Abstract
A proper inflammatory response is critical to the restoration of tissue homeostasis after injury or infection, but how such a response is modulated by the physical properties of the cellular and tissue microenvironments is not fully understood. Here, using H358, HeLa, and HEK293T cells, we report that cell density can modulate inflammatory responses through the Hippo signaling pathway. We found that NF-κΒ activation through the proinflammatory cytokines interleukin-1β (IL-1β) and tumor necrosis factor α (TNFα) is not affected by cell density. However, we also noted that specific NF-κΒ target genes, such as cyclooxygenase 2 (COX-2), are induced much less at low cell densities than at high cell densities. Mechanistically, we observed that the transcriptional coactivators Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) are localized to the nucleus, bind to TEA domain transcription factors (TEADs), recruit histone deacetylase 7 (HDAC7) to the promoter region of COX-2, and repress its transcription at low cell density and that high cell density abrogates this YAP/TAZ-mediated transcriptional repression. Of note, IL-1β stimulation promoted cell migration and invasion mainly through COX-2 induction, but YAP inhibited this induction and thus cell migration and invasion. These results suggest that YAP/TAZ–TEAD interactions can repress COX-2 transcription and thereby mediate cell density–dependent modulation of proinflammatory responses. Our findings highlight that the cellular microenvironment significantly influences inflammatory responses via the Hippo pathway.
Keywords: Hippo pathway, NF-κB, histone deacetylase (HDAC), inflammation, cytokine, cell signaling, Yes-associated protein (YAP), COX-2, TAZ, TEADs
Introduction
Inflammation is the body's primary protective response when tissues are damaged due to infection, injury, or other physical stresses. It is generally an acute response and can be resolved as soon as the offending agents are removed or otherwise countered, after which tissue structure and function are restored to homeostasis (1). Conceivably, cells and tissues integrate the physical properties of their microenvironments, such as cell density, cell geometry, extracellular matrix stiffness, and mechanical stretches, to properly calibrate an inflammatory response (2, 3). However, the failure of this process can lead to excessive tissue damage, making them prone to chronic inflammation (4), which has been linked to numerous chronic diseases, including type 2 diabetes, atherosclerosis, neurodegenerative diseases, and cancer (5, 6).
The NF-κΒ signaling pathway plays an essential role in executing an inflammatory response (7, 8). NF-κΒ is a transcription factor complex that is normally retained in the cytosol of resting cells by virtue of its binding to IκB inhibitory proteins, such as IκBα. In response to a variety of stimuli (e.g. proinflammatory cytokines interleukin-1β (IL-1β)2 and tumor necrosis factor α (TNFα)), transforming growth factor-β–activated kinase 1 (TAK1) and IκB kinase complexes are sequentially activated, leading to IκBα phosphorylation and subsequent ubiquitination and degradation. Consequently, NF-κΒ is freed and translocated into the nucleus where it binds to target genes and induces their expression (9). More than 200 genes are known to be activated by NF-κΒ, including chemokines, cytokines, adhesion molecules, inflammatory mediators, apoptosis inhibitors, and others (10). These genes perform a variety of essential functions. For example, cyclooxygenase-2 (COX-2), an inducible isozyme that catalyzes the first step in the synthesis of prostanoids, mediates an effective inflammatory response, whereas negative regulators of the NF-κΒ signaling pathway (such as IκBα and deubiquitinating enzymes A20 and CYLD) help resolve inflammation and reset the response to latency (11, 12).
The Hippo signaling pathway is critical in the control of organ size and has been implicated in tumorigenesis (13). This pathway includes the mammalian orthologs of Drosophila melanogaster Hippo kinase mammalian STE20-like protein kinases 1 (MST1; also known as STK4) and 2 (MST2; also known as STK3), large tumor suppressors 1 (LATS1) and 2 (LATS2), the transcriptional coactivators Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ; also known as WWTR1), and the TEA domain–containing sequence-specific transcription factors TEAD1–TEAD4 (14–22). Once the Hippo kinase module is activated, the upstream kinases MST1/2 are activated, which in turn phosphorylate and activate LATS1/2 (23, 24). LATS1/2 then phosphorylate YAP and TAZ (25, 26), leading to their cytoplasmic retention (27, 28). When this module is turned off, MST1/2 and LATS1/2 are inactivated, resulting in the dephosphorylation of YAP and TAZ and their translocation into the nucleus where they form active transcriptional complexes with TEADs to initiate target gene expression (22, 29, 30). The Hippo signaling pathway can be modulated by the physical properties of cell and tissue microenvironments (31, 32). For example, low cell density, a high degree of cell spreading, or stiff matrices will render MST1/2 and LATS1/2 inactive, leading to YAP/TAZ activation and their translocation into the nucleus. Conversely, high cell density, a low degree of cell spreading, or soft matrices will result in activation of MST1/2 and LATS1/2, which phosphorylate and inactivate YAP/TAZ (33, 34). Whether the Hippo signaling pathway can transduce physical properties of the cellular and tissue microenvironments to modulate inflammatory responses, however, has not been fully investigated.
Here, we report how the physical properties of the cell microenvironment can affect inflammatory responses. We used cell density as the model and studied its modulatory effects on the cell's proinflammatory responses. Cell density varies in different tissues and organs, which would respond differently to proinflammatory stimuli. In addition, cell density is a critical factor that affects tumor growth in which chronic inflammation is also implicated (35). We found that cell density strongly modulated the inflammatory responses, and YAP/TAZ, the effector proteins of the Hippo pathway, mediated the effects.
Results
Cell density modulates inflammatory responses
Cell density is an important physical property that influences cellular behaviors, but whether it modulates inflammatory responses is not fully understood. We examined its effect on the NF-κB signaling pathway by plating cells at different cell densities and stimulating them with IL-1β or TNFα. Neither the pattern of IκBα degradation and its resynthesis nor the induction of A20, a deubiquitinating enzyme and a negative regulator of NF-κΒ signaling that terminates NF-κΒ activation (11), was affected by cell confluence levels (Fig. S1, A and B), which indicates that cell density did not affect the activation of the NF-κΒ signaling pathway.
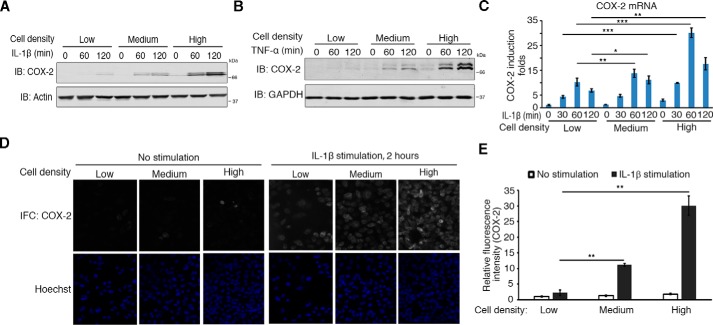
Given the importance of COX-2 in mediating inflammatory responses, we also checked its induction. Protein and mRNA levels of COX-2 both increased with the increment of cell densities (Fig. 1, A–E). However, the expression of COX-1, the constitutively expressed isoform of COXs, was not affected (Fig. S1B). This suggests cell density could modulate COX-2 induction in response to proinflammatory stimulation.
Figure 1.
Cell density modulates COX-2 induction. A and B, cell density modulates the induction of COX-2 at the protein level. H358 cells (about 2 × 105 cells) were seeded onto 10-, 6-, and 3.5-cm plates for low, medium, or high cell densities, respectively, for 6 h and then treated with 10 ng/ml IL-1β (A) or 10 ng/ml TNFα (B) for the times indicated. Cells were then harvested and analyzed by immunoblotting (IB) with the indicated antibodies. C, cell density modulates the induction of COX-2 at the transcriptional level. H358 cells (about 2 × 105 cells) were seeded onto 10-, 6-, and 3.5-cm plates for different cell densities and then treated with IL-1β for the times indicated after 6 h. Cells were then harvested for RNA extraction. The mRNA level of COX-2 was measured by qRT-PCR. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; **, p < 0.01; and ***, p < 0.001, by unpaired, two-tailed Student's t test. D and E, cell density modulates COX-2 induction. H358 cells were cultured on cover glasses in 24-well plates at different densities (2.5 × 104, 7.5 × 104, and 2.25 × 105 cells/well) for 6 h, stimulated with IL-1β for another 2 h, and fixed for immunofluorescence (IFC) staining for COX-2. Nuclei were counterstained with Hoechst (D). Relative mean fluorescence intensities of COX-2 of each cell were determined (E). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. **, p < 0.01, by unpaired, two-tailed Student's t test (E).
Other stress conditions also stimulate COX-2 induction. We used deferoxamine (DFO), a hypoxia-mimicking agent, and serum starvation as stimulators because both are known to induce the expression of COX-2 by activating, respectively, hypoxia-inducible factor-α and p38 signaling (stress-activated protein kinase) (36, 37). We found that both stimulators induced COX-2 expression, and its level was also dependent on cell density (Fig. S1, C and D).
