Abstract
The immune system is characterized by the generation of structurally and functionally heterogeneous immune cells that constitute complex innate and adaptive immunity. This heterogeneity of immune cells results from changes in the expression of genes without altering DNA sequence. To achieve this heterogeneity, immune cells orchestrate the expression and functional status of transcription factor (TF) networks, which can be broadly categorized into 3 classes: pioneer TFs that facilitate initial commitment and differentiation of hematopoietic cells, subset-specific TFs that promote the generation of selected cell lineages, and immune-signaling TFs that regulate specialized function in differentiated cells. Epigenetic mechanisms are known to be critical for organizing the TF networks, thereby controlling immune cell lineage-fate decisions, plasticity, and function. The effects of epigenetic regulators can be heritable during cell mitosis, primarily through the modification of DNA and histone methylation patterns at gene loci. By doing so, the immune system is enabled to mount a selective but robust response to stimuli, such as pathogens, tumor cells, autoantigens, or allogeneic antigens in the setting of transplantation, while preserving the immune cell reservoir necessary for protecting the host against numerous other unexpected stimuli and limit detrimental effect of systemic inflammatory reactions.
Keywords: Dendritic cell development, epigenetic regulation, heterogeneous immune cells, immune system, transcription factor (TF) networks
A hallmark of the immune system is its capability to produce highly diversified immune cells that protect the host against various types of infections and tumors.1–9 For example, dendritic cells (DCs) are professional antigen-presenting cells crucial for eliciting primary T-cell responses.10–12 Based on their surface phenotype, anatomical location, and function, DCs at the steady-state condition are broadly categorized into conventional DCs (cDCs) and plasmacytoid DCs (pDCs).13,14 Under inflammatory conditions, DCs undergo profound changes in their phenotype and functionality.14–18 They present antigenic peptide to trigger antigen-specific T-cell responses.10–12 Dependent on the type of inflammatory stimuli, certain subset(s) of DCs may be preferentially selected to produce special types of cytokines (e.g., interleukin 12 [IL-12], IL-23) and Notch ligands (e.g., DLL1 and DLL4), thereby inducing heterogeneous effector T cells, such as T helper 1 (TH1), TH2, TH17 cells, and cytotoxic T cells (CTLs).19–23 Targeted deletion of a specific subset of DCs or T cells leads to selective impairment of adaptive immunity against the corresponding pathogen(s) and tumor.24–29 Thus, immune cell heterogeneity is designed to selectively protect the host against a specific type of invader(s) while limiting the damage of potentially lethal consequences of systemic immune responses.
Emerging evidence indicates that the heterogeneity of immune cells results from changes in the expression of genes without altering DNA sequence.3,30–32 The difference in gene expression initially arises during development of immune cells. Precise control of gene expression is achieved through epigenetic mechanisms, which facilitate heritable and stable programming of gene transcription while retaining potential to be modified.33–36 The development of DCs and their functional maturation in response to inflammatory stimuli provide a unique cellular model to study epigenetic regulation of the immune system. In this chapter, we will discuss our current understanding of the epigenetic effects on DC development as well as the epigenetic programs involved in the regulation of DC heterogeneity. The epigenetic regulation of T-cell immune responses was previously reviewed in Youngblood et al.,4,37 Yang et al.,38 Russ et al.,39 and He et al.40 and is not discussed here.
TRANSCRIPTION FACTORS AND GENERATION OF DISTINCT DC SUBSETS
Under steady-state condition, DCs develop from hematopoietic stem/progenitor cells (HSPCs) through successive steps of lineage commitment and differentiation: multiple potent progenitors (MPPs) → common DC progenitors (CDPs) → cDCs and pDCs (Fig. 1).13,14,24,26,41–44 Dendritic cells can be induced from monocytes (named monocyte-derived DCs [moDCs]).10,11,14,45,46 In humans, moDCs have been widely used as vaccine adjuvants for the treatment of cancer and chronic infections.14,45 Analysis of gene-targeted mice has identified many critical transcription factors (TFs) in DC development, with some (e.g., PU.1 and Signal transducer and activator of transcription 3 [STAT3]) influencing all DCs and others (e.g., transcription factor 4 [TCF4], which is known as E2–2, inhibitor of DNA binding 2 [ID2], inter-feron regulatory factor 4 [IRF4], interferon regulatory factor 4 [IRF8], and Kruppel-like factor 4 [KLF4]) controlling specific subsets.24,26,41,47–49 For instance, STAT3, which is activated by FLT3 ligand, induces generation of both cDCs and pDCs.50,51 Targeted deletion of either FLT3 ligand or STAT3 causes significantly impaired generation of DCs in vivo.51,52 PU.1 is crucial for DC fate specification. The HSPCs lacking PU.1 show defective DC differentiation potential.53–55 Thus, both PU.1 and STAT3 are known to be pioneer TFs in the regulation of DC commitment and differentiation from MPP.24,41,43
FIGURE 1.
