Abstract
The Nonhomologous end joining (NHEJ) pathway preserves genome stability by ligating the ends of broken chromosomes together. It employs end-processing enzymes, including polymerases, to prepare ends for ligation. We show two such polymerases primarily incorporate ribonucleotides during NHEJ - an exception to the central dogma of molecular biology - both during repair of chromosome breaks made by Cas9, and during V(D)J recombination. Moreover, additions of ribonucleotides, but not deoxynucleotides, effectively promote ligation. Repair kinetics argue ribonucleotide-dependent first strand ligation is followed by complementary strand repair with deoxynucleotides, then by replacement of ribonucleotides embedded in the first strand with deoxynucleotides. Our results indicate as much as 65% of cellular NHEJ has transiently embedded ribonucleotides, which promote flexibility in repair at the cost of more fragile intermediates.
One Sentence Summary
Mammalian NHEJ can resolve a wider variety of chromosome breaks when polymerases introduce ribonucleotides during repair
Nonhomologous end joining (NHEJ) is the primary pathway for repairing chromosomal double-strand breaks (DSBs) in mammals, and is required for genome stability in all cell types as well as assembly of antigen specific receptors by V(D)J recombination in lymphocytes (1). NHEJ employs specialized nucleases and polymerases, including the widely-expressed Pol μ (gene name, Polm) and lymphocyte-specific terminal deoxynucleotidyl transferase (TdT), to modify broken end structures in preparation for ligation (2). Accordingly, loss of Pol μ or TdT results in impaired immune responses (3–6). Loss of the more widely expressed Pol μ additionally interferes with cell growth (7, 8), hematopoiesis (7), and resistance to DNA damage (7–9). Pol μ and TdT notably favor deoxynucleotides 1.4 to 11-fold (depending on nucleotide base) more than ribonucleotides (10). By comparison, other polymerases that maintain DNA genomes (including closely related Pol λ and Pol β) typically incorporate deoxynucleotides several 1000-fold more efficiently than ribonucleotides (11–13). However, it is unknown whether ribonucleotide incorporation occurs during cellular NHEJ, and if ribonucleotide incorporation occurs, if it significantly impacts NHEJ function.
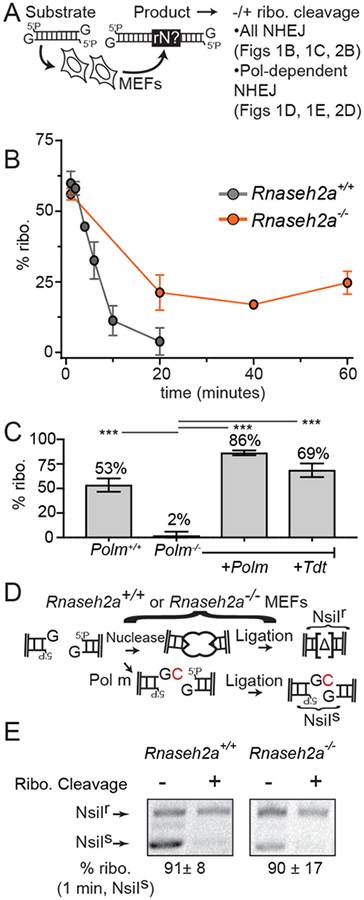
We initially investigated whether ribonucleotides are incorporated during NHEJ after introducing linear DNA substrates into transformed mouse embryonic fibroblasts. We optimized this assay to allow for rapid harvesting of repair products, in anticipation that ribonucleotides were only transiently present. Ribonucleotides embedded in NHEJ products were quantified by assessing the template lost in samples upon cleavage of ribonucleotide containing strands (% ribo.; Fig. 1A, Fig. S1B–C) using validated qPCRs (Fig. S1A, S3A, S4B). We determined embedded ribonucleotides were present in 60% (standard deviation = sd; 4.2%) of NHEJ products (Fig. 1B) when products were assessed within the first minute after electroporation, and were dependent on either Pol μ or TdT (Fig. 1C and Fig. S1, E–F).
Fig. 1: Ribonucleotide incorporation during repair of extrachromosomal substrates.
