Abstract
Nuclear receptor farnesoid X receptor (FXR) functions as the major bile acid sensor coordinating cholesterol metabolism, lipid homeostasis, and absorption of dietary fats and vitamins. Because of its central role in metabolism, FXR represents an important drug target to manage metabolic and other diseases, such as primary biliary cirrhosis and nonalcoholic steatohepatitis. FXR and nuclear receptor retinoid X receptor α (RXRα) form a heterodimer that controls the expression of numerous downstream genes. To date, the structural basis and functional consequences of the FXR/RXR heterodimer interaction have remained unclear. Herein, we present the crystal structures of the heterodimeric complex formed between the ligand-binding domains of human FXR and RXRα. We show that both FXR and RXR bind to the transcriptional coregulator steroid receptor coactivator 1 with higher affinity when they are part of the heterodimer complex than when they are in their respective monomeric states. Furthermore, structural comparisons of the FXR/RXRα heterodimers and the FXR monomers bound with different ligands indicated that both heterodimerization and ligand binding induce conformational changes in the C terminus of helix 11 in FXR that affect the stability of the coactivator binding surface and the coactivator binding in FXR. In summary, our findings shed light on the allosteric signal transduction in the FXR/RXR heterodimer, which may be utilized for future drug development targeting FXR.
Keywords: nuclear receptor, transcription factor, X-ray crystallography, allosteric regulation, signal transduction, metabolism, conformational change, allostery, Farnesoid X receptor, Retinoid X receptor
Introduction
The farnesoid X receptor (FXR, NR1H4)2 is a member of the nuclear receptor (NR) superfamily that plays a key role in the regulation of bile acids, lipid, and glucose metabolisms, as well as anti-inflammatory response and hepatocarcinogenesis inhibition (1, 2). FXR, which is abundantly expressed in liver, kidney, and intestine, has been identified as the main bile acid sensor (3). Over the years, FXR has been demonstrated to be an effective drug target to treat metabolic diseases, because FXR agonist obeticholic acid (OCA or INT747) has been successfully used in primary biliary cirrhosis and nonalcoholic steatohepatitis treatment (4).
FXR belongs to the structurally conserved family of nuclear receptors that function as ligand-regulated transcription factors. It consists of an N-terminal DNA-binding domain, which targets the receptor to specific genes, coupled by a flexible linker to a ligand-binding domain (LBD). The LBD can bind small lipophilic ligands and then serves as the transcriptional switch. Like most NRs, regulation of transcription by FXR is a complex process that relies on the release of corepressors and the ligand-dependent recruitment of coactivator proteins to a surface on the LBD. This surface, usually formed by helices 3, 4, 5, and 12 in FXR, is also called the activation function-2 (AF2) surface. In turn, those coregulatory proteins mediate the interactions with the basal transcriptional machinery, resulting in repression or activation of transcription (5).
FXR operates as a heterodimer with another nuclear receptor named retinoid X receptor-α (RXRα, NR2B1) in vivo (6). RXRα appears to respond to endogenous ligands, such as 9-cis-retinoic acid (9cRA) (7). FXR/RXR heterodimer binds to FXR response element (FXRE), which is mostly a single inverted repeat (IR1) in the promoter region of the target gene (8). RXRs also serve as obligate heterodimer partners for many of the subfamily 1 NRs, including receptors for peroxisome proliferator activators (PPARs), thyroid hormone (TR), retinoic acid (RAR), and vitamin D (9, 10). These different RXR heterodimers can be divided into three classes: permissive, conditional, and nonpermissive RXR heterodimers, based on their different response to RXR ligands (11, 12). Generally, RXR heterodimers that contain PPAR, LXR, and FXR, which can be activated by ligands for either partner in the dimer, are permissive RXR heterodimers. Heterodimer RXR/RAR demonstrates conditional permissiveness, because full response to RXR ligands occurs only in the presence of an RAR agonist. The nonpermissive RXR heterodimers (e.g. RXR/TR, RXR/vitamin D receptor) usually do not respond to RXR ligands (12). Dimerization renders the control of NR function intricate, because the signal across the heterodimer interface provides an exquisite degree of combinatorial control of transactivation (13). Within nuclear receptors, allostery is increasingly recognized as a common regulatory process initiated by DNA, ligands, and coregulators (14). Previous studies have defined an energetic coupled network of amino acids that mediates the allosteric regulation in RXR heterodimers using the statistic coupling analysis (SCA) method. It identifies a signal pathway within the heterodimer connecting the hydrophobic core of the AF2 surface from each receptor, but effects of the mutations in the network vary in different heterodimers (11). More structural information is needed to delineate the SCA network at the atomic level.
To date, a number of FXR–LBD crystal structures have been resolved in complex with a range of distinct ligands, which show that FXR possesses a highly flexible ligand-binding pocket. However, these structures only contained FXR–LBD; thus, it remained unclear whether the presence of RXRα would impact FXR's structure and function. Further understanding of FXR regulation requires a more in-depth knowledge of the interactions between FXR and its binding partner RXRα. Herein, we present crystal structures of FXR–LBD complexed with coactivator and two novel FXR ligands and, for the first time, FXR/RXRα–LBD heterodimer complexes in the presence of novel FXR ligands and natural RXR ligand. In addition, we conducted in vitro studies to determine the transcriptional coactivator steroid receptor coactivator 1 (SRC1) binding affinities for the individual nuclear receptors and receptors in the heterodimer complex. Structure-guided mutagenesis experiments were further used to investigate the signaling between the heterodimers.
Results
Structure determination
The hFXR–LBD was crystallized in the presence of FXR agonists (HNC143 and HNC180) and SRC1 (fragment 741–761) with one LXXLL motif. The purified hFXR/RXRα–LBD heterodimer was crystallized in complex with FXR agonists (HNC143, HNC180, or GW4064), RXR natural ligand 9cRA, and a synthetic peptide derived from the coactivator SRC1–2 (fragment 685–700, the second LXXLL motif in SRC1) containing a single LXXLL motif. Purification of the coexpressed hFXR/RXRα–LBD heterodimer (see “Experimental procedures”) and addition of compounds and SRC1 peptide were critical for the complex crystallization. FXR–LBD/ligand structures were solved by the molecular-replacement method using previously determined FXR–LBD structure as the search probe. The heterodimer complex structures were also determined by molecular replacement method with the FXR and RXRα monomer structures as the searching models in two steps. Electron density maps showed clear features for the respective ligands in the structures, the LXXLL motifs of the SRC1 peptides, and each LBD, except for helix 2 in RXRα and some regions in FXR. The statistics for data collection and structure refinement are summarized in Table 1.
Table 1.
