Abstract
In one more years, we will ‘celebrate’ an exact centenary of the Spanish flu pandemic. With the rapid evolution of the influenza virus, the possibility of novel pandemic remains ever a concern. This review covers our current knowledge of the influenza A virus: on the role of RNA in translation, replication, what is known of the expressed proteins and the protein products generated from alternative splicing, and on the role of base pairing in RNA structure. We highlight the main events associated with viral entry into the cell, the transcription and replication process, an export of the viral genetic material from the nucleus and the final release of the virus. We discuss the observed potential roles of RNA secondary structure (the RNA base-pairing arrangement) and RNA/RNA interactions in this scheme.
Keywords: influenza A virus, RNA and RNA/RNA interactions, viral ribonucleoproteins, RNA viral transcription, RNA viral replication, negative-strand RNA virus
Introduction
The influenza virus belongs to the class known as Orthomyxoviruses [1, 2] and can be divided into three distinct subtypes (influenza A, influenza B or influenza C), depending on its nucleoproteins (NPs) and the antigen determinants of its matrix proteins. The influenza virus is a negative-sense, single-stranded RNA (ssRNA) virus that is segmented into eight (Subtypes A, B) or seven (Subtype C) separate strands. Each segment codes for at least one protein, and based on the extensive studies of the influenza A virus (IAV) in recent years, it seems likely that all subtypes express at least twice as many proteins as its number of segments [3].
Influenza is an envelope virus, where viral nucleocapsid (genome + NPs) is protected by a lipid bilayer derived from the modified host cell’s membrane [2]. The outer layer of the influenza virus is studded with the glycoproteins hemagglutinin (HA) and neuraminidase (NA). On the virion, the HA is found in the form of a homotrimer (HA-trimer), while the NA forms into a homotetramer (NA-tetramer). The HA-trimers are relatively evenly distributed on the surface of the virion in the lipid bilayer, whereas the NA-tetramers appear to localize at one end of the virion, most likely at the exit point of the budding virion [4]. Recently, the distribution of the HA-trimers into larger conglomerates was observed; however, the biological consequences of these processes are not well understood [5].
The IAVs are among the fastest evolving families of viruses [6] and are notable for their ability to rapidly change from one season to the next or even within seasons [7]. Over the course of a few generations, the virus can change sufficiently to become virulent. Like many emerging infectious diseases such as Ebola virus or severe acute respiratory syndrome (SARS) [8], IAV depends on animal reservoirs, where they can reside [9–13] and transfer between species that share the same environment [14]. As one major reservoir involves migratory birds [15], the potential to rapidly spread an infection throughout the world is of major concern and IAV is under constant watch (http://www.who.int/csr/don/archive/year/en/). During the past 100 years, there have been three major epidemics with extensive fatalities [16] and one minor one (mostly because of quick action from the WHO and health practitioners) [17, 18], the Spanish flu (1918, H1N1, 75 million fatalities worldwide), the Asian flu (1957, H2N2, 2 million fatalities), the Hong Kong flu (1968, H3N2, 1 million fatalities) and more recently the swine flu (2009, H1N1, 14 000 fatalities).
Here, we briefly review topics heavily associated with RNA in the IAV, particularly our current knowledge on how the IAV genome is packaged, how the RNA interacts with proteins, what protein products are generated from alternative splicing in the IAV genome and our present knowledge of RNA structure. We also summarize our current understanding of the infection cycle with a focus on how the viral RNA (vRNA) sequences manage to enter the nucleus of the cell, carry out the transcription and replication process and export of the viral genetic material from the nucleus before the final budding and release of the virion. Finally, considering more than one virion can infect a given cell at the same time, it is also possible to shuffle segments between different viral strains in a process known as reassortment. We discuss briefly on the recent discoveries of RNA/RNA interactions between the vRNA segments that help in the packaging of the viral genome. Such interactions would have implications on the likelihood of such reassortment events.
Structure and the viral genome
This section is organized according to the structural features of the viral ribonucleoproteins (vRNP) rather than the numbering of the RNA segments in the genome. Hence, the organizational description of the genome will be with respect to how the virus is built from the individual components rather than the order in which the segments are numbered.
As previously mentioned, IAV consists of a negative-sense, ssRNA genome. The genome is multipartite and contains eight separate segments of various lengths; see Table 1 for details [3, 5, 9, 19–38]. More than 10 major proteins are encoded by these eight segments together with a variety of additional frame-shifted proteins and alternatively spliced products; a short description of what is known of each IAV protein is provided in the corresponding table. Other examples of segmented negative-sense ssRNA viruses are the bi-segmented arenavirus [39, 40], where the segments are called the ‘large segment’ and ‘small segment’, and the tri-segmented bunyavirus [41] (large, medium and small segments).
Table 1.
Influenza a virus RNA segments and the proteins they encode
| RNA segment number | Number of nucleotides | Gene product(s) | Number of amino acids | Function |
|---|---|---|---|---|
| Viral RNA-dependent RNA polymerase (vRdRP)Binds to the ends of each ssRNA segment and synthesizes new copies of the viral RNP. The subunits combine together after import into the nucleus. The vRdRP consists of three main subunits (PB1, PB2 and PA) and peripherally a matrix of NP | ||||
| 1 | 2341 | PB2 | 759 | PB2: Responsible for cap binding. The globular domain is essential for proper association with importin α3 and to a lesser extent α1 and α7 [17, 18] |
| PB2-S1 | 508 | An alternative splicing product of PB2: appears to localize in the mitochondria and inhibit the RIG-I-dependent interferon signaling pathway (humoral immunity) [18] | ||
| 2 | 2341 | PB1 | 757 | PB1: Involved in capturing the cap regions of the host’s mRNA and inserting the primer into the viral mRNA. Holds the polymerase active site and harbors endonuclease activity [17, 20] |
| PB1-F2 | 87–90 | From an alternative reading frame. Can induce apoptosis, regulate host interferon response and modulate susceptibility to bacterial superinfection [19, 20]. May influence intracellular localization of PB1 [21] | ||
| PB1-N40 | N-terminally truncated version of PB1; a product from an in-frame downstream initiation site. The function is unknown, but it might modulate virus-induced pathogenesis [19, 22] | |||
| 3 | 2233 | PA | 716 | Polymerase acidic: functions as an RNA-endonuclease. Cleaves capped RNA fragments off of the host’s pre-RNA to be used as primers for constructing viral mRNA [17, 20] |
| 252 | PA-X | 61 | Frameshifted PA at 191-252 (H7N7), postulated to play an important role in virus replication and shutdown of host innate responses in animal models, but its expression during in vivo infection has not been observed [23, 24] | |
| PA-N155 | 561 | Ribosomal frameshift to AUG start codon at position 155; possible role in viral replication, but function unknown [22] | ||
| PA-N182 | 534 | Ribosomal frameshift to AUG start codon at position 182; function unknown [22] | ||
| 4 | 1778 | HA | 566 | Hemagglutinin: The outer glycoprotein that binds sialic acid of epithelial cells and plays a central role in the fusion process [4, 25] |
| 5 | 1565 | NP | 498 | Nucleoprotein: binds the ssRNA into a large ds (NP protein) helix and serves to regulate the export and import of viral RNPs [8] |
| 6 | 1413 | NA | 454 | Neuraminidase: Helps the virion cut through the mucous coating of epithelial cells. Also thought to be important during the budding process where the newly forming virion breaks away from the host cell [26] |
| 7 | Matrix proteins | |||
| 1027 | M1 | 252 | Full-length structure. Involved in regulating the import and export of the viral RNP. A key regulator for viral assembly, preferentially binding viral RNPs during viral assembly [30] | |
| M2 | 97 | Alternative splice produce. Combines in the form of a tetramer in the viral envelope where it regulates the flow of protons into the viral genome after the capsid has entered the cell and before release of the viral RNPs (endocytosis) [27–29] | ||
| M3 | 9 | Alternative splice product; function unknown [28] | ||
| M4 | 54 | Alternative splice product; function unknown [28] | ||
| M42 | 99 | Alternative splice product; function not fully established; however, it can serve in the place of M2 [28] | ||
| 8 | Nonstructural proteins | |||
| 890 | NS1 | 230 | Full-length structure. Inhibits the interferon-mediated antiviral response [38]. The NS1 protein of IAV serves a critical role in suppressing the production of host mRNAs by inhibiting the 3′-end processing of host pre-mRNAs and consequently blocking the production of host mRNAs, including interferon-β mRNAs. Also involved in the import of the viral RNPs, tends to help hijack the import mechanism using importin alpha [31–33]. | |
| NEP (formerly NS2) | 98 | Important both for the import and export of viral RNPs and mRNA copies to and from the nucleus to the cytosol [34–36] | ||
| NS3 | Function unknown but may be an important protein factor for adaptation to new hosts [31] |
Note: Influenza A viruses have eight gene segments encoding many different proteins, adapted from [38].
