Abstract
Heart failure, coronary artery disease and myocardial infarction are the most prominent cardiovascular diseases contributing significantly to death worldwide. In the majority of situations, except for surgical interventions and transplantation, there are no reliable therapeutic approaches available to address these health problem. Despite several advances that led to the development of biomarkers and therapies based on the renin–angiotensin system, adrenergic pathways, etc, more definitive and consistent biomarkers and specific target based molecular therapies are still being sought. Recent advances in the field of genomic research has helped in identifying non-coding RNAs, including circular RNAs, piRNAs, micro RNAs, and long non-coding RNAs, that play a significant role in the regulation of gene expression and function and have direct impact on pathophysiological mechanisms. This new knowledge is currently being explored with much hope for the development of novel treatments and biomarkers. Circular RNAs and micro RNAs have been described in myocardium and aortic valves and were shown to be involved in the regulation of pathophysiological processes that potentially contribute to cardiovascular diseases. Approximately 32 000 human exonic circular RNAs have been catalogued and their functions are still being ascertained. In the heart, circular RNAs were shown to bind micro RNAs in a specific manner and regulate the expression of transcription factors and stress response genes, and expression of these non-coding RNAs were found to change in conditions such as cardiac hypertrophy, heart failure and cardiac remodelling, reflecting their significance as diagnostic and prognostic biomarkers. In this review, we address the present state of understanding on the biogenesis, regulation and pathophysiological roles of micro and circular RNAs in cardiovascular diseases, and on the potential future perspectives on their use as biomarkers and therapeutic agents.
Keywords: cardiovascular medicine, genetics, ischaemic heart disease, microrna, epigenetics
Introduction
Apart from cancer, cardiovascular disease, in particular heart failure, is still a major health problem across the world that contributes to significant mortality and morbidity, despite the many advances in the understanding of the pathological mechanisms and pharmacological developments.1–4 Inasmuch as the mortality rate for acute heart failure in hospital is 5.6% and 25.7% for 1 year from all causes after hospital admission,5 6 there is a great need for better drugs and ideally suited biomarkers for identifying the disease much earlier.7 Also, heart failure is the leading cause of hospitalisation in people over 65 years of age, across the globe, with about 1 in 5 adults aged over 40 years developing heart failure. This condition has a major impact on society in general.8 9 The number of people dying from acute myocardial ischaemia each year is estimated at approximately 2 million, and 25% of these mortalities occur in the USA alone, emphasising the great socioeconomic and health burden on the society.10
An important precursor event that contributes to cardiac problems is the disturbed homeostasis of the formation of the extracellular matrix (ECM), which is necessary as a support structure for the cardiomyocytes and vasculature. Excessive deposition of ECM and associated cardiac fibrosis occurs under conditions of myocardial infarction mediated cardiac injury, hypertension, etc. Cardiac fibrosis leads to disturbed electrical function of the myocardium and also reduces contractility, thus contributing to disturbed heart function and failure.11 Transformation of fibroblasts into myofibroblasts, which are smooth muscle-like cells with high migratory capacity which secrete large amounts of ECM, is an important event in cardiac fibrosis. This transformation is under the control of several signalling pathways. In particular, both canonical and non-canonical signalling by transforming growth factor-β was found to be critical.12 13 Myofibroblasts, in association with inflammatory cells, aggravate the pathogenesis of fibrosis and lead to heart dysfunction.
The importance of biomarkers in the diagnosis and prognosis of heart failure is studied on several fronts, as they provide insight into the underlying pathological mechanisms and also help in guiding better management of disease. Biomarkers are essential not only for the diagnosis of heart failure, but also to delineate the causative factors of heart failure. Also, some of the biomarkers are useful in evaluating treatment efficacy, in deciding the type and dose of treatments to be used, and as prognostic markers.14
Cardiac injury and the resultant heart failure can result from the haemodynamic and myocardial trophic effects arising from the inappropriate activation of the renin–angiotensin–aldosterone system (RAAS) and sympathetic nervous system. Among the various therapeutic measures for treating heart failure, pharmacological targeting of the RAAS and cardiac natriuretic peptides, along with other neurohormonal systems that regulate cardiovascular pressure/volume homeostasis, have been effectively employed to reduce the symptoms and extend life.1 Thus the recently introduced approaches to block the RAAS as well as degradation of natriuretic peptides with agents such as LCZ 696 (sacubitril/valsartan) has proven effective in reducing all cause mortality and hospital admissions due to heart failure.15 16
It has been recognised that understanding of the molecular events and changes in gene expression and their regulation in myocardial disease is essential for developing effective therapeutics as well as for identifying the biomarkers that guide therapy. Apart from the discovery of many signalling pathways and their disturbances, there has been an explosion of research in the area of non-coding RNAs, such as micro RNA (miRNA), circular RNA (circRNA) and long non-coding RNA (lncRNA), and their role in the control of gene expression and also as critical biomarkers for various diseases.17–20 The present review focuses on the recent studies on the role of miRNA and circRNA in cardiovascular diseases, their development and regulation, and as potential biomarkers.
