Abstract
Ubiquitin (Ub) signaling plays a key regulatory role in nearly every aspect of eukaryotic biology and is initiated by E1 enzymes that activate and transfer Ub to E2 Ub-conjugating enzymes. Despite Ub E1's fundamental importance to the cell and its attractiveness as a target for therapeutic intervention in cancer and other diseases, its only available structural information is derived from yeast orthologs of human ubiquitin-like modifier–activating enzyme 1 (hUBA1). To illuminate structural differences between yeast and hUBA1 structures that might be exploited for the development of small-molecule therapeutics, we determined the first crystal structure of a hUBA1–Ub complex. Using structural analysis, molecular modeling, and biochemical analysis, we demonstrate that hUBA1 shares a conserved overall structure and mechanism with previously characterized yeast orthologs, but displays subtle structural differences, particularly within the active site. Computational analysis revealed four potential ligand-binding hot spots on the surface of hUBA1 that might serve as targets to inhibit hUBA1 at the level of Ub activation or E2 recruitment or that might potentially be used in approaches such as protein-targeting chimeric molecules. Taken together, our work enhances our understanding of the hUBA1 mechanism, provides an improved framework for the development of small-molecule inhibitors of UBA1, and serves as a stepping stone for structural studies that involve the enzymes of the human Ub system at the level of both E1 and E2.
Keywords: ubiquitin, enzyme mechanism, enzyme structure, structure-function, X-ray crystallography, conformational change, protein degradation, adenylation, E1, thioester
Introduction
Ubiquitination is an essential reversible post-translational modification that regulates nearly every cellular process from DNA damage repair to cell cycle progression (1). Modification of a target protein by ubiquitin (Ub)2 can result in several outcomes, including proteasomal degradation, cellular relocalization, and altered protein conformations, interactions, and activities (2). Given the central role ubiquitination plays in so many cellular processes, dysregulation of Ub signaling has been found to be associated with a number of human pathologies such as cancer and neurodegenerative diseases (3). Thus, the ubiquitin–proteasome system provides an attractive target for development of small molecule therapeutics for several diseases with proof of principle provided by proteasome inhibitors already in use in the clinic to treat multiple myeloma and mantle cell lymphoma patients (4).
To prevent dysregulation in healthy cells, conjugation of ubiquitin molecules to target proteins is a specific and highly regulated multistep process involving a three-enzyme cascade (5–8). The first enzyme in the cascade, ubiquitin-activating enzyme E1, activates Ub in a two-step reaction that involves adenylation of the Ub C terminus followed by formation of a thioester bond between the Ub C terminus and the E1 catalytic cysteine (9, 10). The resulting E1∼Ub intermediate next recruits E2-conjugating enzyme, placing the E2 catalytic cysteine in proximity of the E1∼Ub thioester bond to facilitate thioester transfer of Ub from the E1 to E2 catalytic cysteine (11–13). The E2∼Ub thioester intermediate product then dissociates from the E1 enzyme and functions with several families of E3 ubiquitin ligases to conjugate the Ub molecule to a lysine side chain, forming a stable isopeptide bond (14–16). Ub can be attached to target proteins as a single molecule or as polymeric chains linked together through specific lysine residues on Ub, and the nature of these Ub linkages is a major determinant of the biological outcome of ubiquitination (2, 17).
Given its role as the gatekeeper of the Ub conjugation cascade, the E1 enzyme has been intensely studied at the biochemical and structural levels. Previously, structures of yeast model organisms Schizosaccharomyces pombe and Saccharomyces cerevisiae Ub E1, Uba1, alone as well as with Ub and E2 molecules, have been determined (18–23). This has led to an ever-increasing understanding of the Uba1 mechanism for both of its catalytic activities as well as how it interacts with several E2 enzymes. These studies have revealed that Uba1 is a multidomain enzyme that undergoes large conformational changes to perform its dual catalytic activities. At the “base” of the enzyme are the inactive and active adenylation domains (IAD and AAD, respectively) that assemble as a pseudodimer and serve as the rigid body of the enzyme. The AAD binds Ub, ATP, and Mg2+ and harbors the adenylation activity of E1 (18). The cysteine domain is divided into the first and second catalytic cysteine half-domains (FCCH and SCCH, respectively) with the catalytic cysteine residing in the SCCH domain (18). Studies of the E1 for the Ub-like modifier SUMO have shown that the thioester bond formation involves disassembly of the adenylation active site and an ∼130° rotation of the SCCH domain that transits the catalytic cysteine and other structural elements required for catalysis of thioester bond formation into the active site (24, 25). A similar SCCH domain rotation of ∼106° was observed for the S. pombe ortholog of Uba1 (22). Finally, the ubiquitin fold domain, UFD, recruits E2 enzymes in a “distal” conformation and subsequently rotates to a more “proximal” conformation that places the E1 and E2 catalytic cysteines into proximity to facilitate E1–E2 thioester transfer (19). Uba1 orthologs function with tens of different E2s, and a three-residue basic motif at the N-terminal helix of the core of E2s was demonstrated to be important for Ubc2 binding to Uba1 (26). However, Ub E2s share little sequence conservation at their predicted E1-interacting region outside of the three-residue basic motif (19), and the basic motif itself is variably conserved among E2s (26). Altogether, the data suggest that sequence and structural plasticity at the Uba1–E2 interface might underlie the promiscuity of Uba1 and variability in the affinities of E1–E2 interactions. Indeed, previous studies have shown that E1 is able to bind to E2 enzymes via distinct binding modes to accommodate the variability of the E2s (19, 21).
In the context of its attractiveness as a target for small molecule therapeutic intervention in cancer (23, 27), a structural understanding of the Uba1 enzyme and its interactions with Ub and E2 enzymes is crucial for rational approaches to drug design. Herein, we present the crystal structure of a human Uba1–Ub complex, which represents the first structural snapshot of human Uba1 determined to date. Through structural analysis, molecular modeling, and biochemical analysis, we demonstrate that human Uba1 shares a conserved overall structure and mechanism with previously characterized yeast orthologs while noting subtle structural differences in human Uba1, particularly within the active site, that may aid efforts focused on the development of small molecule inhibitors. We also demonstrate that structural plasticity as a mechanism that underlies promiscuity in E1–E2 interactions is a conserved element of the human Ub E1–E2 system. Altogether, our work highlights the utility of structural characterization of orthologous proteins from model organisms as well as the importance of determining structures of bona fide mammalian target protein structures, when feasible.
Results and discussion
Overall structure of a human Uba1–Ub complex
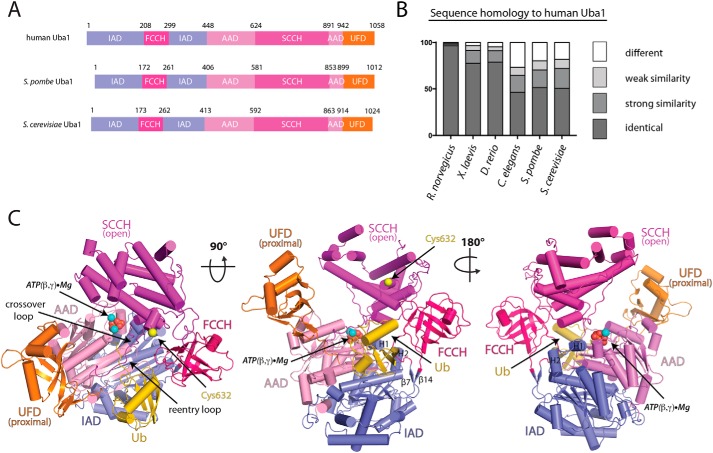
All orthologs of Uba1 share a conserved overall domain organization (Fig. 1A) and have two main functions. First, Uba1 must discriminate against other Ub-like modifiers (Ubls) and specifically recruit and activate Ub in a two-step process involving adenylation of the Ub C terminus followed by formation of a thioester bond between the Uba1 catalytic cysteine and the C-terminal glycine of Ub. Second, Uba1 must recruit E2-conjugating enzymes specific to the Ub system and subsequently catalyze transfer of Ub from the E1 to E2 active-site cysteine to form an E2∼Ub thioester intermediate (E1–E2 thioester transfer or transthiolation). Despite its fundamental importance to cell biology and attractiveness as a target for therapeutic intervention in cancer, the only structures of Uba1 determined to date are of S. cerevisiae and S. pombe orthologs which share ∼50% identity to human Uba1 (Fig. 1B) (18–23). To gain further insights into how human Uba1 (hUba1) performs its functions and to highlight structural differences between yeast and human Uba1 that can potentially guide the development of small molecule therapeutics, we determined the crystal structure of human Uba1 (Fig. 1C). The structure has been refined to 3.14 Å resolution with R/Rfree values of 0.214/0.251 (Table 1). Overall, the electron density maps are of high quality, including for all regions of the structure described throughout (Fig. S1). However, interactions described below, hydrogen bonds in particular, should be considered putative at this modest resolution. The two copies of the hUba1–Ub structure in the asymmetric unit are very similar, superimposing with a root mean square deviation (r.m.s.d.) of 0.86 Å (Fig. S2A), and unless otherwise noted, we describe the complex formed by chains A (hUba1) and B (Ub), which were better ordered.
Figure 1.
Domain organization of Uba1 and overall structure of the human Uba1–Ub complex. A, domain organization of previously crystallized Uba1 orthologs with residue numbers at the domain boundaries listed above. B, overall amino acid sequence homology of select Uba1 orthologs to human Uba1. Strongly similar, weakly similar, and different residues were assigned according to ClustalX. C, hUba1–Ub complex is shown as a cartoon representation with Uba1 domains labeled and color-coded. Ub is colored gold. Only the β- and γ-phosphates of ATP and two accompanying magnesium ions (shown as spheres and labeled “ATP(β,γ)·Mg”) were included in the final model due to poor ordering of the adenine and ribose of ATP. The side chain of the catalytic cysteine of hUba1 (Cys-632) is shown as a yellow sphere.
Table 1.