Statistics of crystallographic data and refinement
| Complex | HNC143–FXR–LBD | HNC180–FXR–LBD | HNC143–FXR/9cRA–RXR | HNC180–FXR/9cRA–RXR | GW4064–FXR/9cRA–RXR |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | F23 | P41212 | P212121 | P43212 |
| Cell dimensions | |||||
| a (Å) | 79.38 | 160.35 | 83.38 | 89.53 | 102.85 |
| b (Å) | 98.77 | 160.35 | 83.38 | 96.13 | 102.85 |
| c (Å) | 119.27 | 160.35 | 161.63 | 114.24 | 109.46 |
| α = β = γ (°) | 90 | 90 | 90 | 90 | 90 |
| Resolution (Å) | 2.88 | 2.60 | 2.10 | 2.95 | 3.05 |
| Rpim | 0.086 (0.507)a | 0.032 (0.267) | 0.024 (0.247) | 0.110 (0.338) | 0.020 (0.274) |
| I/σ | 6.9 (1.6) | 17.2 (3.3) | 18.9 (3.4) | 5.4 (2.0) | 19.4 (3.0) |
| Completeness (%) | 99.9 (100) | 100 (100) | 100 (100) | 98 (98.8) | 99.8 (99.7) |
| Redundancy | 6.2 (6.4) | 10.3 (10.4) | 13.1 (13.5) | 3.5 (3.6) | 13.8 (14.4) |
| Refinement | |||||
| No. of unique reflections | 20,743 | 10,107 | 32,442 | 19,737 | 11,002 |
| Rwork/Rfree (%)b | 21.1/26.2 | 19.6/25.1 | 19.8/24.6 | 22.1/28.2 | 22.5/28.8 |
| No. of atoms | |||||
| Proteins | 3954 | 1982 | 3860 | 7384 | 3638 |
| Ligand/ion | 76 | 64 | 76 | 122 | 58 |
| Water | 4 | 11 | 81 | 17 | 1 |
| Average B-factors | |||||
| Protein | 61.8 | 68 | 43 | 54.9 | 126.3 |
| ligand | 53.504 | 42.99 | 37.1 | 49.5 | 118.1 |
| RMSDc | |||||
| Bond lengths (Å) | 0.0124 | 0.0128 | 0.017 | 0.009 | 0.0118 |
| Bond angles (°) | 1.5924 | 1.6142 | 1.798 | 1.52 | 1.7318 |
| Ramachandran plot | |||||
| Favored (%) | 96.4 | 96.19 | 96.67 | 95.57 | 94.01 |
| Allowed (%) | 2.12 | 2.97 | 3.33 | 3.52 | 5.07 |
| Outliers (%) | 1.48 | 0.85 | 0 | 0.91 | 0.92 |
aThe values in parentheses refer to statistics in the highest resolution bin.
bRfactor = Σ|FP − FPcalc|/ΣFP, where FP and FPcalc are the observed and calculated structure factors, Rfree is calculated from a randomly chosen 5% of reflections that have never been used in refinement, and Rfactor is calculated for the remaining 95% of the reflections.
c RMSD is the root-mean-square deviation from ideal geometry.
Overall structure of the hFXR/RXRα heterodimer
The structures of the hFXR/RXRα LBD heterodimer complexes contain six components: two receptor LBDs, their respective ligands, and two SRC1 peptides (Fig. 1a). The heterodimer complex structure is similar to the previous published RXR LBD heterodimer complexes (15–20). Both LBDs adopted the canonical three-layered α helical sandwich fold, with FXR containing 12 α-helices (H1 to H12) and RXR containing 10 α-helices and 2 β-strands.
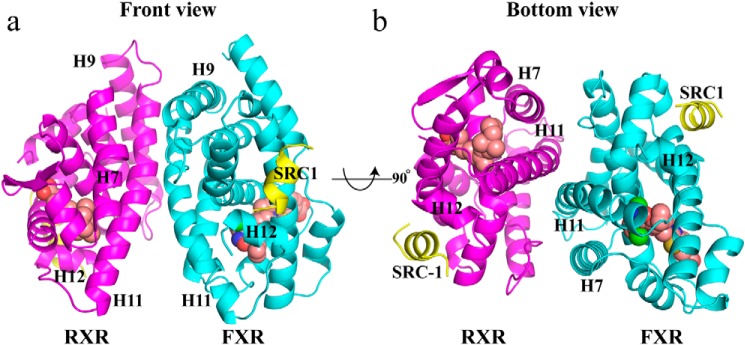
Figure 1.
FXR/RXRα heterodimer structure complexed with HNC143, 9cRA, and SRC1 peptides. a, ribbon diagram of the human FXR/RXRα heterodimer complex with two SRC1 peptides. The SRC1 peptides are in yellow. FXR is cyan, and RXRα is colored magenta. HNC143 in FXR and 9cRA in RXRα are shown in space-filling representation colored by atom type: oxygen as red, nitrogen as blue, sulfur as yellow, chlorine as green, and carbon as pink. b, heterodimer viewed from the bottom of H11, 90° rotation of a.
The FXR and RXRα ligands occupy their respective ligand-binding pockets, and both receptors adopt the active configuration, such that the H12 is folded against the main body of the LBD. This conformation generates a recognition surface (AF2 surface) constituted by mostly hydrophobic residues from H3, H4, and H12 of FXR and RXRα, which allow one SRC1 peptide to bind to each receptor. Two coactivator peptides fold as a two-turn α-helix in the heterodimer complex with the hydrophobic side chains (LXXLL) packed against the agonist-induced FXR and RXRα surfaces.
Binding characteristics of HNC compounds to hFXR
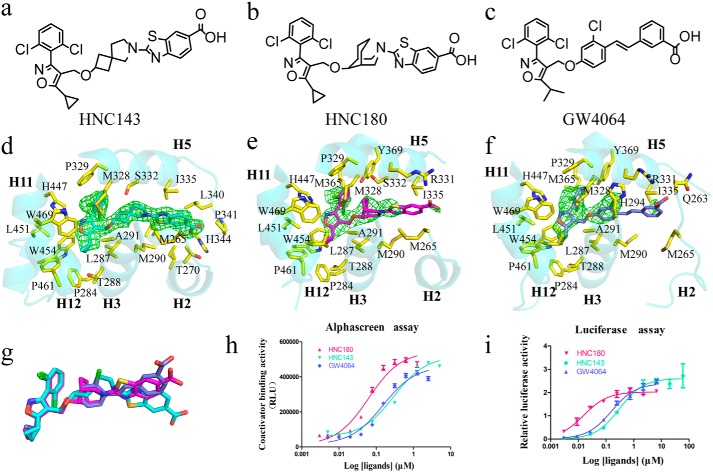
HNC compounds (HNC143 and HNC180) are GW4064 analogues (Fig. 2, a–c), sharing the similar hydrophobic head that can bind in the same position in FXR ligand-binding pocket. Compared with GW4064 and HNC143, HNC180 has a more potent EC50 in a coactivator recruitment assay (Fig. 2h) and is much more active in cell transfection assays (Fig. 2i). Electron density maps allowed unambiguous placement of HNC143, HNC180, and GW4064 into the FXR ligand-binding pocket in the heterodimer structures (Fig. 2, d–f) and monomer structures (data not shown). Overall, these binding modes are similar to that adopted by GW4064 in the monomeric FXR–LBD (21). The nonsteroid ring system of HNC143 or HNC180 is sandwiched between two hydrophobic layers of residues mainly contributed from H3, H5, and H11. The large number of hydrophobic interactions observed between HNC compounds and FXR suggests that these interactions are important for the binding. Structure superposition of HNC143 and HNC180 with GW4064 in the heterodimer structures (Fig. 2g) shows that the tail end of compound HNC143 rotated significantly. This positional difference also exits in the FXR–LBD monomer structures (data not shown). In both FXR–LBD monomer and heterodimer structures with these three ligands, the carboxylate of HNC143 forms a hydrogen bond with His344, leading to stabilization of the loop between H5 and H6, whereas HNC180 and GW4064 interact with Arg331 (Fig. S2). For HNC180 and GW4064-bound structures, the segment between H5 and H6 in FXR–LBD forms a short helix in the monomer structures, whereas it is absent in the heterodimer structure because of a lack of electron density. These results suggest that FXR has considerable conformation dynamics upon binding with different ligands or forming heterodimer with RXR.
Figure 2.
Chemical structure of the compounds and their interactions with FXR. a–c, the two-dimensional structures of HNC143, HNC180, and GW4064. d–f, ligand interactions with FXR–LBD in FXR/RXR heterodimer. The initial electron density maps calculated with σA-weighted Fo − Fc coefficients, before the placement of ligands, are contoured at 3σ, 3σ, and 2σ, respectively. The proteins are represented as cartoons. The side chains of ligand-contacting residues (yellow for carbon, red for oxygen, blue for nitrogen, and green for sulfur) and the ligands (cyan for HNC143, magenta for HNC180, and slate for GW4064) are represented by sticks. Residues lining the binding pocket located within 4 Å of the ligands are shown. g, alignment of the three ligands in the heterodimer structures. h, dose-response curves of the FXR–LBD binding with SRC1 motif in the presence of HNC143, HNC180 and GW4064 as measured by AlphaScreen assays. RLU, relative light units. i, transcriptional activation assay of FXR–LBD with HNC143, HNC180, and GW4064. (EC50: HNC143, 0.306 μm; HNC180, 0.016 μm; GW4064, 0.159 μm.) 293T cells were transiently transfected with pCMV–GAL4–DBD–hFXR–LBD, PGL5, Renilla luciferase reporter plasmids. Relative activity was defined as pGL5–luciferase activity/Renilla luciferase activity. The data are shown as means ± S.D. All experiments were repeated at least three times.
