Abstract
The generation of traction forces and their transmission to the extracellular environment supports the disseminative migration of cells from a primary tumor. In cancer cells, the periodic variation of nuclear stiffness during the cell cycle provides a functional link between efficient translocation and proliferation. However, the mechanical framework completing this picture remains unexplored. Here, the Fucci2 reporter was expressed in various human epithelial cancer cells to resolve their cell cycle phase transition. The corresponding tractions were captured by a recently developed reference-free confocal traction-force microscopy platform. The combined approach was conducive to the analysis of phase-dependent force variation at the level of individual integrin contacts. Detected forces were invariably higher in the G1 and early S phases than in the ensuing late S/G2, and locally colocalized with high levels of paxillin phosphorylation. Perturbation of paxillin phosphorylation at focal adhesions, obtained through the biochemical inhibition of focal adhesion kinase (FAK) or the transfection of nonphosphorylatable or phosphomimetic paxillin mutants, significantly diminished the force transmitted to the substrate. These data demonstrate a reproducible modulation of force transmission during the cell cycle progression of cancer cells, instrumental to their invasion of dense environments. In addition, they delineate a model in which paxillin phosphorylation supports the mechanical maturation of adhesions relaying forces to the substrate.
INTRODUCTION
Beyond their genetic heterogeneity, solid tumors display unique characteristics of stiffness (Beil et al., 2003; Paszek et al., 2005; Kraning-Rush and Reinhart-King, 2012), size, shape, and deformability (Krause and Wolf, 2015). This biophysical fingerprint depends on the composition, architecture, and type of cancer (Mouw et al., 2014). Within neoplastic lesions, cells are embedded in a complex three-dimensional (3D) environment, which contributes overlapping biological and mechanical signals impinging on the metastatic dissemination of cancer seeds. This mechanical interplay is of particular relevance in defining the ability of pervasive cells to migrate in dense interstitial tissues and proliferate in distant body locations (Provenzano et al., 2008; Khamis et al., 2012). In this context, traction forces exerted by cells on the surrounding environment via integrin contacts (i.e., the focal adhesions [FAs]) orchestrate a process of mutual evolution, during which metastatic cells remodel the extracellular matrix (ECM; Wolf et al., 2013; Charras and Sahai, 2014), acquire the ability to repair DNA damage (Kumar et al., 2014), and develop resistance to drugs (Holohan et al., 2013).
A number of studies have evaluated the correlation between cell cycle progression and migration of cancer cells. In particular, the “go or grow” hypothesis postulated that the two activities are mutually exclusive in space and time (Lund-Johansen et al., 1990; Penar et al., 1997; Zhang et al., 1997; Roth et al., 2000). Emerging evidence, however, shows that tumor cells do not defer proliferation for migration (Corcoran and Del Maestro, 2003; Garay et al., 2013). Along this line is the pathological observation that highly proliferative tumors are often likewise invasive (Alves et al., 2011). This correlation is linked to frequent relapses and a very poor prognostic outcome.
The existence of a link between cell proliferation and migration remains elusive, yet a growing number of indications suggest that the key may be found in the way cells generate actomyosin-mediated traction forces and transmit them to the surrounding environment to propel migration (Lauffenburger and Horwitz, 1996). We recently provided evidence that the efficiency of penetrative migration in small interstitial pores varies during the cell cycle progression of cancer cells (Panagiotakopoulou et al., 2016). Nuclear deformability increases immediately after division, a phase in which the biochemical decondensation of chromatin takes place (M/G1 phase). Cells can thus efficiently penetrate small openings due to the reduced physical resistance provided by the nucleus (Cho et al., 2017). Nuclear stiffness increases in the ensuing G1 and S phases (Cramer and Mitchison, 1995; Lesman et al., 2014). This, however, does not impede the pervasion of pores demanding a very large deformation. The same narrow openings become insurmountable obstacles as soon as the cell progresses from the early S to the late S/G2 phase (Panagiotakopoulou et al., 2016).