Crystallographic data collection and refinement statistics
Parentheses indicate statistics for the high-resolution data bin for x-ray data.
| Human Uba1–Ub complex | |
|---|---|
| PDB code | 6DC6 |
| Data Collection | |
| Source | APS 22 ID |
| Wavelength (Å) | 1.00 |
| Resolution limits (Å) | 50–3.14 (3.26–3.14) |
| Space group | P21 |
| Unit cell (Å) a, b, c | 105.0, 70.4, 188.9 |
| Unit cell (°) α, β, γ | 90, 95.2, 90 |
| No. of observations | 133987 |
| No. of reflections | 46012 (4503) |
| Completeness (%) | 94.6 (93.5) |
| Mean I/σI | 8.0 (1.5) |
| CC½ | 0.984 (0.592) |
| Rmergea | 0.139 (0.459) |
| Rpim | 0.088 (0.327) |
| Refinement statistics | |
| Resolution limits (Å) | 41.7–3.14 (3.22–3.14) |
| No. of reflections (work/free) | 45,928/1997 |
| Completeness (%) | 94.5 (92.7) |
| Protein/solvent/ligand atoms | 16,866/0/22 |
| Rcrystb | 0.213 (0.341) |
| Rfree (4.4% of data) | 0.250 (0.391) |
| Bonds (Å)/angles (°) | 0.003/0.554 |
| B-factors: protein/solvent/ligand (Å2) | 90.1/–/54.1 |
| Coordinate error (ML-based) (Å) | 0.50 |
| Ramachandran plot statistics (%) | |
| Favored | 94.1 |
| Allowed | 5.7 |
| Outliers | 0.2 |
| MolProbity score | 1.96–100th percentile (n = 2008, 3.14 Å ± 0.25 Å) |
a Rmerge = Σhkl Σi|I(hkl)i − 〈I(hkl)〉|/ΣhklΣi 〈I(hkl)i〉.
b Rcryst = Σhkl Fo(hkl) − Fc(hkl)|/Σhkl |Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
Analysis of the hUba1–Ub structure shows that, as expected, hUba1 is a modular, multidomain enzyme. As in other E1 structures, the IAD and AAD associate to form a pseudodimeric adenylation domain that serves as the rigid body of the structure (Fig. 1C). Attached to this rigid body are the SCCH domain that is tethered to the AAD by two loops called the crossover and reentry loops, the FCCH domain that is tethered to the AAD by two loops called the β7 and β14 loops, and the UFD, which is tethered to the AAD by a single loop (Fig. 1C). Overall, hUba1 has a “Y” shape with the adenylation domains forming the base of the enzyme, and the SCCH domain and UFD are situated across from each other at the “top” of the enzyme with a large canyon between them that serves to accommodate the E2-conjugating enzyme during E1–E2 thioester transfer (Fig. 1C).
Previous studies have revealed remarkable conformational plasticity in the Uba1 structure that is important for the enzyme to fulfill its functions. Major conformational changes in the flexible loops connecting the FCCH, SCCH, and UFD to the AAD accompany rigid body rotations of these domains that are essential for catalysis of Ub activation and transfer of activated Ub from E1 to E2 (18–23). A comparative analysis of the overall architecture of hUba1 to previously determined structures of S. pombe and S. cerevisiae Uba1 reveals a conformational snapshot of the enzyme that is poised for catalysis of adenylation (Fig. 1C). In the hUba1 structure, Ub engages in contacts with the IAD, AAD, and FCCH domain and the crossover loop, so named because it crosses over the C terminus of Ub as it leads into the SCCH domain. The C-terminal glycine of Ub (Gly-76) is positioned within the hUba1 adenylation active site. The hUba1 SCCH domain is perched on α-helices H1 and H2 of the IAD in an “open” conformation with the catalytic cysteine (Cys-632) separated from the C terminus of Ub by ∼35 Å (Fig. 1C). The SCCH domain also engages in a network of contacts with the FCCH domain that likely contributes to its stabilization in the open conformation. This contrasts structural snapshots of S. pombe Uba1 (22) and the E1 for the Ub-like modifier SUMO that is poised for thioester bond formation (25) where the SCCH domain rotates ∼106–130° relative to the open conformation to adopt a “closed” conformation (Fig. S2B). Closure of the SCCH domain serves to: 1) transit the catalytic cysteine into the active site proximal to the Ub C terminus for thioester bond formation; 2) transit additional structural elements into the Uba1 active site that transform the active site into a thioester bond formation–competent state along with concomitant disassembly of the adenylation active site; and 3) disrupts contact between the SCCH and FCCH domains and the SCCH domain and IAD that facilitate SCCH domain closure along with formation of a new network of contacts that stabilize the closed conformation. Finally, the hUba1 UFD adopts a proximal conformation in which the length of the canyon between the UFD and SCCH domain is at a minimum and, as will be discussed in more detail below, resembles the likely conformation of the hUba1 UFD during E1–E2 thioester transfer.
Human Uba1–Ub interface
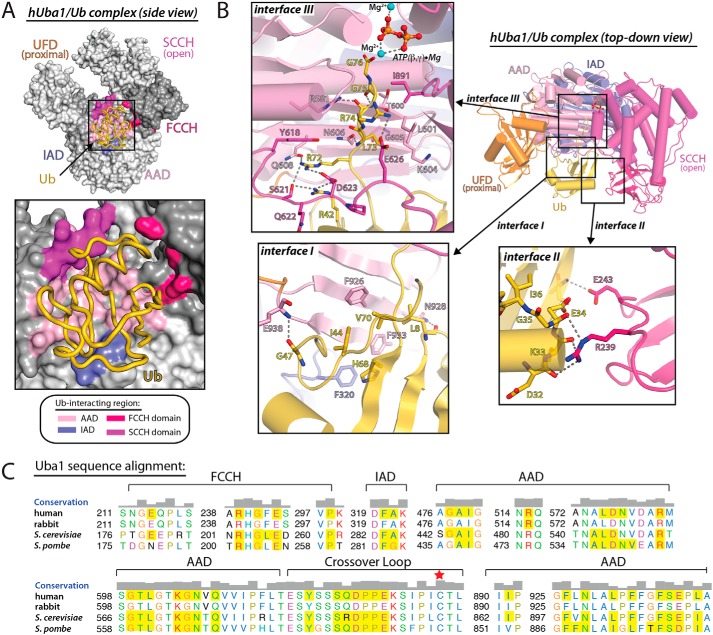
As in other Uba1–Ub structures (18–23), residues from the AAD, IAD, FCCH domain and the crossover loop form the Ub-binding surface of hUba1 (Fig. 2A). A total of ∼3200 Å2 of solvent-accessible surface area is buried upon formation of the hUba1–Ub complex. The hUba1–Ub interface is contiguous, but there are three distinct networks of intermolecular interactions (Fig. 2B) that were previously defined as interfaces 1–3 based on the S. cerevisiae Uba1–Ub structure (18). Interface 1 involves residues from the “bottom” of the globular β-grasp domain of Ub, which predominantly contacts the AAD of Uba1. The Interface 1 network of interactions is centered on the “Ile-44 hydrophobic patch” of Ub (Leu-8, Ile-44, His-68, and Val-70), which engage in contacts with Phe-320, Phe-926, Phe-933, and Ser-937 of hUba1 (Fig. 2B). The importance of the Ile-44 hydrophobic patch of Ub in mediating interaction with hUba1 is demonstrated by a previous study showing that alanine substitutions of Leu-8 and Ile-44 diminish the ability of Ub to be activated by Uba1 (28). At the periphery of Interface 1 are a few hydrogen bond-mediated interactions involving backbone atoms of Ub. On one side of the interface, the backbone carbonyl oxygen of Leu-8 of Ub engages in a hydrogen bond to the side chain of Asn-928 of the AAD, and on the other side, the carbonyl oxygen of Gly-47 engages in a hydrogen bond to the backbone nitrogen atom of Glu-938 of the AAD (Fig. 2B). Interface 2 involves residues from one of the “sides” of the globular β-grasp domain of Ub, which contacts the FCCH domain of hUba1 through hydrogen bond–mediated interactions. Specifically, the backbone carbonyl oxygen atoms of Asp-32, Lys-33, and Glu-34 of Ub engage in hydrogen bonds to the side chain of Arg-239 of the hUba1 FCCH domain (Fig. 2B). The backbone nitrogen atom of Thr-12 of Ub also engages in a hydrogen bond to the side chain of Glu-243 of the FCCH domain.
Figure 2.
Human Uba1–Ub interface involves a conserved network of interactions. A, top, hUba1 is shown as a surface representation with domains labeled and colored various shades of gray. hUba1 residues involved with contacts with ubiquitin are colored according to the domain where they reside (colored as in Fig. 1C). Ub is shown as a worm representation. Bottom, magnified view of the Uba1–Ub interface. B, hUba1–Ub structure is shown as a ribbon representation as in Fig. 1C with magnified views of Interfaces 1–3 interaction networks. Hydrogen bonds are indicated by dashed lines. C, sequence alignment of the Ub-interacting regions of Uba1 from the indicated species. Residue coloring and sequence conservation (shown as a bar graph above the alignment) were determined using default ClustalX parameters. Residues involved in contacts with Ub based on crystal structures are shaded yellow. Residue numbers are indicated to the left, and the domain to which the residues reside are indicated at top of the alignment.
Interface 3 involves residues from the AAD and crossover loop of hUba1, which likely function in guiding the flexible Ub C terminus toward the hUba1 active site and properly positioning the carboxylate of Gly-76 for catalysis. Arg-72 of the Ub C terminus engages in an extensive network of interactions with hUba1, including hydrogen bond and salt bridge interactions with Gln-608 of the AAD and Ser-621 and Asp-623 of the crossover loop (Fig. 2B). Arg-72 of the Ub C terminus also engages in van der Waals contacts with Asn-606 of the AAD and Tyr-618 of the crossover loop of hUba1. The backbone nitrogen atom of Leu-73 of the Ub C terminus engages in a hydrogen bond to the side chain of Asn-606 of the hUba1 AAD and the Leu-73 side chain participates in van der Waals contacts with Leu-601, Lys-604, and Gly-605 of the AAD (Fig. 2B). The importance of the glycine residue at position 605 of the hUba1 AAD in creating space to accommodate Ub C terminus has previously been noted (18, 24, 29). The backbone carbonyl oxygen of Arg-74 of Ub participates in hydrogen bonds with Arg-581 of the hUba1 AAD and the side chain of Arg-74 also engages in putative salt bridge interactions with Glu-626 of the hUba1 crossover loop (Fig. 2B).