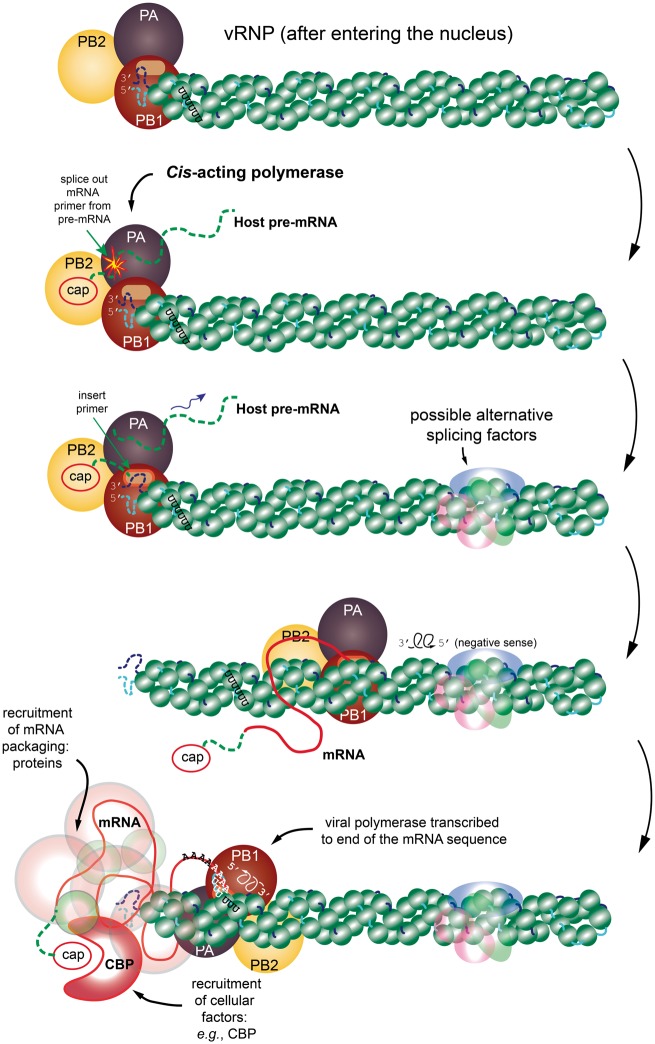
Each of the eight segments is individually packaged into a structurally distinct vRNP particle. A cartoon of a typical vRNP is shown in Figure 1 (top panel). The medium-gray/green beads represent the NP (Segment 5 in Table 1), where a polymer of NP molecules forms a scaffold consisting of a right-handed double-stranded (ds) helix of NPs in an ensiform (needle-like) structure [42–44]. The single-stranded vRNA sequence is represented by the light-gray/cyan lines (on the 5′ half of the sequence) that connects adjacent NP beads together and similarly the blackish/navy-blue lines (on the 3′ half). Thus, the vRNA sequence is left partially exposed as it winds around the NP-scaffolding; as though the NP forms a lining for the vRNA. The NP-scaffold does not play a significant role in protecting the vRNA because the NP-lined vRNA can be cut by ribonucleases [42]. The only part of the vRNA sequence where any dsRNA base pairing occurs is the highly conserved 12 and 13 nucleotide (nt) sequence on the 5′- and 3′-most ends of the vRNA segment, respectively [45]. Likewise, the NP-lined vRNA complex also appears to strongly restrict the formation of RNA secondary structure, though the last section of this review will discuss an emerging nuanced view.
Figure 1.
A cartoon representation of the transcription process. The figure shows the time-dependent and structural relationships of the vRNP and associated viral polymerase, which consists of the subunits PB1, PB2 and PA (from genome Segments 2, 1 and 3, respectively). The viral polymerase binds to the partially complementary 5′ and 3′ termini of the vRNA segment. The progression is from top to bottom. (Top) The vRNP has entered the nucleus of the host cell and has separated from other vRNPs. (Next step) The transcription is initiated by PB2 hijacking a 5′ cap from the host pre-mRNA, followed by endonucleolytic cleavage of the host pre-mRNA by PA. (Next frame) Transcription begins from the 3′ end of the vRNA sequence with the spliced-in primer sequence and cap merged onto the forming mRNA sequence. (Next frame) Transcription continues with the viral polymerase somehow dislodging the NP in its wake to obtain the mRNA transcript. Also depicted faintly are alternative splice factors from the spliceosome that can influence the resulting transcription. (Next frame) While the transcription process is happening, the vRNA and NP tend to recombine back to the rest state. (Final step) The poly-adenylation step is reached where further editing is required of the structure using the host RNA polymerase II system. It is important to remember that during this entire process, various mRNA packaging proteins are being recruited (e.g. the CBP. These are shown as a faint covering of the final mRNA. All this packaging is required for transport to the cytosol for translation into viral proteins. The schematic is inspired from [44] and [42]. The structure of the vRNP is based on the right-handed helix in [42]; however, it is important to remember that [43] reports a left-handed helix. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
An additional component of the vRNP is the viral polymerase. In Figure 1 (top panel, left-hand side), the viral polymerase is a hetrotrimer comprised the following proteins: polymerase acidic (PA; Segment 3), polymerase basic 1 (PB1; Segment 2) and polymerase basic 2 (PB2; Segment 1), with the polymerase located near the 5′-/3′-termini of the vRNA sequence [42]. It is important to note that [43] reports the location of the viral polymerase at the hairpin side (the exact opposite side of the 5′/3′-termini) of the ensiform structure, and the ds helix of NPs has a left-handed twist instead of a right-handed twist. Nevertheless, either way, the nucleocapsid proteins are PB1, PB2, PA and NP, where NP adopts a needle-shaped scaffold for the ssRNA and the viral polymerase is positioned at one end. The viral polymerase is essential for the transcription of the viral mRNA sequences and replication of the viral genome [44].
In the nucleocapsid, electron microscopy and tomography reveal eight rod-shaped vRNPs structures, which are organized inside of the influenza virus in a rolled-up daisy chain pattern called the ‘7 + 1’ structure. The vRNPs make contact with the matrix layer (lipid bilayer) at the budding tip of each of the virions [46]. This cluster also appears to be present in the virion during the fusion process (i.e. when the virus enters the host cell), during the budding process (i.e. when the new virion escapes from the cell) and even on entering the nucleus. Hence, some significant binding interactions are probably present that can be broken only in the nucleus of host cell.
