Abstract
Several strains of Thermus thermophilus were tested in order to detect purine nucleoside synthase activity using pyrimidine nucleosides as the sugar-donor and adenine or hypoxanthine as bases. High productivity values (t =1 hr) were obtained while completely avoiding adenosine-deaminase degradation of the products. N-2-deoxy-ribosyltransferase activity is described for the first time in hyperthermophilic bacteria.
Keywords: Biocatalysis, Nucleoside synthesis, Thermus thermophilus.
Introduction
Thermophiles are a group of microorganisms that grow under extreme temperature conditions. These microorganisms offer useful enzymes to expand the range of reaction conditions suitable for biocatalysis. In this way some interesting enzymes have been described such as esterases, lipases, proteases, alcohol dehydrogenases, etc. [1,2]. Nucleoside analogues are labile and polyfunctional molecules which chemical synthesis requires several protection/deprotection steps [3]. These compounds have a wide range of uses, mainly as antiviral or antitumoral drugs, but also in the treatment of hypertension or inflammatory processes. The one-pot synthesis using nucleoside phosphorylases (NPs) or nucleoside 2’-deoxyribosyltransferases (NdRTs) are an alternative to the chemical synthesis [3,4,5].
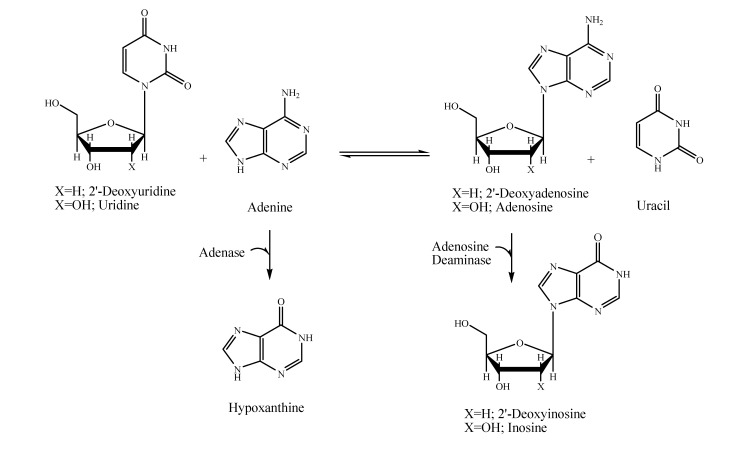
Several mesophile microorganisms have been identified as active for purine nucleoside synthesis, giving relatively good yield values after short reaction times [5,6]. However, the adenosine degradation by adenosine-deaminase (ADA) still remains as a problem for many of them (Scheme 1). In this work, we identify several strains of Thermus thermophilus capable of reaching high yield values, overcoming the ADA problem.
Scheme 1.
Reaction pathways of the adenine nucleoside synthesis and degradation of adenine and adenine nucleosides by ADA.
Results and Discussion
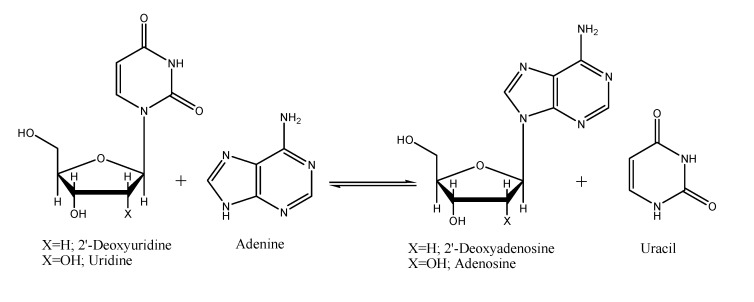
The reactions of 2’-deoxyuridine (X =H) or uridine (X =OH) with hypoxanthine were tested. These reactions yield 2’-deoxyinosine (X =H) or inosine (X =OH), respectively (Scheme 2). Different Thermus thermophilus strains - cultured in optimum conditions - were tested. Better yields and productivity values were obtained with 2’-deoxyuridine (dUrd) than in the case of uridine (Urd) (Table 1). These results seem to indicate that one very active nucleoside 2’-deoxyribosyltransferase or one thymidine nucleoside phosphorylase plus one purine nucleoside phosphorylase are present in these strains.
Scheme 2.
One-pot enzymatic synthesis of adenine nucleosides using uridine or 2’-deoxyuridine as donors of sugar moiety.