The C-terminal diglycine motif of Ub (Gly-75 and Gly-76) is guided toward the hUba1 active site by a small network of interactions with residues from the AAD. The backbone carbonyl oxygen of Gly-75 of Ub participates in hydrogen bonds to both the Arg-581 side chain and the backbone nitrogen atom of Thr-600 of the Uba1 AAD, whereas the backbone nitrogen atom of Gly-75 engages in a hydrogen bond with the backbone carbonyl oxygen of Thr-600 of the hUba1 AAD (Fig. 2B). Gly-75 also engages in van der Waals contacts with the Ile-891 side chain from the SCCH domain of hUba1, further locking it in place. It is also worth noting that the hUba1 crossover loop, which itself is highly flexible, participates in a network of contacts with a 310 helix (g1 helix) and the β3 strand of Ub that properly position it for interactions with the Ub C terminus (Fig. 2B). Here, Arg-42 from the β3 strand of Ub engages in hydrogen bond and salt bridge interactions with Ser-621 and Asp-623 from the hUba1 crossover loop. Asp-39 and Gln-40 of the g1 helix of Ub also participate in van der Waals contacts with Gln-622 and Pro-624 from the hUba1 crossover loop (Fig. 2B). Overall, the hUba1–Ub interface is highly conserved with orthologous Uba1–Ub structures, including the identity of Uba1 residues involved in contacts with Ub (Fig. 2C).
Human Uba1 adenylation active site
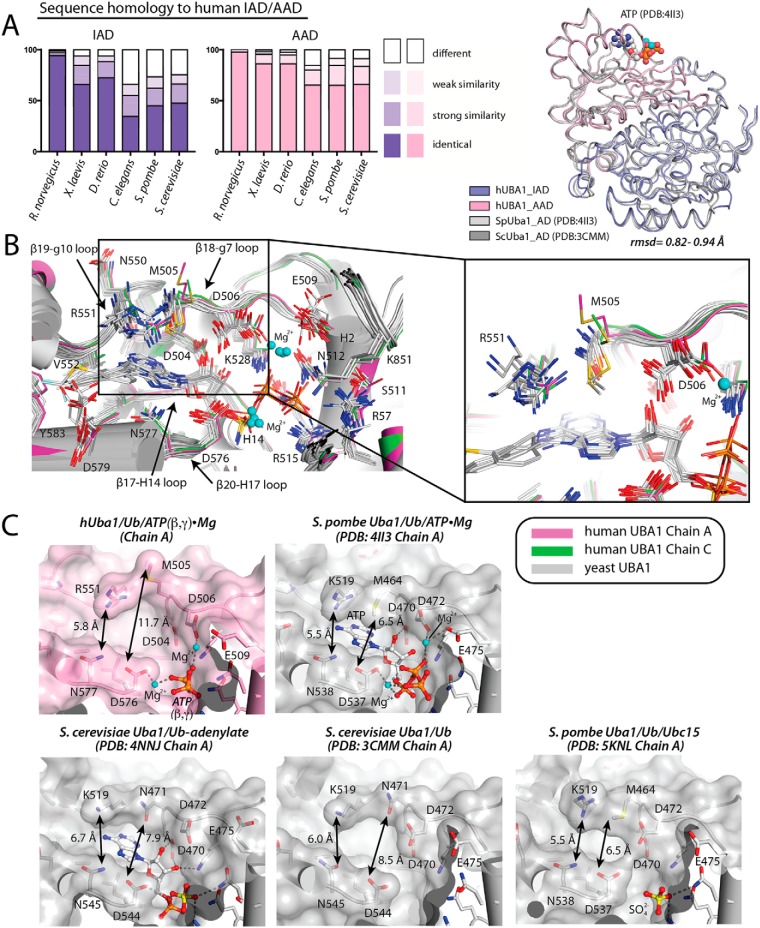
The adenylation active site of hUba1 is formed by structural elements in the IAD and AAD, which both harbor Rossman-like folds that assemble as a pseudodimer (Fig. 3A). Although the IAD is catalytically inactive and does not bind Ub, it harbors residues at it N terminus that are essential for catalysis of adenylation by the AAD. As noted previously, the IAD cannot bind Ub due to the presence of a 4-helix bundle that sterically blocks the surface of the IAD that would interact with Ub (Fig. S3A) (18). The IAD also lacks conservation of residues involved in contacts with ATP and has an arginine at the second glycine position of the “GXGXXG” p-loop at the N terminus of H2 of the AAD that is important for ATP binding and constitutes the oxyanion hole of the active site (Fig. S3B). The IADs of S. pombe and S. cerevisiae share 45–48% identity with that of hUba1; the AADs share 66–68% identity, and the overall adenylation domains superimpose with an r.m.s.d. of 0.82–0.94 Å (516 of 531 eq Cα atoms superimposed) (Fig. 3A).
Figure 3.
Comparison of the human Uba1 active site to other Uba1 orthologs. A, left, amino acid sequence homology of the AAD and IAD of select Uba1 orthologs to human Uba1. Right, superposition of the AAD/IAD of S. pombe and S. cerevisiae Uba1 onto hUba1 with the r.m.s.d. range indicated below the structure (516 of 531 equivalent Cα atoms superimposed). ATP from the S. pombe Uba1–Ub/ATP·Mg structure (PDB code 4II3) is shown as spheres. B, left, adenylation domains of S. pombe and S. cerevisiae Uba1 (shown as cartoon representations and colored gray) from existing Uba1–Ub structures (PDB codes 4II2, 4II3, 3CMM, 4NNJ, 5KNL, 5L6H, 5L6I, and 5L6J) were superimposed onto hUba1. Residues involved in contacts with ATP, Uba1 inhibitor molecules, and magnesium are shown as sticks. Magnesium ions are shown as cyan spheres. Right, magnified view of three regions in the active site that exhibit sequence and conformational variability. C, ATP-binding pockets of the indicated structures of Uba1 are shown as semitransparent surface representations with cofactors and select Uba1 residues involved in contacts with cofactors shown as sticks. Regions of sequence and conformational divergence that result in differences in the shape of the ATP-binding pocket are labeled.
The ATP-binding pocket of hUba1 is formed by residues from H14, H15, and the β17-H14, β18-g7, β19-g10, and β20-H17 loops of the AAD as well as H2 from the IAD (Fig. 3B) (“H” indicates α-helices, and “g” indicates 310 helices). Although ATP and magnesium were included in the crystallization buffer used to determine our hUba1 structure, electron density for the adenine, ribose, and α-phosphate of ATP are both poorly ordered in one copy of hUba1 (chain A), whereas only the ribose is weakly ordered in the other copy (chain B) with no visible electron density for the adenine. In contrast, very strong electron density corresponding to the binding sites for magnesium and β- and γ-phosphates of ATP is evident in both copies of hUba1. Although it is formally possible that this density may belong to magnesium and sulfate ions from the crystallization solution, the near precise structural overlap mentioned above led us to model this electron density as magnesium and β- and γ-phosphates of ATP, while the adenosine and α-phosphate of ATP were not included in the final model (Fig. 3A). The reason for the disordering of adenosine and the α-phosphate of ATP is unclear, but the β- and γ-phosphates of ATP are involved in coordination of two magnesium ions and participate in a similar network of contacts with hUba1 (Fig. 3, B and C, and Fig. S4A) as observed in previous structures of S. pombe and S. cerevisiae Uba1 (19, 20, 23). Specifically, the γ-phosphate of ATP engages in contacts with Arg-57, Arg-515, and Lys-851 of hUba1, and the β-phosphate of ATP engages in contacts with Asp-506, Asn-512, Lys-528, and Asp-576 (Fig. 3B and Fig. S4A). Next, we docked ATP onto our hUba1 structure based on similarity to S. pombe and S. cerevisiae structures bound to ATP, and we found that the α-phosphate of ATP is in position to make contacts with Ala-478 and Asp-576 of hUba1 (Fig. 3B and Fig. S4A). In the model, the ribose of ATP is in position to interact with Gly-475 and Asp-504 of hUba1, and the adenine of ATP is in position to interact with Arg-551, Val-552, Leu-575, and Asn-577 (Fig. 3B and Fig. S4A).
Previous studies have shown that Asp-576 and Lys-528 are critical for ordered substrate binding (with ATP being the preferred leading substrate) and transition state stabilization during Uba1-catalyzed Ub adenylation (30). Analysis of our hUba1–Ub structure and a comparison with other structures of Uba1, SUMO E1, and Nedd8 E1 provide a clue to the structural basis for this observation. As mentioned above, the SCCH domain of Uba1 likely exists in an equilibrium of open (adenylation active) and closed (thioester bond formation active) conformations (22, 25). In the open conformation, the adenylation active site is “assembled” with residues positioned optimally for contacts with ATP·Mg, and in the closed conformation the adenylation active site is “disassembled” with the structural elements harboring these ATP-binding residues either being disordered or suboptimally positioned (25). Binding of ATP·Mg to E1 results in “rigidification” of the active site that includes ordering of helices H1 and H2 from the IAD, which harbors residues involved in contacts with the γ-phosphates of ATP, and remodeling of the “g1 helix” from the AAD of Uba1, which positions residues Asn-512 and Arg-515 for contacts with the β- and γ-phosphates of ATP. Ordering of H1 and H2 is also important because it likely shifts the conformational equilibrium of the SCCH domain toward the open state, since ordering of H1/H2 sterically blocks the SCCH domain from adopting the closed conformation. A recently determined structure of S. pombe Uba1 in which the adenylation active site is disassembled and the SCCH domain has undergone a 106° domain alternation relative to the open conformation (Uba1SCCH_ALT) suggests that there is an ensemble of SCCH domain conformations between the open and closed states (22).