A pivotal role in regulating both cell proliferation and migration is played by cell adhesions to ECM, which are established upon integrin activation (Griffith and Swartz, 2006; Huber-Keener et al., 2012; Bonnans et al., 2014). Integrin contacts are partially lost during cell division (Marchesi et al., 2014) and gradually reassembled by daughter cells upon abscission. Cancer cells feature a greater number of large adhesions to the substrate in the G1 and S phase (Panagiotakopoulou et al., 2016). It is therefore logical to speculate that in these specific phases of the cell cycle, the transmission of cellular forces to the substrate is sufficient to actuate the necessary deformation and squeezing of a relatively stiff nucleus.
Actomyosin-generated contractility is transmitted to the substrate at the level of FAs (Geiger et al., 2009). Mature adhesions connect with actin stress fibers at their cytoplasmic sides (i.e., the adhesion plaque; Kanchanawong et al., 2010). Beside actin dynamics (Choi et al., 2008; Thievessen et al., 2013), master regulators of this mechanical maturation are focal adhesion kinase (FAK) and paxillin (Chrzanowska-Wodnicka and Burridge, 1994; Retta et al., 1996; Frame, 2004; Schlaepfer and Mitra, 2004; Zaidel-Bar et al., 2007; Pasapera et al., 2010; Plotnikov et al., 2012). In particular, FAK-mediated paxillin phosphorylation induces the recruitment of vinculin at the adhesion site, thus reinforcing the connection to the actin cytoskeleton (Kanchanawong et al., 2010). This process may support the transmission of traction forces to the substrate in correspondence with individual integrin contacts, yet a direct demonstration is still missing.
Traction force microscopy (TFM) provides access to cellular forces transmitted to the substrate via FAs. TFM detects the resulting substrate deformations by tracking the displacement of optical landmarks (Roca-Cusachs et al., 2017). To do this, classic TFM approaches, based on continuum hydrogels or silicon substrates, require the acquisition of a reference image (Plotnikov et al., 2014), which renders them incompatible with the analysis of protein activity by immunohistochemistry. Discrete TFM methods do not require this disruptive step, yet introduce significant artifacts in the formation of integrin contacts (Plotnikov et al., 2014). A reference-free continuum TFM method, recently developed in our laboratory, allows the generation of spatially resolved overlapping maps of protein activity and traction forces (Bergert et al., 2016). Here, we exploit this approach to investigate the variation of force transmission to the substrate in cancer cells during their progression along the cell cycle and link it to the corresponding dynamics of FA maturation via FAK-mediated paxillin phosphorylation.
RESULTS AND DISCUSSION
HeLa Fucci2 cells were adopted as a general model of cell cycle progression in human epithelial cancer. Their adhesion to the substrate and migratory behavior in standard culture and in vitro models of interstitial invasion are well established (Miwa et al., 2015; Duan et al., 2017; Chu et al., 2018). The cells stably express the Fucci two-color cell cycle reporter (Sakaue-Sawano et al., 2008), which is characterized by the alternate nuclear expression of hCdt1-mCherry during the G1 phase (red fluorescent channel) and hGem-Azami Green during the S/G2/M phases (green fluorescent channel; Sakaue-Sawano et al., 2008). Calibration of the relative signals from the two fluorescent channels and their correspondence to the cell transition in distinct cell cycle phases were previously reported for these cells. In particular, cells in M/G1 show no fluorescent emission, while the G1, early S, and S/G2 phases are identified by red, orange, or green emissions, respectively (Figure 1A; Panagiotakopoulou et al., 2016).