(A) DNA fragments with 3’G overhangs at ends were introduced into MEFs, and % cellular NHEJ products with embedded ribonucleotides (% ribo.) quantified by comparing amplification efficiency with and without prior cleavage at sites of ribonucleotide incorporation (also Fig. S1C). (B) % ribo. after introduction of substrate into Rnaseh2a+/+ (gray) or Rnaseh2a−/− (orange) MEFs. Data points are the mean of 3 transfections, and error bars represent sd. (C) %ribo. in products recovered after 1 minute. Data points are the mean of 3 transfections, and error bars represent sd. Means were compared in pairs to Polm−/− by ANOVA (***; p < 0.001). (D, E) Digestion of amplified products with NsiI and electrophoresis distinguishes Pol μ-dependent +C products from products with deletions of flanking sequence (Δ). (E) The mean % ribo. in NsiIs products recovered after 1 minute from three independent transfections, ± sd, is noted below gel.
Embedded ribonucleotides in NHEJ products decreased in frequency until almost undetectable after 20 minutes (Fig. 1B, black line). To determine if this reduction was due to replacement of incorporated ribonucleotides with deoxynucleotides (Ribonucleotide excision repair; RER), we employed CRISPR/Cas9 to generate a MEF variant deficient in Rnaseh2a (Fig. S1G), which initiates RER (14). Levels of embedded ribonucleotides in Rnaseh2a-deficient cells were initially equivalent to wild type cells; unlike wild type cells, embedded ribonucleotides were not completely removed in Rnaseh2a-deficient cells, and stabilized at levels approximately two-fold less than was initially observed (Fig. 1B, orange line). Re-expression of RNaseH2A in the Rnaseh2a-deficient variant was sufficient to reduce embedded RNA in NHEJ products to the low levels observed in wild type cells (Fig. S1H).
The substrate used above possessed a single nucleotide 3’ overhang (3’G). Approximately half of repair products require ligation after addition of a single complementary C (+C product), are dependent on both Ku and Pol μ (15), and can be identified by sensitivity to a restriction enzyme (NsiI; Fig. 1D). Sequencing indicates the remaining NsiI resistant products have 1–5 bp deletions of flanking sequence that are at best modestly effected by Polm deficiency (15). Embedded ribonucleotides were present in 91% (sd 8%) of NsiI-sensitive products after 1 minute (Fig. 1E). Similar results were observed when using a different method to detect ribonucleotide-containing products (Fig. S2A), and when using a substrate with a different overhang template (C3’) and Pol μ-dependent added nucleotide (+G product; Fig. S2B). We conclude that most Pol μ- and TdT-dependent NHEJ products contain embedded ribonucleotides, and that the modest preference of these polymerases for addition of deoxynucleotides in vitro (10) is overwhelmed by higher concentrations of ribonucleotides in cells (15).
As also informed by data in subsequent figures, we suggest early products involve one ligated strand only. Subsequent repair of the complementary strand with deoxynucleotides accounts for the two-fold dilution of products with embedded ribonucleotides that is independent of Rnaseh2a (Fig. 1B, orange line), while complete removal of ribonucleotides requires Rnaseh2a-dependent RER.
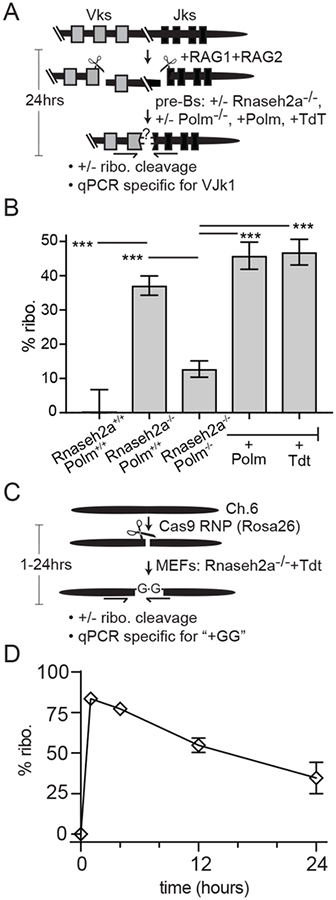
We determined if ribonucleotides are similarly incorporated during repair by NHEJ of chromosomal breaks. We used a pre-B cell line that can be induced to arrest in G1 phase and undergo V(D)J recombination at the immunoglobulin kappa locus (Igk; Fig. 2A and Fig. S3E) (15), since Pol μ is efficiently engaged by the 3’ overhang intermediates in this process(5, 15–17). Embedded ribonucleotides were undetectable 24 hours after induction when cells were proficient in RER. By comparison, 35% of Igk recombination products possessed embedded ribonucleotides in an Rnaseh2a-deficient variant (Fig. 2B and Fig. S3, A–B). This frequency is approximately half of the frequency of Igk products where Pol μ is active (5, 17), consistent with the model proposed above, where only the first strand of a chromosome double strand break is repaired with ribonucleotides. Embedded ribonucleotides were again largely dependent on either Pol μ or TdT (Fig. 2B and Fig. S3, C–D).