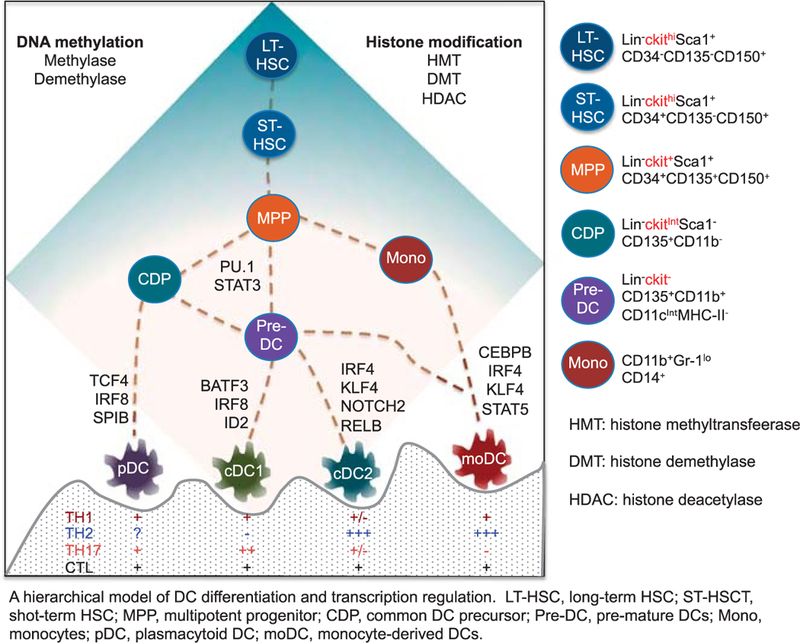
A hierarchical model of DC differentiation and transcription regulation. LT-HSC indicates long-term HSC; ST-HSCT, short-term HSC; MPP, multipotent progenitor; CDP, common DC precursor; Pre-DC, premature DCs; Mono, monocytes.
Dendritic cell subset–specifying TFs are required for committed CDP to become functionally distinct DC lineages.24,26,41,43 For example, HSPC-derived cDCs (CD11c+SeglecH−B220−) can be further classified into 2 classes: cDC1 (CD8α+/CD103+D11b−) and cDC2 (CD8α−CD11b+).24,26,56–58 cDC1 are particularly efficient in cross-presenting exogenous antigens to CD8+ CTLs. cDC1 development requires the expression of TFs, including BATF3, IRF8, and ID2.9,57 BATF3 has a nonredundant role in CD103+ cDC development and a partial effect on inducing CD8α+ DCs in lymph organs.25,26,59 IRF8-deficient animals lack spleen-resident CD8α+ cDCs and nonlymphoid tissue CD103+ cDCs.24,26,27 Functional analysis shows that BATF3 is crucial for cDC1-mediated antitumor activity, whereas IRF8 is also important for CD8+ cDC maturation and IL-12 production that regulates both TH1 and CTL responses.24,25,27
cDC2 are the second branch of cDCs expressing TFs, such as IRF4, KLF4, Neurogenic locus notch homolog protein 2, and RELB. These TFs are important in regulating cDC2 differentiation, survival, and function.57,60 For example, IRF4 is required for cDC2 to prime CD4+ T cells and promote TH17 differentiation in both lung and intestine.61,62 Interestingly, KLF4-expressing cDC2 preferentially promotes TH2 responses to Mansoni infection, but not TH1 and TH17 responses against herpes simplex virus and Toxoplasma gondii infections.29 Thus, cDC2 can be functionally heterogeneous despite their homogeneous expression of surface CD11b.
Plasmacytoid DCs are characterized by their production of high levels of interferon α upon activation.22,63,64 Plasmacytoid DCs are thought to be important for mediating antiviral immune responses and autoimmune diseases.13,22,65 Several TFs are known to regulate pDC differentiation, including TCF4, IRF8, and SPIB.47,48,66 Both TCF and IRF8 are crucial for establishing the pDC gene expression and enhancer state in pDCs.61,67 Furthermore, TCF4 in peripheral pDCs represses the up-regulation of cDC genes.65,67 ID2 is known as a counteracting TF and possesses the ability to reduce TCF4 expression, thereby inhibiting pDC development from hematopoietic progenitor cells (HPCs).65–69 This regulatory loop between TCF4 and ID2 is important for balancing the generation of pDCs and cDCs while maintaining DC plasticity.