Biogenesis of non-coding RNAs
The classic view of central dogma took a different turn when it was revealed that only a fraction of the human genome is transcribed to protein coding mRNAs, and that more than 80% of the genome, consisting of non-protein coding regions, intronic regions and repeat sequences, is transcribed to non-coding RNA. A recent compilation showed that there are about 2000 different miRNAs and nearly 56 000 lncRNAs coded in the human genome.17 21 Much work has shown that many of the non-coding RNAs have important regulatory roles controlling overall gene expression.17 22–24 Cytosol is the main location of miRNAs whereas circRNAs are found both in cytosol and the nucleus. Many of the circRNAs and lncRNAs are classified on the basis of their orientation, such as sense or antisense, unlike the small non-coding RNAs. The presence of circRNAs was also detected in the circulation,25 26 indicating their possible significance as biomarkers for diagnostic and therapeutic evaluation for various diseases, including cardiac diseases.27
Biogenesis of miRNAs
miRNAs are about 22 nucleotides (nt) long and inhibit protein synthesis by annealing with their seed region of 2–8 nt, with the 3’UTR of the target mRNAs, leading to the breakdown of mRNAs.28 29 These are found to vary in their expression and content in different tissues and circulations, under different pathophysiological conditions.1 30 Longer precursor RNAs of up to >10 kb in length, called primary miRNAs, transcribed by RNA polymerase II, give rise to the smaller pre-miRNAs of 70 nt, by the action of RNase III, Drosha and its essential cofactor DGCR8 protein, encoded by DGCR8 (DiGeorge syndrome critical region gene 8). These pre-miRNAs, which are formed in the nucleus and have a stem loop structure, are transported to the cytoplasm by Exportin-5 and Ran-GTP, where they are processed into ~22 nt double-stranded miRNA by Dicer, another RNase III and associated cofactor proteins. The small RNA duplex thus produced in the cytosol forms the RNA induced silencing complex (RISC), with the participation of Argonaute proteins, and other cofactors. As the passenger RNA is separated and degraded, the mature miRNA is formed.31 32 The RNA induced silencing complex with the loaded single-stranded miRNA attaches to the target RNA and directs it for degradation. Nearly half of the known miRNA genes are located in the intergenic regions of the genome. Among the 2000 miRNAs coded by the human genome, those with identical seed regions are grouped into miRNA families because these families target similar groups of transcripts; each miRNA family targets up to 300 mRNA targets.21 22 33 miRNA genes in the intergenic regions occur either in isolation or in clusters, and are under the control of their own promoters. Those in the intronic regions may be co-transcribed with the host gene or may also have their own promoters. On the other hand, exonic miRNA genes overlap exon and intronic regions.34 35
Biogenesis of circRNAs
CircRNAs are from 100 nt to a few kilobases in length and are generally produced from the exon regions as well as the intron, exon-intron and tRNA intron regions of protein coding genes, by the exon or intron circularisation of the 3’ and 5’ ends of the RNA.36 Recently, nearly 32 000 exonic circRNAs have been catalogued.37 There are two models of exon circularisation—in the ‘lariat driven circularisation’ model, skipping of exon(s) of the transcript leads to a lariat that contains exons, followed by spliceosome mediated circularisation38; in the ‘intron pairing driven circularisation’ model, pairing between the complementary motifs present in the intronic regions causes circularisation, and this process is facilitated by the complementary flanking Alu elements or other inverted repeats.39–41 Recent studies also identified exon–intron circRNAs, even though the precise mechanisms of their formation is not clear,42 and also circRNAs from lariat introns.43 Many circRNAs contain two exons, and in the human heart, titin and ryanodine receptor 2 genes were found to produce nearly 50 different circRNAs.44
Functions of circRNAs and miRNAs in the heart
Circular RNAs
RNA-Seq studies revealed the presence of a large number of different circRNAs expressed in cardiac tissues from humans and rodents.44 45 However, relatively fewer circRNAs have been ascribed functions in the heart, and important information regarding their functions, genomic information and splice sites can be found in circBase.46 A prominent change in failing myocardium is a switch to a fetal gene expression programme associated with expression of β-myosin heavy chain, N2BA titin variant, etc.47 48 Interestingly, circRNAs which are highly expressed in neonatal hearts are also elevated in failing hearts although not to the same extent. However, the precise function of the many altered circRNAs in the dilated heart, such as cTTN1, cTTN2, cTTN4 and cTTN5, is not well defined.49 Among the highly expressed circRNAs, those expressed from slc8a1 (ncx1) and rhobtb3 genes, which respectively code for SLC8a1 sodium/calcium exchanger and a Rab9 regulated ATPase involved in endosomes to Golgi transport, are induced during cardiac development.50 Thus altered expression of various circRNAs in failing myocardium is likely important in the overall function of the heart.51
Altered expression of circRNAs in heart disease
Heart related circRNA (HRCR) was the first circRNA found to be repressed in hypertrophic heart and in heart failure in mouse models.52 It was observed that HRCR, like a sponge, binds and inhibits the miRNA miR-223, which was implicated in cardiac hypertrophy.53 miR-223 exerts its effects by blocking the function of ARC (apoptosis inhibitor with CARD domain). On the other hand, HRCR overexpression was found to protect against hypertrophy in a mouse model via blockade of miR-223.52 Apart from HRCR, CDR1AS circRNA, which was originally observed in the brain as a sponge for miR-7, was also found in heart and was elevated following acute myocardial infarction in mouse models. Cardiomyocyte apoptosis due to myocardial infarction caused by ischaemia was found to be prevented by miR-7 and miR-7a, which block expression of proapoptotic proteins PARP and also SP1. Importantly, circRNA CDR1AS binds to miR-7 and miR-7a and aggravates myocardial infarction mediated cardiomyocyte loss.54 However, its role in the pathophysiology of the human heart is yet to be defined.51
It was reported that levels of circRNA_081881 are lowered by 10-fold in blood samples of patients with myocardial infarction.55 CircRNA_081881 is proposed to bind with miR-548, which targets the mRNA of cardiac protective gene PPARγ. Another circRNA, circ-Foxo3, derived from FOXO3 (the gene coding for the FOXO3 transcription factor), present in high amounts in aged hearts, was found to cause cell cycle arrest at G1 to S transition, and lead to cardiac cell senescence. Circ-Foxo3 was shown to antagonise the alleviating effects of ID1, E2F1, FAK, and HIF1α transcription factors on the process of ageing.56 57
Atherosclerotic vascular disease was found to be associated with single nucleotide polymorphisms near the INK4/ARF locus on chromosome 9p21.3. These single nucleotide polymorphisms regulate expression of INK4/ARF via changes in the levels of circRNA cANRIL, which is an antisense transcript derived from the INK4/ARF locus by alternate splicing. Thus cANRIL circRNA is implicated in the pathogenesis of atherosclerosis.58 59
