, this treatment will have the opposite effect, i.e. reducing the levels of Hsp90 will reveal phenotypic diversity and improve adaptive potential [6,15].
Because line-crossing epistasis appears to be prevalent, at least in simulations and in the two studied cases of Hsp90 and H2A.Z [4,5], its impact on evolutionary potential needs to be considered. At minimum, prevalent line-crossing epistasis should make evolutionary trajectories highly contingent on starting genetic backgrounds, the specific mutations that arise, and any environmental factors that impact a gene that has epistatic interactions. This possibility makes it critical to develop more theory addressing how complex-trait evolution proceeds when genetic architectures are highly epistatic and pleiotropic [2,94,95]. One way to directly ask questions about what happens when we replay life’s tape across contexts that subtly differ is by performing laboratory evolution experiments that have sufficient power to identify adaptive mutations and quantify their effects with high resolution [96,97]. Any amount of predictability amidst contingency could be practically useful in identifying the degree to which evolutionary outcomes are context-dependent and whether any trends explain when evolutionary outcomes shift.
7. Conclusion
Decanalizing our thinking on canalization reveals a latent opportunity to gain insights into the panoply of mechanisms that modulate variation in complex traits. In an era when clinicians aim to use personalized genetic information to achieve precision medicine, understanding such mechanisms is key. It is unclear at present whether pessimism or optimism about this endeavor is appropriate. A pessimist might raise these findings of pervasive line-crossing epistasis [4,5] — as well as clear cases of high-order epistasis involving more than pairwise interactions [98–100] — to argue that mutational effects on complex traits are so highly contingent that they are largely unpredictable. An optimist might counter with the regularities that might be at play, such as expected differences between activating and repressing interactions in regulatory networks or the way natural selection prunes variation. Much research, addressing the open questions highlighted above, is needed.
Highlights.
Gene products underlying genetic canalization should buffer the effects of mutations
Putative canalizers buffer some mutations’ effects but potentiate others’ effects
Buffering and potentiation emerge from interactions in gene-regulatory networks
Although epistasis might be common, genetic canalization of complex traits might not
Acknowledgments
We thank Dmitri Petrov for useful discussions. We thank the editors and reviewers for useful comments on an earlier version of this manuscript. This work was supported by National Institutes of Health grant R35GM118170 to MLS, and fellowship F32GM103166 to KAGS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469–469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavlicev M, Wagner GP. Evolutionary Systems Biology: Shifting Focus to the Context-Dependency of Genetic Effects. Integrative Organismal Biology. 2015:91:108. [Google Scholar]
- 3.Hermisson J, Wagner GP. The population genetic theory of hidden variation and genetic robustness. Genetics. 2004;168:2271–2284. doi: 10.1534/genetics.104.029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson JB, Uppendahl LD, Traficante MK, Levy SF, Siegal ML. Histone variant HTZ1 shows extensive epistasis with, but does not increase robustness to, new mutations. PLoS Genet. 2013;9:e1003733. doi: 10.1371/journal.pgen.1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML. Selection Transforms the Landscape of Genetic Variation Interacting with Hsp90. PLoS Biol. 2016;14:e2000465. doi: 10.1371/journal.pbio.2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitesell L, Santagata S, Lin NU. Inhibiting HSP90 to treat cancer: a strategy in evolution. Curr Mol Med. 2012;12:1108–1124. doi: 10.2174/156652412803306657. [DOI] [PubMed] [Google Scholar]
- 8.Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays. 2000;22:1095–1105. doi: 10.1002/1521-1878(200012)22:12<1095::AID-BIES7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 9.Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, et al. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci USa. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi KH. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Mol Ecol. 2013;22:1356–1365. doi: 10.1111/mec.12091. [DOI] [PubMed] [Google Scholar]
- 11.Peuß R, Eggert H, Armitage SAO, Kurtz J. Downregulation of the evolutionary capacitor Hsp90 is mediated by social cues. Proc Biol Sci. 2015;282:20152041. doi: 10.1098/rspb.2015.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabater-Muñoz B, Prats-Escriche M, Montagud-Martínez R, López-Cerdán A, Toft C, Aguilar-Rodríguez J, et al. Fitness Trade-Offs Determine the Role of the Molecular Chaperonin GroEL in Buffering Mutations. Mol Biol Evol. 2015;32:2681–2693. doi: 10.1093/molbev/msv144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohner N. Biology and Evolution of the Mexican Cavefish. Elsevier; 2016. Selection Through Standing Genetic Variation; pp. 137–152. [DOI] [Google Scholar]
- 14.Salathia N, Queitsch C. Molecular mechanisms of canalization: Hsp90 and beyond. J Biosci. 2007;32:457–463. doi: 10.1007/s12038-007-0045-9. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 16.Waddington CH. Canalization of Development and the Inheritance of Acquired Characters. Nature. 1942;150:563–565. doi: 10.1038/150563a0. [DOI] [Google Scholar]