YAP and TAZ mediate the modulation of COX-2 induction by cell density
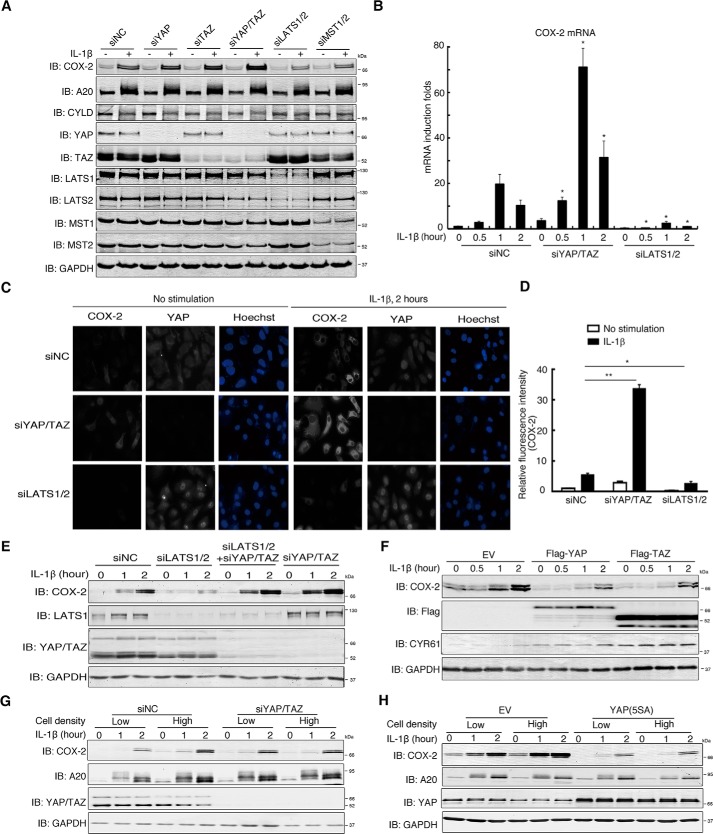
Cell density regulates the Hippo signaling pathway (38), so we investigated whether this pathway might mediate cell density–dependent modulation of COX-2 expression. For the Hippo pathway, inactivation or loss of MST1/2 and LATS1/2 leads to dephosphorylation and nuclear translocation of YAP/TAZ (22). We found that short interfering RNA (siRNA)-mediated knockdown of YAP/TAZ or LATS1/2 did not affect the activation of either ERK1/2 or the NF-κΒ pathways stimulated by IL-1β because the pattern of ERK activation, IκBα degradation and resynthesis, and A20 induction were similar among the control siRNA, siYAP/TAZ, and siLATS1/2 groups (Fig. 2A and Fig. S2A). Compared with control siRNA, however, IL-1β induced more COX-2 at both the protein and mRNA levels upon knockdown of YAP, TAZ, or both together but less COX-2 upon knockdown of MST1/2 or LATS1/2 (Fig. 2, A and B). Immunofluorescence staining also confirmed that, compared with control siRNA, knockdown of YAP/TAZ increased COX-2 induction, whereas knockdown of LATS1/2 decreased it, along with the relocation and accumulation of YAP in the nucleus (Fig. 2, C and D).
Figure 2.
The Hippo pathway mediates the effect of cell density on COX-2 induction. A, Hippo pathway regulates the induction of COX-2 by IL-1β. H358 cells were transfected with the indicated siRNAs and treated with IL-1β for 2 h. Cells were then lysed and analyzed by immunoblotting (IB) with the indicated antibodies. B, knockdown of YAP/TAZ enhances whereas knockdown of LATS1/2 diminishes the transcription of COX-2. H358 cells transfected with control siRNA or siRNA against YAP/TAZ or LATS1/2 were treated with IL-1β for the indicated duration of time. Cells were harvested, and total RNAs were extracted for measurement of COX-2 mRNA level by qRT-PCR. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05 (compared with siNC group at the same induction time point), by unpaired, two-tailed Student's t test. C and D, knockdown of YAP/TAZ enhances COX-2 induction by IL-1β, whereas knockdown of LATS1/2 diminishes it. H358 cells transfected with control siRNA or siRNA against YAP/TAZ or LATS1/2 were cultured onto cover glasses, treated with or without IL-1β for 2 h, and fixed for immunofluorescence staining using the indicated antibodies (C). The relative mean fluorescence intensity (anti-COX-2) of each cell was determined. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; and **, p < 0.01, by unpaired, two-tailed Student's t test (D). E, LATS1/2 regulate COX-2 induction through YAP and TAZ. H358 cells were transfected with siRNAs targeting LATS1/2, YAP/TAZ, or both and then treated with IL-1β for the indicated time. Cells were harvested for immunoblotting with the indicated antibodies. F, overexpression of YAP and TAZ attenuates the induction of COX-2 by IL-1β. HeLa cells were transfected with FLAG-YAP or FLAG-TAZ and treated with IL-1β for the time indicated. Cells were then lysed and analyzed by immunoblotting with the indicated antibodies. G, knockdown of YAP/TAZ abolishes the effect of cell density on COX-2 induction by IL-1β. H358 cells plated at low or high cell density were transfected with control siRNA or siRNAs against YAP and TAZ for 48 h and then stimulated with IL-1β for the time indicated. Cells were then lysed and analyzed by immunoblotting with the indicated antibodies. H, overexpression of YAP(5SA) abolishes the effect of cell density on COX-2 induction by IL-1β. H358 cells plated at low or high cell density were transfected with empty vector (EV) or a plasmid expressing the constitutively active YAP mutant YAP(5SA) for 24 h and then stimulated with IL-1β for the indicated duration of time. Cells were then lysed and analyzed by immunoblotting with the indicated antibodies. NC, negative control.
To exclude the possibility that LATS1/2 might regulate the COX-2 induction independently of YAP/TAZ, we transfected YAP/TAZ, LATS1/2 siRNAs, or both into cells. Compared with control siRNA, knockdown of LATS1/2 led to reduction of COX-2 induction, but this disappeared upon further knockdown of YAP/TAZ, which was similar to that of YAP/TAZ knockdown alone (Fig. 2E). These results suggest the modulation of COX-2 induction by LATS1/2 relied on YAP and TAZ. This was further reinforced by the fact that overexpression of YAP or TAZ led to a marked decrease of IL-1β–stimulated COX-2 induction (Fig. 2F). Remarkably, knockdown of YAP and TAZ or stably expressing YAP(5SA), a constitutively active form of YAP, completely abolished cell density–modulated COX-2 induction by IL-1β stimulation (Fig. 2, G and H). Knockdown of YAP and TAZ also strongly enhanced COX-2 induction by DFO or serum starvation, whereas knockdown of LATS1/2 suppressed it (Fig. S2, B and C). Hydrodynamic tail vein injection of YAP(5SA) caused liver tumors in mice over the course of 3 months (39). Notably, immunohistochemistry of the tumors revealed a negative correlation between the expression of YAP(5SA) and COX-2 (Fig. S2D).
To test for the possibility that YAP/TAZ indirectly inhibit COX-2 induction by inducing other genes to be expressed first, the latter then inhibiting COX-2 induction, we knocked down LATS1/2 in H358 cells and then treated them with cycloheximide (CHX), a translation inhibitor. Although the qRT-PCR results showed that CHX enhanced the induction of COX-2 by IL-1β stimulation (most likely by inhibiting the new synthesis of the IκBα protein), CHX did not block the suppression of COX-2 induction upon LATS1/2 knockdown (Fig. S2E). We also tested the possibility that microRNA could be involved but found that knocking down Dicer, the enzyme responsible for cleaving pre-miRNA into mature miRNA, did not affect the suppression of COX-2 induction upon LATS1/2 knockdown (Fig. S2F). These results suggest that new protein synthesis and microRNAs were irrelevant to the suppression function of YAP/TAZ. Together, these results demonstrate that the Hippo pathway mediated the cell density–dependent modulation of COX-2 induction by different stimulators and that YAP and TAZ furthermore functioned in direct transcriptional suppression.
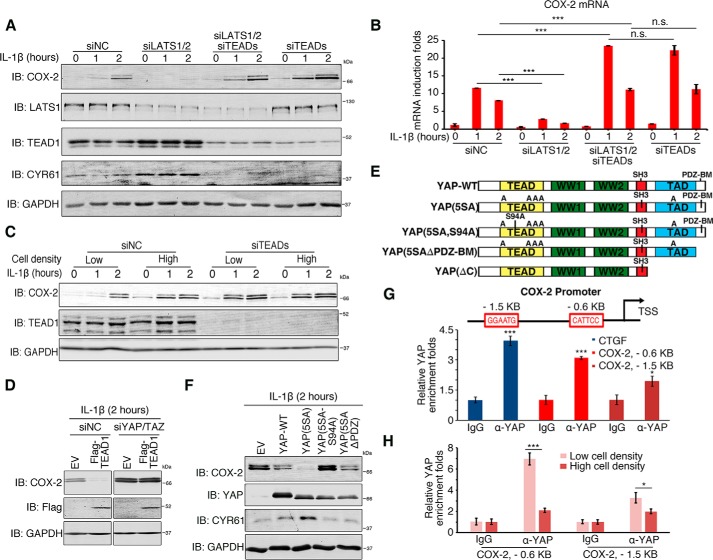
YAP and TAZ require TEADs to function as transcriptional corepressors
YAP and TAZ are transcriptional coactivators that facilitate the transcriptional activators TEADs (TEAD1–4) to induce gene expression (22), and we sought to determine whether the suppressive functions of YAP and TAZ depend on TEADs. Knocking down LATS1/2, which is functionally equivalent to YAP/TAZ activation, diminished COX-2 induction by IL-1β, serum starvation, and hypoxia. This effect, however, was abrogated in TEAD-depleted cells (Fig. 3, A and B, and Fig. S3, A and B). Knockdown of TEAD expression also disrupted the cell density–regulated COX-2 induction stimulated by IL-1β (Fig. 3C). The mRNA level of COX-1 and the knockdown efficiencies were validated using qRT-PCR (Fig. S3C). The mRNA level of connective tissue growth factor validated the dysregulation of the Hippo pathway upon knockdown of LATS1/2 and TEADs (Fig. S3C). Furthermore, overexpression of TEAD1 suppressed IL-1β–stimulated COX-2 induction, but this suppression was abolished when YAP and TAZ were knocked down (Fig. 3D). We therefore concluded that TEADs were needed for the suppression functions of YAP/TAZ.