In heart muscle, genes coding for titin and ryanodine receptor 2 express circRNAs in maximal numbers.51 In hypoxic vascular endothelium, circRNA cZNF292 was found to be induced and similar induction is also suggested in hypoxic myocardium. The significance of cZNF292 became evident in experimental studies showing compromised endothelial function when expression of this circRNA was reduced.60 A recent study described the identification of 1412 aortic valve specific circRNAs with their host genes having valve related functions and signalling pathways, including extracellular matrix–receptor interaction, ErbB signalling and vascular smooth muscle contraction.61
Apart from the changes in the above circRNAs, it was also observed that expression of CAMK2D (calcium/calmodulin dependent protein kinase type II delta chain) circRNAs were reduced in hypertrophic as well as dilated cardiomyopathy.49 Involvement of circRNAs in cardiac fibrosis and cardiomyopathy was also indicated in a mouse model, where it was found that circRNA_000203, which is transcribed from myo9A gene, is elevated in diabetic mouse myocardium and also in angiotensin II induced cardiac fibroblasts. CircRNA_000203 interacts with miR-26b-5p and compromises its antifibrotic activity, by enhancing the fibrotic process via expression of Col1a2 (collagen, type I, a2) and connective tissue growth factor.62
Exosomal circRNAs as biomarkers
Because of the presence of circRNAs in secreted exosomes, at a level that reflects their concentration in cells, exosomal circRNA has been considered an important diagnostic tool for various diseases.50 Thus circulating exosomal hsa_circ_0124644, which is elevated in patients with coronary artery disease,63 was found to be a useful biomarker for the disease. Similarly, the circRNA MICRA (myocardial infarction associated circular RNA), derived from the exon 1 of ZNF609 (zinc finger protein 609), was reported to predict left ventricular dysfunction, following myocardial infarction,25 and also to improve risk classification, indicating the significance of this circRNA as a novel biomarker for myocardial infarction.64
Micro RNAs
Certain miRNA profiling studies conducted between 2008 and 2015 validated the initially identified heart failure related miRNAs from smaller cohorts in larger groups of patients. These profiled and validated miRNAs are classified according to the biological sample (ie, whole blood, plasma, serum, buffy coat, peripheral blood mononuclear cells or cardiac muscle biopsy). Even though there was little consensus among these studies regarding the miRNA signatures in heart failure, the clusters of miRNAs could be used for differentiating heart failure with preserved or reduced ejection fraction.65–73 Many functional and association studies implicated a variety of miRNAs in the regulation of cardiovascular function, and levels of these miRNAs are altered in cardiac remodelling, hypertrophy, fibrosis, apoptosis, hypoxia, cardiomyopathy and heart failure.74 75
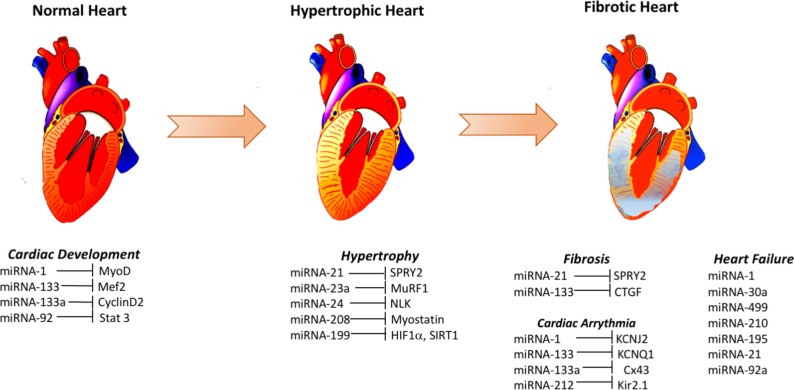
A number of studies indicated the potential use of different miRNAs as biomarkers of heart failure. Elevated levels of miR-1 in the plasma of heart failure patients with acute myocardial infarction,76 miR-195 in the left ventricle biopsy of dilated cardiomyopathy patients,72 miR-30a in the serum of patients with heart failure with reduced ejection fraction71 and miR-499 in the plasma of acute heart failure and acute coronary syndrome patients,76 were among some of the major changes seen (figure 1). A number of miRNAs (eg, miR-1, −124–3 p, −126,–150, −195,–21, −210, −30a, −342–3 p, −423–5 p, −499–5 p, −622 and −92a) were found to be differentially regulated in more than one cohort of heart failure patients, and some of these miRNAs likely target genes related to cardiac hypertrophy and apoptosis (miR-1, miR-30a), cardiac fibrosis (miR-21) and apoptosis signalling pathways (miR-195, miR-499–5 p, miR-92a).1 Therefore, it is possible that some of these differentially regulated miRNAs in heart failure may help in discovering the hitherto unknown molecular mechanisms of pathogenesis and the genes involved and their products.
Figure 1.
Some elevated signatures of micro RNAs (miRNAs) and their targets involved in cardiac disease. Several different miRNAs are altered during cardiac development and during the pathogenesis of hypertrophy, fibrosis, arrhythmia and eventual heart failure. These changes are proposed to be causative or reflective of these pathogenic mechanisms. Here, only some of the elevated miRNAs in normal cardiac development, cardiac hypertrophy, cardiac fibrosis, cardiac arrhythmia and heart failure and their corresponding likely targets are shown.
Altered miRNA expression affecting lipid metabolism in heart disease
Lipid metabolism, in particular cholesterol metabolism and its homeostasis, is important in the pathogenesis of atherosclerosis, which is one of the main causes of coronary heart disease. It is generally believed that a disturbed balance between the low density lipoprotein and high density lipoprotein (HDL) associated cholesterol plays an essential part in promoting atherosclerosis. A critical and rate limiting step in the biosynthesis of cholesterol is catalysed by 3-hydroxy-3-methylglutaryl-CoA synthase-1, and miR-223 inhibits the expression of this enzyme and thus cholesterol biosynthesis. This miR-223 is also known to block the expression of other sterol biosynthetic enzymes, such as methylsterol monooxygenase 1, and also scavenger receptor BI, involved in the uptake of HDL cholesterol.77 ApoB containing low density lipoproteins, whose dysregulation contributes to atherosclerosis, are produced in the liver and their synthesis/assembly is known to be regulated by miR-122, miR-34a and miR-30c.78 Also, liver production of HDL is inhibited by miR-33, which blocks cholesterol efflux to apolipoprotein A1 by interfering with the expression of the adenosine triphosphate binding cassette (ABC) transporter.79 Conditions such as hyperlipidaemia stimulate the arterial endothelial cells to express different adhesion molecules (eg, vascular adhesion molecule-1, intracellular adhesion molecule-1 and E-selectin), which participate in capturing leukocytes on their surfaces, and this process along with the formation of macrophage foam cells are involved in the formation of atherosclerotic plaques. Different miRNAs, such as miR-181b which controls the NF-κB pathway and miR-125a which regulates lipid uptake by macrophages and foam cell formation, play a role in plaque formation.80
Altered miRNAs and peptide hormones in heart disease
Several neurohormone peptides, such as natriuretic peptides, endothelin-1 and angiotensin II, and their cognate receptors, play an important role in cardiovascular function, and perturbation of their function is implicated in the pathogenesis of cardiovascular disease. However, whether miRNAs regulate the expression of these peptides and affect their function is not clearly established. Some evidence from experimental animals indicated that miR-100 targets the 3’UTR of the NPR3 gene, coding for natriuretic peptide receptor 3. Thus elevated levels of miR-100 in heart failure patients may help in maintaining circulating levels of the natriuretic peptides.81 Also, a negative association between miR-125a/b-5p, which binds with the 3’UTR of mRNA coding for prepro-endothelin-1, and endothelin-1 expression was noticed.82
