 A series of 13 phenyl substituted thiosemicarbazones (SB1–SB13) were synthesized and evaluated for their inhibitory potential towards human recombinant monoamine oxidase A and B (MAO-A and MAO-B, respectively) and acetylcholinesterase.
A series of 13 phenyl substituted thiosemicarbazones (SB1–SB13) were synthesized and evaluated for their inhibitory potential towards human recombinant monoamine oxidase A and B (MAO-A and MAO-B, respectively) and acetylcholinesterase.
Abstract
A series of 13 phenyl substituted thiosemicarbazones (SB1–SB13) were synthesized and evaluated for their inhibitory potential towards human recombinant monoamine oxidase A and B (MAO-A and MAO-B, respectively) and acetylcholinesterase. The solid state structure of SB4 was ascertained by the single X-ray diffraction technique. Compounds SB5 and SB11 were potent for MAO-A (IC50 1.82 ± 0.14) and MAO-B (IC50 0.27 ± 0.015 μM), respectively. Furthermore, SB11 showed a high selectivity index (SI > 37.0) for MAO-B. The effects of fluorine orientation revealed that SB11 (m-fluorine) showed 28.2 times higher inhibitory activity than SB12 (o-fluorine) against MAO-B. Furthermore, inhibitions by SB5 and SB11 against MAO-A and MAO-B, respectively, were recovered to near reference levels in reversibility experiments. Both SB5 and SB11 showed competitive inhibition modes, with Ki values of 0.97 ± 0.042 and 0.12 ± 0.006 μM, respectively. These results indicate that SB5 and SB11 are selective, reversible and competitive inhibitors of MAO-A and MAO-B, respectively. Compounds SB5, SB7 and SB11 showed moderate inhibition against acetylcholinesterase with IC50 values of 35.35 ± 0.47, 15.61 ± 0.057 and 26.61 ± 0.338 μM, respectively. Blood–brain barrier (BBB) permeation was studied using the parallel artificial membrane permeation assay (PAMPA) method. Molecular docking studies were carried out using AutoDock 4.2.
Introduction
The role of monoamine oxidases (MAOs) in brain neurochemistry is mainly connected with the oxidative deamination of biogenic amines (BA).1 Amines such as adrenaline, noradrenaline, melatonin and serotonin are predominantly metabolized by MAO-A, whereas benzylamine and phenylethylamine are controlled by MAO-B. Both types of MAO isoforms have common substrates such as dopamine and tyramine.2,3 Selective MAO inhibitors (MAOIs) have a great impact on treating various psychiatric and neurodegenerative disorders by depleting MAO levels in the brain.4 Serotonin, the concentration of which is maintained by inhibitors of MAO-A, shows superior antidepressant activity.5 Conversely, the end-products, hydrogen peroxide and reactive oxygen species (ROS), produced during dopamine metabolism by MAO-B, generate oxidative stress and apoptosis in dopamine producing cells. These highly reactive toxic radicals produce neural toxicity which may be the prime indications for Alzheimer's and Parkinson's diseases (AD & PD).6 Hence, selective MAO-B inhibitors are highly recommended as co-adjuvant therapy for treating AD and PD patients.7,8
The current scenario of PD therapy focuses on restoring the level of dopamine in the brain and thereby curtailing the motor symptoms.9 This therapy is accelerated by the administration of dopamine precursors (L-DOPA), dopamine agonists, catechol-O-methyltransferase (COMT) and MAO-B inhibitors such as selegiline and rasagiline (highly selective and irreversible).10 Molecules which have closer acetylcholinesterase (AChE) and MAO-B affinities are able to limit the neurotoxicity related to sources of ROS in age-related AD diseases.11 In 2011, the molecule ladostigil, a dual inhibitor of both AChE and MAO-B designed by Youdim, entered into the phase II clinical trial for the treatment of AD.12 Considering the complex pathogenetic factors of various neurodegenerative diseases, it is highly appropriate to recommend selective MAO-B inhibitors with cocktail therapy to trigger the multi-targets associated with these diseases.
The MAO-B inhibitors currently in use are the irreversible type, having a covalent bond to the FAD unit of the inhibitor binding cavity (IBC) of the enzyme.13 Disruption of the target, a poor ADME profile and increased duration of action occur due to this irreversible binding. Hence, the development of reversible MAO-B inhibitors has a greater therapeutic value in treating neurodegenerative diseases.14 Some of the evidence also documents that mild symptomatic benefits were gained via the administration of moclobemide (a reversible MAO-A inhibitor) when combined with levodopa for the treatment of PD.15,16 Recently, many small molecules like chalcones, coumarins and chromones have shown considerable potential for the development of MAO-A/MAO-B inhibitors with a highly selective and reversible mode of inhibition.17–19
From a chemical point of view, thiosemicarbazones are thiourea compounds linked with an azomethine scaffold and are the key intermediates for the synthesis of 2-amino-1,3,4-thiadiazoles via a ring chain tautomerism mechanism.20,21 Besides this, thiosemicarbazones are excellent chelators of transition metals such as zinc (Zn), copper (Cu) and iron (Fe), which are potent inhibitors of various carcinogenesis-inducing pathways.22 In the past, many efforts have addressed the development of thiosemicarbazone-based MAOIs.23–29 Moreover, the cyclized form of thiosemicarbazones from chalcones afforded N-thiocarbamoyl pyrazolines, which show a remarkable inhibition profile against MAOs.30–34 The presence of hydrazine units in thiosemicarbazide also afforded a number of hydrazone scaffolds via the acid catalyzed nucleophilic addition mechanism.35 Numerous studies recommend the multi-potent MAO and cholinesterase inhibitors for treating AD and PD.36,37 Accordingly, this work describes the synthesis of phenyl-substituted thiosemicarbazones, the studies on MAO and acetylcholinesterase inhibition, and the kinetics of the inhibition mechanism of MAOs using Lineweaver–Burk plots, the reversibility mode, and blood–brain barrier (BBB) permeation assays. Finally, the lead molecules from the in vitro results were subjected to molecular docking studies to elucidate the binding interactions of both MAO-A and B.
Result and discussion
Chemistry
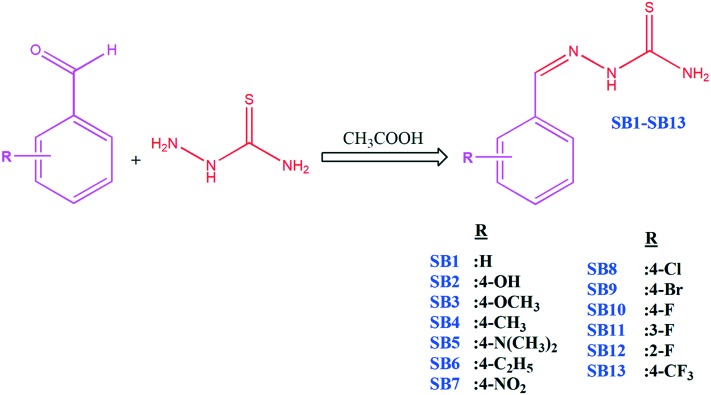
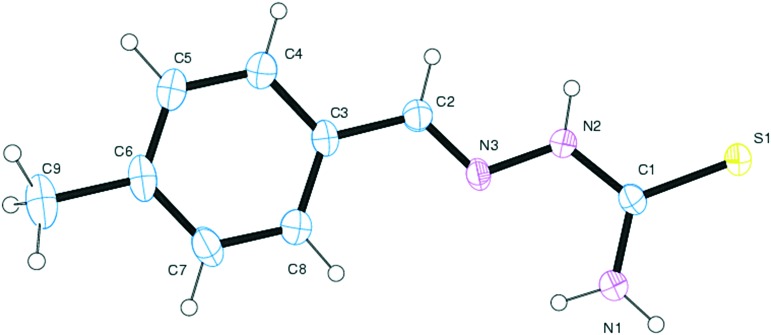
The target aryl thiosemicarbazones were synthesized as presented in Scheme 1. Synthesis was accomplished by the single step reaction between commercially available thiosemicarbazide hydrochloride and various substituted benzaldehydes. In the 1H-NMR spectra, the sharp singlet peak observed between 7.71–7.89 is ascribed to the azomethine (–C[combining low line]H[combining low line] N–) proton. The downfield proton of the NH group attached to the thiocarbamoyl unit is observed in the range between 9.18–9.88. Two broad singlets were found at 6.23–7.25 and 7.23–7.26, corresponding to the terminal NH2 group. 13C NMR spectra displayed carbothioamide groups for SB1–SB13 between δ180.60–184.30. All spectral characterization results are in full agreement with previous literature reports.38–40 The solid state structure of SB4 was ascertained by the single X-ray diffraction technique, and the ORTEP diagram is depicted in Fig. 1. The mass spectra of all fluorinated chalcones showed intensive molecular ions, assisting the structure of the targeted compounds.