Two additional proteins are also known to be lodged in the nucleocapsid: the nonstructural protein 1 (NS1) and 2 [NS2/nuclear export protein (NEP)], both encoded in Segment 8. NS1 plays a critical role in suppression of the 3′-end processing of the host mRNA and blocking the hosts innate immune response stimulated by interferon-α/β [47–49]. NS2, which is also called the NEP, is a multifunctional protein involved in regulating the accumulation of vRNA, antigenomic complementary RNA and viral mRNA synthesized by the vRNA polymerase [9, 50, 51]. The protein is also implicated in the efficient release of budding virions [51, 52]. It seems likely that all of the major expressed proteins are stored in the nucleocapsid to provide additional export, import and viral polymerase functionality, but the exact contents and proportions are unknown.
The virions of the IAV are observed in both a spherical or filamentous shape [4, 53, 54], where in both cases, only one copy of viral genomic appears to be present [54]; the capsid is neither a random collection of vRNPs nor multiple copies of the genome. The virions in which ‘7 + 1’ clusters of vRNP where observed were all spherical [46, 55]. The filamentous structure can sometimes encompass same space as eight spherical virions [46]. One might assume that as the virion can fit in a sphere, it would be the most likely shape of the virus. However, the filamentous structure appears to be the more commonly observed form. It is not presently known exactly what determines whether the virion is spherical or filamentous nor is it clear how the vRNPs combine in the filamentous virion; however, it seems reasonable that the filamentous-shaped virus also preserves this ‘7 + 1’ pattern.
The viral envelop is built from lipids derived from the host membrane and comprises the matrix protein 1 (M1; from Segment 7), which is located at the interface between the nucleocapsid and the lipid envelop. M1 is partly responsible for the curvature of the emerging viroid, supports the arrangement of the eight RNP on the inner surface of the lipid envelope and regulates the import and export of vRNPs into and out of the nucleus. Anchored to the viral envelope, with part of the peptide sequence occupying the border of the lipid and the nucleocapsid, extending through the lipid layer and jutting out from the surface, are two externally oriented glycoproteins: the hemmaglutinin (HA) and the NA. It is important to keep in mind that none of these envelop surface proteins are monomers. The HA forms a homotrimer (HA-trimer) that binds the sialylated glycans located on the surface of epithelial cells and initializes the process of fusion between viral envelope and the host’s endosome. The NA forms a homotetramer (NA-tetramer) and appears to play a major role in the final steps of the budding process. In addition to these glycoproteins, matrix protein 2 (M2, Segment 7) is a small protein that combines into a homotetramer (M2-tetramer) serves as a transmembrane ion channel (a proton pump). HA and NA are the main antigens of IAV and are used to identify various subtypes of IAV, e.g. H3N2 specifies an IAV with the HA belonging to Subtype 3 and the NA also belonging to Subtype 2.
Infection and the role of vRNPs
The IAV infection begins with the virus binding to the surface of an epithelial cell on recognizing the sialic acid glycans. The main sialic acid is N-acetylneuramic acid in mammals and birds [56], typically followed by galactose. These molecules can be connected with an α2,6 bond, which is abundant on the surface of cells in the upper respiratory tract of humans, or with an α2,3 bond, which is found in the digestive tract in birds or in the lungs or bronchial tissue in humans. Although sialylated glycans in epithelial cells are studied mostly for their role in disease, the function of such glycans is diverse, interfacing with signaling proteins and various structural and modulatory roles [57, 58], and in epithelial cells in general, defense against oxidative stress [59] and even nutrition [60, 61]. To defend against infections, epithelial cells secrete mucin (mucous) in the respiratory tract [27, 57, 62]. The major components of mucin are glycans terminated with sialic acid together with various antimicrobials compounds [62]. For the virions to traverse the thick mucosal layer covering the epithelial cells in the lungs, the NA-tetramers on the surface of the influenza A virion must cleave through the sialic acid within the mucous [27]. This allows the HA-trimers of the IAV to interact with the sialylated glycans on the apical cell surface [63], an event known as virus adsorption.
This event is followed by internalization of the virus and the fusion process, which takes advantage of the specific features of HA. Owing to the process of proteolytic cleavage of the HA0 precursor [64], the HA protein is divided into two subunits HA1 and HA2. HA1 forms the globular head that binds to specific glycans on the epithelial cells. HA2 anchors the HA to the viral membrane and facilitates membrane fusion of the virus with the lipid bilayer of the endosomes, where the virus enters an endosome and the fusion peptide (from HA2) interacts with the lipid bilayer. As a result of lowering the pH in the endosome, the HA subunit adopts a low pH conformation [65]. The fusion process also depends on whether the virion is spherical or filamentous. Binding for the spherical virion triggers the internalization into an endosome, where acidification causes the HA-trimers to aggregate or cluster [26, 66–68]. Filamentous virions are, however, too large to fit into a clathrin-coated pit; therefore, they must enter the cell through a process known as macropinocytosis [44, 69–71]; a pathway also used by the Ebola virus and SARS virus [8].
After membrane fusion, the virion remains ensconced within the endosome until the acidification process further dissociates the viral membrane from the vRNPs. Before the endosomes transport the contents to lysosomes for recycling, the acidification process opens the endosome resulting in dissociation of M1 from the vRNPs and releasing the vRNP into the cytosol. After dissociation of M1 and release of the viral RNPs, the vRNPs appears to remain in a single cluster of eight vRNPs (the abovementioned ‘7 + 1’ structure) and enters the nucleus through the nuclear pores still as a clustered unit. This behavior was observed, regardless of whether the virion entered the cell via endocytosis or macropinocytosis. The path to the nucleus appears to be diffusion driven [72].
In the nucleus, the complex of vRNPs separates. The heterotrimer (PA-PB1-PB2) of each vRNP is drawn to the nuclear matrix, where it can bind to RNA polymerase II (Pol II) at the C-terminal domain of the large subunit [73]. By hijacking the host transcriptional machinery, the virus can gain access to many of the host’s mRNA processing proteins (the spliceosome complex), the 5′-capped RNA primer sequences, poly(A) tail processing proteins and the host’s NEPs; this recruitment process is shown in cartoon style in Figure 1. The Pol II provides many of the required tools and raw materials needed to build a properly constructed mRNA for export and translation into proteins. One such process is known as cap-snatching [21], where a short primer sequence at the 5′ end of the host’s mRNA is captured and spliced onto the forming viral mRNA (first two panels of Figure 1). Such a primer sequence include the methylated GTP cap (m7GDP), a 13 nt primer sequence that permits binding of NEPs and recruits the associated cap-binding proteins (CBPs). By binding Pol II, the transport proteins are snatched before the host CBP can finish processing the host pre-mRNAs [36, 73]. The ability of the vRNA polymerase to inhibit Pol II may slow down or block important host antiviral responses [48, 74]. Other factors involved in packaging the mRNA and constructing the poly(A) tail are also required and provided by Pol II (last two panels of Figure 1). M1 and NP also appear to bind to the histones of the nucleosome [36], which may be necessary to anchor the viral polymerase during replication processes.
Each individual vRNP is capable of transcribing its own sequence using the heterotrimer (PA, PB1 and PB2) to accomplish this task. However, replication cannot be carried out by a single vRNA. Therefore, it is important to distinguish the difference between transcription and replication.
Transcription of the vRNA sequence involves obtaining a positive-sense copy of a subsequence of vRNA. Combined with the editing steps provided by the Pol II complex, a positive-sense messenger RNA (mRNA) sequence with full mRNA functionality is generated and subsequently transported to the cytosol and translated into a protein. Multiple splice products of mRNA are produced as a result of using the host spliceosome in alternative splicing. Some of these alternatively spliced products appear to be viable [22, 23, 25, 29, 75]. The general process is shown in cartoon form in Figure 1.