In silico gene expression analysis indicated the possible regulation of genes associated with angiotensin II signalling by miR-132/212 in cardiac fibroblasts.83 Also, negative regulation was reported for expression of AGTR1 (angiotensin II type I receptor) and ANP, by miR-15584 and miR-425,85 respectively, by binding to the 3’UTR of the corresponding mRNAs. Another component of this system, expression of human CYP11B2 (aldosterone synthase), was shown to be downregulated by miR-766, associated with lowered blood pressure.86 Further in silico studies to identify putative miRNA targets in neurohormone signalling pathways implicated in cardiac dysfunction revealed that miRNAs mostly appear to regulate these pathways by lowering the expression of neurohormone receptors and these include endothelin receptors, mineralocorticoid receptor/nuclear receptor subfamily three group C member 2, angiotensin II receptors, natriuretic peptide receptors and corticotropin releasing factor receptors.1
miRNAs and cardiac hypertrophy
The heart normally shows compensatory changes in structural organisation under stress conditions, such as hypertension, ageing and metabolic disturbances, by cardiomyocyte enlargement (cardiac hypertrophy) and also by enhancing extracellular matrix (cardiac fibrosis). Prolonged cardiac hypertrophy leads to a life threatening condition, end stage heart failure with reduced ejection fraction.87 88 The first pro-hypertrophic miRNA was found to be miR-208 in animal studies, showing that deletion of this miRNA was protective against hypertrophy and fibrosis of the heart89 whereas overexpression in mouse hearts led to arrhythmia and pathological cardiac hypertrophy.90 Interestingly, inhibition of miR-208a by a specific antisense oligonucleotide was found to improve cardiac function in a hypertensive heart failure rat model91 and also to protect against diet induced obesity in mice.92
Even though miR-1 is also implicated in cardiac hypertrophy, its precise role remains controversial. On the other hand, miR-133a levels in cardiac tissues were found to be inversely correlated with hypertrophy in animal models of pathological cardiac hypertrophy, exercise induced hypertrophy and also in the hearts of patients with hypertrophic cardiomyopathy or atrial dilatation (figure 1). Inasmuch as miR-133a is also protective against cardiac fibrosis, clinical uses of this miRNA became obvious.80 The pro-autophagic transcription factor FOXO-3, which mediates the cardiomyocyte recycling process, an adaptive protective response against pressure overload in the failing heart, was found to be a target of miR-212 and miR-132, which are overexpressed in murine models of pressure induced cardiac hypertrophy.93 94
Cardiac fibrosis, which is also a major cause of heart failure and myocardial infarction, was found to be associated with elevated levels of miR-21 in the cardiomyofibroblasts in rodent models and human hearts. Since miR-21 targets sprout homologue-1 (Spry1), it was suggested that this miRNA enhances growth factor secretion and fibroblast survival and proliferation, leading to fibrosis.95 Even though some studies showed positive results of ameliorating heart function in rodent models by antagomirs of miR-21, different results were obtained in other studies.96 Besides the above described miR-133a, miR-29, which targets components of fibrosis, including collagens, matrix metalloproteinases, leukaemia inhibitory factor, etc, was also found to be cardioprotective and its levels are found to be reduced in myocardium under stress conditions.97 98
Micro RNAs in the circulation
Even though the proposed function of miRNAs is within the cell, specifically targeting mRNAs, and thus protein synthesis, several miRNAs have also been found extracellularly and in the circulation, and were found to be stable and associated with different types of carrier proteins or exosomes.99 Inasmuch as these circulating miRNAs were found to reflect their corresponding cellular levels, it was proposed that circulating miRNAs can be helpful as potential biomarkers (table 1) for diagnosis or treatment efficacy monitoring, for disease conditions including cardiovascular diseases.75 Currently, blood levels of B-type atrial natriuretic peptides (BNPs) and N-terminal pro-brain natriuretic peptide (NT-proBNP) are considered as sensitive and reliable biomarkers for heart failure, although not highly specific. It has been reported that circulating levels of miR-423–5 p are different in patients having dyspnoea due to heart failure compared with those with causes other than heart failure.100 Apart from miR-423–5 p, changes in other circulating miRNAs have also been described and thus plasma levels of miR-103, miR-142–3 p, miR-30b and miR-342–3 p were found to be lower in patients with acute heart failure68 whereas miR-499 is elevated101 indicating the possibility of combining different miRNAs as panels for the diagnosis of heart disease.75 MiR-29a, which was found to be elevated in cardiac fibrosis, was also found to be elevated in the plasma of hypertrophic cardiomyopathy patients and correlated with left ventricular hypertrophy and cardiac remodelling.102 Also, elevated level of miR-29a in the circulation was shown to be associated with enhanced interventricular septum size, an indicator of fibrosis as well as hypertrophy.103
Table 1.
Circulating micro RNAs as biomarkers for diagnosis, prognosis and treatment response in cardiovascular disease
| miRNA | Biomarker value | Condition | Respective fold change vs controls | Ref |
| miR-22, −92b, −320a, −423–5 p | Diagnosis | Chronic heart failure | Up (1.4; 1.3; 1.2; 1.5) | 73 |
| miR-30c, −146a, −221,–375 | Diagnosis | Chronic heart failure | Down (0.6; 0.7; 0.4; 0.45) | 68 |
| miR-107,–139, −142–5 p | Diagnosis | Chronic heart failure | Down (0.3; 0.2; 0.52) | 75 |
| miR-193b-3p, −193b-5p, −183–3 p, −190a, −211–5 p, −494 | Diagnosis | Chronic heart failure | Down (0.4; 0.5; 0.4; 0.5; 0.45; 0.2) | 69 |
| miR-423–5 p | Diagnosis | Acute heart failure | Up (2.5) | 100 |
| miR-499 | Diagnosis | Acute heart failure | Up (100) | 101 |
| miR-106a-5p, −652–3 p, −18a-5p, −27a-3p, −199a-3p | Diagnosis | Acute heart failure | Down (<0.5 fold, for all) | 105 |
| miR-30b, −103, −142–3 p, −342–3 p | Diagnosis | Acute heart failure | Down (0.4; 0.6; 0.4; 0.5) | 70 |
| miR-30c, −146a | Diagnosis | HFpEF vs HFrEF | Down (0.8; 0.7) | 68 |
| miR-221,–328, −375 | Diagnosis | HFpEF vs HFrEF | Up (1.25; 1.7; 1.4) | 68 |
| miR-29a | Diagnosis | Cardiac fibrosis and hypertrophic cardiomyopathy | Up (3.1) | 102 |
| miR-18a-5p, −652–3 p | Prognosis | Heart failure, 180 day all cause mortality | Down (<0.5 fold, for all) | 105 |
| miR-126, −508–5 p | Prognosis | Cardiovascular death after 2 years | Up (1.33; 10.2) | 104 |
| miR-30d | Prognosis | Acute heart failure, 1 year all cause mortality | Down (0.33) | 106 |
| miR-26b-5p, −29a-3p, −92a-3p, 145–5 p, −30e-5p | Treatment response | 12 months after effective CRT | Up (15.2; 9; 14; 6; 8) | 107 |
| miR-1, −208a/208b, −499 | Treatment response | 3 months after LVAD | Down (0.1; 0.03; 0.014) | 111 |
| miR-1202 | Treatment response | 3 months after LVAD | Down (0.9) | 110 |
CRT, cardiac resynchronisation therapy; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; LVAD, left ventricular assist device.