Notably, one-half of the channel on E1 that threads the Ub C terminus toward the active site is formed by different E1 structural elements in the open, closed, and alternative (alt) SCCH conformations, with the reentry loop forming half of the channel in the open conformation, and the crossover loop forming half of the channel in the closed conformation (Fig. S4B). Analysis of the distribution of B-factors within different conformational snapshots of E1s reveals that residues from the reentry loop have lower B-values in open structures compared with residues from the crossover loop in the closed structure, which implies that the channel itself exhibits less conformational variability in open structures compared with closed (Fig. S4B). Furthermore, in the S. pombe Uba1SCCH_ALT structure, the Uba1 crossover loop overlaps with the binding site for the Ub C terminus, and the channel that threads the Ub C terminus toward the active site is significantly widened (Fig. S4B). Based on this analysis, it is possible that stabilization of the Uba1 SCCH domain in the open conformation upon ATP·Mg binding might account for ordered substrate binding by both rigidifying the Ub C-terminal binding channel in an open state, as suggested previously (30), and by reducing the fraction of Uba1 molecules with intermediate SCCH domain conformations incompatible with Ub binding. Reduced ATP binding and thus a greater degree of conformational variability of the Ub C-terminal binding channel could account for the shift to random substrate addition in Asp-576 and Lys-528 Uba1 mutants, although we acknowledge this is speculative.
Comparison of human and yeast Uba1 adenylation active sites
Whereas the majority of interactions with ATP are conserved between human, S. pombe, and S. cerevisiae based on our hUba1–Ub/ATP(β,γ)/Mg structure and the hUba1–Ub/ATP/Mg model, comparative analysis of human, S. pombe, and S. cerevisiae Uba1 structures shows that there are two primary positions of divergence that slightly alter the shape of the ATP-binding pocket. The first position of divergence is in proximity to the adenine of ATP, where the side chain of Met-505 projects away from ATP in both copies of hUba1 in the structure to widen the ATP-binding pocket significantly compared with S. pombe and S. cerevisiae Uba1 (Fig. 3C). Met-505 of hUba1 is an asparagine in S. cerevisiae Uba1 (Asn-471) that participates in a hydrogen bond with a nitrogen atom from adenine, and although S. pombe has a methionine conserved at this position (Met-464), the side chain projects toward ATP and engages in van der Waals interactions with adenine (PDB codes 4II2 and 4II3) (Fig. 3C). Interestingly, the rotamer of Met-464 of S. pombe Uba1 adopts a similar conformation regardless of whether ATP is present in the active site (PDB codes 4II2 and 4II3 versus 5KNL and 5UM6) (Fig. 3C), which suggests that the conformational difference observed at human Met-505 may not be simply due to the poor ordering of the adenine of ATP in the hUba1 active site. Whereas Arg-551 of hUba1 is conserved in S. pombe Uba1 (Arg-512) and the overall shape of the ATP-binding pocket is similar in this area, S. cerevisiae harbors a lysine residue at this position (Lys-519), and the shorter and less bulkier side chain results in a widening of the ATP-binding pocket in this region (Fig. 3C). Consistent with these structural differences in the active site, the ability of a recently identified Uba1-specific inhibitor (TAK-243) to target S. cerevisiae Uba1 required N471M and K519R mutations to “humanize” the active site (27).
The second position of divergence is at Asp-506 of hUba1. Although Asp-506 is conserved in both S. pombe and S. cerevisiae (Asp-465 and Asp-472, respectively), this residue adopts two distinct conformations in all Uba1 structures that segregate into two populations depending on the substrate/product-binding status of the active site (Fig. 3B). In Uba1 structures harboring ATP (PDB codes 4II2 and 4II3, and this study), the residue corresponding to Asp-506 of hUba1 is involved in coordination of a magnesium ion that contacts the β- and γ-phosphates of ATP and is unique to the Uba1 active site compared with E1s for other Ub-like modifiers (Fig. 3, B and C). In Uba1 structures harboring Ub-adenylate intermediate (PDB code 4NNJ) or Ub-adenylate intermediate mimetics (PDB codes 5L6H and 5L6J), the magnesium ion coordinated by the residue corresponding to Asp-506 of hUba1 is absent, likely due to the absence of the β- and γ- phosphates of ATP. Nevertheless, the side chain of the corresponding aspartate residues adopts a similar conformation as in the ATP-bound structures (Fig. 3, B and C). In contrast, in Uba1 structures lacking ATP or adenylate intermediate (PDB codes 3CMM, 5KNL, and 5UM6), the aspartate side chain projects in the opposite direction and occupies similar structural space as the 2′-hydroxyl group of ATP (Fig. 3, B and C). The only outlier to this pattern is Asp-472 in the structure of S. cerevisiae Uba1 in complex with the inhibitor MLN-4924 (PDB code 5L6I), which notably lacks a hydroxyl group at the 2′ position of the AMP mimetic and thus can accommodate the aspartate side chain in this conformation (Fig. 3, B and C). Based on our structural analysis, it appears that the “resting” conformation of the residue corresponding to Asp-506 of hUba1 in the absence of ATP is projecting toward where the 2′-hydroxyl of the ribose is situated upon ATP binding. The side chain of hUba1 Asp-506 flips toward the β- and γ-phosphates of ATP to adopt an “active” conformation that avoids steric clashes with the 2′-hydroxyl group of the ribose of the incoming ATP molecule and coordinates magnesium to stabilize the negative charges on the β- and γ- phosphates of ATP (Fig. 3, B and C, and Fig. S4C). The question of why Asp-506 needs to undergo such a substrate-dependent structural transition is made apparent when considering the fact that a second acidic residue (Glu-509, human numbering) is in proximity to the side chain of hUba1 Asp-506 in the active conformation and is also involved in magnesium coordination (Fig. 3, B and C, and Fig. S4C). If Asp-506 adopted the active conformation in the absence of magnesium, its side chain would experience electrostatic repulsion with the side chain of Glu-509 (Fig. S4C). Thus, recruitment of magnesium along with ATP not only serves to stabilize the negative charges on the β- and γ-phosphates of ATP but also relieves electrostatic repulsion that would occur between Glu-509 and Asp-506 when Asp-506 is in the active conformation.
Human Uba1 UFD and plasticity in its interactions with Ub E2s
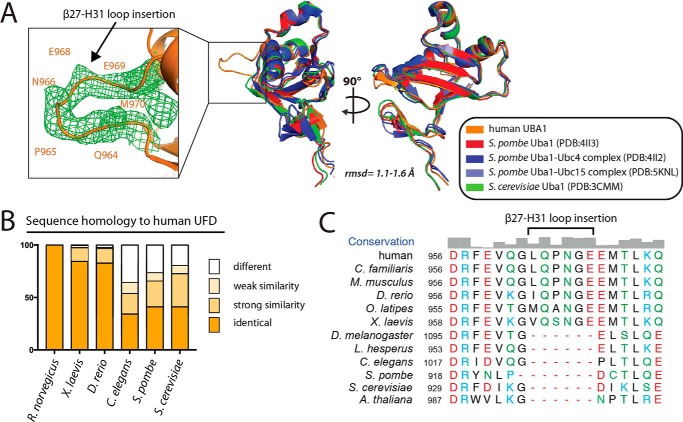
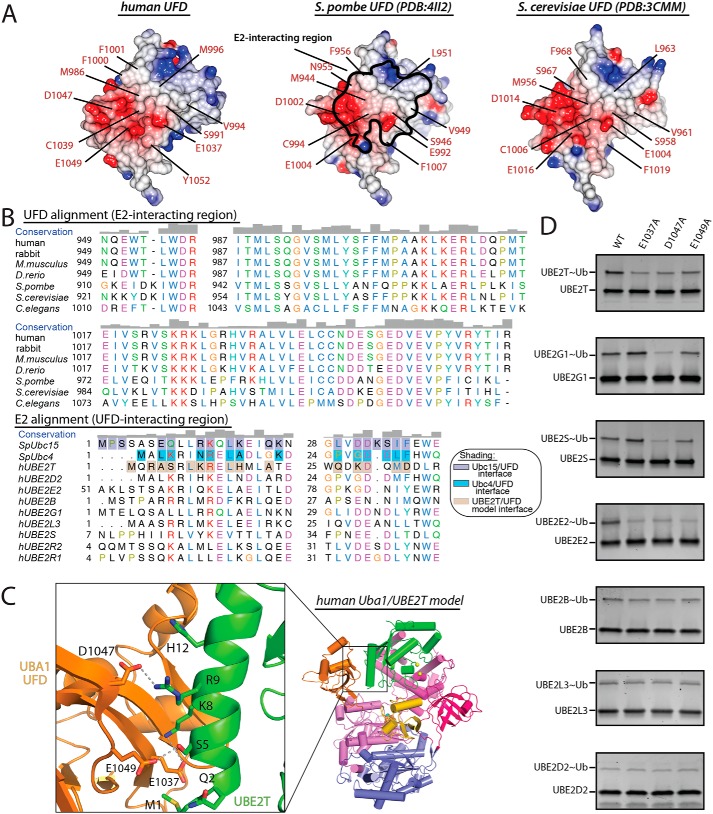
Upon adenylation and formation of a thioester bond between the Uba1 catalytic cysteine and the C-terminal carboxylate of Ub, Uba1 next recruits Ub E2-conjugating enzymes and transfers activated Ub to the E2 catalytic cysteine residue to generate E2∼Ub thioester intermediate (E1–E2 thioester transfer or transthiolation). The UFD of Uba1 is located at the C terminus of Uba1, adopts a β-grasp fold resembling Ub, and is required for the initial recruitment of E2s (26). The UFDs of S. pombe and S. cerevisiae both share only 42% identity with that of hUba1 and superimpose with an r.m.s.d. of 1.1–1.6 Å (110 of 119 equivalent Cα atoms superimposed) (Fig. 4, A and B). The most distinctive structural feature of the hUba1 UFD compared with other structurally characterized Uba1 orthologs is a six-residue insertion in the β27–H31 loop (residues Leu-963 to Glu-968) for which there is clear electron density (Fig. 4, A and C). This insertion in the β27–H31 loop is not conserved in yeast, Caenorhabditis elegans, or Drosophila melanogaster, but the sequence and length of the insertion are highly conserved in all vertebrate Uba1 orthologs we analyzed (Fig. 4C). A functional role that the β27–H31 loop insertion might play is unclear because the E2-binding surface is located on the opposite side of the UFD, although this region is located in proximity to the region of the UFD that facilitates functionally important conformational changes (see below).