Scheme 1. Synthetic route of aryl thiosemicarbazones.
Fig. 1. ORTEP diagram of compound SB4.
Monoamine oxidase inhibition studies
Inhibition profile of aryl substituted thiosemicarbazones
Six different compounds of the derivatives showed high inhibitory activities (more than 50%) against MAO-A or MAO-B, while the other compounds were not effective (Table 1). Compounds SB3, SB5, and SB6 showed efficient inhibition against MAO-A with IC50 values of 4.99 ± 0.25, 1.82 ± 0.14, and 5.98 ± 0.15 μM, respectively. Compounds SB7, SB11, and SB12 were also effectively inhibitory against MAO-B with IC50 values of 1.43 ± 0.014, 0.27 ± 0.015, and 7.61 ± 0.49 μM, respectively. All 6 compounds showed good selectivity, and SB5 and SB11 were the most potent for MAO-A and MAO-B, respectively. Furthermore, SB11 showed a high selectivity index (SI > 37.0) for MAO-B with a low IC50 value (0.27 μM), suggesting it is a good candidate for selective MAO-B inhibition. Considering the structural comparisons, we concluded that based on their IC50 values, the m-fluorine substitution of the compounds (SB11) showed 28.2 times higher inhibitory activity against MAO-B than o-fluorine substitution (SB12) and >37.0 times more potent than p-fluorine (SB10) (Table 1). However, the substituent p-NO2 (SB7) was more effective than the p-fluorine substituent (SB10).
Table 1. Inhibition of recombinant human MAO enzymes and acetylcholinesterase by aryl substituted aryl thiosemicarbazones a .
| Compounds | Residual activity at 10 μM (%) |
IC50 (μM) |
SI b | |||
| MAO-A | MAO-B | MAO-A | MAO-B | AChE | ||
| SB1 | 83.5 ± 0.7 | 75.5 ± 0.7 | >10.0 | >10.0 | >40.0 | — |
| SB2 | 66.0 ± 1.4 | 87.5 ± 3.5 | >10.0 | >10.0 | >40.0 | — |
| SB3 | 37.5 ± 0.7 | 82.1 ±1.4 | 4.99 ± 0.25 | >10.0 | 36.14 ± 0.45 | <0.50 |
| SB4 | 56.5 ± 0.7 | 91.5 ± 2.1 | >10.0 | >10.0 | >40.0 | — |
| SB5 | 16.5 ± 2.1 | 82.3 ± 1.4 | 1.82 ± 0.14 | >10.0 | 35.35 ± 0.47 | <0.18 |
| SB6 | 36.0 ± 5.7 | 86.2 ± 1.4 | 5.98 ± 0.15 | >10.0 | 26.72 ± 0.006 | <0.60 |
| SB7 | 75.5 ± 3.5 | 7.5 ± 0.7 | >10.0 | 1.43 ± 0.014 | 15.61 ± 0.057 | >6.99 |
| SB8 | 76.0 ± 1.4 | 60.5 ± 2.1 | >10.0 | >10.0 | >40.0 | — |
| SB9 | 59.5 ± 0.7 | 82.5 ± 4.9 | >10.0 | >10.0 | 31.12 ± 0.44 | — |
| SB10 | 84.5 ± 3.5 | 86.5 ± 2.1 | >10.0 | >10.0 | 37.93 ± 0.57 | — |
| SB11 | 75.5 ± 2.1 | 3.0 ± 1.4 | >10.0 | 0.27 ± 0.015 | 26.61 ± 0.34 | >37.0 |
| SB12 | 77.5 ± 0.7 | 33.5 ± 2.1 | >10.0 | 7.61 ± 0.49 | 30.84 ± 0.58 | >1.31 |
| SB13 | 92.5 ± 0.7 | 77.5 ± 0.7 | >10.0 | >10.0 | 36.23 ± 0.44 | — |
| Toloxatone | — | — | 0.92 ± 0.016 | >80 | — | <0.012 |
| Lazabemide | — | — | >80 | 0.042 ± 0.0010 | — | >1900 |
| Clorgyline | — | — | 0.0071 ± 0.0003 | 1.69 ± 0.32 | — | 0.0042 |
| Pargyline | — | — | 1.31 ± 0.068 | 0.091 ± 0.005 | — | 14.4 |
| Tacrine | — | — | — | — | 0.23 ± 0.014 | — |
aResults are expressed as means ± standard errors of duplicate experiments. Inhibitory activities for reference compounds of MAO and AChE were measured after preincubation with the enzymes for 30 min and 15 min, respectively.
bSI was expressed for MAO-B by dividing the IC50 value of MAO-A by that of MAO-B.
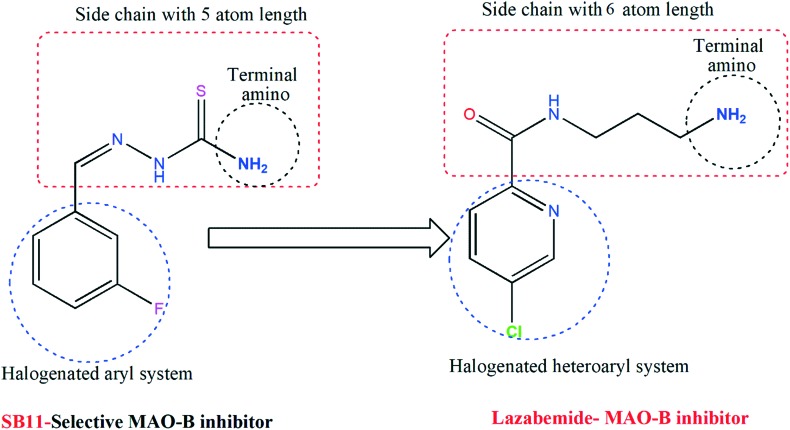
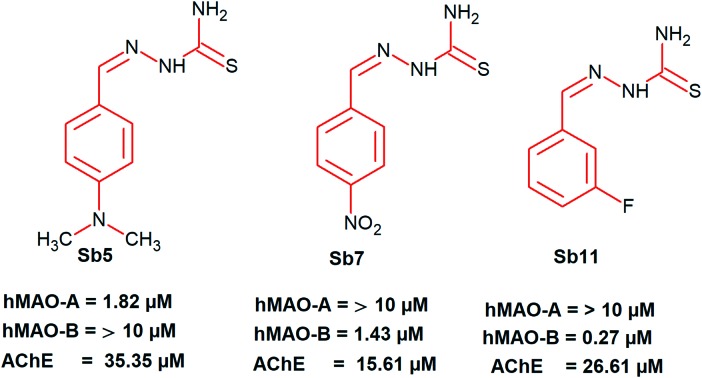
The potency of SB5 for MAO-A (IC50 = 1.82 μM) was lower than that of IM5 (IC50 = 0.30 μM), which is a synthesized imidazole-bearing chalcone derivative of the eleven series and is the most potent for MAO-A reported by our group recently.41 However, the SI value of SB5 (0.18) in this study was >4.2 times greater than that of IM5 (0.75). The potency of SB11 for MAO-B (IC50 = 0.27 μM) was higher than that of IM4 (IC50 = 0.32 μM), which is another derivative and the most potent for MAO-B in the IM series. Similar to SB5, the SI value of SB11 (>37.0) was >11.2 times higher than that of IM4 (3.3). Although the potency of SB11 for MAO-B was 6.4 times lower than that of the marketed drug lazabemide for MAO-B (IC50 = 0.042 μM), the relatively low molecular weight of SB11 (MW = 197.2) is comparable to that of lazabemide (MW = 199.6) and lower than that of IM4 (MW = 288.3); it also has an IC50 value comparable to that of lazabemide in the nanomolar concentration range. The structural features of SB11 were mainly divided as follows: (a) a halogenated aryl system, (b) a side chains with an almost similar length to that of lazabemide, and (c) a terminal amino group at the side chain. Similar types of features are seen in the potent MAO-B inhibitor (lazabemide) and these are depicted in Fig. 2. These structural features are responsible for the design and development of a new class of MAO-B inhibitors.