Monocyte-derived DCs are thought to be inflammatory DCs and are widely used as vaccine adjuvants in humans.10,11,70,71 Upon induction by granulocyte macrophage colony-stimulating factor and IL-4, both HPCs and monocytes may differentiate into moDCs.10 These cells are CD8α−CD11b+ and produce high levels of inducible nitric oxide synthase and arginase, resembling in vivo–generated inflammatory DCs.14,72,73 Transcription factors, including CCAAT/enhancer-binding protein beta (CEBPB), IRF4, KLF4, STAT5, RELB, and CCAAT/enhancer-binding protein alpha, are able to regulate moDC differentiation.24,26,29 Granulocyte macrophage colony-stimulating factor–driven moDC differentiation requires expression of functional IRF4 and CEBPB.56,62,67,74 CEBPB can promote moDC differentiation by counteracting IRF8 effects.67 Notably, KLF4 induces a set of monocyte lineage–associated molecules and is a key switch factor regulating differentiation of monocytes into moDCs.29 The engagement of multiple TFs in the regulation of moDCs implies not only their importance in immune responses, but also their heterogeneity in function and tissue distribution.14
EPIGENETIC PROCESSES IN DCs
Epigenetic mechanisms regulate cell development, identity, and function. This can be achieved by catalyzing histone modifications at promoter and enhancer regions, thereby changing chromatin conformation and altering TF binding. For example, monomethylation of histone H3 lysine 4 (H3K4me1) and acetylation of histone H3 lysine 27 (H3K27ac) mark genomic regions that indicate primed enhancers and active enhancers, respectively.42,49,75–78 Enhancers identified by H3K4me1 and H3K27Ac are associated with genes critical for DC subset specification. For example, pDC and moDCs are distinguished by thousands of differential enhancers.41,43,56 Analysis of H3K4me1 and H3K27Ac modifications reveals a large number of differential sites between pDCs and moDCs. The amount of both H3K4me1 and H3K27ac in pDCs is significantly higher for pDC-specific genes than moDC-specific genes.43,56 Similarly, moDC-specific genes demonstrated significantly higher H3K4me1 and H3K27ac intensity than pDC-specific genes.43,56 Identification of these DC-specific enhancer regions is important for defining the specific effects of epigenetic regulators.
Intriguingly, by systematically mapping more than 180,000 protein-DNA interactions of 25 TFs during moDC response to lipopolysaccharide (LPS), Amit and colleagues found that chromatin marks, including H3K4me3, H3K4me1, and H3K27Ac, are significantly less dynamic compared with changes in expression of TFs.67,79 Binding of H3K4me3 at the promoter regions was remarkably stable during the first 2 hours of LPS response in moDCs.67,79 Similar results are observed for these chromatin marks in pDCs.56,67 These studies suggest that the chromatin landscape of TFs crucial for DC differentiation have been established prior to inflammatory stimulation and perhaps early during subset-specification stage. It is important to examine which chromatin-modifying enzyme(s) play a critical role in modifying these enhancers under steady-state and inflammatory conditions.