Figure 3.
TEADs are required for YAP/TAZ-mediated suppression of COX-2 induction. A and B, lack of TEADs abolishes the suppression of COX-2 induction. H358 cells were transfected with control siRNA or with siRNAs against LATS1/2, TEADs, or both and treated with IL-1β for the indicated duration of time. Cells were then lysed and analyzed by immunoblotting (IB) (A) or by qRT-PCR (B). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. ***, p < 0.001; and n.s., not significant (p > 0.05), by unpaired, two-tailed Student's t test. C, lack of TEADs abolishes the effect of cell density on COX-2 induction. H358 cells plated at low or high densities were transfected with control siRNA or siRNA against TEADs and then stimulated with IL-1β for the indicated time. Cells were harvested and immunoblotted using the indicated antibodies. D, TEAD1 suppression of COX-2 induction depends on YAP/TAZ. HeLa cells were transfected with control siRNA or siRNAs against YAP and TAZ for 48 h followed by transient overexpression of empty vector (EV) or FLAG-tagged TEAD1 for 24 h. Cells were then stimulated with IL-1β for 2 h and harvested for immunoblotting using the indicated antibodies. E, diagram presents YAP mutants used in the study. F, YAP requires the association with TEADs to suppress COX-2 induction. HeLa cells were transfected with YAP WT or its mutants, stimulated with IL-1β, and harvested for immunoblotting with the indicated antibodies. G, YAP binds directly to the COX-2 promoter. Top, diagram showing the COX-2 gene promoter in the vicinity of its transcription start site (TSS). The red boxes indicate the putative TEAD-binding sequences. Bottom, H358 cells stably expressing YAP(5SA) were subjected to ChIP analysis using anti-YAP or IgG, and precipitated DNA was measured by qPCR. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; and ***, p < 0.001, by unpaired, two-tailed Student's t test. H, cell density affects the enrichment of YAP at the COX-2 promoter. H358 cells seeded at low or high cell densities were subjected to ChIP analysis using anti-YAP or IgG, and precipitated DNA was measured by qPCR. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; and ***, p < 0.001, by unpaired, two-tailed Student's t test. NC, negative control; CTGF, connective tissue growth factor.
To further dissect the molecular basis of the YAP/TAZ–TEADs as suppression complexes, we tested three different YAP mutants: YAP(5SA); YAP(5SA,S94A), an inactive form of YAP due to its deficiency in association with TEADs; and YAP(5SAΔPDZ-BM), which lacks the PDZ-binding motif and is also inactive (Fig. 3E) (40). As shown in Fig. 3F, overexpression of YAP or YAP(5SA) inhibited the induction level of COX-2 by IL-1β. However, this inhibitory activity was completely abolished in mutant YAP(5SA,S94A). Surprisingly, although as deficient as YAP(5SA,S94A) at inducing CYR61 (a commonly used target gene of YAP/TAZ), YAP(5SAΔPDZ-BM) was still able to suppress COX-2 induction. In addition, TEAD1(Δ88–101), a TEAD1 deletion form deficient in DNA binding, also failed to suppress COX-2 induction (Fig. S3D). Together, these data suggested that although the binding of YAP/TAZ–TEADs to DNA is essential for their suppression activities, their transcriptional activity may not be.
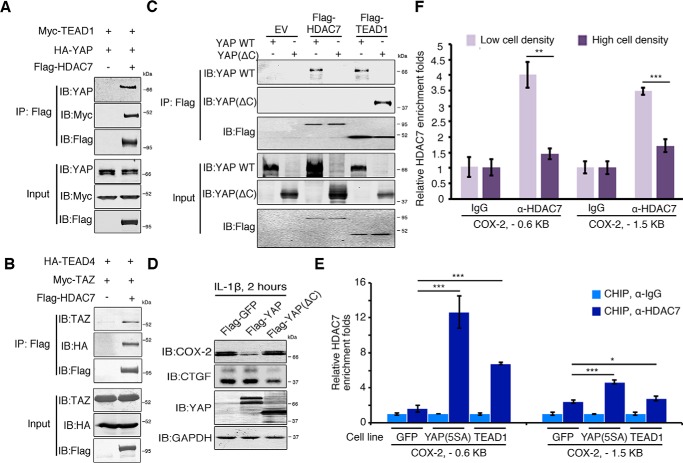
Next, we sought to determine whether YAP/TAZ–TEAD complexes bind directly to the promoter region of COX-2. There are two putative TEAD-binding sequences (GGAATG and CATTCC) in the COX-2 promoter. One is located about 0.6 kb upstream of the transcriptional start site, and the other is about 1.5 kb upstream of it (Fig. 3G). We conducted chromatin immunoprecipitation (ChIP) assays using an anti-YAP1 antibody and observed the enrichment of YAP at both the −0.6 and −1.5 kb sites (Fig. 3G). Importantly, YAP enrichment at the two sites was modulated by cell density: more YAP was recruited at low cell density compared with high cell density (Fig. 3H). These data suggest there was direct binding between YAP/TAZ–TEADs and the COX-2 promoter.
YAP/TAZ–TEADs recruit HDAC7 to the COX-2 promoter to suppress its induction
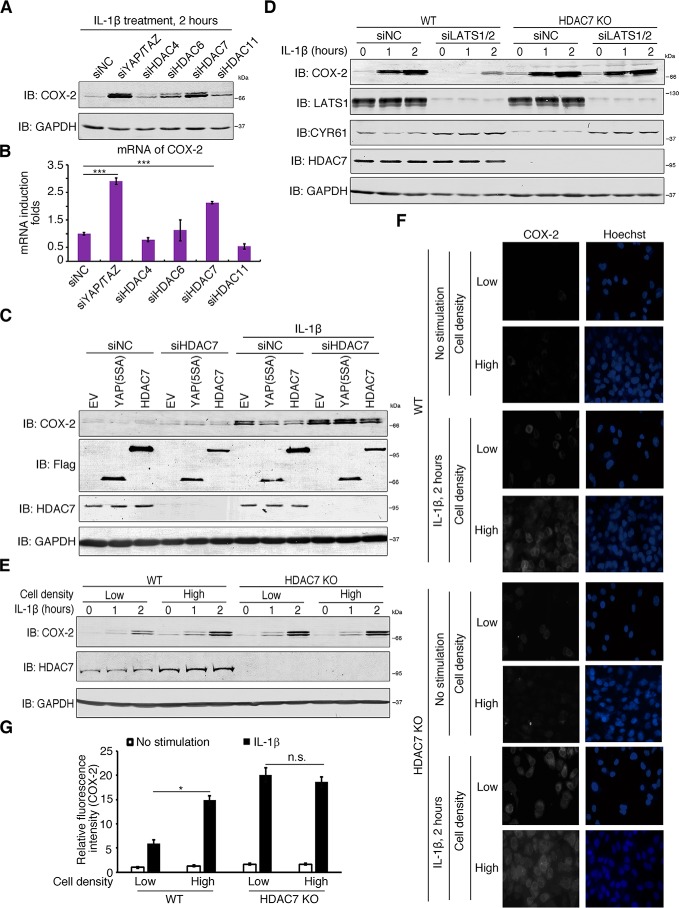
We next tested whether histone deacetylases (HDACs) were involved in the transcriptional suppression activity of the YAP/TAZ–TEAD complexes (41). Treatments with both sodium butyrate and trichostatin A, which specifically inhibit the class I and II mammalian HDACs, restored COX-2 induction by serum starvation in LATS1/2-depleted H358 cells (Fig. S4, A and B). This suggests that HDACs may play a role in the suppression activities of YAP/TAZ–TEAD complexes. Upon overexpression, HDAC4, -6, -7, and -11 diminished COX-2 induction with HDAC7 being the most effective (Fig. S4C). Only knockdown of HDAC7, however, was able to boost the induction of COX-2 by IL-1β stimulation, which was comparable with that of YAP/TAZ depletion (Fig. 4, A and B). The knockdown efficiencies were measured by qRT-PCR (Fig. S4D).