- 17.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 18.Gibson G. Decanalization and the origin of complex disease. Nat Rev Genet. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- 19.Gibson G, Reed LK. Cryptic genetic variation. Curr Biol. 2008;18:R989–90. doi: 10.1016/j.cub.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger D, Bauerfeind SS, Blanckenhorn WU, Schäfer MA. High temperatures reveal cryptic genetic variation in a polymorphic female sperm storage organ. Evolution. 2011;65:2830–2842. doi: 10.1111/j.1558-5646.2011.01392.x. [DOI] [PubMed] [Google Scholar]
- 21.Dammerman KJ, Steibel JP, Scribner KT. Increases in the mean and variability of thermal regimes result in differential phenotypic responses among genotypes during early ontogenetic stages of lake sturgeon (Acipenser fulvescens) Evol Appl. 2016;9:1258–1270. doi: 10.1111/eva.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shama LNS. The mean and variance of climate change in the oceans: hidden evolutionary potential under stochastic environmental variability in marine sticklebacks. Sci Rep. 2017;7:8889. doi: 10.1038/s41598-017-07140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegal ML, Masel J. Hsp90 depletion goes wild. BMC Biol. 2012;10:14. doi: 10.1186/1741-7007-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Visser JAGM, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 25.Paaby AB, Rockman MV. Cryptic genetic variation: evolution’s hidden substrate. Nat Rev Genet. 2014;15:247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 27.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 28.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangster TA, Salathia N, Lee HN, Watanabe E, Schellenberg K, Morneau K, et al. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci USa. 2008;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharloo W. Canalization: Genetic and developmental aspects. Annual Review of Ecology and Systematics. 1991;22:65–93. doi: 10.1146/annurev.es.22.110191.000433. [DOI] [Google Scholar]
- 32.Loison L. Canalization and genetic assimilation: reassessing the radicality of the Waddingtonian concept of inheritance of acquired characters. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Waddington CH. The Strategy of the Genes, a Discussion of Some Aspects of Theoretical Biology, by C. H. Waddington,… With an Appendix [Some Physico-chemical Aspects of Biological Organisation] by H. Kacser,…. 1957. [Google Scholar]
- 34.Takahashi KH. Multiple modes of canalization: links between genetic, environmental canalizations and developmental stability, and their trait-specificity. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Draghi JA. Links between evolutionary processes and phenotypic robustness in microbes. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Zabinsky R, Mason G, Queitsch C, Jarosz D. It’s not magic - Hsp90 and its effects on genetic and epigenetic variation. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badyaev AV. Evolutionary transitions in network controllability reconcile adaptation with continuity of evolution. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Hallgrímsson B, Green RM, Katz J, Fish J, Bernier F, Roseman CC, et al. Developmental-Genetics of Canalization. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegal ML, Leu JY. On the Nature and Evolutionary Impact of Phenotypic Robustness Mechanisms. Annu Rev Ecol Evol Syst. 2014;45:495–517. doi: 10.1146/annurev-ecolsys-120213-091705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schell R, Mullis M, Ehrenreich IM. Modifiers of the Genotype-Phenotype Map: Hsp90 and Beyond. PLoS Biol. 2016;14:e2001015. doi: 10.1371/journal.pbio.2001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 42.Miyata Y, Nakamoto H, Neckers L. The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des. 2013;19:347–365. doi: 10.2174/138161213804143725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci USa. 2013;110:2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USa. 2014;111:E2310–8. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lachowiec J, Lemus T, Borenstein E, Queitsch C. Hsp90 promotes kinase evolution. Mol Biol Evol. 2015;32:91–99. doi: 10.1093/molbev/msu270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 47.Hall DW, Mahmoudizad R, Hurd AW, Joseph SB. Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genet Res (Camb) 2008;90:229–241. doi: 10.1017/S0016672308009324. [DOI] [PubMed] [Google Scholar]
- 48.Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harb Symp Quant Biol. 2009;74:103–108. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- 49.Tomala K, Korona R. Molecular chaperones and selection against mutations. Biol Direct. 2008;3:5. doi: 10.1186/1745-6150-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Zhou L, Prodromou C, Savic V, Pearl LH. HECTD3 Mediates an HSP90-Dependent Degradation Pathway for Protein Kinase Clients. Cell Reports. 2017;19:2515–2528. doi: 10.1016/j.celrep.2017.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420–aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, et al. Exploring genetic suppression interactions on a global scale. Science. 2016;354:aag0839–aag0839. doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 55.Vanhaeren H, Gonzalez N, Coppens F, De Milde L, Van Daele T, Vermeersch M, et al. Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. eLife. 2014;2014:653. doi: 10.7554/eLife.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitesell L, Santagata S, Mendillo ML, Lin NU, Proia DA, Lindquist S. HSP90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proc Natl Acad Sci USa. 2014;111:18297–18302. doi: 10.1073/pnas.1421323111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg SM, Queitsch C. Medicine. Combating evolution to fight disease. Science. 2014;343:1088–1089. doi: 10.1126/science.1247472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 59.Jarosz D. Hsp90: A Global Regulator of the Genotype-to-Phenotype Map in Cancers. Adv Cancer Res. 2016;129:225–247. doi: 10.1016/bs.acr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown AJ, Gow NA. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7:333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 62.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. Genetic architecture of Hsp90-dependent drug resistance. Eukaryotic Cell. 2006;5:2184–2188. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJT, Ji J, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155:1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rendel JM. Canalisation and gene control. Academic Press; 1967. [Google Scholar]