Table 1.
Yield and productivity values in the reaction dUrd or Urd + hypoxanthine, using different T. thermophilus strainsa.
| Strain | 2’-Deoxyinosine synthesis | Inosine synthesis | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells ]b | Productivityc | Yield (%) | [cells ]b | Productivityc | |
| NAR1 | 24 | 10,000 | 12 x 10-5 | 8.5 | 9,883 | 4.3 x 10-5 |
| HB27 | 28 | 6,364 | 22 x 10-5 | 14 | 6,364 | 11 x 10-5 |
| PRQ-16 | 24 | 6,316 | 19 x 10-5 | 11 | 6,111 | 9.0 x 10-5 |
| PRQ-25 | 25 | 6,250 | 20 x 10-5 | 11 | 5,978 | 9.2 x 10-5 |
| B | 24 | 7,059 | 17 x 10-5 | 11 | 7,142 | 7.7 x 10-5 |
| RQ1 | 20 | 10,417 | 9.6 x 10-5 | 8.7 | 10,610 | 4.1 x 10-5 |
| N17 | 21 | 7,500 | 14 x 10-5 | 10.5 | 7,500 | 7.0 x 10-5 |
| HN1.11 | 12 | 3,529 | 17 x 10-5 | 5.3 | 3,786 | 7.0 x 10-5 |
| Fiji3A1 | 16.5 | 2,750 | 30 x 10-5 | 8.3 | 2,767 | 15 x 10-5 |
| CC16 | 23 | 6,053 | 19 x 10-5 | 10 | 5,952 | 8.4 x 10-5 |
| VG7 | 22 | 2,683 | 41 x 10-5 | 13 | 2,708 | 24 x 10-5 |
a Reaction conditions: 65ºC, 5 mM deoxyuridine or uridine, 5 mM hypoxanthine, 30 mM sodium phosphate buffer (pH= 7). Total volume 4 mL; b million cells. mL-1; c µmol.h-1.million cells-1.
In order to test the efficiency of the biocatalysts in the synthesis of 6-aminopurine nucleosides, the synthesis of 2’-deoxyadenosine (X=H) and adenosine (X=OH) (Scheme 2) was studied (Table 2). Productivity values, which are related to the number of cells carrying the reaction, were used to compare results, since each assay was performed from different cultures with different cells concentrations, making overall yield a poor parameter for comparison.
Table 2.
Yield and productivity of the reaction dUrd or Urd + adenine using some selected strains a.
| Strain | 2’-Deoxyadenosine synthesis | Adenosine synthesis | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells] b | Productivity c | Yield (%) | [cells] b | Productivity c | |
| NAR1 | 30 | 10,714 | 14 x 10-5 | 15 | 10,563 | 7.1 x 10-5 |
| HB27 | 44 | 11,000 | 20 x 10-5 | 21 | 10,938 | 9.6 x 10-5 |
| PRQ-16 | 29 | 6,041 | 24 x 10-5 | 15 | 6,000 | 12.5 x 10-5 |
| PRQ-25 | 28 | 5,600 | 25 x 10-5 | 15 | 5,556 | 13.5 x 10-5 |
| B | 38 | 9,500 | 20 x 10-5 | 21 | 9,545 | 11 x 10-5 |
| RQ1 | 28 | 10,769 | 13 x 10-5 | 15 | 10,870 | 6.9 x 10-5 |
| N17 | 27 | 7,105 | 19 x 10-5 | 12 | 6,771 | 9.6 x 10-5 |
| HN1-11 | 15 | 5,769 | 13 x 10-5 | 6.9 | 5,847 | 5.9 x 10-5 |
| Fiji3A1 | 14 | 5,000 | 14 x 10-5 | 6.3 | 5,250 | 6.0 x 10-5 |
| CC16 | 30 | 6,818 | 22 x 10-5 | 14 | 6,667 | 10.5 x 10-5 |
| VG7 | 23 | 4,423 | 26 x 10-5 | 12 | 4,286 | 14 x 10-5 |
a Reaction conditions: 65ºC, 5 mM deoxyuridine or uridine, 5 mM hypoxanthine, 30 mM sodium phosphate buffer (pH= 7). Total volume 4 m; b million cells. mL-1; c µmol.h-1.million cells-1.