Some studies suggest that the differentiation of HPCs into DC lineages is associated with the establishment of hierarchical organization of TF networks. Among them, PU.1 and CEBPB represent the pioneer TF regulating DC lineage commitment, whereas TFs downstream of immune signaling pathways (e.g., activator protein 1 nuclear factor kappa-light-chain-enhancer of activated B cells, and STAT1) are important for mediating DC responses upon inflammatory stimulation.43,56 PU.1 and CEBPB bind tens of thousands of chromatin sites in pDCs and moDCs, respectively.43,67 Intriguingly, more than 70% of other TFs bind in close proximity to these pioneer TFs. In contrast, immune-signaling TFs show stimuli-dependent binding dynamics in DCs.43,55,56,68,79 These observations suggest a critical role of PU.1 and CEBPB in orchestrating the enhancer regions for other TFs in differentiating HPCs, whereas immune-signaling TFs are programmed to regulate short-term stimulatory responses.43,79 Notably, genome-wide ChIP-seq analysis suggests that genes mediating fast immune responses (e.g., activator protein 1 nuclear factor kappa-light-chain-enhancer of activated B cells, and STAT1) have been programmed as early as DC commitment stage.43,79 Similar hierarchical organization of TFs has been shown for other immune cells (such as T and B cells), although the composition of hierarchical TF networks and their epigenetic regulators vary in different cell types.4,32,80–85
EPIGENETIC ORGANIZATION OF HIERARCHICAL TF NETWORKS IN DC PROGENITORS
Recent studies have examined the epigenetic organization of TF networks in DC progenitors.43,56 Genome-wide analysis reveals that certain groups of genes are associated with distinct development stages and subsets of DCs. Dendritic cell commitment (e.g., MPP → CDP transition) is associated with down-regulation of genes (e.g., CCAAT/enhancer-binding protein alpha, GATA2, GFI1, and T-cell acute lymphocytic leukemia protein 1) that restrict hematopoietic cell differentiation from HSPCs49,75,86 and up-regulation of CDP signature genes (e.g., E2F2 and HOXA1) associated with cell cycle.56 Interestingly, pan-DC genes (e.g., FLT3, IRF5, SPIB, and STAT1), which are expressed in both cDCs and pDCs but expressed at low levels in MPP and CDP, are known to regulate the overall generation and immune response of DCs.56 These gene profiles provide the basis for further investigating how DCs organize the hierarchical TF network for their commitment, subset specification, and differentiation.
In this hierarchical TF network regulating DC development, PU.1 may enable nucleosome positioning and local histone modifications to regulate TF binding. PU.1 has a determining function in fate decisions of hematopoiesis, including the establishment of DC lineages.49,53–55,68,75,87,88 ChIP-seq–based analysis of PU.1 and H3K4me1 colocalization shows an increasing overlap of PU.1 binding and H3K4me1 deposition from 20% in MPP to 70% in cDCs.56 This indicates an increased recruitment of PU.1 to enhancer elements during DC development. Furthermore, PU.1 is believed to function as an enhancer factor for many other TFs.67,79 However, a substantial proportion of enhancer regions shows no PU.1 occupancy, such as in MPP and CDP.56 This suggests that PU.1 binding of chromatin likely requires other co–binding factors in DCs and that these PU.1-interaction factors may vary in DCs at distinct stage of commitment and differentiation.
Determining the specific effects of functionally relevant epigenetic enzymes will greatly facilitate our understanding of how epigenetic mechanisms control DC heterogeneity and function through interacting with TFs. Recent studies have identified the capacity of PU.1 that regulates (or interacts with) DNA demethylation (machinery) and direct monocyte differentiation into moDCs.89,90 Monocytes driven to differentiation by granulocyte macrophage colony-stimulating factor and IL-4 are uniquely affected by this phenomenon.89 Interestingly, STAT6 interaction with TET2, a DNA demethylase, is important for moDC differentiation. The effect of STAT6 and TET2 on DNA demethylation is associated with functional PU.1, as evidenced by the observation that silencing PU.1 in monocytes markedly impairs DNA demethylation.87 However, ChIP-seq analysis suggests that PU.1 motifs are significantly enriched in both hypermethylated and hypomethylated genes.91 Thus, PU.1 likely participates in the process of mediating both hyper- and hypo-DNA methylation, thereby influencing DC commitment, differentiation, and subset specification.
EPIGENETIC PROGRAMMING OF DC HETEROGENEITY AND HERITABILITY
Immunological memory provides the host with long-term protection against pathogens. It is well accepted that memory T cells are long-lived, self-renewing cells that are able to rapidly and robustly respond to a specific antigen (pathogen) upon re-encounter.4,32,80,81 Memory B cells function in a similar capacity, by producing high-affinity antibodies that neutralize virus upon secondary exposure.82–85
Interestingly, Netea, Stunnenberg and colleagues have discovered that after an initial priming by inflammatory stimuli monocytes can keep such attack “in memory” during their subsequent differentiation into macrophages.92–95 For example, as compared with unprimed naive macrophages, macrophages derived from monocytes primed with β-glucan short term (24 hours) show an enhanced inflammatory status. This phenomenon is termed “trained immunity.” In contrast, macrophages derived from monocytes that were pretreated with LPS short term produced less pro-inflammatory mediators (e.g., IL-6 and tumor necrosis factor α) upon challenge with certain Toll-like receptor agonist(s).92,93,95 They observed that both mammalian target of rapamycin and hypoxia-inducible factor 1α are important for the establishment of “trained immunity” in monocytes.92 Genome-wide analysis of histone marks (e.g., H3K4me1, H3K4me3, and H3K27ac) and DNase I accessibility identifies that approximately 8000 dynamic regions in monocytes are linked to trained immunity and attenuated inflammatory pathways in differentiated macrophages compared with their naive counterparts. These changes in monocytes have been established within 24 hours after priming by β-glucan or LPS and last more than 5 days after priming.92–94 Thus, epigenetic effects are associated with heritability of immunological memory in innate immune cells. Identifying the precise role of epigenetic regulator(s) in mediating “trained” and attenuated immunity in macrophages will introduce a new epigenetic perspective innate immunity and better define pathophysio-logical roles of both innate and adaptive immunity.