Fig. 2: Ribonucleotide incorporation during repair of chromosomal breaks.
(A, B) SP9 pre-B cells were induced for 24 hours, resulting in expression of RAG1+2 nuclease and introduction of chromosome breaks adjacent to VK and JK coding segments (boxes). % ribo. in VJK coding junctions was measured as in Fig. 1A. (B) Data points are the mean of five independent inductions, and error bars represent sd. Means were compared in pairs as noted by ANOVA (***; p < 0.001). (C, D) Rosa26 locus-targeting Cas9 ribonucleoprotein was introduced into MEFs deficient in Rnaseh2a and expressing TdT. % ribo. was detected as in Fig. 1A using a qPCR specific for the TdT-dependent +GG product (also Fig. S4). (D) Data points are the mean of 3 independent transfections, and error bars represent sd.
We sought to track Polymerase-dependent ribonucleotide incorporation during chromosomal NHEJ earlier than was possible using the V(D)J recombination model, and also to extend analysis to non-lymphoid cells. We directly introduced Rosa26 locus-targeted Cas9 nuclease into Rnaseh2a-deficient MEFs, which allowed for rapid accumulation of repair products (Fig S4D), and thus assessment of ribonucleotides in these products immediately after they were generated. Sequencing of repair products from wild type vs. Polm−/− MEFs confirms Pol μ promotes repair accuracy (Fig. S4A). However, the blunt ends generated by Cas9 engage Pol μ less frequently (16% of all repair) than V(D)J recombination intermediates, and the contribution of Pol μ is distributed over several mostly template-dependent products that cannot be easily distinguished from polymerase-independent repair. We therefore expressed TdT in these cells (Fig. S1F), as this generates a class of repair products - addition of two or more Gs, with no loss of flanking DNA (“+GG products”)—that are abundant (18% of NHEJ products, Table S1), unambiguously polymerase-dependent, and can be detected by a sensitive product-specific qPCR (Fig. 2C–D, Fig. S4B–D). Ribonucleotide incorporation by TdT and Pol μ is not significantly different in cells (Fig. 1C, 2B) and using steady state kinetics in vitro(10), arguing characterization of ribonucleotide incorporation by TdT during NHEJ is directly comparable to Pol μ dependent repair. Embedded ribonucleotides were present in 84% and 77% of +GG NHEJ products 1 and 4 hours after introduction of Cas9 respectively (Fig. 2D), and were reduced twofold in these RER-deficient cells over the next 20 hours. This is consistent with strong favoring of ribonucleotide incorporation by these polymerases for first strand repair, followed by repair of complementary strands with deoxynucleotides. As expected, embedded ribonucleotides were much lower in repair products when cells were proficient in RER (23% after 1 hour; Fig. S4F), and undetectable at a nearby locus where breaks were not induced (whether cells were RER deficient or not; Fig. S4E).