Figure 4.
HDAC7 mediates YAP/TAZ suppression of COX-2 induction. A and B, HDAC7 inhibits COX-2 induction. H358 cells were transfected with siRNAs against the indicated HDACs and treated with IL-1β for 2 h. Cells were harvested for immunoblotting (IB) with the indicated antibodies (A) and qRT-PCR (B) for COX-2 induction. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. ***, p < 0.001, by unpaired, two-tailed Student's t test. C, HDAC7 deficiency abolishes YAP repression of COX-2 induction. HeLa cells were transfected with control siRNA or siRNA against HDAC7 followed by transfection of the indicated plasmids. Cells were then stimulated with IL-1β and harvested for immunoblotting using the indicated antibodies. D, lack of HDAC7 abolishes the Hippo-mediated inhibition of COX-2 induction. WT and HDAC7 KO H358 cells were transfected with control siRNA or siRNA targeting LATS1/2 and then stimulated with IL-1β for the indicated duration of time. Cells were harvested and immunoblotted with the indicated antibodies. E, F, and G, loss of HDAC7 weakens the effect of cell density on COX-2 induction. H358 cells of WT or HDAC7 KO at the same cell numbers (about 2 × 105 cells) were cultured in 10-cm (low cell density) or 3.5-cm (high cell density) plates and treated with IL-1β for the indicated time. Cells were then harvested and immunoblotted with the indicated antibodies (E). WT and HDAC7 KO H358 cells were cultured on cover glasses in 24-well plates at low (2.5 × 104 cells/well) or high (2.25 × 105 cells/well) densities and treated with IL-1β for 2 h. Cells were then fixed and subjected to immunofluorescence staining for COX-2 (F). The relative mean fluorescence intensities of COX-2 staining of single cells were determined. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; and n.s., not significant, by unpaired, two-tailed Student's t test (G). NC, negative control; EV, empty vector.
We then examined whether HDAC7 was involved in the YAP-mediated suppression of COX-2 induction. Overexpression of YAP(5SA) or HDAC7 reduced COX-2 induction levels by IL-1β, but this effect was abolished when HDAC7 was depleted using siRNAs (Fig. 4C). Similarly, knockdown of LATS1/2 reduced IL-1β–stimulated COX-2 induction, but this effect was abolished in HDAC7 knockout (KO) cells (Fig. 4D). More importantly, there was significantly less COX-2 induction at low cell density than at high cell density in wildtype (WT) cells, but this difference was lost in HDAC7 KO cells (Fig. 4, E, F, and G). These data suggest that HDAC7 mediated the suppression activity of the YAP/TAZ–TEAD complexes.
We next examined the interaction between HDAC7 and YAP/TAZ–TEAD complexes to dissect their molecular basis. Coimmunoprecipitation revealed the direct binding of HDAC7 with YAP–TEAD1 or TAZ–TEAD4 complexes (Fig. 5, A and B). Domain mapping suggested the C-terminal TAD domain of YAP was involved in its interaction with HDAC7 because YAP C-terminal deletion (YAP(ΔC)) (Fig. 3E) was not immunoprecipitated by FLAG-HDAC7 (Fig. 5C). Overexpression of YAP(ΔC) also did not suppress the COX-2 induction by IL-1β stimulation (Fig. 5D).
Figure 5.
HDAC7 binds directly to the COX-2 gene promoter. A and B, YAP/TAZ–TEAD complexes interact with HDAC7. 293T cells were cotransfected with Myc-TEAD1, HA-YAP1, and FLAG-HDAC7 (A) or HA-TEAD4, Myc-TAZ, and FLAG-HDAC7 (B). Cell lysates were subjected to immunoprecipitation with M2 beads, and bound proteins were analyzed using the indicated antibodies. C, the C terminus of YAP mediates the interaction of YAP–TEAD complex with HDAC7. 293T cells were transfected with the indicated plasmids, and cell lysates were subjected to immunoprecipitation with M2 beads. Bound proteins were analyzed using the indicated antibodies. D, the C-terminal transcriptional activation domain of YAP is required for its repression function. HeLa cells were transfected with FLAG-YAP or its C-terminal deletion mutant, stimulated with IL-1β for 2 h, and harvested for immunoblotting using the indicated antibodies. E, HDAC7 binds to the COX-2 promoter through YAP–TEAD complex. H358 cells stably expressing GFP, YAP(5SA), or TEAD1 were subjected to ChIP using anti-HDAC7 or IgG, and precipitated DNA was measured by qPCR using primers to amplify the indicated regions surrounding the putative TEAD-binding sites on the COX-2 promoter. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05 and ***, p < 0.001, by unpaired, two-tailed Student's t test. F, cell density regulates HDAC7 recruitment onto the COX-2 promoter. H358 cells grown at low or high cell densities were subjected to ChIP using anti-HDAC7 or IgG, and precipitated DNA was measured by qPCR. n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. **, p < 0.01; and ***, p < 0.001, by unpaired, two-tailed Student's t test. EV, empty vector.
Next, we investigated whether HDAC7 was recruited onto the COX-2 promoter. We generated GFP (as a control)-, YAP(5SA)-, and TEAD1-expressing stable cell lines in H358 cells and performed a ChIP assay using an anti-HDAC7 antibody. HDAC7 was enriched at the two putative TEAD-binding loci of the COX-2 promoter in GFP-expressing cells, and this effect was enhanced in cells with stable overexpression of YAP(5SA) and TEAD1 (Fig. 5E). This suggests that TEAD1 and YAP facilitated the binding of HDAC7 to the COX-2 promoter. More importantly, HDAC7 recruitment to the COX-2 promoter was modulated by cell density in that less HDAC7 was recruited at high cell density than at low cell density (Fig. 5F). Taken together, these data indicate that HDAC7 formed a repressor complex with YAP/TAZ–TEADs that bound to the promoter region of COX-2 and suppressed its transcription.
YAP impairs cellular metastasis mainly via suppression of COX-2 induction
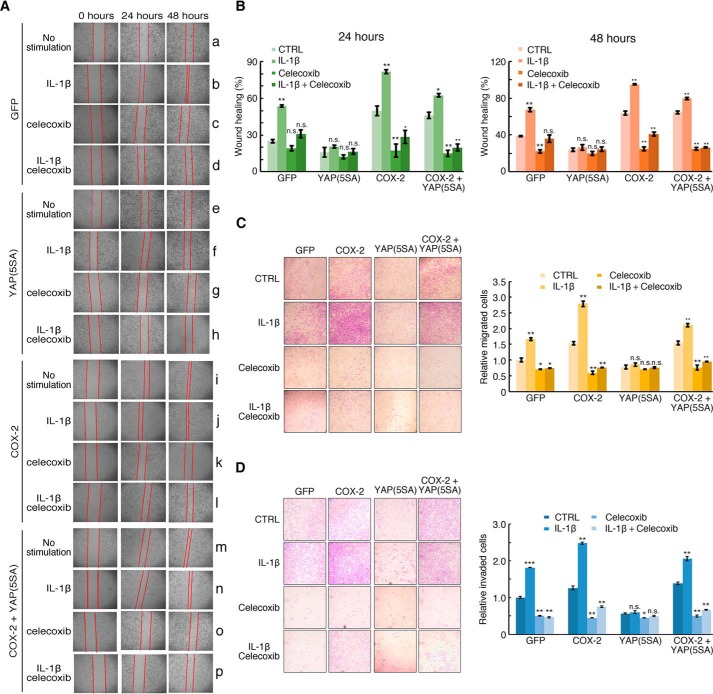
We next investigated the effects of YAP/TAZ-mediated modulation of inflammatory responses. For this, we established H358 cell lines stably expressing GFP (as a control), COX-2, YAP(5SA), or both COX-2 and YAP(5SA) (Fig. S5A). An MTT assay showed that these four cell lines were similar in viability and proliferation, which were also not affected by IL-1β stimulation (Fig. S5B). We then performed wound healing and Transwell assays using these four stable cell lines. In the wound healing assay, IL-1β stimulation significantly elevated the cell migration ability in GFP-expressing cells (Fig. 6A, group b), but not in YAP(5SA)-expressing cells (Fig. 6A, group f). Although stable expression of COX-2 promoted cell migration in the same manner as IL-1β stimulation (Fig. 6A, groups i and j), the effect was not blocked by YAP(5SA) expression (Fig. 6A, groups m and n). Of note, in all cases except YAP(5SA)-expressing cells, the COX-2–specific inhibitor celecoxib significantly blocked the migration of these cell lines (Fig. 6, A, groups d, h, k, l, o, and p, and B). Consistently, the invasion assay in vitro also showed significant suppression activity of YAP(5SA) on the increased capacity of cell invasion by IL-1β stimulation, an effect reversed by the expression of COX-2 (Fig. 6, C and D). Collectively, these data show that IL-1β promoted cell migration through COX-2 induction that was inhibited by YAP via its suppression of COX-2 induction. This suggests that YAP could function as a tumor suppressor by inhibiting the inflammation-induced promotion of cancer cell migration and metastasis.
Figure 6.