Fig. 2. Similarity-based structures of SB11 and a standard MAO-B inhibitor.
Structure–activity relationship (SAR) analysis of MAO inhibition
Changes in the MAO inhibitory potency of the tested aryl thiosemicarbazones could be correlated to the effect of various electron donating and withdrawing groups anchored to the phenyl system. To explore the structure–activity relationship of the target compounds, we initially focused on varying the substituents at the para position of the phenyl system of aryl thiosemicarbazones. Unsubstituted aryl thiosemicarbazones are less effective against both MAO-A and MAO-B with IC50 > 10.0 μM. Modifications in the position and orientation of substituents on the phenyl system result in a shift in this trend. The presence of electron donating groups (EDGs) such as methoxy, dimethylamino and ethyl on the para position of the phenyl system in compounds SB3, SB5 and SB6 significantly contributes to the activity ratio towards MAO-A. Furthermore, introduction of electron donating groups such as hydroxyl and methyl (SB2 & SB4) results in a dramatic decrease in activity. Hence, it is commonly recommended that EDGs with bulky groups on the phenyl system of aryl thiosemicarbazones adapt well in the hydrophobic pocket of the inhibitor binding cavity (IBC) of MAO-A. Shifting of MAO-A selectivity was revealed after the introduction of an electron withdrawing nitro group. The presence of halogens such as chlorine, bromine and fluorine at the para position of aryl thiosemicarbazones had no impact on MAO-B inhibition. In particular, shifting of the fluorine atom to the meta position results in more potent MAO-B inhibition (SB11) with a Ki value of 0.12 ± 0.006 μM. This inhibition constant value was found to be better than that of the standard hMAO-B inhibitor (irreversible type) which was reported previously by our research group.42–50
Kinetics
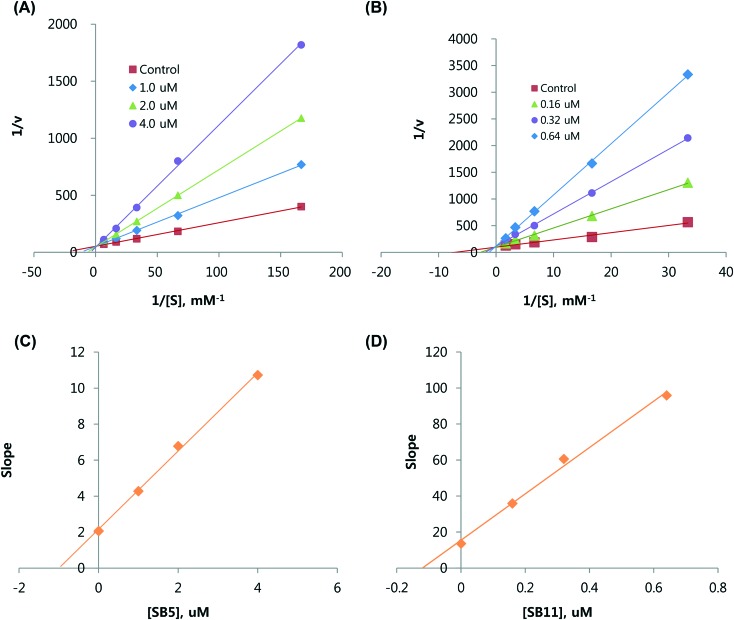
The inhibition modes of SB5 and SB11 for MAO-A and MAO-B, respectively, were analyzed using Lineweaver–Burk plots. The plots for SB5 and SB11 were linear and intersected the y-axis (Fig. 3A and C). The Ki values determined from the secondary plot (the slopes of the Lineweaver–Burk plots vs. the inhibitor concentrations) of MAO-A inhibition by SB5 and MAO-B inhibition by SB11 were 0.97 ± 0.042 and 0.12 ± 0.006 μM, respectively (Table 1, Fig. 3B and D). These results indicate that SB5 and SB11 are selective and reversible competitive inhibitors of MAO-A and MAO-B, respectively.
Fig. 3. Kinetic analyses of inhibition of MAO-A by SB5 (A) and of MAO-B by SB11 (C) using Lineweaver–Burk plots, and their respective secondary plots of slopes vs. inhibitor concentrations of SB5 (B) and SB11 (D).
Reversibility studies
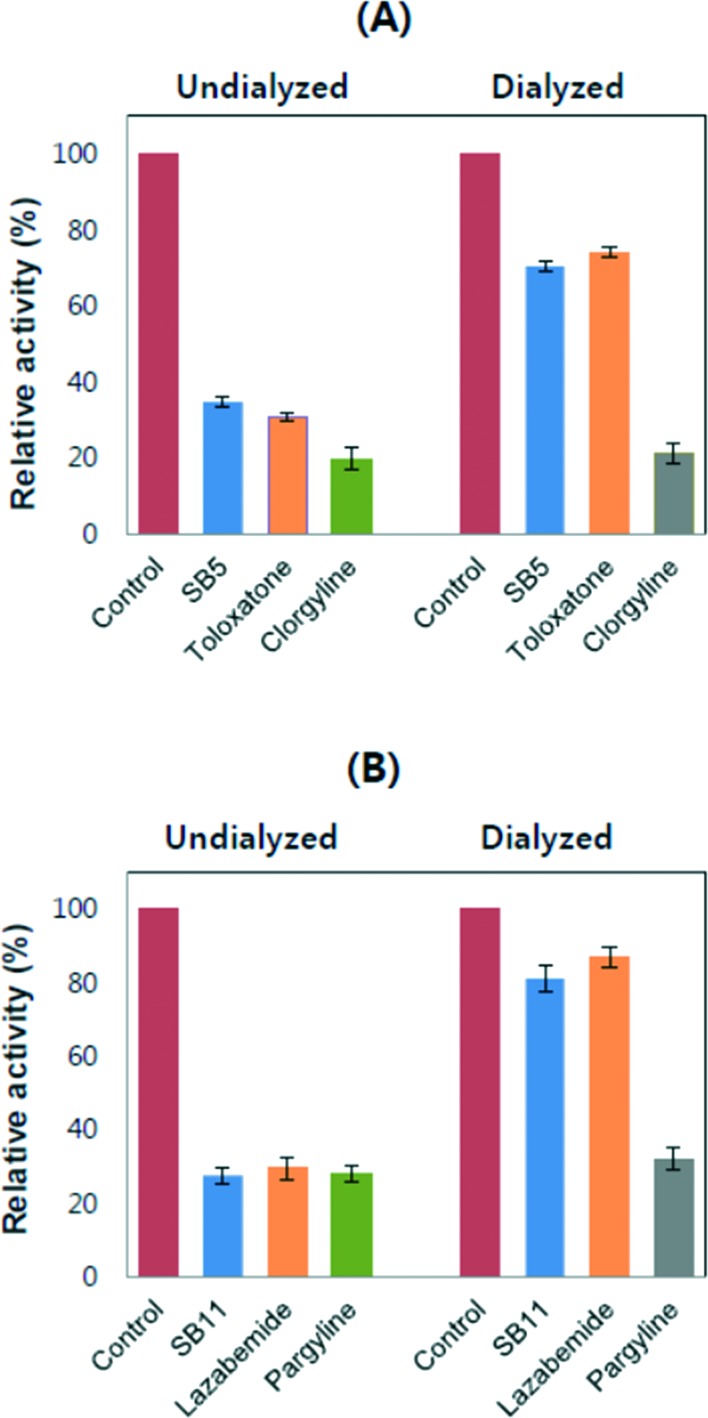
No changes in the residual activities were observed when SB5 and SB11 were preincubated with MAO-A and MAO-B, respectively, for up to 30 min. In the reversibility experiments, the AU and AD values obtained using SB5 for MAO-A were 34.7% and 70.3%, respectively (Fig. 4A). The values for toloxatone (a reversible inhibitor) reference experiments for MAO-A were 30.7% and 74.0%, respectively, and the values for clorgyline (an irreversible inhibitor) were 21.4% and 19.8%, respectively. The inhibition by toloxatone was greatly recovered by dialysis, while inhibition by clorgyline was not recovered. Similar to these results, the activity by SB5 was recovered close to the reversible reference level. The enzymatic activity of MAO-B by SB11 revealed AU and AD values of 27.4% and 81.0%, respectively (Fig. 4B); the values for lazabemide were 29.5% and 86.9%, respectively, and those for pargyline were 27.9% and 32.0%, respectively. MAO-B inhibition by pargyline was not recovered by dialysis, whereas the activity by lazabemide was greatly recovered, similar to the recovery to near reference levels of the inhibitory activity by SB11. These results indicate that analogues SB5 and SB11 are reversible inhibitors of MAO-A and MAO-B, respectively.