In analogy to macrophages, monocytes may be able to imprint the gene programs triggered by prior stimulation into DCs. In support of this idea, recent studies have shown that differentiation of DCs from monocytes involves histone modifications and DNA demethylation.90,91,96 First, there exists a general mechanism that allows immune cells to generate differential enhancer landscapes and stable functional states by chromatin-regulating TFs.49,75 Second, different types of DCs may produce specific patterns of immune responses even when these differential lineages of DCs are activated by the same stimulus. For instance, upon inflammatory stimulation, pDCs produce high levels of interferon α/β. cDC1 produces IL-12, and cDC2 stimulates innate lymphoid cells to produce IL-22.24–27,29,59 In addition, we have recently discovered that triggering of Toll-like receptors induces high levels of the NOTCH ligand DLL4 in both cDCs, which is crucial for TH1 and TH17 differentiation. In contrast, despite inflammatory stimuli, activation of moDCs cannot induce DLL4.72,73,97 Third, IRF8 binds more than 30,000 enhancer regions (e.g., H3K4me1) highly enriched in pDCs. Enforced IRF8 expression leads to down-regulation of IRF4 and CEBPB, thereby directing the epigenetic landscape toward a pDC-specific pattern.67 Together, these observations argue that properties acquired by DCs during the early stages of development may represent a pattern of cellular memory. Identification of the epigenetic mechanisms that establish heritable programs in DC progenitors during differentiation will address whether a “memory machinery” may critically regulate the production of functionally specialized DC subsets.
CONCLUDING REMARKS
Advances in mapping chromatin states in DCs and other immune cells greatly facilitate our understanding of the epigenetic mechanisms that regulate immune cell diversity and immune responses to variable stimuli. However, the precise roles of chromatin-modifying enzymes that control DC development and function remain largely unknown. Future studies should focus on identifying the functional relevance of specific epigenetic regulators in DCs and their progenitors. Results from these studies will better define pathophysiological roles of tumor and inflammation in mediating immune dysfunction under disease conditions.
Our understanding of immune function for DCs is derived mostly from studies of cells developed under steady-state conditions. However, under inflammatory conditions, HPCs may instate the transcriptional programs to reflect their initial encounter of the environmental stimuli. For example, DCs derived from engrafted donor HSPCs display aberrant phenotype and function in mice undergoing allogenic hematopoietic stem cell transplantation.98–101 While cDC2-like cells in the spleen have impaired antigen-presenting function,98,99 CD103+ cDC1 shows augmented capacity to mediate alloreactive T-cell responses.102 We have previously discovered that generation of thymic DCs and DLL4+ DCs from engrafted donor HSPCs is impaired in transplant mice with graft-versus-host disease.73,103 In addition, the tumor microenvironment may have major impact on the production of functionally and phenotypically different DC subsets.27,28,104,105 It will be intriguing to investigate under these inflammatory conditions whether and how epigenetic effects may help immune cells to adopt previously recognized phenotype and functions, thereby influencing overall consequence of immune responses.
Finally, various pharmacological approaches that target these epigenetic regulators have been tested for cancer treatment in clinical trials.35,36,106–109 Data from our studies and others indicate that certain epigenetic regulators may play essential roles in regulating immune cell function and survival capability. For example, EZH2 is essential for promoting the survival and production of effector T cells.90,110 Drugs inhibiting EZH2, which are in clinical trials for treating cancers, might have adverse effects on T-cell immunity against tumor when they are tested for cancer treatment. Similar effects have been shown for DNA methylation inhibitors that can suppress cancer cell proliferation and survival, but also induce the generation of regulatory T cells, which are known to blunt the efficacy of cancer immunotherapy.111 It is likely that new strategies such as systems biology are needed for identifying an optimal pharmacological approach that can maximize the efficacy of immune cells on eliminating tumor cells while directly controlling tumor growth.