YAP inhibits cell migration and invasion through suppression of COX-2 induction. A and B, active YAP disrupts IL-1β–stimulated cell migration by inhibiting COX-2 induction. H358 cells stably expressing GFP (control), YAP(5SA), COX-2, or YAP(5SA) plus COX-2 were generated and used in a wound healing assay. The cells were treated with IL-1β and/or the COX-2 inhibitor celecoxib as indicated and are shown after recovery of 24 or 48 h (A). The frequency of wound recovery is shown (B). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. n.s., not significant (p > 0.05); *, p < 0.05; and **, p < 0.01 (compared with control (CTRL) in each group, respectively), by unpaired, two-tailed Student's t test. C, active YAP inhibits IL-1β–stimulated COX-2 induction to disrupt cell migration in a Transwell assay. The four stable cell lines as in A with the indicated treatment were added to the top chambers. After incubation for 24 h, the migrated cells on the underside of the chamber were fixed, stained with crystal violet, and then examined under a light microscope (left panel). The number of migrated cells was counted from the images of five randomly selected fields (right panel). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. n.s., not significant (p > 0.05); *, p < 0.05; and **, p < 0.01 (compared with control (CTRL) in each group, respectively). D, YAP inhibits IL-1β–induced COX-2 to disrupt cell invasion in a Transwell invasion assay. The invasion ability of H358 stable cell lines as in A was measured by a Transwell invasion assay. Cells were added to the top chambers coated with Matrigel. After incubation for 24 h, the invaded cells were fixed, stained with crystal violet, and then examined under a light microscope (left panel). The number of invasive cells was counted from the images of five randomly selected fields (right panel). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01; and ***, p < 0.001 (compared with control (CTRL) in each group, respectively), by unpaired, two-tailed Student's t test.
YAP suppresses induction of selective genes that play key roles in inflammation and immune responses
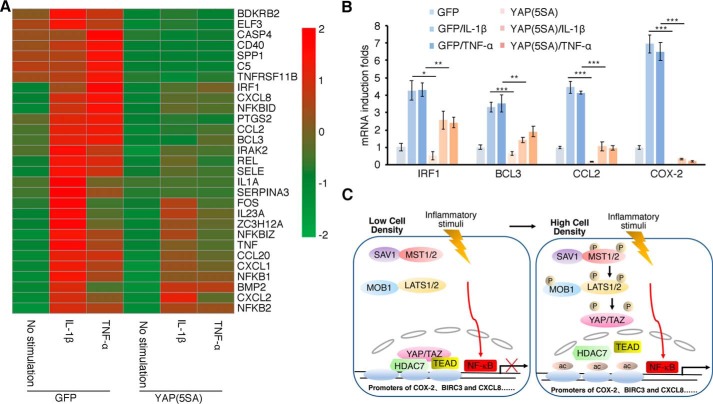
To further understand the genes that might be regulated by the Hippo signaling pathway during proinflammatory responses, we stimulated our GFP- and YAP(5SA)-expressing cells with TNFα or IL-1β and conducted microarray assays to obtain their gene expression profiles. As expected, many of the NF-κB target genes were not affected by YAP(5SA) (Fig. S6A), among which TRAF1, A20, and IκBα were confirmed by qRT-PCR (Fig. S6B). Interestingly, YAP(5SA) negatively regulated a significant number of NF-κΒ target genes (Fig. 7A) as was also the case of COX-2. Some we confirmed by qRT-PCR (Fig. 7B). It is worth noting as well that YAP(5SA) enhanced the induction of a few NF-κΒ target genes, such as IL-6 and PTX3 (Fig. S6, A and B).
Figure 7.
Genes negatively regulated by YAP/TAZ play key roles in regulation of inflammatory and immune responses. A and B, activation of YAP impairs the induction by proinflammatory factors. H358 cells stably expressing GFP or YAP(5SA) were treated with IL-1β or TNFα for 1 h and harvested for extraction of RNA, which was then used for microarray analysis. The heat map is the expression intensity of the selective genes induced by IL-1β or TNFα or suppressed by YAP(5SA). The scale on the right is the log2 -fold changes of corresponding genes among compared groups (A). Some genes shown in A were validated by qRT-PCR analysis (B). n = 3 independent experiments. Data are presented as mean ± S.D. Error bars represent S.D. *, p < 0.05; **, p < 0.01; and ***, p < 0.001, by unpaired, two-tailed Student's t test. C, a proposed model depicts the mechanism of YAP/TAZ–TEAD–HDAC7 complex–mediated negative regulation of transcription. At low cell density, Hippo signaling is off, and YAP/TAZ are located in the nucleus. They form a repressive complex with TEADs and HDAC7 and bind onto the promoter regions of selective NF-κΒ target genes, such as COX-2, to inhibit their transcription. Conversely, high cell density leads to activation of the Hippo pathway, YAP/TAZ hyperphosphorylation, and their cytosolic relocation, which results in initiation of the transcription of COX-2 upon the inflammatory stimulus.
In addition, many other inflammation-related genes were regulated by YAP(5SA), although they are not NF-κΒ target genes, and their expression did not change upon IL-1β or TNFα stimulation. Some of these genes were down-regulated (Fig. S6C), whereas others were up-regulated (Fig. S6D). Notably, for those targeted genes involved in inflammatory responses, the heat map showed that YAP(5SA) down-regulated a greater number of genes than it up-regulated. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated that many of the differentially regulated genes play critical roles in signaling pathways related to antimicrobial responses (Fig. S6E and Table S1). Furthermore, gene ontology analysis indicated that the expression of YAP(5SA) suppressed many genes that play key roles in regulating cell adhesion, cell migration, prostaglandin metabolism, and angiogenesis (Fig. S6F), consistent with the results of the migration and invasion assays (Fig. 6). The microarray analysis further established YAP as a bona fide transcriptional corepressor to regulate inflammatory responses.
Discussion
Inflammation is the body's primary response to infections and injuries. A disproportionate or inappropriately extended response can be detrimental, predisposing the body to chronic diseases (4). Therefore, the cells and tissues involved integrate the physical properties of their microenvironments to confer a response with proper magnitude and duration. Testing the effect of cell density on proinflammatory responses, we found that the same proinflammatory stimuli elicited differing magnitudes of responses, such as the induction of COX-2, when cells were at different density levels: the lower the density, the weaker the response; the higher the density, the stronger the response (Fig. 7C). Based on the microarray data, which indicated that YAP repressed a number of selective target genes of NF-κB, we propose that other related proinflammatory factors could also be regulated by cell densities. It is highly possible that other physical features of the cells and tissues, such as extracellular matrix stiffness, cell geometry, and mechanical stretch, could also exert influence on proinflammatory responses. In fact, several reports suggest that some mechanical forces, such as the rigidity of substrate and osmotic stress, regulate inflammatory responses and cancer invasion (2, 42, 43).
Our study shows that the Hippo pathway is involved in conveying the cell density signals that determined the magnitude of the inflammatory responses. The Hippo pathway closely cross-talks with other signaling pathways involved in development, such as the WNT (44), transforming growth factor-β (45), and Notch signaling pathways (46). We discovered that although it does not influence the activation of NF-κB signaling, the Hippo pathway modulates selective NF-κΒ target genes and therefore is also involved in regulating the inflammation and immune responses. Notably, previous studies have shown the connection between the Hippo pathway and innate immunity from other angles (47–49). Yorkie, the Drosophila homolog of YAP, impeded antimicrobial responses by directly regulating mRNA levels of the IκB homolog Cactus (48). LATS1/2 regulated antitumor immune responses (50). Given that Hippo has been shown to relay physical cues, such as cell polarity, cell–cell contacts, and mechanical and cytoskeleton cues, and chemical signals like growth factors and G protein–coupled receptor ligands into cellular responses (38, 51–53), it would be interesting to study how the Hippo signaling pathway integrates these changes in microenvironment to modulate inflammatory and immune responses. Although the pathophysiological significance of these regulations needs more study, we speculate that the mechanism at least partly controls the magnitude and duration of inflammation of cells and tissues in response to their microenvironmental cues.
It is generally understood that YAP/TAZ function as transcriptional coactivators to facilitate other transcriptional activators, such as TEADs, Runx1/2, and ErbB4 (54, 55). However, we have shown that they could also function as transcriptional repressors. This repressor activity is a direct rather than indirect effect via activating expression of other inhibitory factors. Two previous studies have also reported YAP/TAZ or TEADs to be transcriptional repressors (56, 57). Intriguingly, YAP/TAZ still use TEADs as the important components in repression. Therefore, it seems YAP/TAZ–TEADs alone cannot be simply categorized as activators or repressors. Rather, whether they function as activators or repressors depends on the histone-modifying enzymes they recruit. They will recruit histone acetyltransferases, such as p300 or cAMP-response element–binding protein (CREB)-binding protein, to activate gene transcription (58) but will recruit HDACs, such as HDAC7 (this study), to repress gene transcription. Even more intriguing is that both the activation and suppression of gene transcriptions occur in the same spatiotemporal settings. Why and how YAP/TAZ–TEADs are activators for some genes but repressors for others is a course for future study.