Use of circulating miRNAs for disease prognosis
The prognostic value of circulating miRNAs was evaluated in several studies and it was found that decreased plasma miR-126 is related to cardiovascular mortality in patients with ischaemic heart failure, whereas such a relationship in non-ischaemic heart failure patients was seen with elevated circulating miR-508a-5p.104 Reduced plasma levels of miR-18a-5p and miR-652–3 p were found to be predictive of 180 day cardiovascular death in hospitalised heart failure patients.105 A recent study on 96 acute heart failure patients showed that reduced serum miR-30d levels are predictive of 1 year all cause mortality in these patients.106 Besides being useful as prognostic biomarkers, miRNAs have also been found to respond to treatments and thus reflect treatment efficacy in patients with cardiovascular disease. Thus cardiac resynchronisation therapy was found to be associated with higher circulating levels of miR-26b-5p, miR-145–5 p, miR-92a-3p, miR-30e-5p and miR-29a-3p,107 and also miR-30d108 and miR-30e109 in responding patients. Similarly, it was noted that responsiveness to left ventricular assist device could be assessed by plasma levels of miR-1202,110 whereas elevated circulating miR-483–3 p and decreased miR-208, miR-499 and miR-1 levels were found following implantation of this device due to unloading of the heart.111 On the other hand, anthracycline chemotherapy to cancer patients is known to cause cardiotoxicity and plasma miR‐29b and miR‐499 are found to be acutely elevated following anthracycline therapy with a dose–response relationship.112
Even though these studies emphasise the significance of circulating miRNAs as potential biomarkers for cardiovascular disease, there is a great need for systematic comparison of large panels of miRNAs in larger cohorts of patients with simultaneous comparison with established biomarkers, for diagnosis, prognosis and treatment response. Also, use and validation of miRNAs as biomarkers in assessing the clinical outcome of cardiac diseases is the most important task that needs to be accomplished. In addition, considering that miRNAs can be secreted out of the cell and can be taken up by other cells where they can exert their effects, and also because of their stability in the circulation, it is possible that these can be used as potential therapeutics. However, because of the multiple mRNA targets for many of the miRNAs and also effects in more than one organ, much needs to be worked out for them to be used in clinical practice.
Nevertheless, many studies have proven their potential as therapeutics in cardiac disease models in rodents.113 114 The diagnostic/prognostic abilities of some of the miRNAs have been described by receiver operating characteristic (ROC) curve analyses. Thus while assessing non-invasive biomarkers of graft rejection in heart transplant recipients, it was found that serum levels of miR-10a, miR-31, miR-92a and miR-155, which strongly correlate with their expression in cardiac tissue, significantly discriminated patients with allograft rejection from those without rejection, on the basis of the AUC analyses of the corresponding ROC curves (miR-10a AUC=0.975; miR-31 AUC=0.932; miR-92a AUC=0.989; and miR-155 AUC=0.998, P<0.0001 for all).115 Similarly, ROC curve analysis for miR-499 in acute myocardial infarction patients compared with healthy controls showed an AUC of 0.947, which correlated significantly with circulating cTnT, a well known biomarker.30 Also, for a diagnosis of heart failure, miR-423–5 p was found to show a significant clinically relevant association, with an AUC of 0.91 for ROC curve analysis (95% CI 0.84–0.98).30
Conclusions
The significance of non-coding RNAs, including miRNAs, circRNAs and also lncRNAs in the regulation of various cellular processes and the pathophysiology of the whole organisms is now clearly established. The importance of miRNAs and circRNAs in cardiovascular diseases is well studied and the role of these non-coding RNAs in the disease process and their use as diagnostic and prognostic biomarkers is recognised. Inasmuch as the existing biomarkers are only able to help differentiating the causes and progression of disease, to a limited extent the newly identified miRNAs and circRNAs are making it possible to fill this void. Apart from their use as disease biomarkers, miRNAs have also proven to be useful in evaluating patient response to the available therapies during the course of treatments and also to understand whether a patient will be responsive to a given treatment.116 117 Also, miRNAs are being tested as drugs for cardiovascular diseases in multiple preclinical studies and have proven to be efficacious. However, considering that miRNAs and circRNAs have multiple cellular targets and may show their effects in organs not directly involved in the disease pathogenesis, attempts are being made to improve the in vivo specificity of miRNA mimics. Also, the effectiveness of panels of multiple miRNAs and/or circRNAs is being studied as biomarkers for diagnosis, prognosis and treatment response monitoring. The future holds promise for these biomolecules and their mimics as therapeutics as well as biomarkers for cardiovascular diseases.
Acknowledgments
Author acknowledges support from the First Affiliated Hospital of Xinxiang Medical University.
Footnotes
Contributors: GZ planned the review, collected all of the literature, assembled the information and wrote thereview.
Funding: Beijing Medical and Health Foundation (No:B17246-045); Foundation of He’nan Educational Committee (No:18A320005 and No:19A360032).