As in the case of 6-oxonucleosides (Table 1), the synthesis of 2’-deoxyadenosine (dAdo) leads to better yields than the synthesis of adenosine (Ado) (Table 2). The productivities in dAdo (Table 2) were generally similar or higher compared to those in the synthesis of 2’-deoxyinosine (Table 1) except for strains VG7 and FIJI3A1. In any case, adenine nucleoside degradation products (inosine, 2’-deoxyinosine or hypoxanthine) could not be detected, indicating absence of ADA activity at the tested temperature. From the results of Table 1 and Table 2, we could conclude that the intracellular enzymes of strains show better selectivity versus 6-aminopurine than versus 6-oxopurines. This broad specificity has been described in mesophile bacteria [5,7]. Due to the different biomass production of the strains, productivity values were calculated to select the best biocatalysts. According to this criterion HB27, PRQ-16, PRQ-25, B, Fiji3A1 and VG7 were selected as the most interesting strains. The productivity values are better than those described for wild type mesophile or psychrophile species under the same experimental conditions [5,6,8]. Other workers [9] describe better nucleoside yields but the cell concentration was not given, so we cannot compare the catalytic activity. These productivities, the absence of ADA activity, the thermotolerance and the resistance to extreme conditions make these biocatalysts interesting for further developments. In addition, we must indicate that the cell cultures of each strain – in optimum conditions – are repetitive as we can observe in Table 1 and Table 2.
Similar reactions were performed using thymidine as the sugar-donor nucleoside and adenine or hypoxanthine as acceptor bases to explore the presence of one thymidine-nucleoside phosphorylase (selective versus thymidine and 2’-deoxyuridine (dUrd) compared to uridine (Urd) [11,12,13]). The combination of this enzyme plus a non-specific PNP can give activities similar to the expected for an active NdRT. The obtained productivity values with the selected strains (Table 3) were generally lower than those obtained with dUrd (Table 1 and Table 2) showing a moderated selectivity versus adenine. Therefore, if the strains: i) gave better yields versus adenine than with hypoxanthine; ii) gave better yields with 2’-deoxyribose nucleoside than with ribose nucleoside; iii) gave lower or similar yields using thymidine than using 2’-deoxyuracil, as sugar donors and iv) uridine is recognised as substrate, we could postulate that the strains present an active NdRT or one PyNP plus one PNP selective versus adenine compared to hypoxanthine.
Table 3.
Yield and productivity of the reaction thymidine + adenine or hypoxanthinea.
| Strain | Thymidine + hypoxantine | Thymidine + adenine | ||||
|---|---|---|---|---|---|---|
| Yield (%) | [cells]b | Productivityc | Yield (%) | [cells]b | Productivityc | |
| HB27 | 22 | 10,000 | 11 x 10-5 | 33 | 9,706 | 17 x 10-5 |
| PRQ-16 | 14 | 5,385 | 13 x 10-5 | 21 | 5,526 | 19 x 10-5 |
| PRQ-25 | 15 | 5,357 | 14 x 10-5 | 21 | 5,250 | 20 x 10-5 |
| B | 21 | 8,750 | 12 x 10-5 | 39.5 | 8,977 | 22 x 10-5 |
| Fiji3A1 | 12 | 5,455 | 11 x 10-5 | 16 | 5,333 | 15 x 10-5 |
| VG7 | 11.5 | 3,382 | 17 x 10-5 | 19 | 3,519 | 27 x 10-5 |
aReaction conditions: 65ºC, 5mM thymidine, 5mM adenine or hypoxanthine, 30mM sodium phosphate buffer (pH=7). Total volume 4 mL; b million cells. mL-1; c µmol.h-1.million cells-1.
Other assays were performed for the optimization of reaction parameters such as pH, molar ratio of nucleoside/base and nature of the buffer. HB27 was selected due to the high productivities and the easy culture conditions. To explore the nature of the active enzyme, the same reaction was tested in two buffers. In Tris/HCl buffer 30mM (pH=7) the productivity values experienced an increase of 44% over those obtained in 30mM sodium phosphate buffer (pH=7). This result suggests, in accordance with the literature [1,2,3,4,6], that one NdRT, rather than two NPs, may be the most active enzyme in HB27.