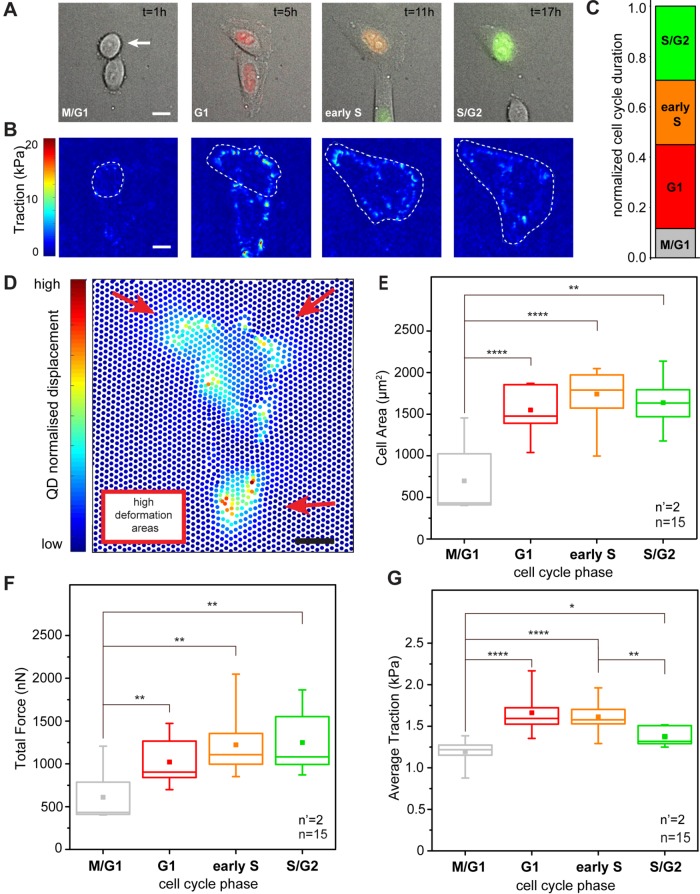
FIGURE 1:
Cell cycle–dependent force transmission in cancer cells. (A) Merged transmission and fluorescence images of a HeLa Fucci2 cell over a complete cell cycle. The corresponding cell cycle phase, as defined by the Fucci2 reporter, is indicated in the bottom left corner of each panel. The elapsed time from the initial cell division is reported in the top right corner. Scale bar is 10 μm. (B) Corresponding maps of actuating traction obtained by cTFM. A white dashed line indicates the basal profile of the cell under analysis. (C) Relative duration of each cell cycle phase. (D) Typical displacement of the QD nanodisc array induced by a HeLa Fucci2 cell in G1. Scale bar is 10 μm. (E) Dynamics of absolute total forces along cell cycle progression. Force is averaged over all tracked cell cycles. (F) Corresponding phase-resolved dynamics of cell spreading and (G) traction transmitted to the substrate along individual cell cycles. *p < 0.05, **p < 0.01, ****p < 0.0001. n = number of cells and n’ = number of independent experiments.
To access the forces imparted by cells to the substrate, HeLa Fucci2 cells were seeded at low density and care was taken to avoid the generation of large clusters, which interfere with the measurement of individual cell contractility (initial seeding density 4 × 104/cm2). To measure the spatial and temporal variation of cellular traction as a function of the cell cycle phase, a red-emitting confocal TFM (cTFM; Bergert et al., 2016) platform was used. The force sensor array was based on a silicone substrate (9:10, CY52-276) yielding an elastic modulus of ∼12.6 kPa, on the surface of which a triangular pattern of red quantum dot (QD) disk-shaped layers of diameter ∼200 nm was printed, with a 2-μm pitch. This configuration provided sufficient spatial resolution and sensitivity to capture deformations induced by single FAs (Bergert et al., 2016). Individual cells were monitored through live-cell microscopy along the entire duration of their cell cycles, from one division to the next. The average duration of these cycles was 20.2 h; therefore a total observation period of 30 h made it possible to monitor a sufficient number of complete progressions. The resulting time lapses yielded stratified information on cell cycle and spreading dynamics and on corresponding deformation of the QD nanodisc array. From the latter, the actuating traction forces were derived by means of a dedicated algorithm (Figure 1B; Bergert et al., 2016).
The average duration of each cell cycle phase was extracted as the first quantitative data layer (Figure 1C). During the recorded cycles, cells spent more time in the G1 phase, while their residence in M/G1 (early stage G1), early S, or late S/G2 phase was shorter. Similarly, phase-resolved values of cell spreading were obtained. The graph in Figure 1F reports the linear increase of basal area upon progression from the M/G1 to the G1 phase. The cell surface increment continued in the ensuing development to reach a plateau in late S/G2.