Heterodimerization and coactivator recruitment
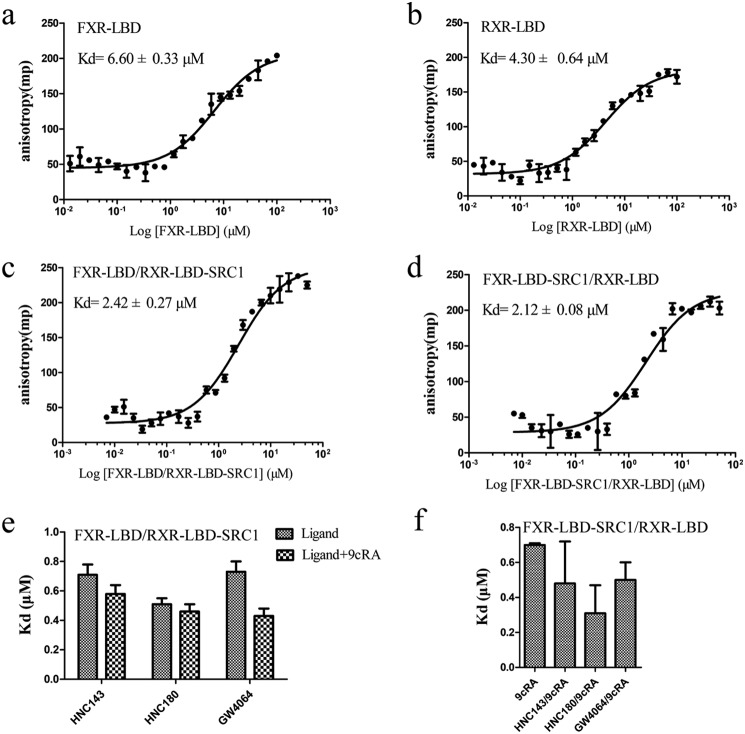
Given the importance of heterodimerization in nuclear receptor signal pathway, we addressed the question whether association of FXR and RXRα modifies the intrinsic ability of the receptors to interact with coregulators. To this end, we purified FXR–LBD and RXRα–LBD with or without fused LXXLL motifs (see “Experimental procedures”) to examine whether nuclear receptor LBD dimerization can influence the coactivator binding. The fused SRC1 to one of the two receptors in the heterodimer was designed to assess the binding of biotin-labeled SRC1 to the nonfused receptor in coactivator binding assay. To validate this approach, FXR/RXR heterodimer fused with two SRC1 peptides was tested in the fluorescence anisotropy assay. The results show that there is no binding between the two-SRC1–fused heterodimer and fluorescein-labeled peptide (Fig. S1). We then examined the SRC1 binding affinity for FXR and RXR in the context of the heterodimer and in response to different ligands (Table S1). The affinity of SRC1676–700 LXXLL motifs binding to the FXR–LBD alone (Kd = 6.60 μm) is lower than binding to the FXR–LBD in the FXR/RXR–LBD heterodimer complex (Kd = 2.42 μm) (Fig. 3, a and c). A similar effect was found in RXRα–LBD (Fig. 3, b and d). Therefore, dimerization enhances the binding affinity of FXR–LBD and RXR–LBD for LXXLL motifs. In addition, contributing to the overall stability of the receptor, FXR agonists or RXR agonist (9cRA) can enhance the SRC1 binding, and the addition of 9cRA or FXR agonists will further increase the binding affinity in FXR/RXR·SRC1 or FXR·SRC1/RXR heterodimer (Fig. 3, e and f). We further tested the EC50 of FXR ligands in coactivator recruitment assay in the context of FXR monomer or FXR/RXR heterodimer, with or without RXR agonist 9cRA (Table 2). The results show that heterodimerization with RXR will improve the EC50 of all these ligands. However, when 9cRA is added, differentiated effects of the FXR compounds were observed. Ligands HNC180 and GW4064 have better EC50 in FXR/RXR–LBD dimer; on the other hand, EC50 of HNC143, INT747, and ivermectin decrease to FXR monomer level or even lower. This result suggested that the higher-order nuclear receptor complex structure improves coactivator binding, and the synergistic effect between the two receptors ligands may have ligand selectivity.
Figure 3.
Characterization of the interaction between coactivator and FXR–LBD, RXR–LBD, and FXR/RXR–LBD. a and b, binding curve of SRC1676–700 peptide to FXR–LBD, RXR–LBD monomer without ligands, measured by fluorescence anisotropy. c and d, binding curve of SRC1676–700 peptide to FXR/RXR–LBD·SRC1 and FXR–LBD·SRC1/RXR–LBD without ligands. The heterodimeric LBD complex consists of FXR–LBD and RXR–LBD fused with SRC1 or vice versa. This allows for the free biotinylated SRC1 peptide to bind only the unfused LBD. e and f, dissociation constants of the SRC1676–700 and FXR–LBD/RXR–LBD·SRC1 complexes (e) and FXR–LBD·SRC1/RXR–LBD (f), obtained from fluorescence anisotropy-based titrations. The experiments were carried out in the presence of FXR agonists (HNC143, HNC180, and GW4064), with or without RXR agonist (9cRA). The values are means ± S.D. of three independent experiments.
Table 2.
EC50 of ligands in the coactivator recruitment assay with monomeric or heterodimeric FXR–LBD
The data are shown as the mean values ± S.D. (n = 3).
| Receptor | EC50 |
||||
|---|---|---|---|---|---|
| HNC143 | HNC180 | GW4064 | INT747 | Ivermectin | |
| μm | |||||
| FXR–LBD | 0.228 ± 0.015 | 0.043 ± 0.002 | 0.131 ± 0.033 | 0.521 ± 0.027 | 0.468 ± 0.049 |
| FXR/RXR-LBD | 0.086 ± 0.005 | 0.032 ± 0.001 | 0.036 ± 0.012 | 0.359 ± 0.008 | 0.396 ± 0.036 |
| FXR/RXR–LBD·9cRA | 0.211 ± 0.030 | 0.049 ± 0.021 | 0.050 ± 0.004 | 0.843 ± 0.159 | 0.726 ± 0.035 |
The asymmetric hFXR–RXRα heterodimer interface
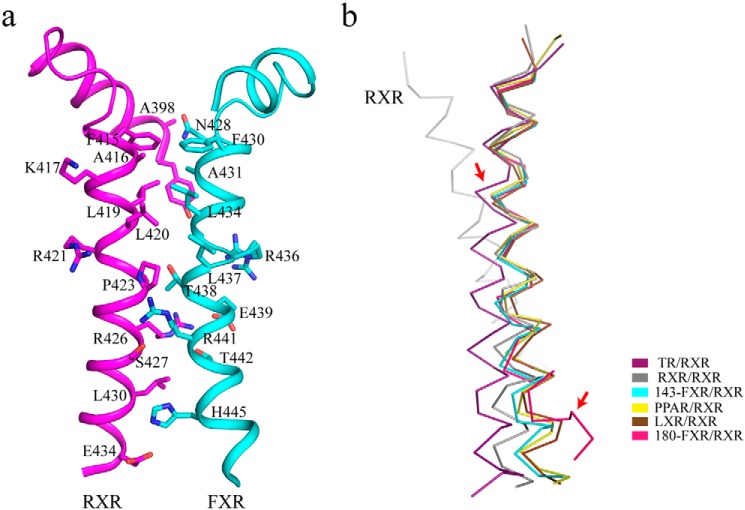
Consistent with other RXR heterodimer structures, FXR and RXRα interact via the conserved asymmetric dimer interface, largely comprised of H11 in each monomer, with additional contacts from H7 and H9, loops L8–9 and L9–10. The interactions consist of an intricate network of hydrophobic and polar interactions (Table S2), with H11 of RXRα and FXR forming a coiled coil structure as shown in Fig. 4a. During the protein purification, we found that the isolated FXR–LBD is mostly monomeric, whereas mixing equivalent FXR and RXR LBDs yields essentially heterodimer (data not shown).