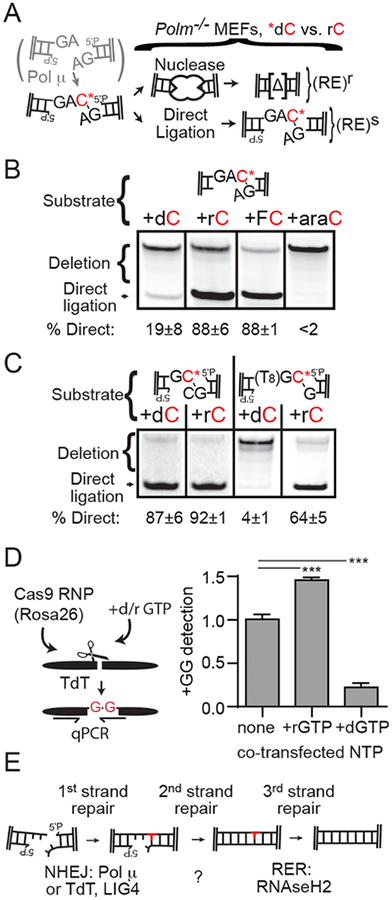
We investigated the consequences of ribonucleotide addition on ligation, the next step of cellular NHEJ. We initially focused on a substrate with 3’ GA overhangs, where 66% of NHEJ occurs by ligation after Pol μ-dependent addition of a single complementary C (Fig. S5A). We made two variants of this substrate with a C already added – one where the added C was a ribonucleotide (+rC), vs. one where it was a deoxynucleotide (+dC) (Fig. 3A). We then introduced these two substrate variants into cells that express neither Pol μ nor TdT, to isolate the effect of the different pre-added nucleotides on ligation, and confirmed repair under these conditions relies on the NHEJ-specific ligase (LIG4; Fig. S5B). Strikingly, only the +rC variant was able to efficiently promote direct ligation in cells, while the +dC variant was largely ineffective (Fig. 3B; reduced over 20-fold, Table 1). Direct ligation was also stimulated when the ribonucleotide 2’OH was substituted with Fluorine (+2’FC), and blocked when the terminal nucleotide was substituted with a ribonucleotide stereoisomer, arabinofuranosylcytidine (+AraC) (Fig. 3B, Table 1). AraC differs from rC only by the orientation of the 2’OH and the favored sugar pucker (rC favors C3’ endo, AraC favors C2’ endo), suggesting a terminal C3’ endo nucleotide is required for stimulation of ligation (Fig. S5C). A terminal ribonucleotide also stimulated LIG4-dependent ligation in vitro, confirming the impact on cellular NHEJ is specific to the ligation step. In contrast, T4 DNA ligase gained no benefit from a terminal ribonucleotide (Fig. S5D). LIG4 may be alone amongst mammalian ligases in the ability to take advantage of added ribonucleotide termini, analogous to the in vitro activity of bacterial Pseudomonas LigD when compared to that of other prokaryotic ligases (18).
Fig. 3: Impact of ribonucleotide termini on the NHEJ ligation step.
(A–C) Termini of NHEJ substrates were varied to be consistent with polymerase-dependent addition of a ribonucleotide vs. a deoxynucleotide, and introduced into MEFs expressing neither Pol μ nor TdT. Sensitivity of amplified products to a diagnostic restriction enzyme (RE) was used to identify examples of direct head-to-tail ligation. (B-C) Substrates with indicated terminal nucleotides were introduced into MEFs expressing neither Pol μ nor TdT. The mean % directly ligated products for three independent transfections, ± sd, is noted below. (D) Deoxy-guanine triphosphate (+GTP), ribo-guanine triphosphate (+rGTP), or an equivalent amount of the relevant salt (“none”), were added to Rosa26 Cas9-sgRNP transfections performed as in Fig. 2C–D, and genomic DNA was harvested after 1 hour. Data are the mean from 4 transfections, and error bars represent sd. Means were compared by ANOVA in pairs to no NTP added (***; p < 0.001). (E) Triple strand break repair model. Pol μ or TdT-dependent ribonucleotide addition is noted in red.
Table 1.
Stimulation of NHEJ repair pathway by a terminal ribonucleotide
| Substrate | Terminal C | Relative Joining efficiency# | % Direct ligation | Ligation Stimulation+ |
|---|---|---|---|---|
| --GC [-- | deoxyC | 1 | 87 ± 6 | 1 |
| --] CG-- | (ribo)C | 1.2 ± 0.7 | 92 ± 1 | 1.3 |
| deoxyC | 1 | 19 ± 8 | 1 | |
| --GAC [-- | (ribo)C | 5.8 ± 1.9 | 88 ± 6 | 26 |
| --] AG-- | 2’fluoroC | 5.4 ± 3.9 | 88 ± 1 | 25 |
| arabinoC | 1.1 ± 0.6 | < 2* | < 0.1* | |
| --GACGC [-- | deoxyC | 1 | 32 ± 3 | 1 |
| --] GCAG-- | (ribo)C | 6.3 ± 3.1 | 88 ± 3 | 17 |
| --TTTTTTTTGC [-- | deoxyC | 1 | 4 ± 1 | 1 |
| --] G-- | (ribo)C | 3.0 ± 0.9 | 64 ± 5 | 48 |
Joining efficiency rC/joining efficiency dC, as measured by qPCR that amplifies all NHEJ products.