Cell-induced displacement of the QD nanodiscs from their original, perfectly-ordered position in the triangular array (through the deformation of the elastic substrate to which the nanodiscs firmly adhered) was automatically captured under each tracked cell and over the entire cell cycle (Figure 1, B and D). A finite element analysis of the resulting vectorial displacement field, based on the constitutive material model of the substrate (Bergert et al., 2016), rendered spatially and temporally resolved maps of the actuating traction forces. Using the overlapping cell profiles, stress magnitudes were integrated under the entire cell surface, yielding the total force intensity exerted by the cell under analysis (Figure 1E). Forces transmitted by HeLa Fucci2 cells increased from the M/G1 phase (699 ± 144 nN) to G1 and lasted through early S (1741 ± 89 nN), in line with previous reports establishing a linear correlation between cell area and force generation (Paszek et al., 2005; Mekhdjian et al., 2017). A slight decrease was instead measured toward the end of the cell cycle, in the late S/G2 phase (1638 ± 101 nN; Figure 1E).The same total force and cell area trends are well captured by measurements along individual cell cycles (Supplemental Figures S1–S4).
To better understand the link between cell spreading and force generation in cell cycle progression, the measured cellular force (Figure 1E) was divided by the cell basal area (Figure 1F) to obtain the average traction (Figure 1G). This derived descriptor corresponds to the density of force exerted by individual cells on the substrate and increased significantly from the M/G1 to the G1 phase (1.126 ± 0.2 kPa to 1.65 ± 0.2 kPa). It persisted upon transition to the S phase (1.54 ± 0.2 kPa) while dropping significantly when cells entered the S/G2 phase. In the S/G2 phase the average traction was only slightly higher than that measured in M/G1 (1.3 ± 0.2 kPa). These results suggest that the cellular machinery supporting force transmission matures during cell cycle progression along with cell spreading, but with different dynamics. The highest average traction is transmitted to the substrate before the cell surface reaches its full extension (Supplemental Figures S1–S4). Finally, no direct correlation between cell shape and exerted traction could be detected in individual cell cycles (Supplemental Figure S5), suggesting that the small and rapid variations of cell shape recorded under our experimental conditions did not significantly impinge on the force generation dynamic.
cTFM provides a reference-free detection of cell-mediated substrate deformations from which a measure of transmitted cell contractility can be obtained directly without the necessity of disruptive cell removal (Bergert et al., 2016). The phase-resolved evolution of average traction obtained from fixed samples (Figure 2A), and thus from different cells, revealed the same temporal variation as that captured by the live cell analysis (Figure 1G). This result has a twofold value. First, it validates cTFM as conducive to endpoint sample processing (i.e., immunostaining) to reveal the level of activity of specific molecules as an additional layer of information. Second, it demonstrates that the G1 traction peak is significant beyond intercellular variability.
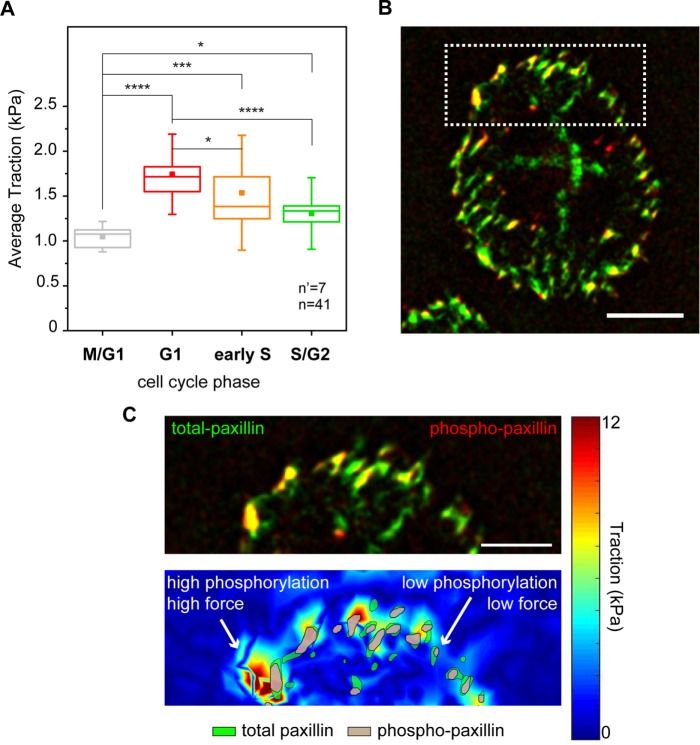
FIGURE 2:
Combined analysis of forces and focal adhesion maturation. (A) Cell cycle–dependent variation of traction forces transmitted to the substrate in fixed samples. *p < 0.05, ***p < 0.001, ****p < 0.0001. n = number of cells and n’ = number of independent experiments. (B) Basal distribution of total (green) and phosphorylated (red) paxillin signals obtained from immunostaining. (C) Corresponding map of actuating stresses (top panel). Magnification of the cell periphery (bottom panel), including a group of individual focal adhesions. Green area outlines the total paxillin profile, and gray area outlines phosphorylated paxillin regions.