Numerous studies have shown that both YAP/TAZ and COX-2 are pro-oncogenic. YAP/TAZ are activated in many human tumors and play a role in the initiation, progression, and even metastasis of some cancers, whereas COX-2 can drive tumorigenesis by producing prostaglandins. These in turn act directly both on cancer and stromal cells, inhibiting apoptosis and enhancing cell migration in the former and promoting neoangiogenesis in the latter (59–61). Increased expression of COX-2 has been found in colon, prostate, lung, breast, pancreas, and gastric tumors and also positively correlates with poor patient prognosis (62–67). A recent report also suggests that tumor-derived COX-2 can facilitate cancer immune evasion (68). It is thus counterintuitive with respect to our findings that active YAP/TAZ inhibit COX-2 expression and consequently inflammatory microenvironment-induced cell migration and invasion (Fig. 6). Nevertheless, a few other studies also implied YAP to be a tumor suppressor (69, 70). Thus, we suggest that YAP/TAZ should not simply be designated as either oncogenes or tumor suppressors. Because the Hippo pathway can be regulated by various physical or biochemical signal inputs, YAP/TAZ could coordinate with other signaling pathways to give an integrated response, be it cell growth and proliferation or growth inhibition. This also suggests that by purposely altering the physical properties of microenvironment, we could be able to put YAP/TAZ into use for therapeutics.
COX-2, one of the key enzymes for biosynthesis of prostaglandins and a crucial regulator of inflammation, angiogenesis, and tumorigenesis (71), is known to be highly inducible at the transcriptional level by various hormones, growth factors, and inflammatory stimuli as well as other stress conditions through the mitogen-activated protein kinase, NF-κB, WNT, and hypoxia-response pathways (72–75). However, few studies have focused on the possibility of its transcriptional inhibition as we have done with YAP and TAZ. This further clarifies the extent of the COX-2 regulatory network and implies a potential therapeutic means to inhibit COX-2 activity.
In summary, we report that the Hippo signaling pathway transduced the cell density signal to modulate proinflammatory responses. YAP/TAZ–TEADs formed novel complexes with HDAC7, which then functioned as transcriptional repressors to modulate the induction of selective genes related to inflammation, immunity, and metastasis. Thus, YAP/TAZ can have a role in tumor suppression. We propose that the Hippo signaling pathway acts as an important inflammatory regulator by transducing the physical properties of the cell and tissue microenvironments. Our study also points out the possibility that we can change the responses of cells and tissues to stimuli by modifying the physical properties of their microenvironments, which can then be exploited as a therapeutic option against inflammatory diseases and cancer.
Materials and methods
Reagents and plasmids
Recombinant human IL-1β and TNFα were purchased from Sino Biological. DFO (mesylate salt), sodium butyrate, and trichostatin A were purchased from Sigma. Celecoxib was from MedChem Express. pCMV-GAL4-YAP and pCMV-GAL4-YAP(ΔC) were kindly provided by Bin Zhao (Life Science Institute, Zhejiang University, China). YAP, TAZ, YAP(5SA), YAP-(5SA,S94A), YAP(5SAΔPDZ-BM), TEAD1, TEAD1(Δ88–101), and HDAC1/3/4/6/7/8/9/10/11 were all constructed in pLX304-puro vectors. cDNAs encoding YAP(5SA) and COX-2 were inserted into pHAGE-CMV-full-EF1a-IRES-Zs Green vectors as lentiviral transfer plasmids.
Cell culture, transfections, and infections
H358, HeLa, and HEK293T cells were obtained from American Type Culture Collection (ATCC). All cell lines were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Gibco), 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C in 5% CO2 (v/v). Lipofectamine 2000 (Invitrogen) was used for plasmid transfection. Transfection of siRNAs was performed using Lipofectamine RNAiMAX (Invitrogen) reagent. The empty vector-, YAP(5SA)-, COX-2–, and YAP(5SA) plus COX-2–expressing H358 cells were generated by recombinant lentivirus-mediated gene transduction. For production of lentiviral supernatants, HEK293T cells were cotransfected with the transfer vectors harboring the indicated genes above together with helper plasmids psPAX2 and pMD2.G (Addgene). Virus stocks were prepared by collecting the media 48 h after transfection. H358 cells were infected with the recombinant viruses in the presence of Polybrene (1 μg/ml) and selected with 1 μg/ml puromycin. H358 HDAC7 knockout cells were obtained by CRISPR genomic editing technology. The guide RNA sequences were cloned into pEP330, which was modified from px330 (Addgene). Plasmids were transfected into H358 cells. Twenty-four hours after transfection, puromycin (0.5 μg/ml) was added into culture medium for selection. The details of guide RNA sequences are provided in Table S2. For cell density manipulation, we generally plated the same number of cells (about 2 × 105) onto 10-, 6-, and 3.5-cm plates for low, medium, or high cell densities, respectively. Alternatively, we plated cells at about 2.5 × 104, 7.5 × 104, and 2.25 × 105 cells/well in 24-well plates to represent different cell confluences, respectively.
Quantitative RT-PCR assay
H358 cells with specified siRNA transfection were lysed, and total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was generated by a PrimeScript RT reagent kit (TaKaRa). Quantitative real-time PCR was performed using SYBR Green (TaKaRa) and the ABI 7500 real-time PCR system (Applied Biosystems). The primer sequences are provided in Table S2.
Western blotting and immunoprecipitation
H358 or HeLa cells transfected with the indicated plasmids were lysed in Buffer A (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, 1 mm DTT, and Complete protease inhibitor mixture) for 10 min on ice and centrifuged at 20,000 × g for 10 min. Cell lysates were analyzed by SDS-PAGE and immunoblotting with antibodies specified in figures. For coimmunoprecipitation studies, HEK293T cells transfected with the indicated plasmids were lysed and subjected to immunoprecipitation using M2-conjugated magnetic beads (Sigma). M2 beads were resolved by 2× SDS loading buffer and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Detailed information of all antibodies used in immunoblotting analysis is provided in Table S3.
Immunofluorescence and microscopy
To visualize the protein expressions of COX-2 and YAP, H358 cells stimulated with or without IL-1β were fixed in 4% paraformaldehyde, permeabilized, blocked in 2% BSA in PBS for 2 h, and incubated sequentially with the indicated primary antibodies and Alexa Fluor–labeled secondary antibodies (Life Technologies) with extensive washing. Slides were then mounted with Vectashield and stained with Hoechst 33342 (Life Technologies). Immunofluorescence images were obtained and analyzed using a Nikon Eclipse Ti inverted microscope. ImageJ was used to quantify the mean fluorescence intensity of each cell.
ChIP
Cells were fixed with 1% paraformaldehyde for 10 min at room temperature with constant shaking. Cells were then neutralized with 125 mm glycine for 5 min at room temperature, harvested with PBS, and resuspended in SDS lysis buffer (1% SDS, 50 mm Tris-Cl, pH 8.1, and 10 mm EDTA). Lysates were sonicated to shear DNA to lengths between 200 and 1000 bp. Lysates were cleared by centrifugation for 20 min at 16,100 × g. The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, and 167 mm NaCl). The immunoprecipitating antibody was added to the supernatant fraction and incubated overnight at 4 °C. Protein A/G–agarose beads (Abmart) were added and incubated for 1 h at 4 °C. The agarose beads were washed and eluted with 1% SDS and 100 mm NaHCO3 overnight at 68 °C. After RNase and Proteinase K treatment, proteins were denatured and removed by phenol-chloroform extraction, and the soluble DNAs were recovered by ethanol precipitation. Oligo sequences used in ChIP assays are available in Table S2.
MTT assay
H358 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS). Cells (1 × 104 cells/ml) in 0.1 ml of medium were placed in each well of 96-well plates. For IL-1β treatment, IL-1β was added into each well as indicated (final concentration, 10 ng/ml). The plate was then placed in the incubator for 24, 48, and 72 h. At the end of each individual incubation time point, 20 μl of the 5 mg/ml MTT solution (Sigma) was added into each sample well. The plate was then incubated again in the dark for 4 h. The supernatant was removed; the crystals were then dissolved in 150 μl/well DMSO. The plate was then gently shaken for 5 min to ensure equal mixing, and absorbance at 570 nm was taken using a spectrometer.
Wound healing assay
H358 cells (5 × 105) were seeded onto 6-well plates. Cells were treated with 10 ng/ml IL-1β and 20 μg/ml celecoxib as indicated. Scratch wounds were made by scraping the cell layer across each culture plate using the tip of 200-μl pipette tips. After wounding, the debris was removed by washing the cells with PBS. Wounded cultures were incubated in serum-free medium for 48 h, and then three fields were randomly picked from each scratch wound and visualized by microscopy to assess cell migration ability. The experiments were performed in triplicates.
Migration and invasion assays
The cell migration assay was performed using Transwells (24-well insert; 8-mm-pore-size polycarbonate membrane) obtained from Corning. H358 cells (5 × 105/ml) in 0.1 ml of medium with 1% FBS were placed in the upper chamber; the lower chamber was loaded with 0.8 ml of medium containing 10% FBS. Cells were treated with IL-1β and celecoxib. The total number of cells that migrated into the lower chamber was counted after 12 h of incubation at 37 °C with 5% CO2. Six visual fields were chosen for counting. The Transwell cell invasion assay was performed using Transwells preloaded with a layer of Matrigel (Sigma-Aldrich) on the upper surface. The rest of the experimental procedure is as same as the cell migration assay.