Figure 4.
UFD of human Uba1 contains a unique loop insertion within a conserved overall structure. A, right, UFDs from the indicated Uba1 structures were superimposed onto the hUba1 UFD with the r.m.s.d. range indicated below the structure (110 of 119 eq Cα atoms superimposed). Left, magnified view of the β27–H31 loop insertion of hUba1 UFD, which is unique to vertebrates, with 2Fo − Fc composite omit map electron density for the insertion shown as green mesh (contoured at 1σ). B, amino acid sequence homology of the UFD from select Uba1 orthologs to human Uba1. C, sequence alignment of the indicated Uba1 orthologs around the β27–H31 loop insertion of the hUba1 UFD. Residues are colored as in Fig. 2C.
Analysis of surface electrostatic representations of Uba1 orthologs shows that overall surface features of the E2-interacting region of hUba1 are similar to S. pombe and S. cerevisiae Uba1, including the presence of a large acidic patch known to be important for E1–E2 thioester transfer activity (Fig. 5A) (18, 19, 21). One unique feature of the Ub conjugation cascade compared with those for other Ubls is that there are a large number of E2s that must interact with either one E1 (lower eukaryotes) or two E1s (mammals) in the Ub system. An elegant kinetic analysis of E1–E2 thioester transfer identified a three-residue basic motif on hA of Ub E2s, mutation of which altered affinity in the predicted fashion (26). The relative but not absolute conservation of this three-residue basic motif across Ub E2s suggested that although important, this motif was not the only contribution to E1 binding and that variability in E2 sequence accounts for variability in affinity through an induced fit model (26). Consistent with this, recent structural studies in S. pombe have demonstrated that the physicochemical properties of E2 (Ubc4 and Ubc15) residues involved in direct contacts with the UFD are variably conserved (Fig. 5B) and that promiscuity in E1–E2 interactions is achieved through structural plasticity at the UFD–E2 interface (19, 21). Similarly, the physicochemical properties of human E2 residues predicted to be involved in contacts with the hUba1 UFD based on E1–E2 crystal structures are also poorly conserved (Fig. 5B).
Figure 5.
Structural plasticity in human Uba1–E2 interactions. A, UFDs from the indicated Uba1 structures are shown as surface electrostatic representations. The E2-interacting region of S. pombe UFD is indicated with black outline, and residues involved in contacts with Ubc4 (PDB code 4II2) and Ubc15 (PDB code 5KNL) are highlighted in the middle panel. The corresponding residues of human and S. cerevisiae UFD are highlighted in the left and right panels. B, top, sequence alignment of the E2-interacting region of UFD, and bottom, the UFD-interacting region of E2s from the indicated species. Residues involved in intermolecular contacts in the Uba1–Ubc15 and Uba1–Ubc4 structures are shaded purple and cyan, respectively. Uba1 and UBE2T residues involved in contacts in the hUba1/UBE2T model are shaded gray. Residues are colored as in Fig. 2C. C, UFD/E2 interface of the hUba1/UBE2T model is shown as a cartoon representation with residues involved in intermolecular interactions shown as sticks. Putative hydrogen bonds are indicated by dashed lines. D, E1–E2 thioester assays of the indicated human E2s and hUba1 variants were performed as described under “Experimental procedures”.
Two questions we next wished to address are whether the conserved acidic patch of the Uba1 UFD is important for E1–E2 interactions in the human Ub system and whether there is structural plasticity in human E1–E2 interactions as observed in the yeast system. To help answer these questions, we first created a model of hUBA1 in complex with UBE2T by docking UBE2T onto hUba1 based on the S. pombe Uba1–Ubc4 interaction and then subjected the model to optimization of rigid body position and side-chain conformation using the RosettaDock server (Fig. 5C) (31). Based on the resulting model, there are three basic residues on UBE2T (Lys-8, Arg-9, and His-12) that are in proximity to three conserved residues within the hUba1 acidic patch (Glu-1037, Asp-1047, and Glu-1049) that can potentially engage in salt bridge or longer range electrostatic interactions at the hUba1–UBE2T interface (Fig. 5C). We mutated these three acidic residues on hUba1 to alanine and tested their ability to transfer Ub to UBE2T in E1–E2 thioester transfer assays performed in the linear region of the reaction with protein concentrations determined based on total protein of apparently homogeneous enzyme preparations. The results of these assays reveal that each of the hUba1 mutants exhibit impaired E1–E2 thioester transfer activity (Fig. 5D). Next, we analyzed a structure-based sequence alignment of human E2s and found that Lys-8, Arg-9, and His-12 of UBE2T are poorly conserved (Fig. 5B), an observation consistent with structural plasticity being a major determinant of promiscuity in human E1–E2 interactions as it is in the yeast system. To test this, we subjected the three hUba1 mutants we analyzed above to E1–E2 thioester transfer assays with a panel of different human E2s (Fig. 5D). Consistent with structural plasticity being a conserved feature of hUba1–UFD interactions, the human E2s we tested exhibited a range of distinct activity signatures with the different hUba1 mutants. At the two extremes were UBE2D2, UBE2L3, and UBE2B, which were not affected by mutation of any of the three acidic Uba1 residues, and UBE2E2, which was similar to UBE2T in that all three Uba1 mutants diminished thioester transfer activity (Fig. 5D). UBE2G1 and UBE2S had patterns in between these extremes. For UBE2G1, only D1047A diminished thioester transfer activity, whereas for UBE2S, both D1047A and E1049A had defective thioester transfer activity (Fig. 5D). Altogether, these results confirm the importance of the Uba1 acidic patch for E1–E2 thioester transfer and demonstrate that there is likely a significant degree of structural plasticity at the human Uba1–E2 interface due to variably conserved E2 residues participating in distinct networks of interactions with the hUba1 UFD, consistent with previous kinetic (26) and structural (18, 19, 21) studies.
Conformational changes in the UFD and modeling a human SCCH domain–E2 interface
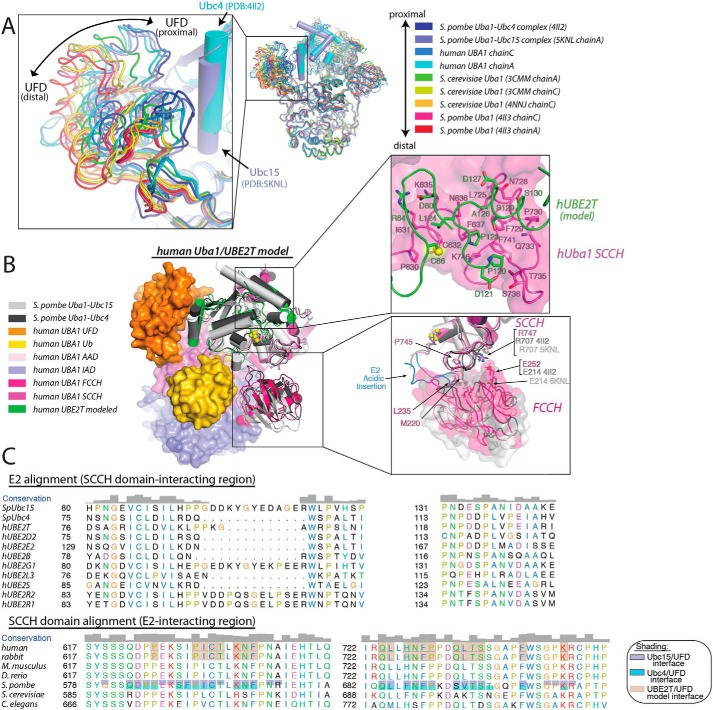
Previous structural studies have demonstrated that the Uba1 UFD adopts a spectrum of conformations that result in differences in the width of the canyon between the UFD and the SCCH domain (18–23). At the two extremes of this spectrum are the distal and proximal conformations in which the distance between the UFD and the Uba1 catalytic cysteine residue on the SCCH domain are at a maximum and minimum, respectively (Fig. 6A and Fig. S5A). Previous modeling experiments suggest that E2 is initially recruited to the Uba1 UFD in distal conformations with the E1 and E2 active sites separated by as much as 25 Å (Fig. S5B) (19). Recruitment of E2 with the UFD in a distal conformation with a larger canyon between the UFD and SCCH domain likely allows the E2 to sample greater conformational space for productive complex formation with the UFD. Subsequent transition of the UFD from the distal to proximal conformation brings the E1 and E2 active sites into proximity, resulting in a network of intermolecular interactions that facilitate nucleophilic attack of the E1∼Ub thioester bond by the E2 catalytic cysteine during E1–E2 thioester transfer.
Figure 6.