ACKNOWLEDGMENT
The authors thank Ms Janaki Purushe for proofreading and fruitful discussion.
This work was supported by the Department of Defense (to Y.Z.) and NationalInstitutes of Health (CA172106–01 and HL127351–01A1 to Y.Z.).
Footnotes
The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- 1.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. [DOI] [PubMed] [Google Scholar]
- 2.Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14(spec no 1):R41–R46. [DOI] [PubMed] [Google Scholar]
- 3.Schuyler RP, Merkel A, Raineri E, et al. Distinct trends of DNA methylation patterning in the innate and adaptive immune systems. Cell Rep. 2016;17: 2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youngblood B, Hale JS, Ahmed R. Memory CD8 T cell transcriptional plasticity. F1000Prime Rep. 2015;7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlach C, Rohr JC, Perié L, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. [DOI] [PubMed] [Google Scholar]
- 6.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalia V, Sarkar S, Gourley TS, et al. Differentiation of memory B and T cells. Curr Opin Immunol. 2006;18:255–264. [DOI] [PubMed] [Google Scholar]
- 8.Buchholz VR, Flossdorf M, Hensel I, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. [DOI] [PubMed] [Google Scholar]
- 9.Miller JC, Brown BD, Shay T, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. [DOI] [PubMed] [Google Scholar]
- 11.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. [DOI] [PubMed] [Google Scholar]
- 12.Zamvil SS, Steinman L. The T lymphocyte in experimental allergic encephalomyelitis. Annu Rev Immunol. 1990;8:579–621. [DOI] [PubMed] [Google Scholar]
- 13.Watowich SS, Liu YJ. Mechanisms regulating dendritic cell specification and development. Immunol Rev. 2010;238:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 2007;7:19–30. [DOI] [PubMed] [Google Scholar]
- 15.Naik SH. Demystifying the development of dendritic cell subtypes, a little. Immunol Cell Biol. 2008;86:439–452. [DOI] [PubMed] [Google Scholar]
- 16.Reizis B, Colonna M, Trinchieri G, et al. Plasmacytoid dendritic cells: one-trick ponies or workhorses of the immune system? Nat Rev Immunol. 2011;11:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuniga EI, McGavern DB, Pruneda-Paz JL, et al. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat Immunol. 2004;5:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlitzer A, Loschko J, Mair K, et al. Identification of CCR9− murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood. 2011;117:6562–6570. [DOI] [PubMed] [Google Scholar]
- 19.Strioga MM, Felzmann T, Powell DJ Jr, et al. Therapeutic dendritic cell–based cancer vaccines: the state of the art. Crit Rev Immunol. 2013; 33:489–547. [DOI] [PubMed] [Google Scholar]
- 20.Palucka K, Banchereau J. Dendritic-cell–based therapeutic cancer vaccines. Immunity. 2013;39:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology. 2013;140:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–1226. [DOI] [PubMed] [Google Scholar]
- 23.Benlahrech A, Duraisingham S, King D, et al. Human blood CD1c dendritic cells stimulate IL-12–independent IFN-gamma responses and have a strikingly low inflammatory profile. J Leukoc Biol. 2015;97: 873–885. [DOI] [PubMed] [Google Scholar]
- 24.Merad M, Sathe P, Helft J, et al. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildner K, Edelson BT, Purtha WE, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy TL, Grajales-Reyes GE, Wu X, et al. Transcriptional control of dendritic cell development. Annu Rev Immunol. 2016;34:93–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmon H, Idoyaga J, Rahman A, et al. Expansion and activation of CD103 (+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity. 2016;44:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laky K, Evans S, Perez-Diez A, et al. Notch signaling regulates antigen sensitivity of naive CD4+ T cells by tuning co-stimulation. Immunity. 2015;42:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tussiwand R, Everts B, Grajales-Reyes GE, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kakaradov B, Arsenio J, Widjaja CE, et al. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017;18:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly AD, Kroeger H, Yamazaki J, et al. A CpG island methylator pheno-type in acute myeloid leukemia independent of IDH mutations and associated with a favorable outcome. Leukemia. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139: 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Kirby JE, Sunwoo H, et al. Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 2016;30: 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russ BE, Olshanksy M, Smallwood HS, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41:853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He S, Tong Q, Bishop DK, et al. Histone methyltransferase and histone methylation in inflammatory T-cell responses. Immunotherapy. 2013;5: 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul F, Amit I. Plasticity in the transcriptional and epigenetic circuits regulating dendritic cell lineage specification and function. Curr Opin Immunol. 2014;30:1–8. [DOI] [PubMed] [Google Scholar]