Microarray analysis
GFP- or YAP(5SA)-transfected H358 cells were treated with 10 ng/ml TNFα, 10 ng/ml IL-1β, or vehicle for 1 h, respectively. Cells were harvested, and total RNAs were isolated. The microarray assay was performed by the service provider BGI, China. RNA samples were assessed for quality and integrity using Eukaryote total RNA Nano (Agilent Technologies). Fluorescent RNA targets were prepared using the OneArray® Amino Allyl aRNA Amplification kit (Phalanx Biotech Group, Taiwan) and Cy5 dyes (Amersham Biosciences) from total RNA samples. Fluorescent targets were hybridized to the Human Whole Genome OneArray with Phalanx hybridization buffer using the Phalanx hybridization system. The slides were scanned by an Agilent G2505C scanner (Agilent Technologies, Santa Clara, CA). The Cy5 fluorescence intensities of each spot were analyzed by GenePix 4.1 software (Molecular Devices). Normalized spot intensities were transformed to gene expression log2 ratios between the control and treatment groups. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE115207.
Statistical analysis
Quantitative data are presented as mean ± S.D. All statistical tests were performed using a Student's t test (unpaired, two-tailed): *, p < 0.05; **, p < 0.01; and ***, p < 0.001. No statistical method was used to predetermine sample size. All experiments were repeated a minimum of three times to ensure reproducibility.
Author contributions
Q. Z., X. H., J. S., L. H., P. X., H. S., L. Z., B. Z., Y.-j. W., and Z. X. conceptualization; Q. Z. and Z. X. data curation; Q. Z. software; Q. Z. formal analysis; Q. Z. and Y.-j. W. validation; Q. Z., X. H., X. X., Y. Z., and L. H. investigation; Q. Z. visualization; Q. Z. and Z. X. writing-original draft; Q. Z., J. C., J. X., and Z. X. writing-review and editing; Z. X. supervision; Z. X. funding acquisition.
Supplementary Material
This work was supported by National Key Research and Development Program of China, Stem Cell and Translational Research Grant 2016YFA0100300 (to Z. X. and Y. W.) and Natural Science Foundation of China Grants 31571445 and 31371416 (to Z. X.) and 31401200 (to X. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Tables S1–S3.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE115207.
- IL
- interleukin
- TNFα
- tumor necrosis factor α
- YAP
- Yes-associated protein
- TAZ
- transcriptional coactivator with PDZ-binding motif
- COX
- cyclooxygenase
- TEAD
- TEA domain transcription factor
- HDAC
- histone deacetylase
- MST
- mammalian STE20-like protein kinase
- LATS
- large tumor suppressor
- DFO
- deferoxamine
- ERK
- extracellular signal-regulated kinase
- CHX
- cycloheximide
- qRT-PCR
- quantitative RT-PCR
- miRNA
- microRNA
- KO
- knockout
- TAD
- transcriptional activation domain
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- FBS
- fetal bovine serum
- qPCR
- quantitative PCR
- PDZ-BM
- PDZ-binding motif.
References
- 1. Kulinsky V. I. (2007) Biochemical aspects of inflammation. Biochemistry 72, 595–607 [DOI] [PubMed] [Google Scholar]
- 2. Abolhassani M., Wertz X., Pooya M., Chaumet-Riffaud P., Guais A., and Schwartz L. (2008) Hyperosmolarity causes inflammation through the methylation of protein phosphatase 2A. Inflamm. Res. 57, 419–429 10.1007/s00011-007-7213-0 [DOI] [PubMed] [Google Scholar]
- 3. Schwartz L., Guais A., Pooya M., and Abolhassani M. (2009) Is inflammation a consequence of extracellular hyperosmolarity? J. Inflamm. 6, 21 10.1186/1476-9255-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nathan C. (2002) Points of control in inflammation. Nature 420, 846–852 10.1038/nature01320 [DOI] [PubMed] [Google Scholar]
- 5. Dubois R. N., Abramson S. B., Crofford L., Gupta R. A., Simon L. S., Van De Putte L. B., and Lipsky P. E. (1998) Cyclooxygenase in biology and disease. FASEB J. 12, 1063–1073 10.1096/fasebj.12.12.1063 [DOI] [PubMed] [Google Scholar]
- 6. Wellen K. E., and Hotamisligil G. S. (2005) Inflammation, stress, and diabetes. J. Clin. Investig. 115, 1111–1119 10.1172/JCI25102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh S., and Karin M. (2002) Missing pieces in the NF-κB puzzle. Cell 109, (suppl.) S81–S96 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- 8. Li Q., and Verma I. M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 10.1038/nri910 [DOI] [PubMed] [Google Scholar]
- 9. Hayden M. S., and Ghosh S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 10.1101/gad.183434.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mantovani A., Allavena P., Sica A., and Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- 11. Lawrence T. (2009) The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1, a001651 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu T., Zhang L., Joo D., and Sun S. C. (2017) NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 17023 10.1038/sigtrans.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharif G. M., Schmidt M. O., Yi C., Hu Z., Haddad B. R., Glasgow E., Riegel A. T., and Wellstein A. (2015) Cell growth density modulates cancer cell vascular invasion via Hippo pathway activity and CXCR2 signaling. Oncogene 34, 5879–5889 10.1038/onc.2015.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harvey K. F., Pfleger C. M., and Hariharan I. K. (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 10.1016/S0092-8674(03)00557-9 [DOI] [PubMed] [Google Scholar]
- 15. Udan R. S., Kango-Singh M., Nolo R., Tao C., and Halder G. (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- 16. Xu T., Wang W., Zhang S., Stewart R. A., and Yu W. (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 17. Creasy C. L., and Chernoff J. (1995) Cloning and characterization of a member of the MST subfamily of Ste20-like kinases. Gene 167, 303–306 10.1016/0378-1119(95)00653-2 [DOI] [PubMed] [Google Scholar]
- 18. Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P. R., Bellen H. J., and Halder G. (2002) Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730 10.1242/dev.00168 [DOI] [PubMed] [Google Scholar]
- 19. Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., and Li Y. (2005) Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675–685 10.1016/j.cell.2004.12.036 [DOI] [PubMed] [Google Scholar]
- 20. Huang J., Wu S., Barrera J., Matthews K., and Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 21. Zhang L., Ren F., Zhang Q., Chen Y., Wang B., and Jiang J. (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., and Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan E. H., Nousiainen M., Chalamalasetty R. B., Schäfer A., Nigg E. A., and Silljé H. H. (2005) The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- 24. Praskova M., Xia F., and Avruch J. (2008) MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321 10.1016/j.cub.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., and Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu C. Y., Zha Z. Y., Zhou X., Zhang H., Huang W., Zhao D., Li T., Chan S. W., Lim C. J., Hong W., Zhao S., Xiong Y., Lei Q. Y., and Guan K. L. (2010) The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TrCP E3 ligase. J. Biol. Chem. 285, 37159–37169 10.1074/jbc.M110.152942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao B., Li L., Tumaneng K., Wang C. Y., and Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev. 24, 72–85 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S., Liu Y., Zheng Y., Dong J., and Pan D. (2008) The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 30. Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., Oren M., Sudol M., Cesareni G., and Blandino G. (2001) Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276, 15164–15173 10.1074/jbc.M010484200 [DOI] [PubMed] [Google Scholar]
- 31. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., and Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 32. Halder G., Dupont S., and Piccolo S. (2012) Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 10.1038/nrm3416 [DOI] [PubMed] [Google Scholar]
- 33. Wada K., Itoga K., Okano T., Yonemura S., and Sasaki H. (2011) Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914 10.1242/dev.070987 [DOI] [PubMed] [Google Scholar]
- 34. Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S. (2013) A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- 35. Basan M., Risler T., Joanny J. F., Sastre-Garau X., and Prost J. (2009) Homeostatic competition drives tumor growth and metastasis nucleation. HFSP J. 3, 265–272 10.2976/1.3086732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaidi A., Qualtrough D., Williams A. C., and Paraskeva C. (2006) Direct transcriptional up-regulation of cyclooxygenase-2 by hypoxia-inducible factor (HIF)-1 promotes colorectal tumor cell survival and enhances HIF-1 transcriptional activity during hypoxia. Cancer Res. 66, 6683–6691 10.1158/0008-5472.CAN-06-0425 [DOI] [PubMed] [Google Scholar]
- 37. Jang B. C., Sanchez T., Schaefers H. J., Trifan O. C., Liu C. H., Creminon C., Huang C. K., and Hla T. (2000) Serum withdrawal-induced post-transcriptional stabilization of cyclooxygenase-2 mRNA in MDA-MB-231 mammary carcinoma cells requires the activity of the p38 stress-activated protein kinase. J. Biol. Chem. 275, 39507–39515 10.1074/jbc.M003224200 [DOI] [PubMed] [Google Scholar]