Human Uba1 UFD adopts the E1–E2 thioester transfer-active proximal conformation. A, right, indicated Uba1 structures were superimposed by their adenylation domains with Uba1 shown as worm representations, and the E2s from the S. pombe Uba1–Ubc4 and Uba1–Ubc15 structures shown as cartoon representations. Left, magnified view of the UFDs of the superimposed structures, with the distal- and proximal-most conformations indicated. B, left, hUba1 from the Uba1/UBE2T model is shown as surface representation, and UBE2T is shown as a cartoon representation (green). Ubc4 (dark gray) from the S. pombe Uba1–Ubc4 structure and Ubc15 (light gray) and the FCCH domain (light gray) from Uba1–Ubc15 structure are also shown as cartoon representations for comparison. Top right, SCCH domain–UBE2T interface from the hUba1–UBE2T model is shown with Uba1 shown as semitransparent surface. hUba1 and UBE2T regions at the SCCH domain–UBE2T interface are shown as cartoon representations with residues involved in direct interactions shown as sticks. Bottom right, magnified view of the region around the FCCH/SCCH domain interface of the hUba1–UBE2T model and the S. pombe Uba1–Ubc15 structure. The SCCH domain is shown as semitransparent surface. Residues involved in conserved interactions between the SCCH and FCCH domain are shown as sticks. These interactions are disrupted in the Uba1–Ubc15 structure due to a rotation of the FCCH domain. C, top, sequence alignment of the SCCH domain-interacting region of E2s, and bottom, E2-interacting region of SCCH domains from the indicated species. Residues involved in intermolecular contacts in the Uba1–Ubc4 and Uba1–Ubc15 structures are shaded cyan and purple, respectively. Uba1 and UBE2T residues involved in contacts in the hUba1/UBE2T model are shaded gray.
The only Uba1 structures to date where the UFD has been observed in the proximal conformation are S. pombe Uba1–Ubc4 and Uba1–Ubc15 complexes that were trapped in a conformation resembling the tetrahedral intermediate formed during E1–E2 thioester transfer by specifically cross-linking their catalytic cysteine residues (Fig. 6A) (19, 21). These structures revealed that the transition from distal to proximal UFD conformation occurs by a bending motion primarily at the first β-strand of the β-grasp fold of the UFD (β27) (Fig. S5C). All Uba1 structures previously determined in the absence of E2 adopt a distal conformation (Fig. 6A) (18–20, 22, 23). Interestingly, analysis of the hUba1 structure reveals that both copies of the UFD in the asymmetric unit adopt a proximal conformation even in the absence of E2 and that the hUba1 UFD achieves the proximal conformation primarily through bending that occurs between residues Asp-956 and Val-960 around the β27 strand of the UFD, similar to yeast Uba1 structures (Fig. S5C). Although both copies of the UFD are engaged in crystal contacts, their crystal packing environments are completely different (Fig. S5D). Thus, it is unclear whether crystal packing or a difference in the equilibrium of the proximal and distal UFD conformations in hUba1 might underlie the observed proximal UFD conformation in the absence of E2.
To determine whether contacts between E2 and the hUba1 and SCCH domain and active site are conserved between the yeast and human systems, we again analyzed our hUba1/UBE2T model generated using Rosetta. Analysis of the hUba1/UBE2T model shows that a hydrophobic patch on the hUba1 SCCH domain formed by Phe-637, Leu-725, Phe-729, Phe-741, and the aliphatic portion of the Lys-746 side chain are engaged by the side chains of Pro-123 and Ala-126 of UBE2T (Fig. 6B). The hydrophobic patch is well conserved in all Uba1 orthologs and engages in a similar set of interactions with E2 in the S. pombe Uba1–Ubc4 and Uba1–Ubc15 complex structures (Fig. 6C). In the hUba1/UBE2T model, Leu-124 of UBE2T is also positioned proximal to the active site and engages in van der Waals contacts with the catalytic cysteine residues of both hUba1 and UBE2T as well as Ile-631 of hUba1 (Fig. 6B). The importance of this network of contacts has been demonstrated in the S. pombe system where mutations of Ubc4 at positions that correspond to Pro-123 and Leu-124 of UBE2T (Pro-118 and Leu-119) significantly diminish the E1–E2 thioester transfer activity of Ubc4 (19). Taken together, the data suggest that the UFD–E2 interface is poorly conserved resulting in considerable plasticity in the nature of its interactions, whereas the E2–SCCH domain interface is much more highly conserved, particularly around the active site.
Also related to E2 recruitment by Uba1 is an additional set of intramolecular interactions between the SCCH and FCCH domains that are observed in the hUba1–Ub structure. In the hUba1–Ub structure, the FCCH and SCCH domains are closely packed together where a small network of interactions, including a salt bridge between Glu-252 of the FCCH domain and Arg-747 of the SCCH domain, and a network of van der Waals contacts between Met-220 and Leu-235 of the FCCH domain and Pro-745 of the SCCH domain take place (Fig. 6B). This network of interdomain contacts, as well as the relative orientation of the FCCH and SCCH domains, are well-conserved in S. pombe and S. cerevisiae structures determined in the absence of E2 (Fig. S6A) (18–20, 22, 23) and may help stabilize the SCCH domain in the open conformation that is required to achieve the adenylation active conformation of Uba1. These interdomain contacts are also conserved in the S. pombe Uba1–Ubc4 complex structure (19), although they are disrupted in the structure of the S. pombe Uba1–Ubc15 complex (21) due to the FCCH and SCCH domains rotating away from each other (Fig. 6B and Fig. S6, A and B). This rotation is the result of the distinct E1-binding mode of Ubc15 that places along acidic insertion in proximity to the Ubc15 active site directly between the FCCH and SCCH domains (21). Based on sequence and structural homology, it is likely that the FCCH and SCCH domains of hUba1 are able to exhibit conformational variability in order to accommodate human E2s with acidic loop insertions that might utilize a similar Uba1-binding mode to S. pombe Ubc15.
Computational analysis of ligand-binding hot spots on human Uba1
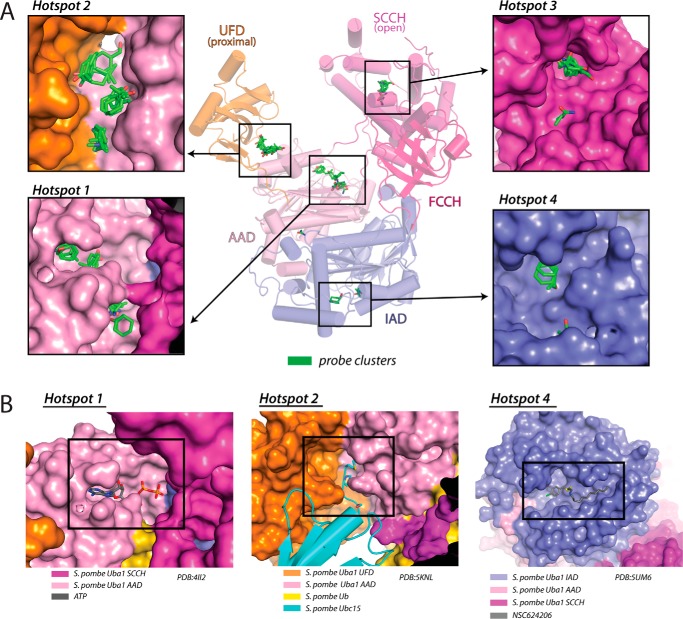
The Ub-proteasome system (UPS) is a validated target for therapeutic intervention in cancer as highlighted by the clinical success of proteasome inhibitors in treating patients with multiple myeloma and mantle cell lymphoma (4). This has resulted in great interest in targeting other components of the UPS for small molecule inhibition, including Uba1 (23, 27). Whereas knowledge of the three-dimensional structures of drug targets can facilitate the development of small molecule therapeutics, the only Uba1 orthologs structurally characterized to date (S. pombe and S. cerevisiae) share only ∼50% sequence identity and 70% sequence similarity with hUba1. Thus, we subjected our hUba1 structure, which represents the first determined structure of a mammalian Uba1 ortholog, to FTMap analysis (32) to identify ligand-binding hot spots that could potentially be used as a framework for development of novel small molecule therapeutics.
FTMap docks a diverse set of 16 small organic molecules containing various functional groups onto a protein surface, clusters the probes, and ranks the clusters on the basis of their average energy (32). FTMap analysis of our hUba1 structure resulted in the identification of four prominent hot spots (HS 1–4) (Fig. 7A). The surface on Uba1 and the E1s for other Ubls that has been targeted most successfully to date is the ATP-binding pocket, and not surprisingly, the highest scoring hot spot identified on hUba1 by FTMap (HS1) corresponds to the ATP-binding site (Fig. 7, A and B). As expected, FTMap analysis of previously determined yeast Uba1 structures shows that the ATP-binding pocket (HS1) is identified as a ligand-binding hot spot in all yeast structures we analyzed (Fig. S7A).
Figure 7.
Ligand-binding hot spots on human Uba1 predicted by FTmap analysis. A, center, hUba1 structure is shown as a cartoon representation as in Fig. 1C with the four top-scoring probe clusters identified by FTMap analysis shown as sticks and their corresponding binding pockets (HS1–4) on hUba1 labeled. Insets, magnified views of the four top-scoring probe clusters shown as sticks, with the corresponding hUba1-binding pockets shown as surface representations. B, left, S. pombe Uba1–Ubc4/Ub/ATP·Mg structure (PDB code 4II2) is shown as a surface representation with ATP shown as sticks in the same orientation as the HS1 inset in A. HS1 identified by FTMap analysis corresponds to the ATP-binding pocket of Uba1. Center, the S. pombe Uba1–Ubc15/Ub structure (PDB code 5KNL) is presented in the same orientation as HS2 in A with Uba1 shown as surface representation and Ubc15 shown as cartoon. The extended N terminus of Ubc15, which partially occupies HS2, is shown as sticks. Right, S. pombe Uba1–NSC624206 inhibitor complex structure (PDB code 5UM6) is presented in the same orientation as HS4 in A with hUba1 shown as a surface representation and NSC624206 shown as sticks.