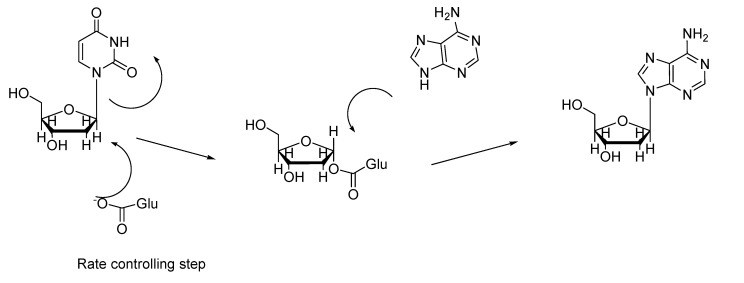
In Scheme 3 we show the reaction mechanism for the 2’-deoxynucleoside synthesis catalyzed by NdRT [3,5]. We can see that phosphate ion is not necessary for the catalytic process. This is the fundamental difference with NPs, which produce 1-α-ribose-phosphate [5,11,14,15].
Scheme 3.
Reaction mechanism of the N-2-deoxyribosyltranferase catalysis.
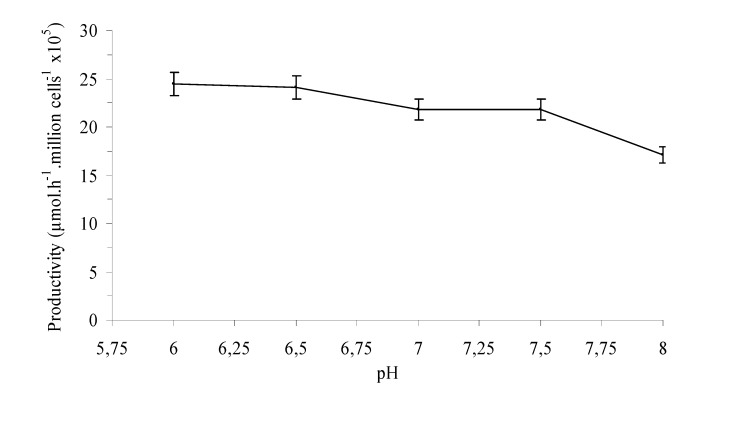
The results in Figure 1 indicate that better productivities are obtained at pH <7.5. Similar optimum pH values were recommended in the case of NdRTs from Lactobacillus helveticus (6 <pH <6.5 [16]) or Lactococus, Streptoccocus, Aeroccoccus and Leuconostoc genera (pH =6.5 [17]). Therefore, these results seem to support the NdRT hypothesis.
Figure 1.
Influence of pH in the productivity of deoxyadenosine by T. thermophilus HB27. Standard reaction conditions: 65ºC, 5mM deoxyuridine, 5mM adenine, 30mM sodium phosphate buffer (pH=7). Total reaction volume 4 mL.
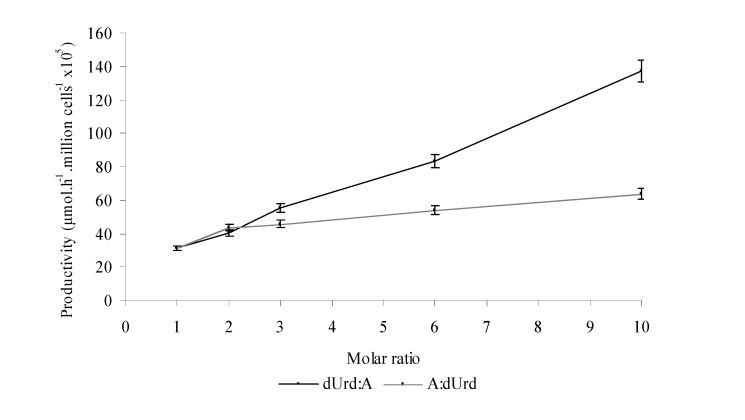
In Figure 2 we show that an excess in dUrd dramatically increases the productivity values. Similar results have been described [5,9,16,18,19], suggesting that the rate controlling step is the first one (Scheme 3). The excess of adenine moderately increases the yield. Taking into the account the mechanism of NdRT [14,15] we could only explain this result by assuming that the greater the concentration of dUrd is, the higher the concentration of active intermediate (1-α-glutamyl-ribose , Scheme 3) is, and therefore the yield increases. Contrarily, if the dUrd concentration is the same, the concentration of the reaction intermediate is constant and so an increase in the adenine concentration does not dramatically increase the yield as observed (Figure 2). We must indicate that an increase in the adenosine concentration does not lead to hypoxanthine formation.
Figure 2.