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Wong LL, Wang J, Liew OW, Richards AM, Chen YT. MicroRNA and heart failure. Int J Mol Sci 2016;17:502 10.3390/ijms17040502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, Falk V, González-Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GM, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P. Authors/Task Force Members Document Reviewers. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail 2016;18:891–975. 10.1002/ejhf.592 [DOI] [PubMed] [Google Scholar]
- 3. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Colvin MM, Drazner MH, Filippatos G, Fonarow GC, Givertz MM, Hollenberg SM, Lindenfeld J, Masoudi FA, McBride PE, Peterson PN, Stevenson LW, Westlake C. Writing Committee Members. 2016 ACC/AHA/HFSA focused update on new pharmacological therapy for heart failure: an update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation 2016;134:e282–e293. 10.1161/CIR.0000000000000435 [DOI] [PubMed] [Google Scholar]
- 4. Vardeny O, Claggett B, Packer M, Zile MR, Rouleau J, Swedberg K, Teerlink JR, Desai AS, Lefkowitz M, Shi V, McMurray JJ, Solomon SD. Prospective Comparison of ARNI with ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure (PARADIGM-HF) Investigators. Efficacy of sacubitril/valsartan vs. enalapril at lower than target doses in heart failure with reduced ejection fraction: the PARADIGM-HF trial. Eur J Heart Fail 2016;18:1228–34. 10.1002/ejhf.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Targher G, Dauriz M, Laroche C, Temporelli PL, Hassanein M, Seferovic PM, Drozdz J, Ferrari R, Anker S, Coats A, Filippatos G, Crespo-Leiro MG, Mebazaa A, Piepoli MF, Maggioni AP, Tavazzi L. ESC-HFA HF Long-Term Registry investigators. In-hospital and 1-year mortality associated with diabetes in patients with acute heart failure: results from the ESC-HFA Heart Failure Long-Term Registry. Eur J Heart Fail 2017;19:54–65. 10.1002/ejhf.679 [DOI] [PubMed] [Google Scholar]
- 6. Roger VL. Epidemiology of heart failure. Circ Res 2013;113:646–59. 10.1161/CIRCRESAHA.113.300268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richards AM, Frampton CM. Can circulating biomarkers identify heart failure patients at low risk? Eur J Heart Fail 2015;17:1213–5. 10.1002/ejhf.438 [DOI] [PubMed] [Google Scholar]
- 8. Alla F, Zannad F, Filippatos G. Epidemiology of acute heart failure syndromes. Heart Fail Rev 2007;12:91–5. 10.1007/s10741-007-9009-2 [DOI] [PubMed] [Google Scholar]
- 9. Stewart S, MacIntyre K, Hole DJ, Capewell S, McMurray JJ. More ’malignant' than cancer? Five-year survival following a first admission for heart failure. Eur J Heart Fail 2001;3:315–22. 10.1016/S1388-9842(00)00141-0 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Pan X, Fan Y, Hu X, Liu X, Xiang M, Wang J. Dysregulated expression of microRNAs and mRNAs in myocardial infarction. Am J Transl Res 2015;7:2291–304. [PMC free article] [PubMed] [Google Scholar]
- 11. Berk BC, Fujiwara K, Lehoux S. ECM remodeling in hypertensive heart disease. J Clin Invest 2007;117:568–75. 10.1172/JCI31044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leask A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ Res 2010;106:1675–80. 10.1161/CIRCRESAHA.110.217737 [DOI] [PubMed] [Google Scholar]
- 13. Squires CE, Escobar GP, Payne JF, Leonardi RA, Goshorn DK, Sheats NJ, Mains IM, Mingoia JT, Flack EC, Lindsey ML. Altered fibroblast function following myocardial infarction. J Mol Cell Cardiol 2005;39:699–707. 10.1016/j.yjmcc.2005.07.008 [DOI] [PubMed] [Google Scholar]
- 14. Schmitter D, Cotter G, Voors AA. Clinical use of novel biomarkers in heart failure: towards personalized medicine. Heart Fail Rev 2014;19:369–81. 10.1007/s10741-013-9396-5 [DOI] [PubMed] [Google Scholar]
- 15. Menendez JT. The mechanism of action of LCZ696. Card Fail Rev 2016;2:40–6. 10.15420/cfr.2016:1:1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vardeny O, Miller R, Solomon SD. Combined neprilysin and renin-angiotensin system inhibition for the treatment of heart failure. JACC Heart Fail 2014;2:663–70. 10.1016/j.jchf.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 17. Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res 2014;42:D98–D103. 10.1093/nar/gkt1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, Singha RS, Malakar AK, Chakraborty S. Interplay between miRNAs and human diseases. J Cell Physiol 2018;233:2007–18. 10.1002/jcp.25854 [DOI] [PubMed] [Google Scholar]
- 19. Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans 2018;46:11–21. 10.1042/BST20170037 [DOI] [PubMed] [Google Scholar]
- 20. Gomes CPC, Spencer H, Ford KL, Michel LYM, Baker AH, Emanueli C, Balligand JL, Devaux Y; Cardiolinc network. The function and therapeutic potential of long non-coding RNAs in cardiovascular development and disease. Mol Ther Nucleic Acids 2017;8:494–507. 10.1016/j.omtn.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinberg RA. The dark side of the genome. Technology Review 1991;94:45–51. [Google Scholar]
- 25. Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, Burkhardt R, Thiery J, Wagner DR, Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol 2016;68:1247–8. 10.1016/j.jacc.2016.06.040 [DOI] [PubMed] [Google Scholar]
- 26. Qian Z, Liu H, Li M, Shi J, Li N, Zhang Y, Zhang X, Lv J, Xie X, Bai Y, Ge Q, Ko EA, Tang H, Wang T, Wang X, Wang Z, Zhou T, Gu W. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine 2018;27:18–26. 10.1016/j.ebiom.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haque S, Harries LW. Lorna Harries. Circular RNAs (circRNAs) in health and disease. Genes 2017;8:353 10.3390/genes8120353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cathcart P, Lucchesi W, Ottaviani S, De Giorgio A, Krell J, Stebbing J, Castellano L. Noncoding RNAs and the control of signalling via nuclear receptor regulation in health and disease. Best Pract Res Clin Endocrinol Metab 2015;29:529–43. 10.1016/j.beem.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 29. Li M, Zhang J. Circulating MicroRNAs: potential and emerging biomarkers for diagnosis of cardiovascular and cerebrovascular diseases. Biomed Res Int 2015;2015:1–9. 10.1155/2015/730535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duggal B, Gupta MK, Naga Prasad SV. Potential role of microRNAs in cardiovascular disease: are they up to their hype? Curr Cardiol Rev 2016;12:304–10. 10.2174/1573403X12666160301120642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 2005;11:1753–61. 10.1261/rna.2248605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004;10:1957–66. 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14:1902–10. 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol 2010;222:540–5. 10.1002/jcp.21993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126–39. 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 36. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 2016;6:34985 10.1038/srep34985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015;10:103–11. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 39. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014;159:134–47. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 41. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 42. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–64. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 44. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103–7. 10.1016/j.yjmcc.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 45. Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD, Li PY, Richards AM, Foo RS. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:cvw250–309. 10.1093/cvr/cvw250 [DOI] [PubMed] [Google Scholar]