- 42.Mildner A, Schönheit J, Giladi A, et al. Genomic characterization of murine monocytes reveals C/EBPbeta transcription factor dependence of Ly6C− cells. Immunity. 2017;46:849–862, e847. [DOI] [PubMed] [Google Scholar]
- 43.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. [DOI] [PubMed] [Google Scholar]
- 44.Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naik SH, Metcalf D, van Nieuwenhuijze A, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7:663–671. [DOI] [PubMed] [Google Scholar]
- 46.Geissmann F, Manz MG, Jung S, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh HS, Cisse B, Bunin A, et al. Continuous expression of the transcription factor e2–2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cisse B, Caton ML, Lehner M, et al. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laouar Y, Welte T, Fu XY, et al. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–912. [DOI] [PubMed] [Google Scholar]
- 52.Karsunky H, Merad M, Cozzio A, et al. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson KL, Perkin H, Surh CD, et al. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J Immunol. 2000;164:1855–1861. [DOI] [PubMed] [Google Scholar]
- 54.Anderson KL, Smith KA, Conners K, et al. Myeloid development is selectively disrupted in PU.1 null mice. Blood. 1998;91:3702–3710. [PubMed] [Google Scholar]
- 55.Carotta S, Dakic A, D’Amico A, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. [DOI] [PubMed] [Google Scholar]
- 56.Lin Q, Chauvistré H, Costa IG, et al. Epigenetic program and transcription factor circuitry of dendritic cell development. Nucleic Acids Res. 2015;43: 9680–9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price JD, Tarbell KV. The role of dendritic cell subsets and innate immunity in the pathogenesis of type 1 diabetes and other autoimmune diseases. Front Immunol. 2015;6:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlitzer A, Sivakamasundari V, Chen J, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015; 16:718–728. [DOI] [PubMed] [Google Scholar]
- 59.Toubai T, Sun Y, Luker G, et al. Host derived CD8+ dendritic cells are required for induction of optimal graft-versus-tumor responses after experimental allogeneic bone marrow transplantation. Blood. 2013;121: 4231–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price JD, Hotta-Iwamura C, Zhao Y, et al. DCIR2+ cDC2 DCs and Zbtb32 restore CD4+ T-cell tolerance and inhibit diabetes. Diabetes. 2015;64: 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura T, Tailor P, Yamaoka K, et al. IFN regulatory factor-4 and −8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. [DOI] [PubMed] [Google Scholar]
- 62.Vander Lugt B, Khan AA, Hackney JA, et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol. 2014;15:161–167. [DOI] [PubMed] [Google Scholar]
- 63.Piccioli D, Tavarini S, Borgogni E, et al. Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood. 2007;109:5371–5379. [DOI] [PubMed] [Google Scholar]
- 64.Gilliet M, Boonstra A, Paturel C, et al. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med. 2002; 195:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15:471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spits H, Couwenberg F, Bakker AQ, et al. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre-DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J Exp Med. 2000;192: 1775–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bornstein C, Winter D, Barnett-Itzhaki Z, et al. A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin States. Mol Cell. 2014;56:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy KM. Transcriptional control of dendritic cell development. Adv Immunol. 2013;120:239–267. [DOI] [PubMed] [Google Scholar]
- 69.Masson F, Minnich M, Olshansky M, et al. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013; 190:4585–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Burg SH, Arens R, Ossendorp F, et al. Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer. 2016;16:219–233. [DOI] [PubMed] [Google Scholar]
- 71.Melief CJ, van Hall T, Arens R, et al. Therapeutic cancer vaccines. J Clin Invest. 2015;125:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mochizuki K, Meng L, Mochizuki I, et al. Programming of donor T cells using allogeneic δ-like ligand 4–positive dendritic cells to reduce GVHD in mice. Blood. 2016;127:3270–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mochizuki K, Xie F, He S, et al. Delta-like ligand 4 identifies a previously uncharacterized population of inflammatory dendritic cells that plays important roles in eliciting allogeneic T cell responses in mice. J Immunol. 2013;190:3772–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herzig Y, Nevo S, Bornstein C, et al. Transcriptional programs that control expression of the autoimmune regulator gene Aire. Nat Immunol. 2017; 18:161–172. [DOI] [PubMed] [Google Scholar]
- 75.Adams D, Altucci L, Antonarakis SE, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. [DOI] [PubMed] [Google Scholar]