Fig. 4. Reversibility of MAO enzymes by aryl substituted thiosemicarbazones. MAO-A and MAO-B were inhibited at approximately 2 × IC50 by SB5 (A) and SB11 (B), respectively, and the activities were recovered by dialysis experiments against 100 mM sodium phosphate (pH 7.2) before measuring the residual activities. Concentrations of inhibitors and references used: SB5, 3.6 μM; toloxatone, 2.0 μM; clorgyline, 0.014 μM; SB11, 0.54 μM; lazabemide, 0.08 μM; pargyline, 0.20 μM.
Acetylcholinesterase inhibition
As presented in Table 1, it was noted that all the compounds are moderate and less potent than the reference compound tacrine for AChE inhibition. Considering the type of substitution at the phenyl ring, the inhibitory potency against AChE clearly favored the electron withdrawing nitro group at the para position of the phenyl system (SB7, IC50 15.61 ± 0.057 μM). According to the data, the presence of a chlorine or hydroxyl group is not crucial for imparting inhibitory potential against AChE in aryl thiosemicarbazones. All the fluorinated thiosemicarbazones showed moderate AChE inhibition. The ortho- and meta-substituted analogues SB12 and SB11 show slightly higher AChE inhibitory activities compared to the para-substituted analogue SB10, likely being MAO-B inhibition.
Blood–brain barrier (BBB) permeation assay
An essential requirement for successful CNS drugs is the ability to cross the BBB, which is determined using the parallel artificial membrane permeation assay (PAMPA). According to the limits established by Di et al., BBB permeation test compounds are classified as follows:51CNS + (high BB permeation predicted) : Pe(×10–6 cms–1) – >4.00CNS – (high BB permeation predicted) : Pe(×10–6 cms–1) – >2.00
Table 2 shows the permeability results of the PAMPA-BBB assay of commercial drugs and the 6 top ranked aryl thiosemicarbazones. Our results indicate that all the tested thiosemicarbazones are capable of crossing the BBB to target the MAO-A and MAO-B enzymes in the central nervous system (CNS), which is consistent with our design strategy.
Table 2. Results of PAMPA-BBB assay of aryl thiosemicarbazones and commercial drugs.
| Compounds a | Bibliography | Experimental | Prediction |
| Pe b (× 10–6 cms–1) | Pe c (× 10–6 cms–1) | ||
| SB3 | — | 11.24 ± 0.44 | CNS+ |
| SB5 | — | 10.14 ± 0.56 | CNS+ |
| SB6 | — | 09.44 ± 0.65 | CNS+ |
| SB7 | — | 12.78 ± 0.45 | CNS+ |
| SB11 | — | 13.12 ± 0.54 | CNS+ |
| SB12 | — | 10.33 ± 0.22 | CNS+ |
| Testosterone | 17.0 | 17.33 ± 0.12 | CNS+ |
| Progesterone | 9.3 | 08.13 ± 0.42 | CNS+ |
| Dopamine | 0.2 | 0.21 ± 0.01 | CNS– |
| Hydrocortisone | 1.8 | 1.71 ± 0.02 | CNS– |
aCompounds were dissolved in DMSO to a concentration of 5 mg mL–1 and diluted with PBS/EtOH (70 : 30). The final concentration of the compound was 100 μg mL–1.
bTaken from ref. 51.
cValues are expressed as the mean ± SEM of three independent experiments.
Molecular docking
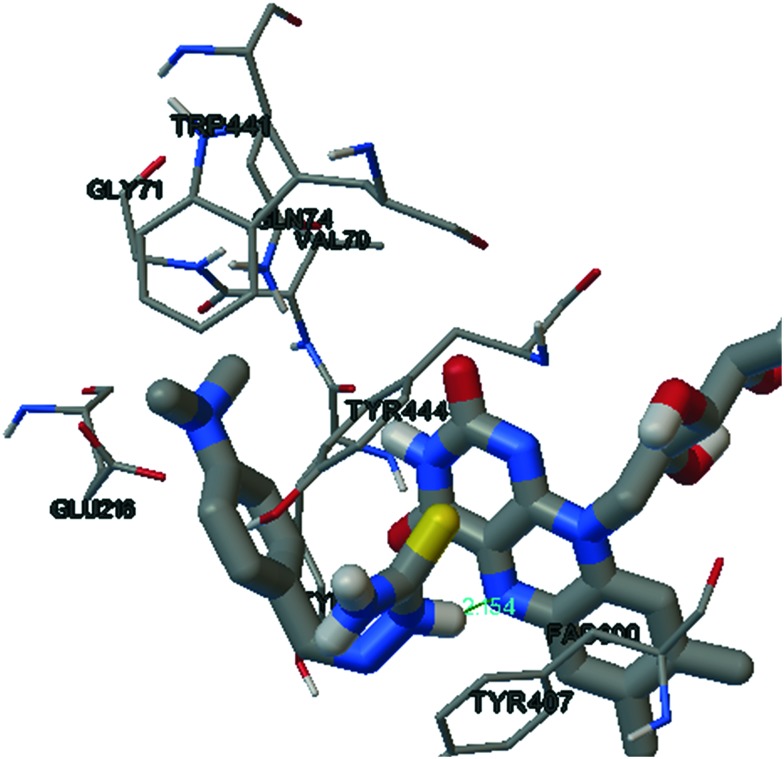
From the in vitro results, it is evident that compounds SB5 and SB11 show a good inhibitory profile towards MAO-A and MAO-B, respectively, in the micro molar range. We therefore attempted to investigate the hypothetical binding modes of the respective compounds in the IBC of isoenzymes. Of the 50 runs in the docking methodology, we selected the highest binding energy in the largest cluster for the hypothetical binding pose. The binding mode of SB5 (an hMAO-A inhibitor) is shown in Fig. 5. The presence of imino nitrogen and a terminal amino group in SB5 contributes significant hydrogen bonding interactions with the TYR444 and the N5 atom of the flavin adenine dinucleotide (FAD) unit of MAO-A, respectively. SB5 adopts an ‘L’ type configuration in which the aryl thiosemicarbazone unit is accommodated by the facing FAD unit with a hydrogen bond of distance 2.15 Å surrounded by the aromatic cage of TYR444 and TYR407. The position and close proximity towards the FAD unit of SB5 may enhance its binding energy towards MAO-A.
Fig. 5. SB5 in the active site of MAO-A.