Figure 4.
The FXR/RXRα heterodimer interface. a, intermolecular interactions mediated by the helices 11 of RXRα and FXR. Interacting residues on the heterodimer interface are labeled. b, alignment of different RXRα heterodimers and conformational variations of RXRα partner. Several NRs (colored as indicated) (TR/RXR: PDB code 4ZO1; PPAR/RXR: PDB code 1FM6; and LXR/RXR: PDB code 1UHL) were superimposed over the highly conserved N-terminal portion of helix 11 in RXR–LBD homodimer (PDB code 1MZN), corresponding to residues 413–427 in RXRα. TR and FXR H11 shifts are showed by red arrows.
Comparing the structurally available RXR heterodimers with RXR homodimer, RXR partners differ in the degree of bending and orientation of H11, even though they share similar electrostatic and polar contacts in the heterodimer interface. In the nonpermissive TR/RXR heterodimer, TRβ H11 has a marked shift, resulting in a rotation of H11 and H5 in RXRα and then disruption of the active conformation of RXRα (20). Although the backbone of the N terminus of H11 in the permissive RXR partners superimpose well with the RXR homodimer, noticeable shifts exist in the C terminus of H11 (Fig. 4b). The differences in the heterodimer interface (especially H11) between the permissive and nonpermissive RXR heterodimer suggest that receptor/ligand-specific bending of H11 also contributes to the cross-dimer signaling.
Role of helix 11 in cross-dimer signaling
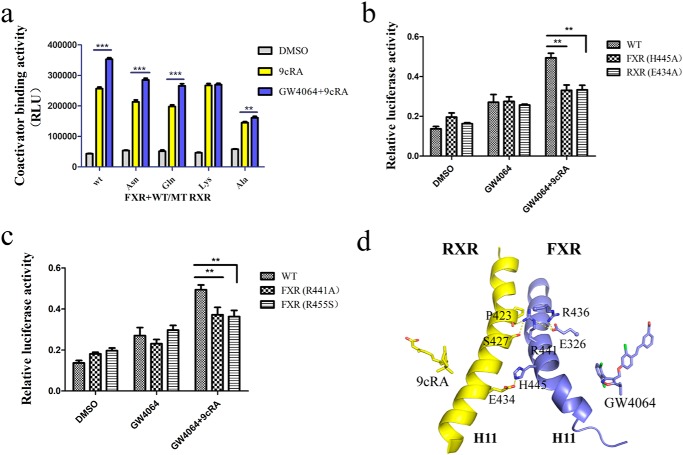
Previous studies have shown that an electrostatic tethering across the dimer interface exists in most RXR heterodimers in the plane of ligand, suggesting a conduit for structural information from the ligand to the partner receptor (22). In our FXR/RXR heterodimer structures, residue Glu434 in RXRα can form a hydrogen bond across the dimer interface with His445 in FXR (Fig. 5d). We then tested several RXR–LBD Glu434 mutations (E434N, E434Q, E434K, and E434A) with WT FXR–LBD in the coactivator binding assay, using RXRα agonist 9cRA and FXR agonist GW4064. The results showed that RXR mutants could affect the signal transduction between the heterodimer (Fig. 5a). When Glu434 in RXR was mutated to Ala, the FXR agonist could hardly increase the ability of RXR to recruit SRC1. Using FXR and RXR mutants, cell-based transfection assay demonstrated that both full-length FXR H445A and RXR E434A mutations decreased the synergistic effect of 9cRA and GW4064 (Fig. 5b). These results indicated that the interaction between the RXR Glu434 and FXR His445 in the heterodimer interface is essential for the synergistic effect. We then looked at whether His445 in FXR is conserved in other RXR partners. Structure-based sequence alignment showed that the presence of basic amino acid (His) at this particular position is unique for FXR. In other nuclear receptors, the residue that forms the electrostatic tethering with Glu434 in RXR corresponds to the position one turn following the His445 in H11 of FXR (Fig. S3).
Figure 5.
The role of helix 11 in modulating the allosteric signal. a, FXR–LBD·SRC1/RXR–LBD (WT RXR–LBD and Glu434 mutants) proteins were used in coactivator recruitment assay, and FXR agonist GW4064 (1 μm) and RXR agonist 9cRA (1 μm) were added. RLU, relative light units. b and c, 293T cells were cotransfected with full-length WT FXR or FXR mutant expression plasmids, WT RXR or RXR mutant expression plasmids, FXR-responsive luciferase reporter (pGL3-IR1), and Renilla reporter plasmids. The cells were treated with DMSO, RXR agonist 9cRA (1 μm), and FXR agonist GW4064 (1 μm) for 24 h. Relative activity was defined as pGL3-luciferase activity/Renilla activity. The values are means ± S.D. of three independent experiments. **, p < 0.001; ***, p < 0.0001 (Student's t test). d, signal transduction pathway from the FXR/RXR heterodimer interface. H11 of FXR and RXR in GW4064-bound heterodimer are shown in cartoon; interacting residues on the heterodimer interface are shown in sticks.
Using SCA, Ranganathan and co-workers (11) identified a network of 27 coevolved amino acids that are energetically coupled and mediate allosteric signaling in the RXR heterodimers. The mutagenesis study showed that FXR E326A and the corresponding mutations in other permissive partners LXR, PPAR, and Nurr1 exhibit different transcriptional activity in RXR heterodimer. In our GW4064–FXR/9cRA–RXR structure, residue Glu326 in FXR forms salt bridges with and stabilizes Arg441 and Arg436, which participate in the formation of heterodimer interface (Fig. 5d). The E326A mutation would disrupt the interaction network and hinder the allosteric signal transduction from RXR, thereby decreasing the transcriptional activity of FXR. Furthermore, we generated other FXR mutations (R441A and R455S) that belong to the SCA network; each of them decreased the synergistic effect of the two receptor agonists in the cell transactivation assay (Fig. 5c). Taken together, these findings are directly in line with previous findings found in other RXR heterodimers: there are two patches of complementary clustered residues for signal transduction through H11 in FXR–RXR heterodimer. One is through the physically connected AF2 hydrophobic core of the receptor across the dimer interface in the SCA network. The other one connects the dimer interface via an electrostatic tethering across H11 at the plane of the ligands (Fig. S4).