Replication means making a complete unedited copy of the original viral genome (with possible mutations introduced by replication ‘errors’). In the first step of replication, the vRNA sequence uses the original viral polymerase as an anchor and recruits a second free viral polymerase (in trans) to build a positive-sense copy of the vRNA sequence (vRNA → cRNA). This is a complete positive-sense strand copy of the original segment. The final result is a complete vRNP that contains only the cRNA sequence. This new cRNA must be again replicated to obtain a negative-sense copy of the sequence (cRNA → ccRNA = vRNA), where ccRNA is the newly created complement of cRNA (which will serve as a vRNA for next generations of virus). This freshly made vRNA sequence must also have access to newly synthesized copies of M1, PA, PB1, PB2 and NP; hence, these proteins (generated by the transcribed mRNA products) must be imported back into the nucleus. The PA + PB1 + PB2 must assemble into a new viral polymerase, which together with the nascent NPs and vRNA is used to form the new vRNP [20, 44, 69].
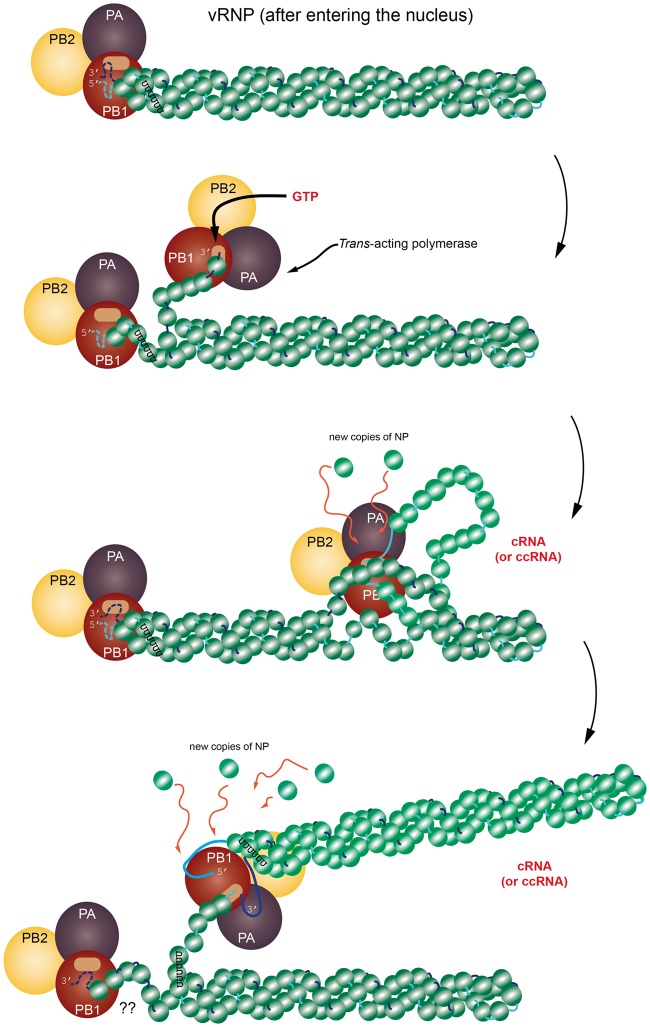
Currently, how exactly the selection between transcription and replication is determined is not completely clear. Nevertheless, based on the images of the RNPs [42, 43], there are some indications as to how the process works. One possible explanation is that the selection is influenced by the different proportions of M1 and NP [76]. In this respect, all processes are essentially transcription and are regulated by these ratios and other factors to establish whether the product will be mRNA or cRNA/ccRNA. Another view [50, 51] is that transcription involves a single viral polymerase, the so-called cis-acting polymerase, which binds somewhere in the vicinity of the promoter region (near the 3′ end of the RNA sequence) and copies the sequence moving in the 3′–5′ direction [42] and possibly responding to splicing factors, as illustrated in Figure 1 (middle). On the other hand, replication of vRNA requires transcription by an independent viral polymerase transcribing starting from the 3′-most end of the negative-sense viral ssRNA sequence—the so-called trans-acting polymerase [42] (Figure 2).
Figure 2.
A cartoon representation of the replication process showing the time-dependent relationship between the vRNP and trans-acting viral polymerase (PB1, PB2 and PA). Note that this model is still under investigation and the possibility remains that replication is also done in cis. The progression of the figure is from top to bottom. (Top) The vRNP has entered the nucleus of the host cell and has separated from other vRNPs. Perhaps at this stage, the vRNP has become anchored to chromatin, though this is not specified. (Next step) The 3′ terminus of the vRNA segment somehow unwinds and the trans-acting viral polymerase captures this free segment and begins copying the sequence in the 3′–5′ direction, making an antisense copy of whatever template is present. (Next frame) The process continues with the viral polymerase somehow unwinding the vRNA + NPs, all the while recruiting new NPs for the copy. (Next frame) When the replication is complete, the structure is eventually released. There are several steps in the trans-acting model that are not fully determined yet. The schematic is inspired from [44] and [42]. The structure of the vRNP is based on the right-handed helix in [42]; however, it is important to remember that [43] reports a left-handed helix. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
Transcription (viral mRNA production) is required to generate new copies of the viral proteins for creating a new virion. Several additional editing steps must be done before a viable mRNA can be exported and translated; the mRNA must have a properly constructed 5′-end on the nucleic acid sequence, a cap with a 5′ untranslated region (5′-UTR) and a poly-adenylated [poly(A)] tract inserted at the 3′-end of the sequence. The viral polymerase catalyzes not only RNA transcription but also cleaves the host cell’s mRNA and splices it onto the forming viral mRNA using cap-snatching by way of the PA and PB2 subunits [21]. To do this, the host Pol II and resident spliceosome factors become involved in this transcription process. For example, although M1 is translated from the unspliced mRNA, both M2 and M42 are the spliced variants. Similarly, NS1 is the unspliced version, while the NEP (NS2) is the spliced variant [44]. Additional known splice variants that have been identified in the viral genome are listed in Table 1. The Pol II is also captured to recruit its capping machinery, which provides the required cap and 13 nt primer sequence at the 5′-end of the viral transcript using PA and PB2 (a process shown as the second and third steps from the top in Figure 1). The mimicry must be complete, so that the viral mRNA is processed in the same way as the host’s mRNA. Midway along the vRNA sequence in Figure 1 (middle), one can see an image of various splice factors that introduce the possibility of alternative splicing in the transcription. Just like the host’s own mRNA, by the end of this process, the viral mRNA is probably coated with the host’s exon-junction-core proteins and associated interacting proteins, the cap-binding complex, poly(A) binding proteins and most likely other important proteins needed for export to the cytosol and translation into proteins by the host’s ribosomal RNA [77].
Some of the proteins generated by this transcription process must find their way back to the nucleus to complete the replication process. Nuclear import of the nascent copies of M1, NP, PA, PB1 and PB2 (produced from the viral mRNA) requires the special actin-activated proteins—importin β and various induced fit supplementary proteins—importins α1, α3, α5 and α7 [47, 78]; a list of proteins and their nuclear import proteins is given in Table 2 [20]. In essence, the importin β cannot by itself import the viral proteins and requires NS1 and, possibly, other alternatively spliced units of the viral mRNA transcripts to induce sufficient fit to allow the import of M1, NP and PB2. PA and PB1 are imported together with Ran-binding protein 5 (RanBP5)—a member of the importin β family. This PA + PB1 complex must then associate with PB2. NEP is small enough that it is able to pass freely through the nuclear pores. This complexity may seem like a wild ride ‘around Robin Hood’s barn’; yet, it shows that this simple virus (with <20 proteins) has evolved a sophisticated strategy to circumvent its host.
Table 2.