Interestingly, HS2 is located at the junction between the UFD and AAD at a region of Uba1 that is in proximity to the N-terminal helix of E2s (hA) (Fig. 7A). E2s differ in the length of their N-terminal regions preceding hA with the shortest being Ubc4/UbcH5b that has one amino acid preceding hA and longer N-terminal extensions of ∼250 amino acids for UBE2Q1/2 (Fig. S7B). Although HS2 is empty in the structure of S. pombe Uba1–Ubc4 with the exception of solvent molecules (PDB code 4II2), the slightly extended N terminus of Ubc15 (five residues preceding hA; PDB code 5KNL) partially occupies HS2 in the S. pombe Uba1–Ubc15 structure (Fig. 7B). Thus, HS2 may represent the surface of Uba1 that engages the E2s with extended N termini preceding hA. This raises the possibility that HS2 could be exploited to specifically target interactions with E2s harboring extended N termini. FTMap analysis of previously determined yeast Uba1 structures identified HS2 as a prominent hot spot only in the structure of S. pombe Uba1 with the UFD in the proximal conformation (PDB codes 4II2) (Fig. S7A). This is likely due to the HS2 pocket not being fully formed when the UFD adopts distal conformations resulting in its positioning farther away from the AAD.
HS3 is formed by residues from α-helices H19, H20, H22, H23, and H25 on the SCCH domain at a region far from the catalytic cysteine and the E2-interacting surface (Fig. 7A). Thus, barring some unexpected allosteric effect, targeting this site would not have an obvious functional effect on Uba1 activity. This is also the case for HS4, formed by residues from the β5 strand, H7, the β4–H5 loop, and the H7–H8 loop at the bottom of the IAD, far from the active site and Ub-binding site of Uba1 (Fig. 7A). Interestingly, HS4 was partially occupied by the Uba1 inhibitor NSC624206 in a recently determined crystal structure of S. pombe Uba1 (PDB code 5UM6) (Fig. 7B) (22), and although this interaction was not involved in the inhibitory mechanism of the molecule, it further validates the results of the FTMap analysis. Targeting HS1 and HS2 could be a way to perturb the adenylation and E2-binding activity of hUba1, and with the rapid development of protein-targeting chimeric molecule approaches (33) even HS3 and HS4, which would not appear to have an obvious functional consequence of small molecule binding, could potentially be co-opted for proteolytic targeting of hUba1.
Conclusions
In this study, we have presented a crystal structure of hUba1, the first mammalian Ub E1 structure to date. Previous structures of S. pombe and S. cerevisiae Uba1 have been invaluable for providing insights into the mechanisms of Uba1 catalytic activities and have laid the foundation for understanding Uba1 promiscuity in its interactions with tens of E2 ubiquitin-conjugating enzymes. This work builds on that foundation by providing a consistent but subtly distinct structure of human Uba1. Comparative analysis of the hUba1 structure to previous Uba1 structures highlights two key facts. First, the structural characterization of orthologous proteins can provide meaningful insights to mammalian protein structure and mechanism, and second, the subtle differences in ortholog identity can alter druggable sites of the protein of interest.
To the first conclusion, we have shown that the overall structure of hUba1 is similar to previously solved structures of Uba1, and by utilizing modeling and biochemical analysis, we have shown that the structural basis for mechanisms of Uba1 catalytic activities and E2 promiscuity are conserved from the yeast model organisms to human. Specifically, the Uba1–Ub interaction networks for Ub adenylation that are observed in this structure are highly conserved; the ATP-binding pocket of hUba1 is conserved with the notable exceptions reviewed below; the UFD rigid body structure is highly conserved except for a β27–H31 loop insertion that is well ordered in the structure but has no known biological function; and the SCCH/FCCH domains are highly conserved compared with other Uba1 structures without E2 enzymes.
In our comparative analysis, we identified subtle structural changes in the ATP-binding pocket of hUba1 compared with yeast Uba1 that confirm previous studies of inhibitor specificity. Specifically, we have provided a structural basis for the necessity of N471M and K519R mutations to render S. cerevisiae Uba1 susceptible to a hUba1 inhibitor, TAK-243 (27), due to Met-505 positioning to widen the ATP-binding pocket near the adenine-binding site and the bulkier side chain of Arg-551 in hUba1. This finding highlights the need for structural characterization of mammalian target proteins when feasible for more successful target-directed drug development. In the same vein, we have utilized FTMap to identify ligand-binding hot spots on hUba1 and identified four pockets for future drug development efforts.
Experimental procedures
Cloning and protein expression
DNA fragment encoding human UBA1 residues 49–1058 was cloned into BamHI/NotI sites of vector pFastBac-HTB with an N-terminal TEV-cleavable His6 tag. Catalytic cysteine Cys-632 was mutated to alanine for crystallization. Point mutations were introduced using PCR-based mutagenesis. The DNA fragment encoding WT human ubiquitin (residues 1–76) was inserted into vector pET-29 to introduce an N-terminal TEV-cleavable His6 tag. Human Ub E2s were amplified from the human cDNA library and inserted into pET28 (UBE2B and UBE2L3), pET29 (UBE2S), and pMTTH (UBE2D2, UBE2E2, UBE2G1, and UBE2T) vectors.
hUba1 was expressed using the Bac-to-Bac Baculovirus Expression System as described previously (34). High titer recombinant baculoviruses were used to infect Spodoptera frugiperda (Sf9) cells at a cell density of 2 × 106 cells/ml cultured in Sf-900 II SFM medium (ThermoFisher Scientific). Cells were harvested after 72 h infection and stored at −80 °C before further use. All other proteins in this study were expressed by inducing E. coli strain BL21 (DE3) codon plus (Stratagene) transformed with plasmid carrying target cDNA. All large-scale cultures were grown at 37 °C in baffled flasks to an A600 of 1.0, at which point the flasks were placed into ice water to cold-shock the cells. After 30 min, isopropyl β-d-1-thiogalactoside was added to a final concentration of 0.2 mm, and the cultures were shaken at 18 °C for an additional 18 h.
Protein purification
Bacterial and insect cell pellets were harvested, resuspended, and lysed by sonication in lysis buffer (20 mm Tris-HCl, pH 8.0, 350 mm NaCl, 20 mm imidazole, 2 mm 2-mercaptoethanol (βME)). All lysates were cleared by centrifugation and applied to columns containing nickel-nitrilotriacetic acid Superflow resin (Qiagen) by gravity. After elution in buffer containing 20 mm Tris-HCl, pH 8.0, 350 mm NaCl, 250 mm imidazole, 2 mm βME, proteins with TEV-cleavable His6 tags were processed by adding TEV protease at a ratio of 1:100 (w/w) and incubating overnight at 4 °C. All proteins used for crystallization and biochemical experiments were subjected to gel filtration (Superdex 200 or Superdex 75 based on protein size; Amersham Biosciences). hUba1, UBE2T, UBE2B, and UBE2D2 were subsequently applied to an anion-exchange column (Mono Q; Amersham Biosciences) in 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 2 mm βME buffer and eluted with a linear gradient of 50–350 mm NaCl over 20 column volumes. Fractions containing purified proteins of interest were pooled, concentrated to 2–15 mg/ml, and snap-frozen in liquid nitrogen.
Crystallization and data collection
hUba1 was purified as described above at a final concentration of 170 μm in 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2 mm βME. Ub (340 μm), MgCl2 (5 mm), and ATP (1 mm) were added prior to sparse-matrix screening in Intelli-Plate (Art Robbins Instruments) (400-nl sitting drop vapor diffusion format) at 18 °C. Diffraction quality crystals of the hUba1–Ub/ATP·Mg were grown by mixing 0.9 μl of protein sample (supplemental with ATP and MgCl2) with 0.9 μl of crystallization buffer, and 0.2 μl of Silver Bullet Reagent C1 (Hampton Research), by hanging drop vapor diffusion at 12 °C. Crystallization buffer contains 0.1 m MgSO4, 0.1 m Tris-HCl, pH 8.0, 20% w/v PEG 4000. Crystals were flash-frozen in liquid nitrogen in cryoprotectant composed of mother liquor supplemented with 5 mm MgCl2, 1 mm ATP, and 20% ethylene glycol.
A complete X-ray diffraction data for the hUba1–Ub/ATP·Mg complex was collected at the Advanced Photon Source (Argonne, IL), SER-CAT beamline 22-ID. All data were indexed, integrated, and scaled using HKL2000. The hUba1–Ub/ATP·Mg crystal belongs to P21 with unit cell dimensions a = 105.0, b = 70.4, c = 188.9 Å, and β = 95.2°. There are two hUba1–Ub/ATP·Mg complexes per asymmetric unit and the crystal has a solvent content of 57.6%.
Structural determination and refinement
A complete dataset for the hUba1–Ub/ATP·Mg crystals was collected to a resolution of 3.14 Å. The program Sculptor was used to generate an hUba1 model based on the coordinates of S. cerevisiae Uba1 (PDB code 3CMM). The program PHASER (35) was used to find a molecular replacement solution using the coordinates for two copies of the hUba1 Sculptor model and human Ub (PDB code 1UBQ). The model was refined to R/Rfree values of 0.214/0.251 via iterative rounds of refinement and rebuilding using PHENIX (36) and COOT (37). The initial round of refinement involved rigid body fitting of the AAD/IAD, FCCH, SCCH, and UFD of hUba1 (domain boundaries defined in Fig. 1A) and Ub. This was followed by several iterative rounds of manual model building in COOT and refinement in PHENIX using reciprocal space and individual B-factor refinements, with torsion-angle NCS restraints and optimization of X-ray/stereochemistry and X-ray/ADP weights applied. Composite omit electron density maps were used to guide model building, and TLS refinement was applied during the final rounds of refinement with TLS groups defined using phenix.find_tls_groups. Electron density for the β- and γ-phosphates of ATP and two associated magnesium ions for each copy of hUba1 were clear after the initial round of refinement and were manually placed into the electron density after several additional rounds of model building and refinement. The final hUba1–Ub/ATP·Mg model contains amino acids 1–76 from both copies of Ub (chains B and D), amino acids 49–803 and 821–1057 from copy A of Uba1, and amino acids 49–802 and 816–1057 from copy C of Uba1. The adenine, ribose, and α-phosphate groups of the ATP molecules associated with both copies of hUba1 were poorly ordered and therefore not included in the final model. The final model has good geometry, with 94.0, 5.8, and 0.2% of residues in the favored, allowed, and disallowed regions of Ramachandran space, respectively. All molecular graphic representations of the structures were generated using PyMOL (38) and CCP4mg (39). Structure alignments were performed using the program Superpose in CCP4 software suite (40).