Direct ligation product was undetectable
Ligation stimulation=Relative joining efficiency multiplied by %Direct ligation rC/%Direct ligation dC
We addressed if end structure context impacted whether ribonucleotide additions stimulated ligation. An added ribonucleotide was required for direct ligation in cells whenever the opposite strand was mostly mispaired or gapped (Fig. 3C, Table 1), end structures where Pol μ and TdT are uniquely active during NHEJ (15, 17). By comparison, direct ligation is similarly efficient for ribonucleotide vs. deoxynucleotide additions in contexts where other polymerases are more active - mostly complementary overhangs - both in cells (Fig. 3C, Table 1), and in vitro (10, 19). The class of end structures where Pol μ and TdT uniquely contribute to cellular NHEJ thus correlates well with the class of end structures where ribonucleotides are required for direct ligation. Moreover, deoxynucleotide additions in these contexts is associated with frequent deletion of both the added nucleotide and flanking DNA (Fig. S5, E–F), and repair was less-efficient (Table 1). Our results imply ribonucleotide addition is required for biological activity of Pol μ and TdT.
We sought to more directly address if polymerase function during cellular NHEJ relies on ribonucleotide addition. We introduced breaks in the chromosome with Cas9, and assessed whether introduction of high concentrations of deoxynucleotide triphosphate affected accumulation of the TdT-dependent repair products (“+GG”) characterized in Fig. 2C–D. Introduction of excess dGTP impaired accumulation of these products 4-fold, relative to parallel experiments with unperturbed nucleotide pools. In contrast, introduction of excess rGTP modestly stimulated +GG product recovery, consistent with the already high cellular rGTP pools in unperturbed cells (Fig. 3D). Similar results were also obtained using two methods that more generally measure NHEJ-dependent short insertions and deletions (Fig. S6, A–B). NHEJ was also impaired upon introduction of AraGTP (Fig. S6), in accord with the inability of additions of this ribonucleotide stereoisomer to promote repair (Fig. 3B).
We show that Pol μ and TdT preferentially add ribonucleotides (Fig 1, 2), contribute to repair of a specific subset of end structures (15), the same subset of end structures requires ribonucleotide additions for efficient LIG4-mediated repair (Fig. 3), and LIG4 may be uniquely able to take advantage of added nucleotides. Within all mammalian DNA metabolism, only the synthetic enzymes specific to NHEJ appear to cooperate in this unusual manner. Our results suggest this reflects a coevolution of these enzymes to better repair damaged or mispaired ends, a central problem to this pathway.
Our results also have more general relevance. Ribonucleotides destabilize DNA genomes unless removed by RER(20, 21), and when incorporated during NHEJ pose special problems for the RER pathway. We show safe accommodation of ribonucleotide-containing intermediates during NHEJ is most likely enabled by three sequential and coupled strand-break repair reactions (Fig. 3E); repair of the first strand with ribonucleotides, repair of the second strand with deoxynucleotides, and RNaseH2A-dependent excision of the ribonucleotides embedded during first strand repair. This model explains the two-fold dilution of embedded ribonucleotides that is independent of RER/RNaseH2A (Fig. 1B). Alternative models – where ribonucleotides are incorporated in both strands, or if RNaseH2 incises the first strand before second strand repair is complete – risk re-breakage of the chromosome. Additionally, the transient nature of the ribonucleotide embedded intermediate (t1/2 is likely less than 5 minutes; Fig. 1B) suggests that all three strand break repair reactions are coupled together, possibly by physical interactions between pathway components.
Pol μ is widely expressed and participates in as little as 16% (e.g. Fig. S4A) to as much as 66% (e.g. Fig S5A) of repair, depending on end structure (15). In lymphocytes, either TdT or Pol μ is active in 65% of NHEJ events required for V(D)J recombination (5, 17). The “triple stand break repair” model proposed here (Fig. 3E) is thus relevant to a large fraction of mammalian NHEJ, and is a fundamental departure from the previously accepted model. It is probably relevant to NHEJ in other species (e.g. yeast and bacteria) (18, 22, 23) as well. Our work further indicates ribonucleotide incorporation is required if mammalian Pol μ and TdT are to be effective in promoting long-term cellular proliferative capacity, the development of adaptive immunity, and radioresistance (3–9).