List of viral proteins that enter the nucleus and importin factors that are used, adapted from [20]
| Protein | Import co-proteins |
|---|---|
| PB2 | Importin α1, α3, α5, α7, β |
| PB1 | (PB1+PA)+RanBP5 |
| PA | |
| NP | Importin α1, α3, α5, α7, β |
| vRNP | Importin α1, α5, β |
As the vRNPs enter the nucleus as a cluster, and yet the whole process of vRNA synthesis must begin with the viral polymerase transcribing in trans on a subsegment of the vRNP template (initially, the replication of cRNA), this means that there must be some free viral polymerase (i.e. PA + PB1 + PB2) available to initiate this process. One possible initial source is that some free viral polymerases are already stored inside the nucleocapsid. Later in the infection cycle, these viral polymerases are replaced with fresh copies.
In forming the viral cRNA, the NP coating is either somehow removed or used with the polymerase heterotrimer to produce the cRNA transcript, where the cRNA is assembled into a RNP (cRNA with the polymerase and scaffold of NPs) that is the template used to replicate the negative-strand sequence (ccRNA; the new copy of vRNA). Both cRNA and ccRNA bind the viral polymerase heterotrimer (PA-PB1-PB2). Hence, cRNA and ccRNA have the NP complex coating onto which the newly synthesized polymerase heterotrimer can bind. As both cRNA and its replication product ccRNA bind the viral polymerase, nuclear export requires a way to distinguish between them and export only the replicated vRNPs. The apparent factor determining this is the binding of M1 to the vRNPs, and not the cRNPs [31, 76]. The export of the newly created vRNPs is further mediated by NEP that directly interacts with NEPs CRM1 (chromosomal maintenance 1, also known as exportin 1) [19, 51, 79], where NEP may enlarge the nuclear pores [80]. Somewhere during the transit (in the cytosol) or somewhere near the point where the individual vRNPs arrive at the cellular membrane, the vRNPs manage to group into the eight segments that form the genome in the 7 + 1 pattern [46]. The selective aggregation of the vRNPs may be because of mutual interactions, proteins factors or RNA-RNA interactions [81], where the latter appears to have some support (see next section). However, the precise secondary structure and the interactions between neighboring vRNPs are unknown.
At the end of the cycle, the vRNP cluster with M1 sets into the budding site, the HA-trimers and NA-tetramers position themselves at the apical positions. Evenly distributed on the surface of the budding virion, M1 and M2 also become positioned. Based on the structure in [4, 53], the NA-tetramer appears on only one side, perhaps, to help cleave the budding virion. How this assembly mechanism might lead to organization of HA and NA into complete, budding virions and how it leads to the selective incorporation of viral genome segments will require further investigation [82]. Other proteins such as NEP and NS1 would be buried within the nucleocapsid. The envelop layer encloses the viral material enabling the virion to leave the infected cell [28, 52, 54, 71].
The role of RNA secondary structure
The vRNA is wrapped up in a scaffold composed of the NP proteins [42, 43]; therefore, only a few examples of known RNA base-pair formation have been reported for the packaged vRNPs. One observed interaction occurs at the terminal ends of each vRNA segment. In all eight vRNA gene segments, the terminal 13 and 12 nt fragments at the 5′- and 3′-most ends of the vRNA segments are conserved and known to form the vRNA promoter. This region also exhibits short hairpins of RNA secondary structure [45].
The second observed interactions were inferred from the fact that the majority of virions contain exactly one complete genome packed in the daisy chain structures [9, 46, 81, 83]. This daisy chain connectivity has long suggested that RNA/RNA interactions occur between the vRNPs that help bind them together. Recent observations have shown that there appear to be some distinct RNA/RNA interactions between different vRNPs [83, 84]. In particular, one study [84] revealed that a vRNA hairpin from Segment 2 (the PB1 Segment) and a vRNA hairpin from Segment 8 (the NP Segment) appear to mutually combine via the complementary base-pair sequences of the loops. (Two RNA hairpins bound together by complementary sequences found in their loops are called a ‘kissing loop’ [85, 86].) The evidence for the kissing loop is supported by sequence alignment methods and in vitro experiments involving disrupting the complementary sequences and measuring the relevant fractions of products via native agarose gel electrophoresis [84]. Hence, the linking of the segments is most likely because of RNA/RNA interactions between adjacent segments. However, these interaction networks of kissing loops also appear to be strain-dependent [84]. This suggests that for a particular subtype of IAV to jump between species, not only does the successful subtype has to acquire specific mutations in HA, access effective nuclear import/export factors and produce proteins that successfully block host innate immunity but, additionally, the subtype requires a proper connectivity between vRNPs. This might partly explain why reassortment of various subtypes is not a common occurrence and how the strains manage to remain unique.
Kobayashi et al. [87] attempted to identify the actual RNA secondary structure (hairpins) in Segment 7 (the M Segment) and test their stability at different temperatures. (RNA secondary structure differs from protein secondary structure in that it shows the base pairing between diverse parts of the RNA sequence or between different RNA sequences; therefore, more topology information can be inferred from RNA secondary structure data.) In this analysis, the functionality of conserved stem-loop structures was directly tested using reverse genetic experiments to introduce synonymous mutations designed to disrupt secondary structures predicted at three locations. These disruptions tended to attenuate the infectivity of the recombinant virus. The disruption of the stem-loop structure at nucleotide positions 219–240, 950–964 and 967-994 of Segment 7 (M Segment) showed a significant temperature dependence, which is characteristic of decrease in RNA stability in these regions. This suggests that the conserved secondary structures predicted in Segment 7 are involved in the production of infectious viral particles during IAV replication.
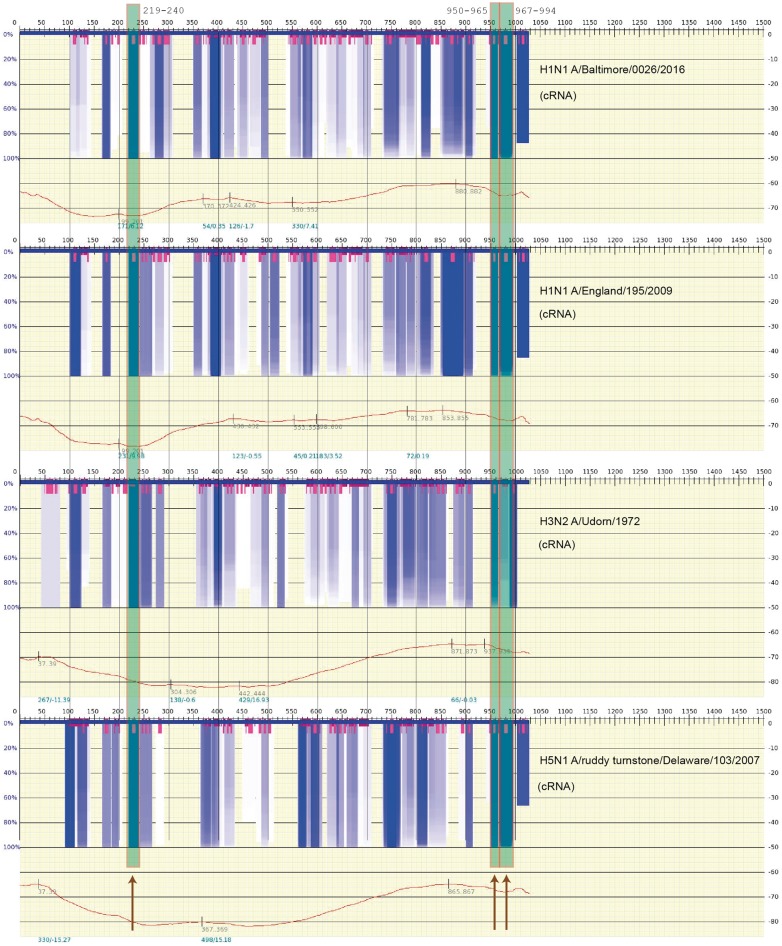
The abovementioned studies strongly suggest that base pairing in the vRNP may play a significant role in the mutual binding of the vRNP structures to one another. The data from Kobayashi et al. is stable. In Figure 3, an actual scan of the free energy and variation in optimal and suboptimal structures of Segment 7 (Segment M) is shown for four different viral strains in the following order: A/Baltimore/0026/2016(H1N1), A/England/195/2009(H1N1), A/udorn/1972(H3N2) and A/ruddy turnstone/DE/509531/2007(H5N1). Sequences of this segment were obtained from the influenza database at https://www.fludb.org [88]. The spectra represent scanning for RNA secondary structure over a window size of 200 nt and, based on the predicted suboptimal structures obtained in the scan, detecting regions where a particular secondary structure motif of RNA repeatedly appears in many (or most) of the optimal and suboptimal structures. The very dark gray/blue boxes represent regions of stable RNA secondary structure that are largely independent of the scanning window size, and segments of highly variable secondary structure are shown by their lack of any color. (The very short gray/magenta boxes indicate the center of the hairpin in the stable (gray/blue) regions of the spectra.) Covering the regions at loci 219–240, 950–964 and 967–994 of Segment 7 reported by Kobayashi et al. [87], Figure 3 shows three long semi-opaque gray/green boxes (also marked by the dark arrows, bottom) overlapping these respective fragments. The scan was performed using vswindow; an expanded application of vs_subopt [89] (http://www.rna.it-chiba.ac.jp/∼vsfold/vs_subopt/) with default settings (window size 200 nt, Kuhn length 4 nt), and the spectrum is generated using Genepoem [90] (http://www.rna.it-chiba.ac.jp/∼vswindow/cgi-bin/index.cgi). Modifying the settings—such as changing the scanning window length or the Kuhn length (a measure of the stiffness of the RNA [89])—generates similar results. As the RNP (RNA wrapped in NPs) is largely exposed [42], the stable RNA secondary structure is also likely to appear in the RNP complex.