Influence of deoxyuridine/adenine ratio. Standard reaction conditions: 65ºC, 5mM deoxyuridine, 5mM adenine, 30mM sodium phosphate buffer (pH=7). Total reaction volume 4 mL.
Several results hint towards the possibility of a NdRT involved in the reactions tested, such as the increased productivity when the reaction is carried in tris-HCl buffer without the presence of phosphate, which is a substrate in the NP reaction, or the apparent acidophility of the process. However, whole, living cells are a complex system with many variables which should be taken into the account. Also, it has been previously reported the existence of NPs in some of the tested strains, such as HB8 [20]. Now, cloning and isolation experiments are in progress in HB27 and PRQ-25 and VG7.
Experimental
Materials and Microorganisms
Nucleosides and purine bases were from Sigma-Aldrich (USA). HPLC solvents were from Scharlab (Spain) and the buffer reagents from Aldrich (Germany). Different strains of Thermus thermophilus were used in this work: NAR1, HB27, PRQ-16. PRQ-25, B, RQ1, Fiji3A1, HN1.11, CC16, VG7 and NR-17. Cells were grown in TB culture medium [0.8 % peptone (w/v), 0.4 % yeast extract (w/v) and 0.3 % NaCl (w/v) in Milli-Q grade water, adjusted to pH 7.5 with NaOH] at 65 ºC [7] for 16 h under shaking at 150 rpm in Erlenmeyer flasks. After growing, the culture broth was centrifuged for 15 min at 10,000 g. The cells were harvested and washed in 10 mL of 30 mM sodium phosphate buffer (pH =7) and then re-centrifuged. The pellet was directly used in the purine nucleoside synthesis test reactions.
Standard synthesis of purine nucleosides
Cells were obtained from 20 mL of culture broth and resuspended 4 mL of reaction mixture (30 mM sodium phosphate buffer, pH=7, sugar-donor nucleoside 5 mM and sugar-acceptor base 5 mM). The reactions were performed at 65ºC under shaking for 1 hour. Samples were obtained and filtered to be immediately analysed by HPLC.
Synthesis in Tris/HCl buffer
Cells were obtained from 20 mL of culture broth and resuspended with 4 mL of reaction mixture (30mM Tris/HCl buffer (pH=7)). Then 2’-deoxyuridine (5 mM) and adenine (5 mM) were added. The reaction was performed at 65ºC under shaking for 1 hour. Samples were filtered to be immediately analysed by HPLC.
Sample analysis
The samples were analyzed by HPLC using water/methanol (90:10 v/v) as the mobile phase and a flow rate of 1.2 mL·min-1. The Agilent 1100 Series HPLC was equipped with an UV detector (set at 254 nm) and a C18 apolar column (0.46 x 15 cm, 5 µm), Teknokroma, Barcelona, (Spain).
Acknowledgements
The research has been supported by a grant from Comunidad Autónoma de Madrid (CAM) Project S-0505/PPQ/0344. One of the authors, Marcos Almendros, thanks the CAM a predoctoral fellow. We also thank Milton S. da Costa for providing most of the strains used in this work. The authors thank to Mr. Davies the critical revision of the manuscript.
Footnotes
Sample availability: Not available.