- 38. Zhao B., Li L., Wang L., Wang C. Y., Yu J., and Guan K. L. (2012) Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54–68 10.1101/gad.173435.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo X., Zhao Y., Yan H., Yang Y., Shen S., Dai X., Ji X., Ji F., Gong X. G., Li L., Bai X., Feng X. H., Liang T., Ji J., Chen L., Wang H., and Zhao B. (2017) Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 31, 247–259 10.1101/gad.294348.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimomura T., Miyamura N., Hata S., Miura R., Hirayama J., and Nishina H. (2014) The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity. Biochem. Biophys. Res. Commun. 443, 917–923 10.1016/j.bbrc.2013.12.100 [DOI] [PubMed] [Google Scholar]
- 41. Shahbazian M. D., and Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 10.1146/annurev.biochem.76.052705.162114 [DOI] [PubMed] [Google Scholar]
- 42. Sorokin L. (2010) The impact of the extracellular matrix on inflammation. Nat. Rev. Immunol. 10, 712–723 10.1038/nri2852 [DOI] [PubMed] [Google Scholar]
- 43. Tse J. M., Cheng G., Tyrrell J. A., Wilcox-Adelman S. A., Boucher Y., Jain R. K., and Munn L. L. (2012) Mechanical compression drives cancer cells toward invasive phenotype. Proc. Natl. Acad. Sci. U. S. A. 109, 911–916 10.1073/pnas.1118910109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park H. W., Kim Y. C., Yu B., Moroishi T., Mo J. S., Plouffe S. W., Meng Z., Lin K. C., Yu F. X., Alexander C. M., Wang C. Y., and Guan K. L. (2015) Alternative Wnt signaling activates YAP/TAZ. Cell 162, 780–794 10.1016/j.cell.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B. M., Dembowy J., Yaffe M. B., Zandstra P. W., and Wrana J. L. (2008) TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837–848 10.1038/ncb1748 [DOI] [PubMed] [Google Scholar]
- 46. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., and Brummelkamp T. R. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 47. Nowell C. S., Odermatt P. D., Azzolin L., Hohnel S., Wagner E. F., Fantner G. E., Lutolf M. P., Barrandon Y., Piccolo S., and Radtke F. (2016) Chronic inflammation imposes aberrant cell fate in regenerating epithelia through mechanotransduction. Nat. Cell Biol. 18, 168–180 10.1038/ncb3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu B., Zheng Y., Yin F., Yu J., Silverman N., and Pan D. (2016) Toll receptor-mediated Hippo signaling controls innate immunity in Drosophila. Cell 164, 406–419 10.1016/j.cell.2015.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Q., Meng F., Chen S., Plouffe S. W., Wu S., Liu S., Li X., Zhou R., Wang J., Zhao B., Liu J., Qin J., Zou J., Feng X. H., Guan K. L., et al. (2017) Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 19, 362–374 10.1038/ncb3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moroishi T., Hayashi T., Pan W. W., Fujita Y., Holt M. V., Qin J., Carson D. A., and Guan K. L. (2016) The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 167, 1525–1539.e17 10.1016/j.cell.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen C. L., Gajewski K. M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., Tao C., and Halder G. (2010) The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 15810–15815 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu F. X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H., Fu X. D., Mills G. B., and Guan K. L. (2012) Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rauskolb C., Sun S., Sun G., Pan Y., and Irvine K. D. (2014) Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143–156 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Komuro A., Nagai M., Navin N. E., and Sudol M. (2003) WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 278, 33334–33341 10.1074/jbc.M305597200 [DOI] [PubMed] [Google Scholar]
- 55. Yagi R., Chen L. F., Shigesada K., Murakami Y., and Ito Y. (1999) A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang S. W., and Eberhardt N. L. (1996) TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP. J. Biol. Chem. 271, 9510–9518 10.1074/jbc.271.16.9510 [DOI] [PubMed] [Google Scholar]
- 57. Kim M., Kim T., Johnson R. L., and Lim D. S. (2015) Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282 10.1016/j.celrep.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 58. Stein C., Bardet A. F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schübeler D., and Bauer A. (2015) YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 11, e1005465 10.1371/journal.pgen.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams C. S., Tsujii M., Reese J., Dey S. K., and DuBois R. N. (2000) Host cyclooxygenase-2 modulates carcinoma growth. J. Clin. Investig. 105, 1589–1594 10.1172/JCI9621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lu L., Li Y., Kim S. M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M. J., Lee J. S., and Johnson R. L. (2010) Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U.S.A. 107, 1437–1442 10.1073/pnas.0911427107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R., and Camargo F. D. (2011) Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782–795 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sheehan K. M., Sheahan K., O'Donoghue D. P., MacSweeney F., Conroy R. M., Fitzgerald D. J., and Murray F. E. (1999) The relationship between cyclooxygenase-2 expression and colorectal cancer. JAMA 282, 1254–1257 10.1001/jama.282.13.1254 [DOI] [PubMed] [Google Scholar]
- 63. Cianchi F., Cortesini C., Bechi P., Fantappiè O., Messerini L., Vannacci A., Sardi I., Baroni G., Boddi V., Mazzanti R., and Masini E. (2001) Up-regulation of cyclooxygenase 2 gene expression correlates with tumor angiogenesis in human colorectal cancer. Gastroenterology 121, 1339–1347 10.1053/gast.2001.29691 [DOI] [PubMed] [Google Scholar]
- 64. Kim H. S., Youm H. R., Lee J. S., Min K. W., Chung J. H., and Park C. S. (2003) Correlation between cyclooxygenase-2 and tumor angiogenesis in non-small cell lung cancer. Lung Cancer 42, 163–170 10.1016/S0169-5002(03)00290-3 [DOI] [PubMed] [Google Scholar]
- 65. Tucker O. N., Dannenberg A. J., Yang E. K., Zhang F., Teng L., Daly J. M., Soslow R. A., Masferrer J. L., Woerner B. M., Koki A. T., and Fahey T. J. 3rd (1999) Cyclooxygenase-2 expression is up-regulated in human pancreatic cancer. Cancer Res. 59, 987–990 [PubMed] [Google Scholar]
- 66. Yip-Schneider M. T., Barnard D. S., Billings S. D., Cheng L., Heilman D. K., Lin A., Marshall S. J., Crowell P. L., Marshall M. S., and Sweeney C. J. (2000) Cyclooxygenase-2 expression in human pancreatic adenocarcinomas. Carcinogenesis 21, 139–146 10.1093/carcin/21.2.139 [DOI] [PubMed] [Google Scholar]
- 67. Huang S. P., Wu M. S., Shun C. T., Wang H. P., Hsieh C. Y., Kuo M. L., and Lin J. T. (2005) Cyclooxygenase-2 increases hypoxia-inducible factor-1 and vascular endothelial growth factor to promote angiogenesis in gastric carcinoma. J. Biomed. Sci. 12, 229–241 10.1007/s11373-004-8177-5 [DOI] [PubMed] [Google Scholar]
- 68. Zelenay S., van der Veen A. G., Böttcher J. P., Snelgrove K. J., Rogers N., Acton S. E., Chakravarty P., Girotti M. R., Marais R., Quezada S. A., Sahai E., and Reis e Sousa C. (2015) Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270 10.1016/j.cell.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ehsanian R., Brown M., Lu H., Yang X. P., Pattatheyil A., Yan B., Duggal P., Chuang R., Doondeea J., Feller S., Sudol M., Chen Z., and Van Waes C. (2010) YAP dysregulation by phosphorylation or ΔNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene 29, 6160–6171 10.1038/onc.2010.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu S. J., Hu J. Y., Kuang X. Y., Luo J. M., Hou Y. F., Di G. H., Wu J., Shen Z. Z., Song H. Y., and Shao Z. M. (2013) MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin. Cancer Res. 19, 1389–1399 10.1158/1078-0432.CCR-12-1959 [DOI] [PubMed] [Google Scholar]
- 71. Wang D., and DuBois R. N. (2004) Cyclooxygenase 2-derived prostaglandin E2 regulates the angiogenic switch. Proc. Natl. Acad. Sci. U.S.A. 101, 415–416 10.1073/pnas.0307640100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schmedtje J. F. Jr., Ji Y. S., Liu W. L., DuBois R. N., and Runge M. S. (1997) Hypoxia induces cyclooxygenase-2 via the NF-κB p65 transcription factor in human vascular endothelial cells. J. Biol. Chem. 272, 601–608 10.1074/jbc.272.1.601 [DOI] [PubMed] [Google Scholar]
- 73. Greenhough A., Smartt H. J., Moore A. E., Roberts H. R., Williams A. C., Paraskeva C., and Kaidi A. (2009) The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 30, 377–386 10.1093/carcin/bgp014 [DOI] [PubMed] [Google Scholar]
- 74. Howe L. R., Crawford H. C., Subbaramaiah K., Hassell J. A., Dannenberg A. J., and Brown A. M. (2001) PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J. Biol. Chem. 276, 20108–20115 10.1074/jbc.M010692200 [DOI] [PubMed] [Google Scholar]
- 75. Cha Y. I., Solnica-Krezel L., and DuBois R. N. (2006) Fishing for prostanoids: deciphering the developmental functions of cyclooxygenase-derived prostaglandins. Dev. Biol. 289, 263–272 10.1016/j.ydbio.2005.10.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.