Figure 3.
A scan of the thermodynamic stability of the cRNA secondary structure (variability of the structures) for the influenza A viral genome (M, Segment 7) where the largely universally conserved segments reported in [87] are indicated by the long semitransparent gray/green boxes (also marked by dark arrows, bottom). The graph spectrum was generated using Genepoem [90] and calculated using vs_subopt/vswindow [89] using default settings (window size 200 nt, Kuhn length 4 nt). The very dark gray/blue boxes indicate regions of highly stable RNA secondary structure (not only hairpins but even a region of stability). The very short gray/magenta boxes indicate regions where a stable loop is present. The particular strains that were calculated are shown on the right side of each free energy spectrum. The sequences were obtained from the influenza database at https://www.fludb.org [88]. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
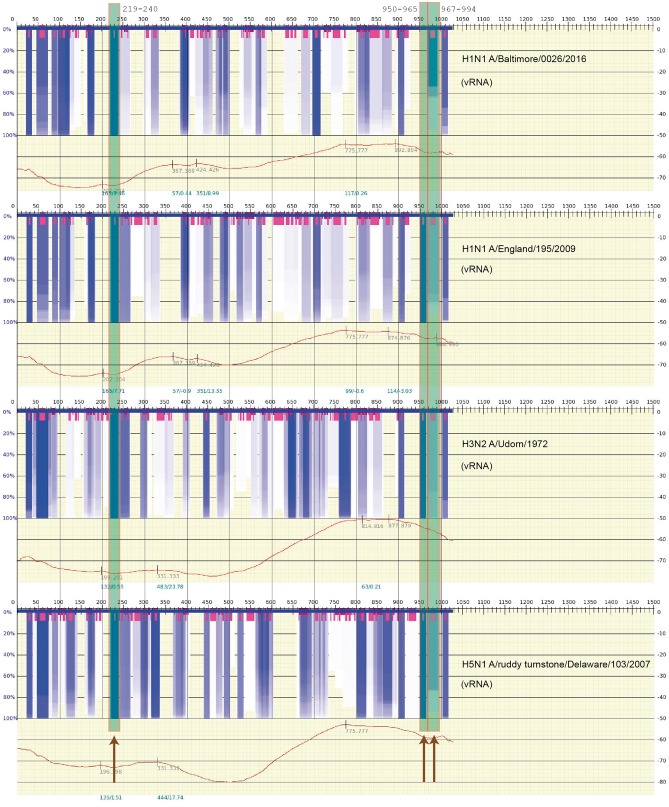
However, it is important to note that the calculations by Kobayashi et al. were for the cRNA sequence. Therefore, in Figure 4, the results for the actual vRNA sequence are shown. Regions at positions 950–964 and 967–994 appear to be weaker (represented in Figure by less intense gray/dark-blue), indicating that there is more variation in possible suboptimal structures for these regions of the vRNA. However, the region at position 219–240 appears prominently in both Figures 3 and 4. Perhaps, these variations in RNA stability help to distinguish the cRNA from vRNA in the steps toward nuclear export.
Figure 4.
The same calculation as Figure 3 of the thermodynamic stability of the vRNA (negative strand) secondary structures. The details of the figure are explained in the caption of Figure 3. The settings and sequences used in the calculation are the same as in Figure 3. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
The somewhat regular pattern of gray/dark-blue bands in strong secondary structure appears on all of the segments of cRNA and vRNA. However, Figures 3 and 4 also suggest that these gray/blue bands are somewhat strain-dependent—as also stressed in reports such as Gavazzi et al. [84] and Kobayashi et al. [87].
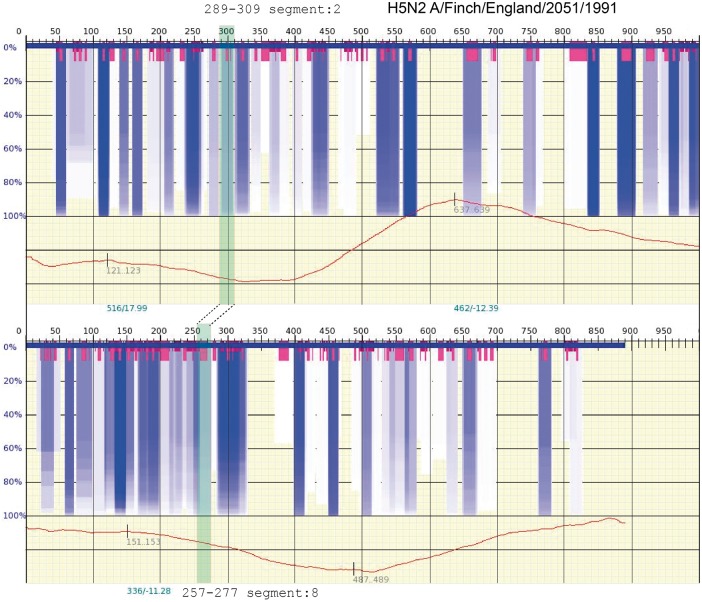
The results from Kobayashi et al. only look at Segment 7; they do not specify where these hairpins might bind to other segments of the vRNP. However, the kissing loop reported in Gavazzi et al. [84] can also be examined in this way. Figure 5 shows a scan of Segments 2 and 8 for the same corresponding vRNA sequence reported in [84], strain A/finch/England/2051/1991(H5N2). The same default settings are used as above for vswindow/Genepoem. Likewise, in Figure 5, the two connected semi-opaque gray/green boxes is used to indicate the corresponding sequence ranges reported in Gavazzi et al. [84], and the dashed lines indicate the connection between Segment 2 and Segment 8. Though both Segment 2 and Segment 8 are clearly visible in the spectrum—indicating that the structures are relatively stable—the intensity of the gray/blue in these regions is less than some other cases in the same spectrum. This lower intensity gray/blue indicates that there is a greater degree of variation in the RNA secondary structure in these locations. In line with the observation that the linking together of vRNPs is the result of kissing hairpins, this may suggest that there are many such contacts regularly dispersed along the vRNA segments that act like a bar code that defines how the pieces should be fit together with one another and identify the vRNPs specific to a particular strain. Though this may diminish some of the long-held fears of facile reassortment of the common flu with such strains as H5N1, it also could explain how individual strains evolve and compete with one another too.