- 46. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666–70. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015;16:126 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev 2007;12(3-4):331–43. 10.1007/s10741-007-9034-1 [DOI] [PubMed] [Google Scholar]
- 49. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE, Pinto YM. RBM20 regulates circular RNA production from the titin gene. Circ Res 2016;119:CIRCRESAHA.116.309568–1003. 10.1161/CIRCRESAHA.116.309568 [DOI] [PubMed] [Google Scholar]
- 50. Li M, Ding W, Sun T, Tariq MA, Xu T, Li P, Wang J. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. Febs J 2018;285:220–32. 10.1111/febs.14191 [DOI] [PubMed] [Google Scholar]
- 51. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, Dimmeler S, Squire I. Cardiolinc network. Circular RNAs in heart failure. Eur J Heart Fail 2017;19:701–9. 10.1002/ejhf.801 [DOI] [PubMed] [Google Scholar]
- 52. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF, Pf L. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602–11. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 53. Taïbi F, Metzinger-Le Meuth V, Massy ZA, Metzinger L. miR-223: an inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta 2014;1842:1001–9. 10.1016/j.bbadis.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 54. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP, Ym S, Xp J. The Circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016;11:e0151753 10.1371/journal.pone.0151753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang F, Zhu P, Guo J, Liu X, Wang S, Wang G, Liu W, Wang S, Ge N. Circular RNAs in thoracic diseases. J Thorac Dis 2017;9:5382–9. 10.21037/jtd.2017.10.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017;38:1402–12. 10.1093/eurheartj/ehw001 [DOI] [PubMed] [Google Scholar]
- 57. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846–58. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One 2009;4:e5027 10.1371/journal.pone.0005027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res 2015;117:884–90. 10.1161/CIRCRESAHA.115.306319 [DOI] [PubMed] [Google Scholar]
- 61. Chen J, Wang J, Jiang Y, Gu W, Ni B, Sun H, Gu W, Chen L, Shao Y. Identification of circular RNAs in human aortic valves. Gene 2018;642:135–44. 10.1016/j.gene.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 62. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342 10.1038/srep40342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918 10.1038/srep39918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 2017;17:33–6. 10.1016/j.ijcha.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Melman YF, Shah R, Das S. MicroRNAs in heart failure: is the picture becoming less miRky? Circ Heart Fail 2014;7:203–14. 10.1161/CIRCHEARTFAILURE.113.000266 [DOI] [PubMed] [Google Scholar]
- 66. Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail 2016;18:457–68. 10.1002/ejhf.495 [DOI] [PubMed] [Google Scholar]
- 67. Vogel B, Keller A, Frese KS, Leidinger P, Sedaghat-Hamedani F, Kayvanpour E, Kloos W, Backe C, Thanaraj A, Brefort T, Beier M, Hardt S, Meese E, Katus HA, Meder B. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J 2013;34:2812–23. 10.1093/eurheartj/eht256 [DOI] [PubMed] [Google Scholar]
- 68. Watson CJ, Gupta SK, O’Connell E, Thum S, Glezeva N, Fendrich J, Gallagher J, Ledwidge M, Grote-Levi L, McDonald K, Thum T. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail 2015;17:405–15. 10.1002/ejhf.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, Chong JP, Ng JY, Chen YT, Chan MM, Chen Z, Yeo PS, Ng TP, Ling LH, Sim D, Leong KT, Ong HY, Jaufeerally F, Wong R, Chai P, Low AF, Lam CS, Jeyaseelan K, Richards AM. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail 2015;17:393–404. 10.1002/ejhf.223 [DOI] [PubMed] [Google Scholar]
- 70. Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail 2013;15:1138–47. 10.1093/eurjhf/hft078 [DOI] [PubMed] [Google Scholar]
- 71. Fukushima Y, Nakanishi M, Nonogi H, Goto Y, Iwai N. Assessment of plasma miRNAs in congestive heart failure. Circ J 2011;75:336–40. 10.1253/circj.CJ-10-0457 [DOI] [PubMed] [Google Scholar]
- 72. Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y, Komuro I. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 2013;113:322–6. 10.1161/CIRCRESAHA.113.301209 [DOI] [PubMed] [Google Scholar]
- 73. Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail 2012;14:147–54. 10.1093/eurjhf/hfr155 [DOI] [PubMed] [Google Scholar]
- 74. Lai KB, Sanderson JE, Izzat MB, Yu CM, Cm Y. Micro-RNA and mRNA myocardial tissue expression in biopsy specimen from patients with heart failure. Int J Cardiol 2015;199:79–83. 10.1016/j.ijcard.2015.07.043 [DOI] [PubMed] [Google Scholar]
- 75. Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, Melillo G, Rigolini R, Costa E, Crea F, Capogrossi MC, Napolitano M, Martelli F. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics 2010;42:420–6. 10.1152/physiolgenomics.00211.2009 [DOI] [PubMed] [Google Scholar]
- 76. Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, Erlinge D. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord 2013;13:12 10.1186/1471-2261-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, Sethupathy P, Remaley AT. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA 2014;111:14518–23. 10.1073/pnas.1215767111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhou L, Irani S, Sirwi A, Hussain MM. MicroRNAs regulating apolipoprotein B-containing lipoprotein production. Biochim Biophys Acta 2016;1861:2062–8. 10.1016/j.bbalip.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 79. Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328:1570–3. 10.1126/science.1189862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dangwal S, Schimmel K, Foinquinos A, Xiao K, Thum T. Noncoding RNAs in heart failure. Handb Exp Pharmacol 2017;243:423–45. [DOI] [PubMed] [Google Scholar]
- 81. Wong LL, Wee ASY, Lim JY, Ng JYX, Chong JPC, Liew OW, Lilyanna S, Martinez EC, Ackers-Johnson MA, Vardy LA, Armugam A, Jeyaseelan K, Ng TP, Lam CSP, Foo RSY, Richards AM, Chen Y-T. Natriuretic peptide receptor 3 (NPR3) is regulated by microRNA-100. J Mol Cell Cardiol 2015;82:13–21. 10.1016/j.yjmcc.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 82. Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, Yuan W, Rui Y-C. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens 2010;28:1646–54. 10.1097/HJH.0b013e32833a4922 [DOI] [PubMed] [Google Scholar]
- 83. Eskildsen TV, Schneider M, Sandberg MB, Skov V, Brønnum H, Thomassen M, Kruse TA, Andersen DC, Sheikh SP. The microRNA-132/212 family fine-tunes multiple targets in angiotensin II signalling in cardiac fibroblasts. J Renin Angiotensin Aldosterone Syst 2015;16:1288–97. 10.1177/1470320314539367 [DOI] [PubMed] [Google Scholar]