Supplementary Material
Acknowledgments
We thank Stephanie Nick McElhinny, Jody Havener, Thomas Kunkel, R. Scott Williams, Qi Zhang, and Ramsden lab members for help guiding the experiments described in this work, Dr. Luis Blanco for providing MEFs, Dr. Yung Chang for providing SP9 cells, and Dr. Eric Hendrickson for providing HCT116 cells.
Funding
This work was supported by NCI R01CA097096 (D.A.R.), ACS PF-14-0438-01-DMC (J.M.P.), NCI F31CA203156 (M.P.C.), T32GM007092 (M.E.L.) and T32GM119999 (A.J.L.).
Footnotes
Competing Interests
J.M.P. is currently an employee at New England Biolabs, a manufacturer and vendor of some of the molecular biology reagents used in this work.
Data and Materials Availability
All materials and raw data are available upon request.
References
- 1.Taccioli GE et al. , Impairment of V(D)J recombination in double-strand break repair mutants. Science 260, 207–210 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Waters CA, Strande NT, Wyatt DW, Pryor JM, Ramsden DA, Nonhomologous end joining: A good solution for bad ends. DNA Repair (Amst.) 17, 39–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komori T, Okada A, Stewart V, Alt FW, Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 261, 1171–1175 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D, Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science 261, 1175–1178 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA, Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19, 203–211 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Cabaniols JP, Fazilleau N, Casrouge A, Kourilsky P, Kanellopoulos JM, Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J. Exp. Med 194, 1385–1390 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucas D et al. , Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair. PLoS genetics 5, e1000389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chayot R, Danckaert A, Montagne B, Ricchetti M, Lack of DNA polymerase μ affects the kinetics of DNA double-strand break repair and impacts on cellular senescence. DNA Repair (Amst.) 9, 1187–1199 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Schimmel J, Kool H, van Schendel R, Tijsterman M, Mutational signatures of nonhomologous and polymerase theta-mediated end-joining in embryonic stem cells. The EMBO journal 36, 3634–3649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nick McElhinny SA, Ramsden DA, Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol 23, 2309–2315 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavanaugh NA, Beard WA, Wilson SH, DNA polymerase beta ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem 285, 24457–24465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JA et al. , A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. Journal of molecular biology 395, 282–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosavi RA, Moon AF, Kunkel TA, Pedersen LC, Bebenek K, The catalytic cycle for ribonucleotide incorporation by human DNA Pol λ. Nucleic Acids Res 40, 7518–7527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JS, Lujan SA, Kunkel TA, Processing ribonucleotides incorporated during eukaryotic DNA replication. Nature reviews. Molecular cell biology 17, 350–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryor JM et al. , Essential role for polymerase specialization in cellular nonhomologous end joining. Proceedings of the National Academy of Sciences of the United States of America 112, E4537–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlissel MS, Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol. Cell. Biol 18, 2029–2037 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nick McElhinny SA et al. , A gradient of template dependence defines distinct biological roles for family × polymerases in nonhomologous end joining. Mol. Cell 19, 357–366 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Shuman S, Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3’-OH monoribonucleotide. J. Biol. Chem 283, 8331–8339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon AF et al. , Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res 45, 9138–9148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nick McElhinny SA et al. , Genome instability due to ribonucleotide incorporation into DNA. Nature Chemical Biology 6, 774–781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reijns MAM et al. , Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA, Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem 280, 20051–20058 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Della M et al. , Mycobacterial Ku and Ligase Proteins Constitute a Two-Component NHEJ Repair Machine. Science 306, 683–685 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Lucas D et al. , Polymerase mu is up-regulated during the T cell-dependent immune response and its deficiency alters developmental dynamics of spleen centroblasts. European journal of immunology 35, 1601–1611 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Uphoff CC, Drexler HG, Detection of Mycoplasma contamination in cell cultures. Curr Protoc Mol Biol 106, 28.4.1–14 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Battaglia M, Pozzi D, Grimaldi S, Parasassi T, Hoechst 33258 staining for detecting mycoplasma contamination in cell cultures: a method for reducing fluorescence photobleaching. Biotech Histochem 69, 152–156 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Brinkman EK, Chen T, Amendola M, van Steensel B, Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42, e168–e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.