Figure 5.
Similar to Figures 3 and 4, a calculation of thermodynamic stability of the vRNA (negative strand) secondary structure for the kissing loop regions for H5N2 in Segments 2 and 8, as reported in [83]. The cRNA sequence was obtained from https://www.fludb.org and converted to vRNA. The details of the spectrum are explained in the caption of Figure 3, and the settings used in the calculations are the same as Figures 3 and 4. The very dark gray/blue boxes indicate regions of highly stable RNA secondary structure and the very short gray/magenta boxes indicate regions where stable hairpin loops are present. The connected-semitransparent gray/green boxes indicate the band where the kissing loop hairpins are found in Segments 2 and 8 of the strain, and a dotted line is shown to indicate the respective interaction between these two vRNP segments. (A colour version of this figure is available online at: https://academic.oup.com/bfg)
Key Points
In this work, we summarize our current understanding of the structural aspects of transcription and replication.
We also outline the current knowledge of existing IAV proteins and the role of alternative splicing and frameshift in their formation. The potential functions of these proteins, if known, are also discussed.
We discuss the observed, potential roles of RNA secondary structure and RNA/RNA interactions in this scheme the formation and clustering of viral ribonucleoproteins.
Funding
WKD, ML, DP were supported by grants from the Polish National Science Centre (grant numbers 2014/15/B/ST6/05082 and 2013/09/B/NZ2/00121); ML, DP were funded by Foundation for Polish Science (TEAM to D.P.). DP was supported by grant 1U54DK107967-01 ‘Nucleome Positioning System for Spatiotemporal Genome Organization and Regulation’ within 4DNucleome NIH program.
Wayne Dawson is a contributing researcher at the Bio-information Lab in Yayoi campus at the University of Tokyo and also associated with Center of New Technologies (CeNT) in Warsaw, Poland. His research is primarily directed to RNA structure prediction and structural biology and recently focused on chromatin structure.
Michał Łaźniewski is a researcher at the Laboratory of Functional and Structural Genomics at the University of Warsaw in the Center of New Technologies (CeNT) and Department of Physical Chemistry in the Faculty of Pharmacy at the Medical University of Warsaw. His research is in bioinformatics and structural biology with a focus on medical applications using bioinformatics.
Dariusz Plewczyński is a professor and head of the Laboratory of Functional and Structural Genomics at the University of Warsaw in the Center of New Technologies (CeNT) in Warsaw, Poland and also the affiliated with the Faculty of Mathematics and Information Science at Warsaw University of Technology in Warsaw. His main expertise covers computational genomics, biostatistics and bioinformatics. He is actively applying computational modeling to various interdisciplinary problems in Human Genomics.
References
- 1. Zheng W, Olson J, Vakharia V, et al. The crystal structure and RNA-binding of an orthomyxovirus nucleoprotein. PLoS Pathog 2013;9:e1003624.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Couch RB. Orthomyxoviruses In: Baron S. (ed). Medical Microbiology. Galveston, TX: University of Texas Medical Branch at Galveston, 1996. https://www.ncbi.nlm.nih.gov/books/NBK7627/ [PubMed] [Google Scholar]
- 3. Yamayoshi S, Watanabe M, Goto H, et al. Identification of a Novel Viral Protein Expressed from the PB2 Segment of Influenza A Virus. J Virol 2015;90:444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wasilewski S, Calder LJ, Grant T, et al. Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine 2012;30:7368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiong X, Corti D, Liu J, et al. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci USA 2015;112:9430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith GJ, Bahl J, Vijaykrishna D, et al. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci USA 2009;106:11709–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC. Influenza antiviral drug resistance: question and answer. https://www.cdc.gov/flu/about/qa/antiviralresistance.htm (29 April 2017, date last accessed).
- 8. Mingo RM, Simmons JA, Shoemaker CJ, et al. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J Virol 2015;89:2931–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ortin J, Martin-Benito J.. The RNA synthesis machinery of negative-stranded RNA viruses. Virology 2015;479–480:532–44. [DOI] [PubMed] [Google Scholar]
- 10. Guan Y, Chen H, Li K, et al. A model to control the epidemic of H5N1 influenza at the source. BMC Infect Dis 2007;7:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M, Veit M.. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein Cell 2016;7:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bettencourt LM, Ribeiro RM.. Real time bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS One 2008;3:e2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolzoni L, Real L, De Leo G.. Transmission heterogeneity and control strategies for infectious disease emergence. PLoS One 2007;2:e747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009;325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garske T, Clarke P, Ghani AC.. The transmissibility of highly pathogenic avian influenza in commercial poultry in industrialised countries. PLoS One 2007;2:e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis 2006;12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain S, Kamimoto L, Bramley AM, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med 2009;361:1935–44. [DOI] [PubMed] [Google Scholar]
- 18. Duque V, Vaz J, Mota V, et al. Clinical manifestations of pandemic (H1N1) 2009 in the ambulatory setting. J Infect Dev Ctries 2011;5:658–63. [DOI] [PubMed] [Google Scholar]
- 19. Boulo S, Akarsu H, Ruigrok RW, et al. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res 2007;124:12–21. [DOI] [PubMed] [Google Scholar]
- 20. Hutchinson EC, Fodor E.. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 2012;30:7353–8. [DOI] [PubMed] [Google Scholar]
- 21. Dias A, Bouvier D, Crepin T, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009;458:914–18. [DOI] [PubMed] [Google Scholar]
- 22. Wise HM, Foeglein A, Sun J, et al. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 2009;83:8021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muramoto Y, Noda T, Kawakami E, et al. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol 2013;87:2455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khurana S, Chung KY, Coyle EM, et al. Antigenic fingerprinting of antibody response in humans following exposure to highly pathogenic H7N7 Avian Influenza Virus: evidence for Anti-PA-X antibodies. J Virol 2016;90:9383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 2012;337:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivanovic T, Harrison SC.. Distinct functional determinants of influenza hemagglutinin-mediated membrane fusion. Elife 2015;4:e11009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cohen M, Zhang XQ, Senaati HP, et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 2013;10:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts KL, Leser GP, Ma C, et al. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J Virol 2013;87:9973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wise HM, Hutchinson EC, Jagger BW, et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog 2012;8:e1002998.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Busath DD. Influenza A M2: channel or transporter? In: Leitmannova L, Iglič A (eds). Advances in Planar Lipid Bilayers and Liposomes. Burlington: Academic Press, 2009, 161–201. [Google Scholar]
- 31. Chaimayo C, Hayashi T, Underwood A, et al. Selective incorporation of vRNP into influenza A virions determined by its specific interaction with M1 protein. Virology 2017;505:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Selman M, Dankar SK, Forbes NE, et al. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg Microbes Infect 2012;1:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Falcon AM, Marion RM, Zurcher T, et al. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J Virol 2004;78:3880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuo RL, Li LH, Lin SJ, et al. Role of N terminus-truncated NS1 proteins of influenza A virus in inhibiting IRF3 activation. J Virol 2016;90:4696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Neill RE, Talon J, Palese P.. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J 1998;17:288–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Robles I, Akarsu H, Muller CW, et al. Interaction of influenza virus proteins with nucleosomes. Virology 2005;332:329–36. [DOI] [PubMed] [Google Scholar]
- 37. Kohno Y, Muraki Y, Matsuzaki Y, et al. Intracellular localization of influenza C virus NS2 protein (NEP) in infected cells and its incorporation into virions. Arch Virol 2009;154:235–43. [DOI] [PubMed] [Google Scholar]