- 84. Yang Y, Zhou Y, Cao Z, Tong XZ, Xie HQ, Luo T, Hua XP, Wang HQ. miR-155 functions downstream of angiotensin II receptor subtype 1 and calcineurin to regulate cardiac hypertrophy. Exp Ther Med 2016;12:1556–62. 10.3892/etm.2016.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, Spagnolli E, Martinez A, Ryan A, Tainsh LT, Kim S, Rong J, Huan T, Freedman JE, Levy D, Miller KK, Hata A, del Monte F, Vandenwijngaert S, Swinnen M, Janssens S, Holmes TM, Buys ES, Bloch KD, Newton-Cheh C, Wang TJ. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest 2013;123:3378–82. 10.1172/JCI67383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Maharjan S, Mopidevi B, Kaw MK, Puri N, Kumar A. Human aldosterone synthase gene polymorphism promotes miRNA binding and regulates gene expression. Physiol Genomics 2014;46:860–5. 10.1152/physiolgenomics.00084.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Braunwald E. Heart Failure. JACC Heart Fail 2013;1:1–20. 10.1016/j.jchf.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 88. Bisping E, Wakula P, Poteser M, Heinzel FR. Targeting cardiac hypertrophy: toward a causal heart failure therapy. J Cardiovasc Pharmacol 2014;64:293–305. [DOI] [PubMed] [Google Scholar]
- 89. van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a MicroRNA. Science 2007;316:575–9. 10.1126/science.1139089 [DOI] [PubMed] [Google Scholar]
- 90. Callis TE, Pandya K, Seok HY, Tang R-H, Tatsuguchi M, Huang Z-P, Chen J-F, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang D-Z. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009;119:2772–86. 10.1172/JCI36154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011;124:1537–47. 10.1161/CIRCULATIONAHA.111.030932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac MicroRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012;149:671–83. 10.1016/j.cell.2012.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078 10.1038/ncomms2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007;13:619–24. 10.1038/nm1574 [DOI] [PubMed] [Google Scholar]
- 95. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JTR, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980–4. 10.1038/nature07511 [DOI] [PubMed] [Google Scholar]
- 96. Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 2010;120:3912–6. 10.1172/JCI43604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008;105:13027–32. 10.1073/pnas.0805038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang Y, Huang X-R, Wei L-H, Chung ACK, Yu C-M, Lan H-Y. miR-29b as a therapeutic agent for angiotensin II-induced cardiac fibrosis by targeting TGF-β/Smad3 signaling. Mol Ther 2014;22:974–85. 10.1038/mt.2014.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res 2010;106:1035–9. 10.1161/CIRCRESAHA.110.218297 [DOI] [PubMed] [Google Scholar]
- 101. Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499–506. 10.1161/CIRCGENETICS.110.957415 [DOI] [PubMed] [Google Scholar]
- 102. Roncarati R, Viviani Anselmi C, Losi MA, Papa L, Cavarretta E, Da Costa Martins P, Contaldi C, Saccani Jotti G, Franzone A, Galastri L, Latronico MV, Imbriaco M, Esposito G, De Windt L, Betocchi S, Condorelli G. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol 2014;63:920–7. 10.1016/j.jacc.2013.09.041 [DOI] [PubMed] [Google Scholar]
- 103. Derda AA, Thum S, Lorenzen JM, Bavendiek U, Heineke J, Keyser B, Stuhrmann M, Givens RC, Kennel PJ, Schulze PC, Widder JD, Bauersachs J, Thum T. Blood-based microRNA signatures differentiate various forms of cardiac hypertrophy. Int J Cardiol 2015;196:115–22. 10.1016/j.ijcard.2015.05.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Qiang L, Hong L, Ningfu W, Huaihong C, Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. Int J Cardiol 2013;168:2082–8. 10.1016/j.ijcard.2013.01.160 [DOI] [PubMed] [Google Scholar]
- 105. Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MA, Liu LC, van der Harst P, Pinto YM, de Boer RA, Meyer S, Teerlink JR, O’Connor CM, Metra M, Davison BA, Bloomfield DM, Cotter G, Cleland JG, Mebazaa A, Laribi S, Givertz MM, Ponikowski P, van der Meer P, van Veldhuisen DJ, Voors AA, Berezikov E. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail 2016;18:414–23. 10.1002/ejhf.332 [DOI] [PubMed] [Google Scholar]
- 106. Xiao J, Gao R, Bei Y, Zhou Q, Zhou Y, Zhang H, Jin M, Wei S, Wang K, Xu X, Yao W, Xu D, Zhou F, Jiang J, Li X, Das S. Circulating miR-30d predicts survival in patients with acute heart failure. Cell Physiol Biochem 2017;41:865–74. 10.1159/000459899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Marfella R, Di Filippo C, Potenza N, Sardu C, Rizzo MR, Siniscalchi M, Musacchio E, Barbieri M, Mauro C, Mosca N, Solimene F, Mottola MT, Russo A, Rossi F, Paolisso G, D’Amico M. Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur J Heart Fail 2013;15:1277–88. 10.1093/eurjhf/hft088 [DOI] [PubMed] [Google Scholar]
- 108. Melman YF, Shah R, Danielson K, Xiao J, Simonson B, Barth A, Chakir K, Lewis GD, Lavender Z, Truong QA, Kleber A, Das R, Rosenzweig A, Wang Y, Kass D, Singh JP, Das S. Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. Circulation 2015;131:2202–16. 10.1161/CIRCULATIONAHA.114.013220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sardu C, Paolisso G, Marfella R. Letter by Sardu et al Regarding Article, "Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study". Circulation 2016;133:e388 10.1161/CIRCULATIONAHA.115.018205 [DOI] [PubMed] [Google Scholar]
- 110. Morley-Smith AC, Mills A, Jacobs S, Meyns B, Rega F, Simon AR, Pepper JR, Lyon AR, Thum T. Circulating microRNAs for predicting and monitoring response to mechanical circulatory support from a left ventricular assist device. Eur J Heart Fail 2014;16:871–9. 10.1002/ejhf.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, Mihailovic A, Sauer M, Ji R, Ramarathnam A, Totary-Jain H, Williams Z, Tuschl T, Schulze PC. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA 2014;111:11151–6. 10.1073/pnas.1401724111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Leger KJ, Leonard D, Nielson D, de Lemos JA, Mammen PP, Winick NJ. Circulating microRNAs: potential markers of cardiotoxicity in children and young adults treated with anthracycline chemotherapy. J Am Heart Assoc 2017;6:e004653 10.1161/JAHA.116.004653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Simonson B, Das S. MicroRNA therapeutics: the next magic bullet? Mini Rev Med Chem 2015;15:467–74. 10.2174/1389557515666150324123208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Martinez-Fernandez A. MicroRNA therapy for the failing heart. Circ Cardiovasc Genet 2014;7:393–4. 10.1161/CIRCGENETICS.114.000687 [DOI] [PubMed] [Google Scholar]
- 115. Duong Van Huyen JP, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, Iserin F, Rouvier P, François A, Vernerey D, Loyer X, Leprince P, Empana JP, Bruneval P, Loupy A, Jouven X. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J 2014;35:3194–202. 10.1093/eurheartj/ehu346 [DOI] [PubMed] [Google Scholar]
- 116. de Gonzalo-Calvo D, Vea A, Bär C, Fiedler J, Couch LS, Brotons C, Llorente-Cortes V, Thum T. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: a novel tool for personalized medicine? Eur Heart J. 2018 doi: 10.1093/eurheartj/ehy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Park KM, Teoh JP, Wang Y, Broskova Z, Bayoumi AS, Tang Y, Su H, Weintraub NL, Kim IM. Carvedilol-responsive microRNAs, miR-199a-3p and -214 protect cardiomyocytes from simulated ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol 2016;311:H371–H383. 10.1152/ajpheart.00807.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]