- 76.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008; 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boyle AP, Davis S, Shulha HP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. [DOI] [PubMed] [Google Scholar]
- 79.Garber M, Yosef N, Goren A, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gattinoni L, Lugli E, Ji Y, et al. A human memory T cell subset with stem cell–like properties. Nat Med. 2011;17:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clybouw C, Fischer S, Auffredou MT, et al. Regulation of memory B-cell survival by the BH3-only protein Puma. Blood. 2011;118:4120–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minton K B cell memory: a second chance for antibodies. Nat Rev Immunol. 2015;15:131. [DOI] [PubMed] [Google Scholar]
- 84.Kugelberg E B cell memory: making sense in humans. Nat Rev Immunol. 2015;15:133. [DOI] [PubMed] [Google Scholar]
- 85.Leavy O Immune memory: sequential evolution of B cell memory. Nat Rev Immunol. 2016;16:72–73. [DOI] [PubMed] [Google Scholar]
- 86.Olsson A, Venkatasubramanian M, Chaudhri VK, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016; 537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de la Rica L, Rodríguez-Ubreva J, García M, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DeRyckere D, Mann DL, DeGregori J. Characterization of transcriptional regulation during negative selection in vivo. J Immunol. 2003; 171:802–811. [DOI] [PubMed] [Google Scholar]
- 89.Vento-Tormo R, Company C, Rodríguez-Ubreva J, et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 2016;17:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Ulm A, Somineni HK, et al. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenetics Chromatin. 2014;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klug M, Schmidhofer S, Gebhard C, et al. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013;14:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng SC, Quintin J, Cramer RA, et al. mTOR- and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014; 345:1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. [DOI] [PubMed] [Google Scholar]
- 95.Novakovic B, Habibi E, Wang SY, et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell. 2016;167: 1354–1368, e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pacis A, Tailleux L, Morin AM, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015;25: 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meng L, Bai Z, He S, et al. The notch ligand DLL4 defines a capability of human dendritic cells in regulating TH1 and TH17 differentiation. J Immunol. 2016;196:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leveque-El Mouttie L, Koyama M, Le Texier L, et al. Corruption of dendritic cell antigen presentation during acute GVHD leads to regulatory T-cell failure and chronic GVHD. Blood. 2016;128:794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wikstrom ME, Fleming P, Kuns RD, et al. Acute GVHD results in a severe DC defect that prevents T-cell priming and leads to fulminant cytomegalovirus disease in mice. Blood. 2015;126:1503–1514. [DOI] [PubMed] [Google Scholar]
- 100.Markey KA, Koyama M, Kuns RD, et al. Immune insufficiency during GVHD is due to defective antigen presentation within dendritic cell subsets. Blood. 2012;119:5918–5930. [DOI] [PubMed] [Google Scholar]
- 101.Markey KA, MacDonald KP, Hill GR. Recipient plasmacytoid DCs are not required to prime allogeneic T-cell responses after BMT. Blood. 2009; 113:6038–6039. [DOI] [PubMed] [Google Scholar]
- 102.Koyama M, Cheong M, Markey KA, et al. Donor colonic CD103+ dendritic cells determine the severity of acute graft-versus-host disease. J Exp Med. 2015;212:1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Y, Hexner E, Frank D, et al. CD4+ T cells generated de novo from donor hemopoietic stem cells mediate the evolution from acute to chronic graft-versus-host disease. J Immunol. 2007;179:3305–3314. [DOI] [PubMed] [Google Scholar]
- 104.Kranz LM, Diken M, Haas H, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016; 534:396–401. [DOI] [PubMed] [Google Scholar]
- 105.Laoui D, Keirsse J, Morias Y, et al. The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat Commun. 2016;7:13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012; 492:108–112. [DOI] [PubMed] [Google Scholar]
- 107.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. [DOI] [PubMed] [Google Scholar]
- 108.Chen S, Shi Y. A new horizon for epigenetic medicine? Cell Res. 2013;23:326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tuma RS. Targeted epigenetic therapies: the next frontier? J Natl Cancer Inst. 2010;102:1824–1825. [DOI] [PubMed] [Google Scholar]
- 110.He S, Xie F, Liu Y, et al. The histone methyltransferase Ezh2 is a crucial epigenetic regulator of allogeneic T-cell responses mediating graft-versus-host disease. Blood. 2013;122:4119–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi J, Ritchey J, Prior JL, et al. In vivo administration of hypomethylating agents mitigate graft-versus-host disease without sacrificing graft-versus-leukemia. Blood. 2010;116:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]