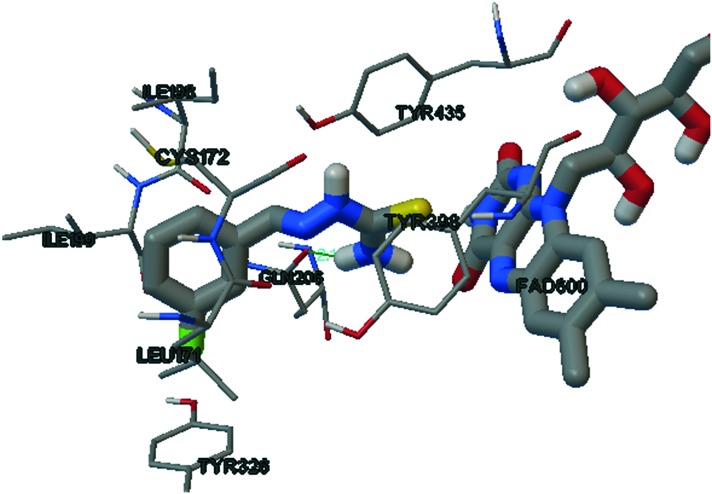
The binding mode of the potent MAO-B inhibitor SB11 is shown in Fig. 6. The entrance cavity of MAO-B leading to the substrate cavity is hydrophobic in nature.52 ILE199 and TYR326 are the side chains responsible for the separation and fusion between the entrance and substrate cavity, depending on the nature of the bound inhibitor.53–56 The meta substituted fluorine of the phenyl system of SB11 is efficiently accommodated in the entrance cavity of MAO-B. This lipophilic environment enhances the binding affinity of SB11 towards the IBC of MAO-B. The terminal amino group of thiosemicarbazone shows significant hydrogen bonding with GLN206 and the electron rich thiocarbamoyl group of SB11 surrounded by the aromatic cage of TYR398 and TYR435 nearer to the FAD unit.
Fig. 6. SB11 in the active site of MAO-B.
Conclusions
To summarize our results, various aryl thiosemicarbazones with different electron donating and withdrawing environments were synthesized, characterized and evaluated for their MAO inhibitory and blood brain barrier permeation potential. The representative compounds SB5 and SB11 were potent for MAO-A and MAO-B, respectively, with a reversible and competitive mode of inhibition, having IC50 values of 1.82 ± 0.14 and 0.27 ± 0.015 μM, respectively. The results explicate that the nature and orientation of groups on the aryl system of the title scaffold can bestow a significant selectivity profile on both MAO-A and MAO-B. The SAR revealed that the presence of an electron donating bulky group produces good selectivity towards MAO, and at the same the time an electron withdrawing nitro group in the same position shifts the selectivity to MAO-B. The presence of a halogen at the para position of the phenyl ring has no impact on MAO inhibition, but the shifting of fluorine to the meta position dramatically results in good MAO-B inhibition with a high selectivity index. Compounds SB5, SB7 and SB11 show moderate inhibition against acetylcholinesterase with IC50 values of 35.35 ± 0.47, 15.61 ± 0.057 and 26.61 ± 0.338 μM, respectively. PAMPA studies revealed that the representative molecules are able to cross the blood–brain barrier, which is a pre-requisite of CNS drug design for the treatment of various neurodegenerative and psychiatric disorders. Molecular modelling studies identified that the presence of the imino nitrogen and terminal amino group of SB5 contributed significant hydrogen bonding interactions with the TYR444 and N5 atom of the flavin adenine dinucleotide (FAD) unit of MAO-A, and the m-fluorine of the phenyl system of SB11 was efficiently accommodated in the entrance cavity of the MAO-B. Also, the terminal amino group of thiosemicarbazone formed significant hydrogen bonding with GLN206 of MAO-B. This study has thus provided new insights into the SARs of various aryl thiosemicarbazone compounds towards MAO inhibition and could possibly afford new attractive and more promising multi-targeted ligands for the treatment of AD and PD.
Experimental
Chemistry
A mixture of thiosemicarbazide hydrochloride and substituted benzaldehyde in the presence of catalytic acetic acid was stirred for 3–4 hours. The resultant mixture was refluxed for 4–5 hours and poured onto crushed ice. The formed solid was washed with water until it was free from the acid, filtered and crystallized with ethanol. The following 13 phenyl substituted thiosemicarbazones (SB1–SB13) were obtained:
(2E)-2-Benzylidenehydrazine-1-carbothioamide (SB1)
Yellowish white; yield: 76%; m.p.: 126–128 °C. 1H NMR (400 MHz, CDCl3) δ: 6.40 (s, 1H, NH2), 7.23 (s, 1H, NH2), 7.44–7.42 (m, 3H, J = 8 Hz, Ar–H), 7.66–7.64 (d, 2H, J = 8 Hz, Ar–H), 7.86 (s, 1H, –CH N–), 9.60 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 182.6 (C S), 142.8 (CH N), 134.3 (Ar–C1), 131.0 (Ar–C4), 129.3 (Ar–C2 & Ar–C6), 128.6 (Ar–C3 & Ar–C5). ESI-MS (m/z): calculated-179.24, observed-361.23.
(2E)-2-[(4-Hydroxyphenyl)methylidene]hydrazine-1-carbothioamide (SB2)
Pale yellow; yield: 63%; m.p.: 210–212 °C. 1H NMR (400 MHz, CDCl3) δ: 5.21 (s, IH, Ar–OH) 6.77, (s, 1H, NH2), 6.94–6.92 (d, 2H, J = 8 Hz, Ar–H), 7.28 (s, 1H, NH2), 7.66–7.64 (d, 2H, J = 8 Hz, Ar–H), 7.77 (s, 1H, –CH N–), 9.56 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.3 (C S), 160.2 (Ar–C4), 142.37 (CH N), 130.3 (Ar–C2 & Ar–C6), 125.2 (Ar–C1), 118.2 (Ar–C3 & Ar–C5). ESI-MS (m/z): calculated-195.24, observed-195.23.
(2E)-2-[(4-Methoxyphenyl)methylidene]hydrazine-1-carbothioamide (SB3)
Yellowish; yield: 78%; m.p.: 150–152 °C. 1H NMR (400 MHz, CDCl3) δ: 3.84 (s, 3H, OCH3), 6.37 (s, 1H, NH2), 6.92–6.90 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.60–7.58 (d, 2H, J = 8 Hz, Ar–H), 7.83 (s, 1H, –CH N–), 9.66 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.4 (C S), 161.9 (Ar–C4), 145.2 (CH N), 129.8 (Ar–C2 & Ar–C6), 128.9 (Ar–C1), 119.3 (Ar–C3 & Ar–C5), 55.4 (OCH3). ESI-MS (m/z): calculated-209.26, observed-209.25.
(2E)-2-[(4-Methylphenyl)methylidene]hydrazine-1-carbothioamide (SB4)
White; yield: 82%; m.p.: 155–157 °C. 1H NMR (400 MHz, CDCl3) δ: 2.38 (s, 3H, CH3), 6.46 (s, 1H, NH2), 7.21–7.19 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.54–7.52 (d, 2H, J = 8 Hz, Ar–H), 7.88 (s, 1H, –CH N–), 9.88 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.6 (C S), 145.3 (CH N), 142.1 (Ar–C4), 132.2 (Ar–C1), 131.7 (Ar–C2 & Ar–C6), 131.4 (Ar–C3 & Ar–C5), 21.6 (CH3). ESI-MS (m/z): calculated-193.26, observed-193.25.
(2E)-2-{[4-(Dimethylamino)phenyl]methylidene}hydrazine-1-carbothioamide (SB5)
Yellowish; yield: 81%; m.p.: 190–192 °C. 1H NMR (400 MHz, CDCl3) δ: 3.03 (s, 6H, N(CH3)2), 6.23 (s, 1H, NH2), 6.68–6.66 (d, 2H, J = 12 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.52–7.49 (d, 2H, J = 12 Hz, Ar–H), 7.71 (s, 1H, –CH N–), 9.18 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.7 (C S), 151.8 (Ar–C4), 141.4 (CH–N), 132.0 (Ar–C2 & Ar–C6), 123.5 (Ar–C1), 114.4 (Ar–C3 & Ar–C5), 41.9 (N–(CH3)2). ESI-MS (m/z): calculated-222.30, observed-222.29.