Ligand effect in the heterodimer interface and helix 11 conformation
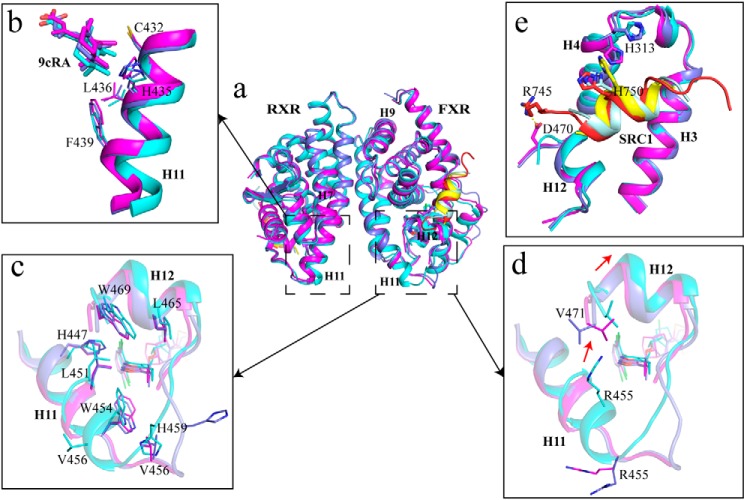
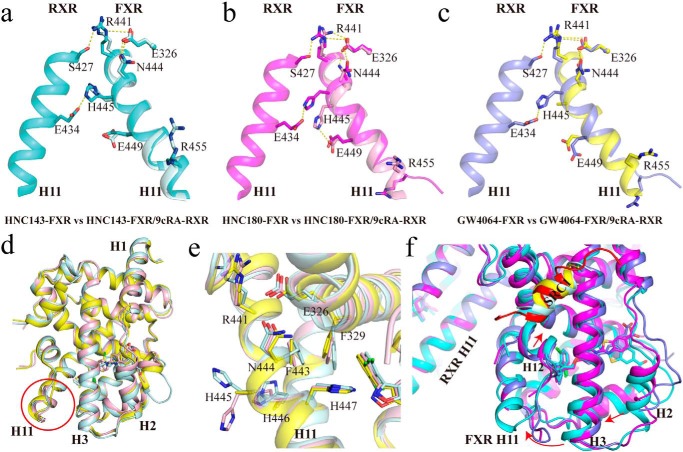
We then addressed the question of whether the dimerization interface could be altered depending on the type of bound ligands, thereby providing a structure basis for the allosteric communication between RXR heterodimers and diverse gene activation profiles (23). In our three FXR–RXR heterodimer structures, all RXRα agonists are 9cRA (Fig. S5). Structure superposition of these complexes showed that the RXR parts superpose well (Fig. 6a), with root-mean-square deviations being 0.460 Å (GW4064–FXR/9cRA–RXR versus HNC143–FXR/9cRA–RXR) and 0.457 Å (HNC180–FXR/9cRA–RXR versus HNC143–FXR/9cRA–RXR) over RXRα Cα atoms. One significant difference is observed in the C terminus of H11 of RXR in HNC143–FXR/9cRA–RXR heterodimer, among which movement of residues (Cys432, His435, Leu436, and Phe439) induce positional change of the ligand 9cRA correspondingly (Fig. 6b). The most significant difference is located in the C terminus of H11 of FXR, with the notable structural change starting from Asn444 and His445. Compared with HNC143 bound heterodimer, the main chain hydrogen bonds formed by Asn444–Glu448 and His445–Glu449 are lost in HNC180- and GW4064-bound heterodimers (Fig. S6). These two pairs of hydrogen bonds are necessary for maintaining a stable helix structure; thus, losing those bonds will trigger the C terminus of helix 11 to turn into a loop. Furthermore, residues His447, Leu451, Trp454, Leu465, and Trp469 shift toward the bound compounds HNC180 or GW4064 (Fig. 6c), leading to a closer ligand/receptor interaction. The change from helix conformation to loop also orientates residue Leu441 to the hydrophobic core, and hydrophilic residue His459 becomes solvent-exposed. It is also worth mentioning that the position of Arg455 changed significantly in the C terminus of H11 in these heterodimeric structures with different FXR agonists bound. As shown in Fig. 6d, Arg455 in HNC143-bound FXR pushed against the C terminus of H12, potentially rendering the H12 unstable in HNC143–FXR/9cRA–RXR heterodimer. Thus, it may explain why the coactivator recruitment ability of HNC143 is lower than that of HNC180, because a stable AF2 surface is necessary for coactivator recruitment. In addition, five more residues of coactivator SRC1 were identified in HNC180–FXR/9cRA–RXR than in HNC143- or GW4064-bound heterodimer, resulting in more interactions between FXR (His313 in H4 and Asp470 in H12) and SRC1 in HNC180–FXR/9cRA–RXR complex (Fig. 6e).
Figure 6.
The ligand-induced structural change in the C terminus of helix 11. a, alignment of the three FXR/RXR heterodimers. Cyan, heterodimer with HNC143; magenta, heterodimer with HNC180; slate, heterodimer with GW4064; pale cyan, SRC1 in HNC143 complex; red, SRC1 in HNC180 complex; yellow, SRC1 in GW4064 complex. b, shift in C terminus of H11 in RXR induced by heterodimerization with FXR and corresponding change in the position of RXR ligands 9cRA. c and d, the details of the difference on H11 and H12 with different ligands in FXR. e, coactivator binding interface (AF2 surface) of FXR in three heterodimers.
Interestingly, H11 is almost identical in the monomeric FXR structures with different ligands (HNC143, HNC180, and GW4064) bound. Compared with the monomer structures, the C terminus of H11 in FXR in the heterodimer changed significantly in the HNC180- and GW4064-bound FXR/RXR heterodimers, but not in HNC143-bound heterodimer (Fig. 7, a–d, Fig. S7). Further analysis of these six structures revealed changes in a network of residues. In HNC180–FXR monomer structure, His445 forms a hydrogen bond with Glu449, but when heterodimerized with RXR, this interaction within FXR is replaced by a hydrogen bond between FXR His445 and RXR Glu434, and at the same time FXR Arg441 forms a hydrogen bond with RXR Ser427. These changes induce significant movement of FXR residues Arg441, Asn444, and His445, disrupting the hydrogen-bond network (Asn444–Glu448 and His445–Glu449) within FXR. Similar scenario is also presented in the GW4064–FXR/9cRA–RXR structure. However, in the HNC143–FXR monomer structure, His445 does not interact with Glu449, whereas in heterodimeric structure with RXR, no significant changes are observed for residues Asn444 and His445 in FXR and overall H11. Interestingly, the superposition of these three ligand-bound FXR–LBD monomer structures reveals only subtle changes induced by the different ligands, with the most notable change occurred in the side chain of His445 (Fig. 7e).
Figure 7.
Ligand effect on the heterodimer interface and helix 11 conformation. a–c, overlay of FXR–LBD and FXR/RXR–LBD. Helix 10/11 is shown in pale cyan for HNC143–FXR–LBD and in cyan for HNC143–FXR/9cRA–RXR (a), in pink for HNC180–FXR–LBD and in magenta for HNC180–FXR/9cRA–RXR (b), and in yellow for GW4064–FXR–LBD and in slate for GW4064–FXR/9cRA–RXR(c). d, alignment of the three ligand-bound FXR–LBD monomer structures show the conserved C terminus of FXR H11. e, ligand-induced side-chain difference in H11 within three different ligand-bound monomeric FXR–LBD. f, the conformation change induced by the tail of FXR ligands. Alignment of three different FXR ligand-bound FXR/RXR heterodimers via superposition on RXR and colored as in a–c. Arrows show the shifts of H2, H3, H11, H12, and SRC1 in FXR.
When superposing the three FXR/RXR heterodimers via RXR, we observed that different orientations of the ligand tails in FXR ligand-binding pocket induce a shift in H2, H3, and H11 and then affect the location of AF2, potentially influencing the affinity for the coactivator binding to the heterodimer (Fig. 7f). Therefore, the change of side chain His445 in these compound-bound complexes, combined with distinct conformation of FXR ligands, results in different conformations for H11 of FXR in the heterodimer. Taken together, these structural evidences demonstrated that different FXR ligands can exert unique influence on H11 and then affect the stability of H11 and H12 of FXR when dimerized with RXR, thereby modulating the recruitment of coactivator and transactivation of target genes.
Discussion
The liver is a vital organ with critical functions in metabolism. FXR is an important transcriptional regulator in liver. Except for regulating bile acid cholesterol and lipid and glucose metabolism, recent studies found that FXR has much broader role in liver regeneration, energy balance, and postnatal liver maturation (24–26). These functions indicate that FXR is an attractive therapeutic target for liver diseases. Structural information of FXR will greatly contribute to the rational drug design.