- 65.Rendel JM, Sheldon BL, Finlay DE. Canalisation of development of scutellar bristles in Drosophila by control of the scute locus. Genetics. 1965;52:1137–1151. doi: 10.1093/genetics/52.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barad O, Rosin D, Hornstein E, Barkai N. Error minimization in lateral inhibition circuits. Sci Signal. 2010;3:ra51–ra51. doi: 10.1126/scisignal.2000857. [DOI] [PubMed] [Google Scholar]
- 67.Matsui T, Lee JT, Ehrenreich IM. Genetic suppression: Extending our knowledge from lab experiments to natural populations. Bioessays. 2017;39:1700023. doi: 10.1002/bies.201700023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hodgkin J. Genetic suppression. WormBook. 2005:1–13. doi: 10.1895/wormbook.1.59.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kidd PB, Young MW, Siggia ED. Temperature compensation and temperature sensation in the circadian clock. Proc Natl Acad Sci USa. 2015;112:E6284–92. doi: 10.1073/pnas.1511215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonze D, Halloy J, Goldbeter A. Robustness of circadian rhythms with respect to molecular noise. Proceedings of the National Academy of Sciences. 2002;99:673–678. doi: 10.1073/pnas.022628299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
- 73.Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69:7619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 75.Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oster H, Yasui A, van der Horst GTJ, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2011;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stelling J, Gilles ED, Doyle FJ. Robustness properties of circadian clock architectures. Proceedings of the National Academy of Sciences. 2004;101:13210–13215. doi: 10.1073/pnas.0401463101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oster H, Baeriswyl S, van der Horst GTJ, Albrecht U. Loss of circadian rhythmicity in aging mPer1−/−mCry2−/− mutant mice. Genes Dev. 2003;17:1366–1379. doi: 10.1101/gad.256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 81.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proceedings of the National Academy of Sciences. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 83.Siegal ML, Bergman A. Waddington’s canalization revisited: developmental stability and evolution. Proceedings of the National Academy of Sciences. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wagner A. Does Evolutionary Plasticity Evolve? Evolution. 1996;50:1008–1023. doi: 10.1111/j.1558-5646.1996.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 85.Azevedo RBR, Lohaus R, Srinivasan S, Dang KK, Burch CL. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature. 2006;440:87–90. doi: 10.1038/nature04488. [DOI] [PubMed] [Google Scholar]
- 86.Siegal ML, Promislow DEL, Bergman A. Functional and evolutionary inference in gene networks: does topology matter? Genetica. 2007;129:83–103. doi: 10.1007/s10709-006-0035-0. [DOI] [PubMed] [Google Scholar]
- 87.Fierst JL, Phillips PC. Modeling the evolution of complex genetic systems: the gene network family tree. J Exp Zool B Mol Dev Evol. 2015;324:1–12. doi: 10.1002/jez.b.22597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilke CO. Selection for fitness versus selection for robustness in RNA secondary structure folding. Evolution. 2001;55:2412–2420. doi: 10.1111/j.0014-3820.2001.tb00756.x. [DOI] [PubMed] [Google Scholar]
- 89.Bloom JD, Lu Z, Chen D, Raval A, Venturelli OS, Arnold FH. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 2007;5:29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wagner A. Robustness and evolvability: a paradox resolved. Proc Biol Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meiklejohn CD, Hartl DL. A single mode of canalization. Trends in Ecology & Evolution. 2002;17:468–473. doi: 10.1016/S0169-5347(02)02596-X. [DOI] [Google Scholar]
- 92.Wagner GP, Booth G, Bagheri-Chaichian H. A Population Genetic Theory of Canalization. Evolution. 1997;51:329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 93.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siegal ML. Crouching variation revealed. Mol Ecol. 2013;22:1187–1189. doi: 10.1111/mec.12195. [DOI] [PubMed] [Google Scholar]
- 95.Pavlicev M, Cheverud JM. Constraints Evolve: Context Dependency of Gene Effects Allows Evolution of Pleiotropy. Annu Rev Ecol Evol Syst. 2015;46:413–434. [Google Scholar]
- 96.Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS, Sherlock G. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkataram S, dunn B, Li Y, agarwala A, chang J, Ebel ER, et al. Development of a Comprehensive Genotype-to-Fitness Map of Adaptation-Driving Mutations in Yeast. Cell. 2016;166:1585–1596.e22. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor MB, Ehrenreich IM. Higher-order genetic interactions and their contribution to complex traits. Trends Genet. 2015;31:34–40. doi: 10.1016/j.tig.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor MB, Ehrenreich IM. Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast. PLoS Genet. 2014;10:e1004324. doi: 10.1371/journal.pgen.1004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor MB, Ehrenreich IM. Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions. PLoS Genet. 2015;11:e1005606. doi: 10.1371/journal.pgen.1005606. [DOI] [PMC free article] [PubMed] [Google Scholar]