(2E)-2-[(4-Ethylphenyl)methylidene]hydrazine-1-carbothioamide (SB6)
White; yield: 84%; m.p.: 125–126 °C. 1H NMR (400 MHz, CDCl3) δ: 1.24–1.22 (t, 3H, J = 8 Hz, CH3), 2.70–2.68 (q, 2H, J = 8 Hz, CH2), 6.47 (s, 1H, NH2), 7.24–7.22 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.57–7.55 (d, 2H, J = 8 Hz, Ar–H), 7.89 (s, 1H, –CH N–), 9.92 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.2 (C S), 145.99 (CH N), 137.3 (Ar–C4), 130.5 (Ar–C1), 129.8 (Ar–C2 & Ar–C6), 128.8 (Ar–C3 & Ar–C5), 33.5 (CH2), 14.8 (CH3). ESI-MS (m/z): calculated-207.29, observed-207.00.
(2E)-2-[(4-Nitrophenyl)methylidene]hydrazine-1-carbothioamide (SB7)
Turmeric yellow; yield: 82%; m.p.: 220–222 °C. 1H NMR (400 MHz, CDCl3) δ: 6.23 (s, 1H, NH2), 6.68–6.66 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.52–7.49 (d, 2H, J = 12 Hz, Ar–H), 7.71 (s, 1H, –CH N–), 9.20 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 183.9 (C S), 150.2 (Ar–C4), 143.4 (CH N), 136.9 (Ar–C1), 131.3 (Ar–C2 & Ar–C6), 124.5 (Ar–C3 & Ar–C5). ESI-MS (m/z): calculated-224.23, observed-224.22.
(2E)-2-[(4-Chlorophenyl)methylidene]hydrazine-1-carbothioamide (SB8)
Pale white; yield: 83%; m.p.: 175–177 °C. 1H NMR (400 MHz, CDCl3) δ: 7.25 (s, 1H, NH2), 7.40–7.38 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.59–7.57 (d, 2H, J = 12 Hz, Ar–H), 7.78 (s, 1H, –CH N–), 9.33 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 184.3 (C S), 142.9 (CH N), 137.3 (Ar–C4), 131.3 (Ar–C1), 130.6 (Ar–C2 & Ar–C6), 130.5 (Ar–C3 & Ar–C5). ESI-MS (m/z): calculated-213.68, observed-213.67.
(2E)-2-[(4-Bromophenyl)methylidene]hydrazine-1-carbothioamide (SB9)
White; yield: 85%; m.p.: 140–142 °C. 1H NMR (400 MHz, CDCl3) δ: 6.39 (s, 1H, NH2), 7.26 (s, 1H, NH2), 7.56–7.54 (d, 4H, J = 8 Hz, Ar–H), 7.76 (s, 1H, –CH N–), 9.29 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 182.0 (C S), 142.9 (CH N), 131.2 (Ar–C1), 130.3 (Ar–C2 & Ar–C6), 130.2 (Ar–C3 & Ar–C5), 121.6 (Ar–C4).ESI-MS (m/z): calculated-258.13, observed-258.13.
(2E)-2-[(4-Fluorophenyl)methylidene]hydrazine-1-carbothioamide (SB10)
White; yield: 82%; m.p.: 120–122 °C. 1H NMR (400 MHz, CDCl3) δ: 6.38 (s, 1H, NH2), 7.40–7.38 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.59–7.57 (d, 2H, J = 12 Hz, Ar–H), 7.78 (s, 1H, –CH N–), 9.33 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 180.6 (C S), 160.3 (Ar–C4), 142.2 (CH N), 131.3 (Ar–C1), 130.6 (Ar–C2 & Ar–C6), 125.6 (d, JC–F = 62 Hz, Ar–C3 & Ar–C5). ESI-MS (m/z): calculated-197.23, observed-197.23.
(2E)-2-[(3-Fluorophenyl)methylidene]hydrazine-1-carbothioamide (SB11)
Pinkish white; yield: 79%; m.p.: 155–157 °C. 1H NMR (400 MHz, CDCl3) δ: 6.39 (s, 1H, NH2), 7.26 (s, 1H, NH2), 7.56–7.54 (m, 4H, J = 8 Hz, Ar–H), 7.76 (s, 1H, –CH N–), 9.29 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 182.2 (C S), 166.2 (Ar–C3), 140.1 (CH N), 136.6, (Ar–C5), 134.3 (Ar–C1), 125.3 (Ar–C6), 122.9 (d, JC–F = 64 Hz, Ar–C4), 121.6 (d, JC–F = 68 Hz, Ar–C2). ESI-MS (m/z): calculated-197.23, observed-197.23.
(2E)-2-[(2-Fluorophenyl)methylidene]hydrazine-1-carbothioamide (SB12)
Yellowish grey; yield: 82%; m.p.: 121–123 °C. 1H NMR (400 MHz, CDCl3) δ: 1H NMR (400 MHz, CDCl3) δ: 6.38 (s, 1H, NH2), 7.12–7.10 (m, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.65–7.63 (m, 2H, J = 8 Hz, Ar–H), 7.85 (s, 1H, –CH N–), 9.66 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 182.1 (C S), 164.1 (Ar–C2), 143.6 (CH N), 134.3, (d, JC–F = 68 Hz, Ar–C4), 130.2 (Ar–C6), 125.2 (Ar–C5), 122.6 (Ar–C1), 121.5 (Ar–C3). ESI-MS (m/z): calculated-197.23, observed-197.22.
(2E)-2-{[4-(Trifluoromethyl)phenyl]methylidene}hydrazine-1-carbothioamide (SB13)
Grey; yield: 79%; m.p.: 135–137 °C. 1H NMR (400 MHz, CDCl3) δ: 1H NMR (400 MHz, CDCl3) δ: 6.44 (s, 1H, NH2), 7.11–7.09 (d, 2H, J = 8 Hz, Ar–H), 7.26 (s, 1H, NH2), 7.37–7.35 (d, 2H, J = 12 Hz, Ar–H), 7.87 (s, 1H, –CH N–), 9.83 (s, 1H, N–NH–C S). 13C-NMR (100 MHz, CDCl3) δ: 181.4 (C S), 160.3 (Ar–C4), 142.1 (CH N), 138.4 (Ar–C1), 135.6 (Ar–C4), 129.2 (Ar–C2 & Ar–C6), 124.3 (q, JC–F = 278 Hz, Ar–C3 & Ar–C5), 123.6 (CF3). ESI-MS (m/z): calculated-247.24, observed-247.23.