In this study, we provide a structural and biochemical characterization of farnesoid X receptor in complex with its obligate heterodimer partner RXRα. In agreement with previous studies, especially a recent FXR/RXR heterodimer structure with known FXR ligand WAY-362450 (27), we observed that FXR/RXR heterodimer has the similar heterodimer interface with other RXR heterodimers. Our structural data provide an allosteric mechanism in which communications across the dimer interface, together with the two respective agonists, enhance the affinity of FXR and RXR for binding to individual LXXLL motifs. The increase in binding affinity for coactivators in nuclear receptor complexes versus individual receptors has also previously been noted for RAR, CAR (constitutive androstane receptor), and PXR (19, 28, 29).
It has previously been predicted that receptors whose C termini lie at the same position as PPARγ should be permissive for activation by RXR ligands, because Lys431 in RXR will form a salt bridge with the C terminus of the AF2 in the partner receptor (15). FXR, having one residue shorter than PPARγ at the C terminus, is a RXR permissive partner. However, in our structures, the RXR did not shift to accommodate this one residue difference as was predicted. Therefore, the permissiveness for activation by RXR ligands is not solely determined by the position of partner's C terminus. There should be other mechanisms to determine the permissiveness or nonpermissiveness. When superposing RXR heterodimer structures using the conserved RXR H11 as the reference, we found that the orientation of H11 in heterodimer partner varies by receptors, and the permissive partner have more bending than nonpermissive partners. This can allow permissive heterodimers to sense both receptor ligands, whereas nonpermissive heterodimers only respond to partner ligands.
Previous studies have shown that the C terminus of helix 11 could be differentially positioned by distinct ligands, thereby controlling the packing of helix 12 in the agonist conformer in NR monomer, especially in PPARγ (30). In our structures, we first identified that the C terminus of the H11 in FXR can have different conformation when dimerized with RXR, and the change is ligand-specific. The C terminus of H11 of FXR in the HNC180/GW4064 heterodimer becomes a loop, instead of α-helix, and brings a closer interaction with ligand. This change has not been found in other RXR heterodimers. We speculate that this is a new regulation mechanism in RXR heterodimers. Through the conformational change in H11 of FXR, the synergistic effect of the two receptor ligands can be modulated.
Here, we also demonstrated that heterodimerization can improve the EC50 of FXR agonists in the coactivator recruitment assay. However, the synergic effect only exists in specific ligand pairs. This is due to the diverse effect different ligands could exert on the heterodimer interface. Drug screening efforts have identified small-molecule receptor modulators using only individual LBDs, usually not considering that allosterically acting ligands could be identified if the entire receptor complex is used in the screening. Although the structure for the ligand–receptor complexes are well studied, it is still impossible to precisely predict the correlation between ligand structure and physiological function. Recently, Gabler et al. (31) have developed imatinib and its optimized derivatives as first-in-class allosteric FXR modulators without RXR activity. This allosteric FXR modulation could open a new avenue to gene-selective FXR modulators. Our study presents a very intricate regulation mechanism in ligand-induced FXR/RXR heterodimer signaling.
In summary, our results provide structure insights into the FXR/RXR heterodimer with different combination of ligands. Particularly, the modulation through the conformational change in helix 11 of FXR in the heterodimer will greatly extent our understanding on the RXR heterodimer signal transduction. Importantly, further analysis of allosteric effects on different gene transcription profiles, combined with structural information of nuclear receptor/ligand interactions, will provide a unique opportunity for rational drug design.
Experimental procedures
Compounds
HNC143 and HNC180 were synthesized based on the published patent WO2017118294. 1H NMR spectra and mass spectra data (Fig. S8) confirm the structure correctness and purity of the compounds. GW4064 was purchased from Selleck. INT747 was purchased from MedChem Express. Ivermectin and 9cRA were purchased from sigma.
Protein expression and purification
The RXR–LBD (residues 225–462) was expressed from the expression vector pET21a (Novagen) without a His tag or from vector pET28a (Novagen) with an N-terminal His6 tag, and the FXR–LBD (225-472) was expressed as a N-terminal His6 fusion protein from the expression vector pET24a (Novagen). For FXR–RXRα–LBD heterodimer protein production, Rosetta 2 cells transformed with two expression plasmids (RXR-pET21a and FXR-pET24a) were grown in LB broth at 37 °C to an A600 of ∼1.0 and induced with 0.1 mm isopropyl β-d-thiogalactopyranoside for 16 h when cooled down at 16 °C. The cells were harvested, resuspended in 80 ml of extraction buffer A (20 mm Tris, pH 8.0, 150 mm NaCl, 10% glycerol, 1 mm tris-(2-carboxyethyl) phosphine hydrochloride) per 1 liter of cells, and passed three times through a French press with pressure set at 1000 pa. The lysate was centrifuged at 12,000 rpm for 40 min, and the supernatant was loaded on a source Q column. The column was equilibrated with buffer B (20 mm Tris, pH 8.0, 5 mm NaCl, 10% glycerol, 1 mm tris-(2-carboxyethyl) phosphine hydrochloride), and protein samples were eluted with 100 ml of buffer C (20 mm Tris, pH 8.0, 1 m NaCl, 10% glycerol, 1 mm tris-(2-carboxyethyl) phosphine hydrochloride) using gradient. The FXR/RXRα–LBD heterodimer was further purified by a Superdex 200 column in buffer D (25 mm Tris, pH7.2, 150 mm NaCl, 5 mm DTT, 0.5 mm EDTA). To prepare the protein–ligand–coactivator complex, we added a 4-fold molar excess of SRC1–2 (685–700) peptide; 4-fold molar excess of HNC143, HNC180, or GW4064; and 4-fold molar excess of 9cRA to the purified heterodimer, followed by filter concentration to 8.8 mg/ml.
For coactivator recruitment assays, RXRα–LBD·SRC1 (RXR residue 225–462) and FXR–LBD·SRC1 (FXR residue 225–472) fusion proteins were designed as a human SRC1 coactivator fragment (residue 678–700, SSHSSLTERHKILHRLLQEGSPS) fused in the C terminus of the two receptors, using a 5-residue glycine–serine linker (GGSGG). The purification procedure was the same as described above. The SRC1 fused plasmids were produced by PCR. The primer sequences used are listed are as follows: pET24a–FXR–LBD·SRC1 sense, 5′-TAAGAAGGAGATATACATATGAAAATTGGTCATCATCATCATCATCAT-3′, and antisense, 5′-TGGTGGTGCTCGAGTGCGGCCGCCTATGAGGGGCTACCCTCCTGTAAGAGCCGGTGTAGAATTTTATGCCGTTCTGTCAATGAGCTATGAGAAGATCCTCCAGATCCACCCTGCACGTCCCAGATTTC-3′; and pET21a–RXRα–LBD·SRC1 sense, 5′-TAAGAAGGAGATATACATATGAGCGCCAACGAGGACATG-3′, and antisense, 5′-ACGGAGCTCGAATTCGGATCCCTATGAGGGGCTACCCTCCTGTAAGAGCCGGTGTAGAATTTTATGCCGTTCTGTCAATGAGCTATGAGAAGATCCTCCAGATCCACCAGTCATTTGGTGCGGCGC-3′. The reverse complementary sequences of the coding sequence of linker (GGSGG) and SRC1 peptide (residue 678–700, SSHSSLTERHKILHRLLQEGSPS) in the antisense primer are underlined.
Crystallization and data collection
The HNC143–9cRA FXR/RXR heterodimer crystals were grown in hanging drops that contained 1 μl of the protein-ligand complex and 1 μl of well buffer that contained 25% PEG 4K, 200 mm ammonium sulfate, and 0.1 m trisodium citrate, pH 5.6, at 20 °C. HNC180/GW4064–9cRA FXR/RXR heterodimer crystals were grown in 0.1 m sodium acetate trihydrate, pH 7.0, 12% PEG3350 at 4 °C. Crystals grew to ∼100–500 μm in dimension in 2–3 weeks and diffracted to in the range between 2.1 and 3.05 Å resolution. The crystals were frozen with liquid nitrogen prior to data collection.