- 38. Lamb RA, Krug RM.. Orthomyxoviridae: the viruses and their replication. Fields Virol 2001;4:1487–531. [Google Scholar]
- 39. Pfau CJ. Arenaviruses In: Baron S. (ed). Medical Microbiology. Galveston, TX: University of Texas Medical Branch at Galveston, 1996. https://www.ncbi.nlm.nih.gov/books/NBK8193/ [PubMed] [Google Scholar]
- 40. Briese T, Paweska JT, McMullan LK, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 2009;5:e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shope RE. Bunyaviruses In: Baron S. (ed). Medical Microbiology. Galveston, TX: University of Texas Medical Branch at Galveston, 1996. https://www.ncbi.nlm.nih.gov/books/NBK8004/ [PubMed] [Google Scholar]
- 42. Moeller A, Kirchdoerfer RN, Potter CS, et al. Organization of the influenza virus replication machinery. Science 2012;338:1631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arranz R, Coloma R, Chichon FJ, et al. The structure of native influenza virion ribonucleoproteins. Science 2012;338:1634–7. [DOI] [PubMed] [Google Scholar]
- 44. Fodor E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol 2013;57:113–22. [DOI] [PubMed] [Google Scholar]
- 45. Catchpole AP, Mingay LJ, Fodor E, et al. Alternative base pairs attenuate influenza A virus when introduced into the duplex region of the conserved viral RNA promoter of either the NS or the PA gene. J Gen Virol 2003;84:507–15. [DOI] [PubMed] [Google Scholar]
- 46. Fournier E, Moules V, Essere B, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res 2012;40:2197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hanke L, Knockenhauer KE, Brewer RC, et al. The antiviral mechanism of an influenza A virus nucleoprotein-specific single-domain antibody fragment. MBio 2016;7:e01569–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garcia-Sastre A. Antiviral response in pandemic influenza viruses. Emerg Infect Dis 2006;12:44–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Das K, Aramini JM, Ma LC, et al. Structures of influenza A proteins and insights into antiviral drug targets. Nat Struct Mol Biol 2010;17:530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. York A, Fodor E.. Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell. RNA Biol 2013;10:1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paterson D, Fodor E.. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog 2012;8:e1003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rossman JS, Lamb RA.. Influenza virus assembly and budding. Virology 2011;411:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Calder LJ, Wasilewski S, Berriman JA, et al. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci USA 2010;107:10685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chlanda P, Schraidt O, Kummer S, et al. Structural analysis of the roles of influenza A virus membrane-associated proteins in assembly and morphology. J Virol 2015;89:8957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhirnov OP, Manykin AA.. Abnormal morphological vesicles in influenza A virus exposed to acid pH. Bull Exp Biol Med 2015;158:776–80. [DOI] [PubMed] [Google Scholar]
- 56. Nicholls JM, Lai J, Garcia JM.. Investigating the interaction between influenza and sialic acid: making and breaking the link In: von Itzstein M. (ed). Influenza Virus Sialidase—a drug discovery target. Basel: Springer Basel AG, 2012, 31–45. [Google Scholar]
- 57. Varki A. Sialic acids in human health and disease. Trends Mol Med 2008;14:351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Crocker PR. Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signalling. Curr Opin Struct Biol 2002;12:609–15. [DOI] [PubMed] [Google Scholar]
- 59. Iijima R, Takahashi H, Namme R, et al. Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett 2004;561:163–6. [DOI] [PubMed] [Google Scholar]
- 60. Vimr ER, Kalivoda KA, Deszo EL, et al. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 2004;68:132–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sprenger N, Duncan PI.. Sialic acid utilization. Adv Nutr 2012;3:392S–7S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Linden SK, Sutton P, Karlsson NG, et al. Mucins in the mucosal barrier to infection. Mucosal Immunol 2008;1:183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012;486:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harrison SC. Viral membrane fusion. Nat Struct Mol Biol 2008;15:690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Carr CM, Chaudhry C, Kim PS.. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA 1997;94:14306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dobay MP, Dobay A, Bantang J, et al. How many trimers? Modeling influenza virus fusion yields a minimum aggregate size of six trimers, three of which are fusogenic. Mol Biosyst 2011;7:2741–9. [DOI] [PubMed] [Google Scholar]
- 67. Floyd DL, Ragains JR, Skehel JJ, et al. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci USA 2008;105:15382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hamilton BS, Whittaker GR, Daniel S.. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses 2012;4:1144–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hutchinson EC, Fodor E.. Transport of the influenza virus genome from nucleus to nucleus. Viruses 2013;5:2424–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rossman JS, Leser GP, Lamb RA.. Filamentous influenza virus enters cells via macropinocytosis. J Virol 2012;86:10950–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. de Vries M, Herrmann A, Veit M.. A cholesterol consensus motif is required for efficient intracellular transport and raft association of a group 2 HA from influenza virus. Biochem J 2015;465:305–14. [DOI] [PubMed] [Google Scholar]
- 72. Martin K, Helenius A.. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 1991;65:232–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vreede FT, Chan AY, Sharps J, et al. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 2010;396:125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thomas PG, Keating R, Hulse-Post DJ, et al. Cell-mediated protection in influenza infection. Emerg Infect Dis 2006;12:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Firth AE, Jagger BW, Wise HM, et al. Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol 2012;2:120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heldt FS, Frensing T, Reichl U.. Modeling the intracellular dynamics of influenza virus replication to understand the control of viral RNA synthesis. J Virol 2012;86:7806–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Singh G, Pratt G, Yeo GW, et al. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 2015;84:325–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pumroy RA, Ke S, Hart DJ, et al. Molecular determinants for nuclear import of influenza A PB2 by importin alpha isoforms 3 and 7. Structure 2015;23:374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nguyen KT, Holloway MP, Altura RA.. The CRM1 nuclear export protein in normal development and disease. Int J Biochem Mol Biol 2012;3:137–51. [PMC free article] [PubMed] [Google Scholar]
- 80. Muhlbauer D, Dzieciolowski J, Hardt M, et al. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J Virol 2015;89:6009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fournier E, Moules V, Essere B, et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 2012;30:7359–67. [DOI] [PubMed] [Google Scholar]
- 82. Chen BJ, Leser GP, Jackson D, et al. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol 2008;82:10059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gavazzi C, Isel C, Fournier E, et al. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic Acids Res 2013;41:1241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gavazzi C, Yver M, Isel C, et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci USA 2013;110:16604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sulc P, Romano F, Ouldridge TE, et al. A nucleotide-level coarse-grained model of RNA. J Chem Phys 2014;140:235102.. [DOI] [PubMed] [Google Scholar]
- 86. Martinez HM, Maizel JV Jr, Shapiro BA.. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J Biomol Struct Dyn 2008;25:669–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kobayashi Y, Dadonaite B, van Doremalen N, et al. Computational and molecular analysis of conserved influenza A virus RNA secondary structures involved in infectious virion production, RNA. Biol 2016;13:883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang Y, Aevermann BD, Anderson TK, et al. Influenza research database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 2017;45:D466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dawson W, Takai T, Ito N, et al. A new entropy model for RNA: part III. Is the folding free energy landscape of RNA funnel shaped? J Nucl Acids Invest 2014;5:2652. [Google Scholar]
- 90. Nakamura S. A novel virtual spectrometry: visualized regulatory motifs on ADM, rPol{beta} and CD83 mRNAs in human-friendly manners. J Biochem 2009;146:251–61. [DOI] [PubMed] [Google Scholar]