Monoamine oxidase inhibition studies
Enzyme assays
Chemicals and enzymes were used as described previously. MAO activities were assayed by the continuous method using 0.06 mM kynuramine for MAO-A and 0.3 mM benzylamine for MAO-B as substrates, and reaction rates were expressed as absorbance change per min. The Km values of kynuramine and benzylamine obtained in this study were 0.040 mM and 0.15 mM, respectively, and the substrate concentrations used were 1.5 × and 2.0 × Km values, respectively.57
Analysis of inhibitory activities and enzyme kinetics
The inhibitions of MAO-A or MAO-B activities by the 13 compounds were primarily analyzed at a concentration of 10 μM. The IC50 values were then determined for 6 compounds showing more than 50% inhibitory activity, along with the reference compounds for reversible and irreversible inhibitors. Two potent compounds, SB5 for MAO-A and SB11 for MAO-B, were further investigated for time-dependent inhibition, kinetic studies for assessing the inhibition types, and Ki values of the compounds, as previously described.58
Analysis of reversibility of the inhibitors
Reversibility experiments for the potent inhibitors were performed using the dialysis method, including reference compounds for reversible and irreversible inhibitors, as previously described. The experiments were conducted using 3.6 μM SB5 for MAO-A and 0.54 μM SB11 for MAO-B in 100 mM sodium phosphate (pH 7.2) after preincubation for 30 min. Residual activities for undialyzed and dialyzed experiments were measured, and the relative activities for undialyzed (AU) and dialyzed (AD) experiments were calculated by comparing with each control without an inhibitor. The reversibility pattern was determined by comparing the relative AU and AD values.59
Acetylcholinesterase inhibition
AChE inhibitory activity was assayed using the method developed by Ellman et al., with slight modifications. The reaction was assayed for 10 min at 412 nm using 0.2 U ml–1 of AChE (Electrophorus electricus, type VI-S, Sigma) in 0.5 ml reaction mixture of 50 mM sodium phosphate (pH 7.5), in the presence of 0.5 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and 0.5 mM acetylthiocholine iodide (ACTI). For the measurement of inhibitory activity, each inhibitor (and tacrine as a reference) was preincubated for 15 min with the enzyme prior to addition of DTNB and ACTI.60
Blood–brain barrier (BBB) assay
The 6 top ranked synthesized thiosemicarbazones and known commercial drugs were dissolved in DMSO at a final concentration of 5 mg mL–1, followed by appropriate dilution with a 70 : 30 mixture of phosphate buffered saline solution and ethanol (PBS/EtOH) to give a final concentration of 25 μg mL–1. The filter membrane in the donor microplate was coated with polar brain lipid (PBL) dissolved in dodecane (4 μg mL–1, 20 mg mL–1). A total of 200 μL of diluted solution and 300 μL of PBS/EtOH (70 : 30) were added to the donor and the acceptor wells, respectively. The donor filter plate was carefully placed on the acceptor plate, and the sandwich system was kept at 25 °C for 16 h. The donor plate was carefully removed, and the concentrations of the compounds and the commercial drugs in the acceptor, donor and reference wells were measured with a UV plate reader.51
Molecular docking
AutoDock 4.2 software was employed for molecular docking studies of the lead molecules.61 Preparation Wizard of Maestro-8.4 (Schrodinger LLC) was used to prepare the protein. Crystallographic models 2BXR (hMAO-A) and 2BYB (hMAO-B) were downloaded from www.rcsb.org.62 Ligands were prepared through the PRODRG webserver (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg).63 Grid preparation was performed and the docking parameters were prepared on the basis of a reported method.64
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea, which is funded by the Ministry of Education (2017R1D1A3B03028559 granted to H. Kim).
Footnotes
†Electronic supplementary information (ESI) available. CCDC 1852751. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8md00399h
References
- Tripton K. F. Cell Biochem. Funct. 1986;4:79–87. doi: 10.1002/cbf.290040202. [DOI] [PubMed] [Google Scholar]
- Ramsay R. R. Curr. Pharm. Des. 2013;19:2529–2539. doi: 10.2174/1381612811319140004. [DOI] [PubMed] [Google Scholar]
- Kumar B., Sheetal, Mantha A. K., Kumar V. RSC Adv. 2016;6:42660–42683. [Google Scholar]
- Carradori S., Secci D., Bolasco A., Chiment P., D' Asceenzio M. Expert Opin. Ther. Pat. 2012;22:909–939. doi: 10.1517/13543776.2012.698613. [DOI] [PubMed] [Google Scholar]
- Mathew B., Suresh J., Mathew G. E., Rasheed S. A., Vilapurathu J. K., Jayaraj P. Curr. Enzyme Inhib. 2015;11:108–115. [Google Scholar]
- Youdim M. B., Bakhle Y. S. Br. J. Pharmacol. 2006;147:S287–S296. doi: 10.1038/sj.bjp.0706464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdim M. B., Edmondson D., Tripton K. F. Nat. Rev. Neurosci. 2006;7:295–309. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- Riederer P., Muller T. Expert Opin. Drug Metab. Toxicol. 2017;13:233–240. doi: 10.1080/17425255.2017.1273901. [DOI] [PubMed] [Google Scholar]
- Mathew B., Dev S., Joy M., Mathew G. E. ChemistrySelect. 2017;2:11645–11652. [Google Scholar]
- Binda C., Milczek E. M., Bonivento D., Wang J., Mattevi A., Edmondson D. E. Curr. Top. Med. Chem. 2011;11:2788–2796. doi: 10.2174/156802611798184355. [DOI] [PubMed] [Google Scholar]
- Pisani L., Catto M., Leonetti F., Nicolotti O., Stefanachi A., Campagna F., Carotti A. Curr. Med. Chem. 2011;18:4568–4587. doi: 10.2174/092986711797379302. [DOI] [PubMed] [Google Scholar]
- Youdim M. B. H., Curr M. B. H. Alzheimers Res. Ther. 2006;3:541–550. doi: 10.2174/156720506779025288. [DOI] [PubMed] [Google Scholar]
- Carradori S., Silvestri R. J. Med. Chem. 2015;85:6717–6732. doi: 10.1021/jm501690r. [DOI] [PubMed] [Google Scholar]
- Binda C., Li M., Hubaìlek F., Restelli N., Edmondson D. E., Mattevi A. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdim M. B., Weinstock M. NeuroToxicology. 2004;25:243–250. doi: 10.1016/S0161-813X(03)00103-7. [DOI] [PubMed] [Google Scholar]
- Sieradzan K., Channon S., Ramponi C., Stern G. M., Lees A. J., Youdim M. B. J. Clin. Psychopharmacol. 1995;15:51S–59S. doi: 10.1097/00004714-199508001-00010. [DOI] [PubMed] [Google Scholar]
- Mathew B., Haridas A., Suresh J., Mathew G. E., Ucar G., Jayaprakash V. Cent. Nerv. Syst. Agents Med. Chem. 2016;16:120–136. doi: 10.2174/1871524915666151002124443. [DOI] [PubMed] [Google Scholar]
- Matos M. J., Vina D., Vazque-Rodrigues S., Uriarte E., Santana L. Curr. Top. Med. Chem. 2012;12:2210–2239. doi: 10.2174/156802612805220002. [DOI] [PubMed] [Google Scholar]
- Mathew B., Mathew G. E., Petzer J. P., Petzer A. Comb. Chem. High Throughput Screening. 2017;20:522–532. doi: 10.2174/1386207320666170227155517. [DOI] [PubMed] [Google Scholar]
- Kasayuki Uda S. J. Heterocycl. Chem. 1979;16:1273–1278. [Google Scholar]