E1–E2 thioester transfer assay
The E1–E2 thioester transfer assays were performed at 25 °C as described previously (19, 21). All thioester transfer assays were performed with 2.5 nm E1, 500 nm E2, 5 μm Ub, 5 mm MgCl2, 200 μm ATP, 20 mm Hepes, pH 7.5, 50 mm NaCl for 120 s. Reactions were terminated through the addition of nonreducing urea SDS-PAGE buffer and subjected to SDS-PAGE, 150 V constant, at 4 °C. The gels were stained with Sypro Ruby (Bio-Rad) and visualized with a ChemiDoc MP (Bio-Rad).
FTMap analysis of human Uba1
The FTmap server (http://ftmap.bu.edu/)3 was used to identify potential ligand-binding hot spots on the surface of hUba1. The program was run with default parameters. Thirteen hot spots were predicted, and the numbers of probe clusters at these sites were 14, 13, 11, 10, 8, 8, 7, 6, 3, 3, 3, 2, and 2, from the highest-ranked to the lowest-ranked hot spot. The 13 clusters localized to five binding pockets (HS1–5) on hUba1. HS1 partially overlaps with the ATP-binding site and contains 13 + 8 + 7 + 3 = 31 clusters. HS2 partially overlaps with the E2 N-terminal binding region and contains 11 + 10 + 8 = 29 clusters. HS3 in SCCH domain contains 14 + 3 + 3 = 20 clusters. HS4 contains 6 + 2 = 8 clusters. HS5, which only contained two clusters, was not included in further analysis.
Author contributions
Z. L., K. M. W., J. H. A., and S. K. O. formal analysis; Z. L. validation; Z. L., L. Y., and S. K. O. investigation; Z. L., J. H. A., and S. K. O. visualization; Z. L., L. Y., and S. K. O. methodology; Z. L., K. M. W., L. Y., and S. K. O. writing-original draft; K. M. W. and S. K. O. writing-review and editing; L. Y. data curation; S. K. O. conceptualization; S. K. O. supervision; S. K. O. funding acquisition; S. K. O. project administration.
Supplementary Material
Acknowledgments
This work was based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by National Institutes of Health Grant P41 GM103403 from NIGMS. X-ray diffraction data were collected at SER-CAT 22-ID and NE-CAT 24-ID-E beamlines at the Advanced Photon Source, Argonne National Laboratory. The Pilatus 6M detector on 24-ID-C beam line is funded by National Institutes of Health ORIP HEI Grant S10 RR029205. This work used resources of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The X-ray crystallography facility used for this work was supported by the Office of the Vice President for Research at the Medical University of South Carolina. The liquid handling robot used was purchased via National Institutes of Health Shared Instrumentation Award S10 RR027139-01.
This work was supported in part by National Institutes of Health Grants R01 GM115568 (to S. K. O.), by Hollings Cancer Center's Ruth L. Kirschstein NRSA T32 CA193201 (to J. H. A.), and by NCI F30CA216921 (to K. M. W.). This work was also supported in part by a Hollings Cancer Center Postdoctoral Fellowship (to Z. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7.
The atomic coordinates and structure factors (code 6DC6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- Ub
- ubiquitin
- Ubl
- ubiquitin-like modifier
- AAD
- active adenylation domain
- IAD
- inactive adenylation domain
- UFD
- ubiquitin fold domain
- FCCH
- first catalytic cysteine half-domain
- SCCH
- second catalytic cysteine half domain
- PDB
- Protein Data Bank
- TEV
- tobacco etch virus
- βME
- 2-mercaptoethanol
- r.m.s.d.
- root mean square deviation
- UPS
- Ub-proteasome system
- h
- human.
References
- 1. van der Veen A. G., and Ploegh H. L. (2012) Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 10.1146/annurev-biochem-093010-153308 [DOI] [PubMed] [Google Scholar]
- 2. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 3. Popovic D., Vucic D., and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 4. Mattern M. R., Wu J., and Nicholson B. (2012) Ubiquitin-based anticancer therapy: carpet bombing with proteasome inhibitors vs. surgical strikes with E1, E2, E3, or DUB inhibitors. Biochim. Biophys. Acta 1823, 2014–2021 10.1016/j.bbamcr.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schulman B. A., and Harper J. W. (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 10.1038/nrm2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Streich F. C. Jr., and Haas A. L. (2010) Activation of ubiquitin and ubiquitin-like proteins. Subcell Biochem. 54, 1–16 10.1007/978-1-4419-6676-6_1 [DOI] [PubMed] [Google Scholar]
- 7. Streich F. C. Jr., and Lima C. D. (2014) Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 10.1146/annurev-biophys-051013-022958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cappadocia L., and Lima C. D. (2018) Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 10.1021/acs.chemrev.6b00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haas A. L., and Rose I. A. (1982) The mechanism of ubiquitin-activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 10. Haas A. L., Warms J. V., Hershko A., and Rose I. A. (1982) Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257, 2543–2548 [PubMed] [Google Scholar]
- 11. Haas A. L., Bright P. M., and Jackson V. E. (1988) Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J. Biol. Chem. 263, 13268–13275 [PubMed] [Google Scholar]
- 12. Hershko A., Heller H., Elias S., and Ciechanover A. (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 13. Pickart C. M., and Rose I. A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581 [PubMed] [Google Scholar]
- 14. Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 15. Buetow L., and Huang D. T. (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spratt D. E., Walden H., and Shaw G. S. (2014) RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem. J. 458, 421–437 10.1042/BJ20140006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yau R., and Rape M. (2016) The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 10.1038/ncb3358 [DOI] [PubMed] [Google Scholar]
- 18. Lee I., and Schindelin H. (2008) Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 10.1016/j.cell.2008.05.046 [DOI] [PubMed] [Google Scholar]
- 19. Olsen S. K., and Lima C. D. (2013) Structure of a ubiquitin E1–E2 complex: insights to E1–E2 thioester transfer. Mol. Cell 49, 884–896 10.1016/j.molcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schäfer A., Kuhn M., and Schindelin H. (2014) Structure of the ubiquitin-activating enzyme loaded with two ubiquitin molecules. Acta Crystallogr. D Biol. Crystallogr. 70, 1311–1320 10.1107/S1399004714002910 [DOI] [PubMed] [Google Scholar]
- 21. Lv Z., Rickman K. A., Yuan L., Williams K., Selvam S. P., Woosley A. N., Howe P. H., Ogretmen B., Smogorzewska A., and Olsen S. K. (2017) S. pombe Uba1–Ubc15 structure reveals a novel regulatory mechanism of ubiquitin E2 activity. Mol. Cell 65, 699–714,e6 10.1016/j.molcel.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lv Z., Yuan L., Atkison J. H., Aldana-Masangkay G., Chen Y., and Olsen S. K. (2017) Domain alternation and active site remodeling are conserved structural features of ubiquitin E1. J. Biol. Chem. 292, 12089–12099 10.1074/jbc.M117.787622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Misra M., Kuhn M., Löbel M., An H., Statsyuk A. V., Sotriffer C., and Schindelin H. (2017) Dissecting the specificity of adenosyl sulfamate inhibitors targeting the ubiquitin-activating enzyme. Structure 25, 1120–1129.e3 10.1016/j.str.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 24. Lois L. M., and Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 10.1038/sj.emboj.7600552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olsen S. K., Capili A. D., Lu X., Tan D. S., and Lima C. D. (2010) Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 10.1038/nature08765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tokgöz Z., Siepmann T. J., Streich F. Jr, Kumar B., Klein J. M., and Haas A. L. (2012) E1–E2 interactions in ubiquitin and Nedd8 ligation pathways. J. Biol. Chem. 287, 311–321 10.1074/jbc.M111.294975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hyer M. L., Milhollen M. A., Ciavarri J., Fleming P., Traore T., Sappal D., Huck J., Shi J., Gavin J., Brownell J., Yang Y., Stringer B., Griffin R., Bruzzese F., Soucy T., et al. (2018) A small-molecule inhibitor of the ubiquitin-activating enzyme for cancer treatment. Nat. Med. 24, 186–193 10.1038/nm.4474 [DOI] [PubMed] [Google Scholar]
- 28. Singh R. K., Kazansky Y., Wathieu D., and Fushman D. (2017) Hydrophobic patch of ubiquitin is important for its optimal activation by ubiquitin-activating enzyme E1. Anal. Chem. 89, 7852–7860 10.1021/acs.analchem.6b04194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L. Jr, Holton J. M., and Schulman B. A. (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 10.1016/S1097-2765(03)00452-0 [DOI] [PubMed] [Google Scholar]
- 30. Tokgöz Z., Bohnsack R. N., and Haas A. L. (2006) Pleiotropic effects of ATP. Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J. Biol. Chem. 281, 14729–14737 10.1074/jbc.M513562200 [DOI] [PubMed] [Google Scholar]
- 31. Lyskov S., Chou F. C., Conchúir S. Ó., Der B. S., Drew K., Kuroda D., Xu J., Weitzner B. D., Renfrew P. D., Sripakdeevong P., Borgo B., Havranek J. J., Kuhlman B., Kortemme T., Bonneau R., et al. (2013) Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS ONE 8, e63906 10.1371/journal.pone.0063906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozakov D., Grove L. E., Hall D. R., Bohnuud T., Mottarella S. E., Luo L., Xia B., Beglov D., and Vajda S. (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 10, 733–755 10.1038/nprot.2015.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deshaies R. J. (2015) Protein degradation: Prime time for PROTACs. Nat. Chem. Biol. 11, 634–635 10.1038/nchembio.1887 [DOI] [PubMed] [Google Scholar]
- 34. Yuan L., Lv Z., Atkison J. H., and Olsen S. K. (2017) Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI. Nat. Commun. 8, 211 10.1038/s41467-017-00272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 38. Delano W. (2002) The PyMOL Molecular Graphics System, Version 1.862, DeLano Scientific, San Carlos, CA [Google Scholar]
- 39. McNicholas S., Potterton E., Wilson K. S., and Noble M. E. (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.