The diffraction data were collected in BL17U1, BL18U1, and BL19U1 Beamlines at Shanghai Synchrotron Radiation Facility of China. The diffraction data were indexed, reduced, merged, and scaled with MOSFLM (32) and AIMLESS (33, 34) from the CCP4 suite of programs (35).
Structure determination and refinement
The FXR monomer structures were determined by molecular replacement with the MOLREP software from CCP4 package (35), using the apo FXR structure (PDB code 3DCT) as the initial search model. For HNC143–FXR–LBD structure, the molecular replacement solution was obtained with a correlation coefficient (CC) of 0.44 and an R factor of 0.81, with the second unrelated peak having a CC of 0.31 and an R factor of 0.87. For HNC180–FXR–LBD, the solution was obtained with a CC of 0.78 and an R factor of 0.36, with the second unrelated peak having a CC of 0.42 and an R factor of 0.58. The FXR/RXR heterodimer structures were determined by molecular replacement with MOLREP in two steps. The apo FXR structure (PDB code 3DCT) was used as the search model in the first step. After obtaining the correct solution for FXR, the apo RXR structure (PDB code 1FBY) was used to locate the position of RXR, whereas FXR was fixed. For HNC143–FXR/9cRA–RXR, in the first step, we obtained top solution with a CC of 0.46 and an R factor of 0.55 in the searching for FXR, with the second unrelated peak having a CC of 0.37 and an R factor of 0.60. In the second step searching for RXR, the top solution has a CC of 0.65 and an R factor of 0.45, with the second unrelated peak having a CC of 0.39 and an R factor of 0.58. The other two heterodimer structures were determined by molecular replacement using the HNC143–FXR/9cRA–RXR as search model. For HNC180–FXR/9cRA–RXR, the solution was obtained with a CC of 0.68 and an R factor of 0.38, and the second unrelated peak had a CC of 0.36 and an R factor of 0.56. For GW4064–FXR/9cRA–RXR, the solution was obtained with a CC of 0.65 and an R factor of 0.50, and the second unrelated peak had a CC of 0.41 and an R factor of 0.66. Model building and refinement was progressed with REFMAC (36) and COOT (37). For HNC143 heterodimer complex structure, the model refinement was optimized using PDB-REDO (38). The statistics of the structure refinement are summarized in Table 1.
Fluorescence anisotropy assay
A fluorescence anisotropy assay was conducted based on fluorescence signal differences between free and protein-bound fluorescein-labeled peptide. A fluorescein-labeled SRC1 peptide (5-FAM-CPSSHSSLTERHKILHRLLQEGSPS), residues 676–700, was synthesized. The reaction system was composed of 50 nm FAM-SRC1 peptide, assay buffer (25 mm HEPES, pH 7.5, 100 mm NaCl, 5% glycerol, and 1 mm DTT), and various concentrations of protein. Ligands were added at the concentrations 3–6-fold of the receptors. The mixture was incubated for 1 h at room temperature and then examined by 2104 Envision multilabel reader (PerkinElmer Life Sciences) using FITC FP dual module with an excitation filter of FITC FP 480 and an emission filter of FITC FP P-pol535 and S-pol535. Curve fitting was carried out using Prism 5.0 (GraphPad). Dissociation constants (Kd) were calculated by fitting to a nonlinear regression, one-site saturation binding model.
Coactivator recruitment assay
The binding affinity of the compound to FXR–LBD was determined by AlphaScreen assays using a hexahistidine detection kit from PerkinElmer Life Sciences. The experiments were conducted with 100 nm receptor LBD and 50–200 nm biotinylated SRC1–2 peptides in the presence of 10 μg/ml donor and acceptor beads in a buffer containing 50 mm MOPS, 50 mm NaF, 0.05 mm CHAPS, and 0.1 mg/ml BSA, all adjusted to pH 7.4. Protein, peptide, ligand, and beads were added together in the buffer, followed by incubation for 2 h in the dark. The signals were measured on a plate reader (PerkinElmer Life Sciences). Statistical analyses were performed with GraphPad Prism.
The peptide with an N-terminal biotinylation is listed below
SRC1–2 (SRC1 residue 676–699; SPSSHSSLTERHKILHRLLQEGSP), FXR, and RXR mutant expression plasmids were generated by site-mutation PCR (from Guangzhou IGE Biotech) and verified by DNA sequencing. Statistical analyses were performed with GraphPad Prism.
Cell culture and Luciferase reporter assay
HEK 293T cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum (Gibco), and cultured at 37 °C in 5% CO2 atmosphere. The cells were plated in 96-well plates at 1.5 × 104/well, 25 ng/well pCMV–GAL4—DBD–hFXR–LBD expression vector, 25 ng/well PGL5 luciferase, and 5 ng/well Renilla luciferase reporter plasmids were transiently transfected into cells using 0.17 μl/well Lipofectamine 2000 (Invitrogen). 8 h after transfection, the cells were treated with FXR agonists in fresh Dulbecco's modified Eagle's medium. Compounds were dissolved in DMSO and added in the concentrations indicated. Luciferase activities were measured after additional 24 h using a Dual-Luciferase reporter assay system (Promega). Relative activity was defined as pGL5–luciferase activity/Renilla luciferase activity. All experiments were repeated at least three times. Statistical analyses were performed with GraphPad Prism.
For the full-length receptor cotransfection assay, 293T cells were transfected with an IR1 (pGL3–FXRE–luc) luciferase reporter plasmid, expression plasmids for FXR, RXRα (pCDNA3.1–hFXR and pCDNA3.1–hRXRα), and Renilla using Lipofectamine 2000. The other procedures were the same as mentioned above.
Author contributions
N. W. and J. L. conceptualization; N. W., Q. Z., J. X., and J. Z. data curation; N. W. formal analysis; N. W. writing-original draft; N. W. and J. L. writing-review and editing; J. Z. and J. L. supervision; J. L. funding acquisition; J. L. project administration.
Supplementary Material
Acknowledgments
We thank the staff at Beamlines 17U1, 18U1, and 19U1 of the Shanghai Synchrotron Radiation Facility of China for support in the diffraction data collection. We also gratefully thank the Guangzhou Branch of the Supercomputing Center of the Chinese Academy of Sciences for support.
This work was supported by National Key Research and Development Program Grant 2017YFA0504104, National Natural Science Foundation of China Grant 31770817, Guangdong Science and Technology Department Grant 2015A030312017, Guangdong Provincial Key Laboratory of Biocomputing Grant 2016B030301007, and National R&D Infrastructure and Facility Development Program of China, “Fundamental Science Data Sharing Platform” Grant DKA2017-12-02-22. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S8.
- FXR
- farnesoid X receptor
- RXR
- retinoid X receptor
- NR
- nuclear receptor
- PPAR
- peroxisome proliferator activator receptor
- TR
- thyroid hormone receptor
- RAR
- retinoic acid receptor
- LBD
- ligand-binding domain
- AF2
- activation function-2
- 9cRA
- 9-cis-retinoic acid
- FXRE
- FXR response element
- IR1
- single inverted repeat
- SCA
- statistic coupling analysis
- SRC1
- steroid receptor coactivator 1
- PDB
- Protein Data Bank
- CC
- correlation coefficient.