- Zelenin K. N., Kuzenetsova O. R., Alekseyev V. V., Terentyev P. B., Ovacharenko V. V. Tetrahedron. 1993;49:1257–1270. [Google Scholar]
- Yu Y., Kalinowski D. S., Kovacevic Z., Siafakas A. R., Jansson P. J., Stefani C. J. Med. Chem. 2009;52:5271–5294. doi: 10.1021/jm900552r. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Pandaey B. R., Barthwal J. P., Kishor K., Bhargava K. P. Res. Commun. Chem. Pathol. Pharmacol. 1978;22:291–300. [PubMed] [Google Scholar]
- Agarwal D. K., Pandaey B. R. Res. Commun. Chem. Pathol. Pharmacol. 1979;26:525–531. [Google Scholar]
- Chimenti F., Maccioni E., Secci D., Bolasco A., Chimenti A., Granese A. J. Med. Chem. 2008;51:4874–4880. doi: 10.1021/jm800132g. [DOI] [PubMed] [Google Scholar]
- Tripathi R. K. P., Goshain O., Ayyannan S. R. ChemMedChem. 2013;8:462–474. doi: 10.1002/cmdc.201200484. [DOI] [PubMed] [Google Scholar]
- Tripathi R. K. P., Rai G. K., Ayyannan S. R. ChemMedChem. 2016;11:1145–1160. doi: 10.1002/cmdc.201600128. [DOI] [PubMed] [Google Scholar]
- Tripathi R. K. P., Sasi M., Gupta S. K., Krishnamurthy S., Ayyannan S. R., Enzyme Inhib J. Med. Chem. 2018;33:37–57. doi: 10.1080/14756366.2017.1389920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatello R., Mazzone S., Castelli F., Mazzone P., Raciti G., Mazzone G. Pharmazie. 1994;49:272–276. [PubMed] [Google Scholar]
- Gokhan N., Yesilada A., Ucar G., Erol K., Bilgin A. A. Arch. Pharm. 2003;336:362–371. doi: 10.1002/ardp.200300732. [DOI] [PubMed] [Google Scholar]
- Ucar G., Gokhan N., Yesilada A., Bilgin A. A. Neurosci. Lett. 2005;382:327–331. doi: 10.1016/j.neulet.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Yabanoglu S., Ucar G., Gokhan S., Salgin U., Yesilada A., Bilgin A. A. J. Neural Transm. 2007;114:769–773. doi: 10.1007/s00702-007-0686-8. [DOI] [PubMed] [Google Scholar]
- Secci D., Carradori S., Bolasco A., Bizzarri B., D'Ascenzio M., Maccioni E. Curr. Top. Med. Chem. 2012;12:2240–2257. doi: 10.2174/156802612805220057. [DOI] [PubMed] [Google Scholar]
- Mathew B., Suresh J., Anbazhagan S., Mathew G. E. Cent. Nerv. Syst. Agents Med. Chem. 2013;13:195–206. doi: 10.2174/1871524914666140129122632. [DOI] [PubMed] [Google Scholar]
- Mathew B., Suresh J., Ahasan M. J., Mathew G. E., Usman D., Subramanyan P. N. Infect. Disord.: Drug Targets. 2015;15:76–88. doi: 10.2174/1871526515666150724104411. [DOI] [PubMed] [Google Scholar]
- Vina D., Matos M. J., Yanez M., Santana L., Uriarte E. Med. Chem. Commun. 2012;3:213–218. [Google Scholar]
- Geldenhuys W. J., Ko K. S., Stinneft H., Van der Schyf C. J. Med. Chem. Commun. 2011;2:1099–1103. [Google Scholar]
- Soares N. A., Almeida M. A., Marins-Goulart C., Chaves O. A., Echevarria A., de Oliveria M. C. C. Bioorg. Med. Chem. Lett. 2017;27:3546–3550. doi: 10.1016/j.bmcl.2017.05.057. [DOI] [PubMed] [Google Scholar]
- Subhashree G. R., Haribabu J., Saranya S., Yuvaraj P., Krishnan D., Karvenbu R. J. Mol. Struct. 2017;1145:160–169. [Google Scholar]
- Liciano P., Moraes C. B., Alcantara L. M., Franco C. H., Pascoalino B., Freitas-Junior L. H. Eur. J. Med. Chem. 2018;146:423–434. doi: 10.1016/j.ejmech.2018.01.043. [DOI] [PubMed] [Google Scholar]
- Sasidharan R., Baek S. C., Manju S. L., Kim H., Mathew B. Biomed. Pharmacother. 2018;106:8–13. doi: 10.1016/j.biopha.2018.06.064. [DOI] [PubMed] [Google Scholar]
- Mathew B., Ucar G., Ciftci S. Y., Baysal I., Suresh J., Mathew G. E., Vilapurathu J. K., Moosa N. A., Pullarottil N., Viswam L., Haridas A., Fathima F. Lett. Org. Chem. 2015;12:605–613. [Google Scholar]
- Mathew B., Mathew G. E., Ucar G., Baysal I., Suresh J., Vilapurathu J. K., Prakasan A., Suresh J. K., Thomas A. Bioorg. Chem. 2015;62:22–29. doi: 10.1016/j.bioorg.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Mathew B., Mathew G. E., Ucar G., Baysal I., Suresh J., Mathew S., Haridas A., Jayaprakash V. Chem. Biodiversity. 2016;13:1046–1052. doi: 10.1002/cbdv.201500367. [DOI] [PubMed] [Google Scholar]
- Mathew B., Ucar G., Mathew G. E., Mathew S., Purapurath P. K., Moolayil F., Mohan S., Gupta S. V. ChemMedChem. 2016;11:2649–2655. doi: 10.1002/cmdc.201600497. [DOI] [PubMed] [Google Scholar]
- Mathew B., Haridas A., Ucar G., Baysal I., Adeniyi A. A., Soliman M. E. S., Joy M., Mathew G. E., Lakshmanan B., Jayaprakash V. Int. J. Biol. Macromol. 2016;91:680–695. doi: 10.1016/j.ijbiomac.2016.05.110. [DOI] [PubMed] [Google Scholar]
- Mathew B., Haridas A., Ucar G., Baysal I., Joy M., Mathew G. E., Lakshmanan B., Jayaprakash V. ChemMedChem. 2016;11:1161–1171. doi: 10.1002/cmdc.201600122. [DOI] [PubMed] [Google Scholar]
- Sasidharan R., Manju S. L., Ucar G., Baysal I., Mathew B. Arch. Pharm. 2016;349:627–637. doi: 10.1002/ardp.201600088. [DOI] [PubMed] [Google Scholar]
- Mathew B., Mathew G. E., Ucar G., Mathew G. E., Nafna E. K., Lohidakshan K. L., Suresh J. Int. J. Biol. Macromol. 2017;104:1321–1329. doi: 10.1016/j.ijbiomac.2017.05.162. [DOI] [PubMed] [Google Scholar]
- Mathew B., Ucar G., Raphael C., Mathew G. E., Joy M., Machaba K. E., Soliman M. E. S. ChemistrySelect. 2017;2:11113–11119. [Google Scholar]
- Di L., Kerns E. H., Fan K., McConnell O. J., Carter G. T. Eur. J. Med. Chem. 2003;38:223–232. doi: 10.1016/s0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Mathew B., Adeniyi A. A., Dev S., Joy M., Ucar G., Mathew G. E., Pillay A. S., Soliman M. E. S. J. Phys. Chem. B. 2017;121:1186–1203. doi: 10.1021/acs.jpcb.6b09451. [DOI] [PubMed] [Google Scholar]
- Novaroli L., Daina A., Favre E., Bravo J., Carotti A., Leonetti F. J. Med. Chem. 2006;49:6264–6272. doi: 10.1021/jm060441e. [DOI] [PubMed] [Google Scholar]
- Legoabe L., Kruger J., Petzer A., Bergh J. J., Petzer J. P. Eur. J. Med. Chem. 2011;46:5162–5174. doi: 10.1016/j.ejmech.2011.08.036. [DOI] [PubMed] [Google Scholar]
- Lan S. J., Pan L. F., Xie S. S., Wang X. B., Kong L. Y. Med. Chem. Commun. 2015;6:592–600. [Google Scholar]
- Lan S. J., Zang T., Liu Y., Zahng Y., Hou J. W., Xie S. S. Med. Chem. Commun. 2017;8:471–478. [Google Scholar]
- Baek S. C., Lee H. W., Ryu H. W., Kang M. G., Park D., Kim S. H. Bioorg. Med. Chem. Lett. 2018;28:584–588. doi: 10.1016/j.bmcl.2018.01.049. [DOI] [PubMed] [Google Scholar]
- Suresh J., Baek S. C., Ramakrishnan S. P., Kim H., Mathew B. Int. J. Biol. Macromol. 2018;108:660–664. doi: 10.1016/j.ijbiomac.2017.11.159. [DOI] [PubMed] [Google Scholar]
- Lee H. W., Choi H., Nam S. J., Fenical W., Kim H. J. Microbiol. Biotechnol. 2017;27:785–790. doi: 10.4014/jmb.1612.12025. [DOI] [PubMed] [Google Scholar]
- Ellman G. L., Courtney K. D., Andres Jr. V., Feather-Stone R. M. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Colibus L., Li M., Binda C., Lustig A., Edmondson D. E., Mattevi A. Proc. Natl. Acad. Sci. U. S. A. 2005;102(36):12684–12689. doi: 10.1073/pnas.0505975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf A. W., van Aalten D. M. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Mathew B., Suresh J., Anbazhagan S., Dev S. Biomed. Aging Pathol. 2014;4:297–301. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.