References and Notes
- 1.Atomi H. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 2005;9:166–173. doi: 10.1016/j.cbpa.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Domínguez A., Fuciños P., Rua M. L, Pastrana L, Longo M.A., Sanromán M.A. Stimulation of novel thermostable extracellular lipolytic enzyme in cultures of Thermus sp. Enzyme Microb. Thechnol. 2007;40:187–194. doi: 10.1016/j.enzmictec.2006.09.006. [DOI] [Google Scholar]
- 3.Mikhailopulo I.A. Biotechnology of nucleic acid constituents- State of the art and perspectives. Curr. Org. Chem. 2007;11:317–338. doi: 10.2174/138527207780059330. [DOI] [Google Scholar]
- 4.Prasad A.K., Trikha S., Parmar V.S. Nucleoside synthesis mediated by glycosyl-transferring enzymes. Biorg. Chem. 1999;27:135–154. doi: 10.1006/bioo.1998.1127. [DOI] [Google Scholar]
- 5.Condezo L.A., Fernandez-Lucas J., Garcia-Burgos C.A., Alcántara A.R., Sinisterra J.V. Enzymatic synthesis of modified nucleosides. In: Patel R.N., editor. Biocatalysis in the Pharmaceutical and Biotechnology Industrie. CRC Press; Boca Raton, FL, USA: 2006. pp. 401–423. Chapter 14. [Google Scholar]
- 6.Fernandez-Lucas J., Condezo L.A., Martinez-Lagos F., Sinisterra J.V. Synthesis of 2’-deoxyribosylnucleosides using new 2’-deoxyribosyltransferase microorganisms producers. Enzyme Microb. Technol. 2007;40:1147–1155. [Google Scholar]
- 7.Bzowska A., Kulikowska E., Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharm. Ther. 2000;88:349–425. doi: 10.1016/S0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 8.Medici R., Lewkowicz E.S., Iribarren A.M. Microbial synthesis of 2,6-diaminopurine nucleosides. J. Mol. Catal. B-Enzym. 2006;39:40–44. doi: 10.1016/j.molcatb.2006.01.024. [DOI] [Google Scholar]
- 9.Utagawa T. Enzymatic preparation of nucleoside antibiotics. J. Mol. Catal. B Enzym. 1999;6:215–222. [Google Scholar]
- 10.Ramírez-Arcos S., Fernandez-Herrero L.A., Berenguer J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta. 1998;1396:215–227. doi: 10.1016/S0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- 11.Pugmiere M.J., Ealick S.E. Structural analysis reveal two distinct families of nucleoside phosphorylases. Biochem. J. 2002;361:1–25. doi: 10.1042/0264-6021:3610001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmerman M., Seidenberg J. Thymidine phosphorylase and nucleoside deoxy-ribosyltransferase in normal and malignal tissues. J. Biol. Chem. 1964;239:2618–2621. [PubMed] [Google Scholar]
- 13.Fukushima M., Suzuki N., Emura T., Yano S., Kazuno H., Tada Y., Yamada Y., Asao T. Structure and activity of specific inhibitors of thymidine phosphorylase to potentiate the function of antitumor 2'-deoxyribonucleosides. Biochem. Pharmacol. 2000;59:1227–1236. doi: 10.1016/S0006-2952(00)00253-7. [DOI] [PubMed] [Google Scholar]
- 14.Holguin J., Cardinaud R., Salemnik C.A. Trans-N-doxyriboxylase: substrate specificity studies. Purine base acceptors. Eur. J. Biochem. 1975;54:515–520. doi: 10.1111/j.1432-1033.1975.tb04164.x. [DOI] [PubMed] [Google Scholar]
- 15.Danzin C., Cardinaud R. Deoxyribosyltransfer catalysis with trans-N-deoxyriboxylase: kinetic study of purine (pyrimidine) to pyrimidine (purine) trans-N-deoxyriboxylase. Eur. J. Biochem. 1976;62:365–372. doi: 10.1111/j.1432-1033.1976.tb10168.x. [DOI] [PubMed] [Google Scholar]
- 16.Cardinaud R., Holguin J. Nucleoside deoxyribosyltransfrerase II from Lactobacillus helveticus. Biochim. Biophys. Acta. 1979;568:339–347. doi: 10.1016/0005-2744(79)90301-2. [DOI] [PubMed] [Google Scholar]
- 17.Chawdhri R.F., Hutchinson D.W., Richards A.O’L. Nucleoside deoxyriboxyltransferase and inosine phosphorylase activity in lactic acid bacteria. Arch. Microbiol. 1991;155:409–411. [Google Scholar]
- 18.Trelles J.A., Fernandez M., Lewkowicz E.S., Iribarrren A.M., Sinisterra J.V. Purine nucleoside synthesis from uridine using immobilized Enterobacter gergoviae CECT 875 whole cells. Tetrahedron Lett. 2003;44:2605–2609. doi: 10.1016/S0040-4039(03)00225-9. [DOI] [Google Scholar]
- 19.Fernandez-Lucas J., Condezo L.A., Quezada M.A., Sinisterra J.V. Low-temperature synthesis of 2’-deoxyadenosine using immobilized psychrotrophic microorganisms. Biotechnol. Bioeng. 2008;100:213–222. doi: 10.1002/bit.21756. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu K., Kunishima N. Purification, crystallization and preliminary X-ray diffraction study on pyrimidine nucleoside phosphorylase TTHA1771 from Thermus thermophilus HB8. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2007;63:308–310. doi: 10.1107/S1744309107009980. [DOI] [PMC free article] [PubMed] [Google Scholar]