References
- 1. Lee F. Y., Lee H., Hubbert M. L., Edwards P. A., and Zhang Y. (2006) FXR, a multipurpose nuclear receptor. Trends Biochem. Sci. 31, 572–580 10.1016/j.tibs.2006.08.002 [DOI] [PubMed] [Google Scholar]
- 2. Wang Y. D., Chen W. D., Moore D. D., and Huang W. (2008) FXR: a metabolic regulator and cell protector. Cell Res. 18, 1087–1095 10.1038/cr.2008.289 [DOI] [PubMed] [Google Scholar]
- 3. Wang H., Chen J., Hollister K., Sowers L. C., and Forman B. M. (1999) Endogenous bile acids are ligands for the nuclear receptor FXR BAR. Mol. Cell 3, 543–553 10.1016/S1097-2765(00)80348-2,10.1016/S1097-2765(00)80484-0 [DOI] [PubMed] [Google Scholar]
- 4. Fiorucci S., Cipriani S., Mencarelli A., Baldelli F., Bifulco G., and Zampella A. (2011) Farnesoid X receptor agonist for the treatment of liver and metabolic disorders: focus on 6-ethyl-CDCA. Mini. Rev. Med. Chem. 11, 753–762 10.2174/138955711796355258 [DOI] [PubMed] [Google Scholar]
- 5. Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., and Yamamoto K. R. (1998) Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12, 3343–3356 10.1101/gad.12.21.3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., Mcmorris T., Lamph W. W., Evans R. M., and Weinberger C. (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81, 687–693 10.1016/0092-8674(95)90530-8 [DOI] [PubMed] [Google Scholar]
- 7. Chambon P. (1996) A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954 10.1096/fasebj.10.9.8801176 [DOI] [PubMed] [Google Scholar]
- 8. Schuetz E. G., Strom S., Yasuda K., Lecureur V., Assem M., Brimer C., Lamba J., Kim R. B., Ramachandran V., Komoroski B. J., Venkataramanan R., Cai H., Sinal C. J., Gonzalez F. J., and Schuetz J. D. (2001) Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome p450. J. Biol. Chem. 276, 39411–39418 10.1074/jbc.M106340200 [DOI] [PubMed] [Google Scholar]
- 9. Mangelsdorf D. J., and Evans R. M. (1995) The RXR heterodimers and orphan receptors. Cell 83, 841–850 10.1016/0092-8674(95)90200-7 [DOI] [PubMed] [Google Scholar]
- 10. Kliewer S. A., Umesono K., Noonan D. J., Heyman R. A., and Evans R. M. (1992) Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358, 771–774 10.1038/358771a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shulman A. I., Larson C., Mangelsdorf D. J., Ranganathan R. (2004) Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116, 417–429 10.1016/S0092-8674(04)00119-9 [DOI] [PubMed] [Google Scholar]
- 12. Forman B. M., Umesono K., Chen J., and Evans R. M. (1995) Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81, 541–550 10.1016/0092-8674(95)90075-6 [DOI] [PubMed] [Google Scholar]
- 13. Leblanc B. P., and Stunnenberg H. G. (1995) 9-cis Retinoic acid signaling: changing partners causes some excitement. Genes Dev. 9, 1811–1816 10.1101/gad.9.15.1811 [DOI] [PubMed] [Google Scholar]
- 14. Fernandez E. J. (2018) Allosteric pathways in nuclear receptors: potential targets for drug design. Pharmacol. Ther. 183, 152–159 10.1016/j.pharmthera.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gampe R. T. Jr., Montana V. G., Lambert M. H., Miller A. B., Bledsoe R. K., Milburn M. V., Kliewer S. A., Willson T. M., and Xu H. E. (2000) Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell 5, 545–555 10.1016/S1097-2765(00)80448-7 [DOI] [PubMed] [Google Scholar]
- 16. Bourguet W., Vivat V., Wurtz J. M., Chambon P., Gronemeyer H., and Moras D. (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5, 289–298 10.1016/S1097-2765(00)80424-4 [DOI] [PubMed] [Google Scholar]
- 17. Svensson S., Ostberg T., Jacobsson M., Norström C., Stefansson K., Hallén D., Johansson I. C., Zachrisson K., Ogg D., and Jendeberg L. (2003) crystal structure of the heterodimeric complex of the LXRα and RXRβ ligand binding domains in a fully agonistic conformation. EMBO J. 22, 4625–4633 10.1093/emboj/cdg456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suino K., Peng L., Reynolds R., Li Y., Cha J. Y., Repa J. J., Kliewer S. A., and Xu H. E. (2004) The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol. Cell 16, 893–905 10.1016/S1097-2765(04)00727-0,10.1016/j.molcel.2004.11.036 [DOI] [PubMed] [Google Scholar]
- 19. Wallace B. D., Betts L., Talmage G., Pollet R. M., Holman N. S., and Redinbo M. R. (2013) Structural and functional analysis of the human nuclear xenobiotic receptor PXR in complex with RXRα. J. Mol. Biol. 425, 2561–2577 10.1016/j.jmb.2013.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kojetin D. J., Matta-Camacho E., Hughes T. S., Srinivasan S., Nwachukwu J. C., Cavett V., Nowak J., Chalmers M. J., Marciano D. P., Kamenecka T. M., Shulman A. I., Rance M., Griffin P. R., Bruning J. B., and Nettles K. W. (2015) Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat. Commun. 6, 8013 10.1038/ncomms9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akwabi-Ameyaw A., Bass J. Y., Caldwell R. D., Caravella J. A., Chen L., Creech K. L., Deaton D. N., Jones S. A., Kaldor I., Liu Y., Madauss K. P., Marr H. B., McFadyen R. B., Miller A. B., Navas F., III, et al. (2008) Conformationally constrained farnesoid X receptor (FXR) agonists: naphthoic acid-based analogs of GW 4064. Bioorg. Med. Chem. Lett. 18, 4339–4343 10.1016/j.bmcl.2008.06.073 [DOI] [PubMed] [Google Scholar]
- 22. Nettles K. W., and Greene G. L. (2005) Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 67, 309–333 10.1146/annurev.physiol.66.032802.154710 [DOI] [PubMed] [Google Scholar]
- 23. Downes M., Verdecia M. A., Roecker A. J., Hughes R., Hogenesch J. B., Kast-Woelbern H. R., Bowman M. E., Ferrer J. L., Anisfeld A. M., Edwards P. A., Rosenfeld J. M., Alvarez J. G., Noel J. P., Nicolaou K. C., and Evans R. M. (2003) A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell 11, 1079–1092 10.1016/S1097-2765(03)00104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim K. H., and Moore D. D. (2017) Regulation of liver energy balance by the nuclear receptors farnesoid X receptor and peroxisome proliferator activated receptor α. Dig. Dis. 35, 203–209 10.1159/000450912 [DOI] [PubMed] [Google Scholar]
- 25. Li G., and Guo L. G. (2015) Farnesoid X receptor, the bile acid sensing nuclear receptor, in liver regeneration. Acta Pharm. Sin. B 5, 93–98 10.1016/j.apsb.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng L., Piekos S. C., Guo G. L., and Zhong X. B. (2017) Role of farnesoid X receptor in the determination of liver transcriptome during postnatal maturation in mice. Nucl. Receptor. Res. 4, 101308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng W., Lu Y., Tian S., Ma F., Wei Y., Xu S., and Li Y. (2018) Structural insights into the heterodimeric complex of the nuclear receptors FXR and RXR. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pogenberg V., Guichou J. F., Vivat-Hannah V., Kammerer S., Pérez E., Germain P., de Lera A. R., Gronemeyer H., Royer C. A., and Bourguet W. (2005) Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J. Biol. Chem. 280, 1625–1633 10.1074/jbc.M409302200 [DOI] [PubMed] [Google Scholar]
- 29. Wright E., Vincent J., Fernandez E. J. (2007) Thermodynamic characterization of the interaction between CAR/RXR and SRC-1 peptide by isothermal titration calorimetry. Biochemistry 46, 862–870 10.1021/bi061627i [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bruning J. B., Parent A. A., Gil G., Zhao M., Nowak J., Pace M. C., Smith C. L., Afonine P. V., Adams P. D., Katzenellenbogen J. A., and Nettles K. W. (2010) Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 6, 837–843 10.1038/nchembio.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gabler M., Kramer J., Schmidt J., Pollinger J., Weber J., Kaiser A., Löhr F., Proschak E., Schubert-Zsilavecz M., and Merk D. (2018) Allosteric modulation of the farnesoid X receptor by a small molecule. Sci. Rep. 8, 6846 10.1038/s41598-018-25158-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 34. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 38. Joosten R. P., Long F., Murshudov G. N., and Perrakis A. (2014) The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 10.1107/S2052252514009324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.