Hence, the total (total-pax) and phosphorylated paxillin (phospho-pax) distributions were visualized in cells contacting the cTFM substrate, providing a direct map of adhesions, paxillin activation, and resulting transmitted forces (Figure 2B). A first, qualitative analysis indicated that large FAs featuring prominent paxillin phosphorylation colocalized with regions of high transmitted force, while smaller adhesions with correspondingly lower phospho-pax levels were found in regions of low force transmission (Figure 2C).
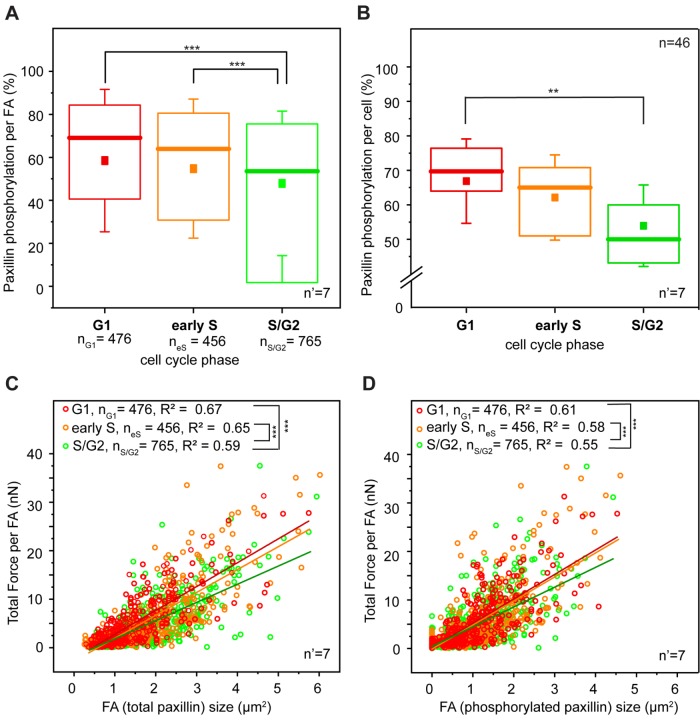
Quantitative large-scale analysis confirmed this observation. HeLa cells do not display resolvable paxillin accumulation at the basal surface during M/G1 (Panagiotakopoulou et al., 2016), consistent with the absence of mature integrin contacts in this phase (Moes et al., 2011). In the ensuing phases, FAs were mostly detected at the cell periphery (Figure 2B). Here, higher levels of phospho-pax correlated linearly with the size of individual adhesions (Supplemental Figure S6), thus linking FA growth to their mechanical maturation (Bonnans et al., 2014; Paszek et al., 2005). Notably, phospho-pax levels were significantly higher in the G1 phase than for FAs of the same size detected in later phases of the cell cycle (Figure 3, A and B). Both the sizes of individual FAs and the corresponding phospho-pax levels were proportional to the amount of force exerted locally on the substrate (Figure 3, C and D). Yet the correlation slope was significantly steeper for cells in the G1 and early S phases. Thus, in the S/G2 phase, individual FAs of the same size exerted relatively less force.
FIGURE 3:
Paxillin phosphorylation and force transmission. (A) Cell cycle–dependent variation of paxillin phosphorylation at focal adhesion sites. (B) Global variation of paxillin phosphorylation during the cell cycle progression of HeLa Fucci2 cells. **p < 0.01, ***p < 0.001. Correlation between force transmitted by individual focal adhesions and corresponding total focal adhesion size (C) and size of phosphorylated paxillin signal (D). Red, yellow, and green dots represent individual focal adhesions detected in G1, early S, and S/G2 cells, respectively. Lines of corresponding colors represent the linear fit to each data set. For panels A, C, and D, n = number of individual focal adhesions from seven independent experiments. For panel B, n = number of cells and n’ = number of independent experiments.
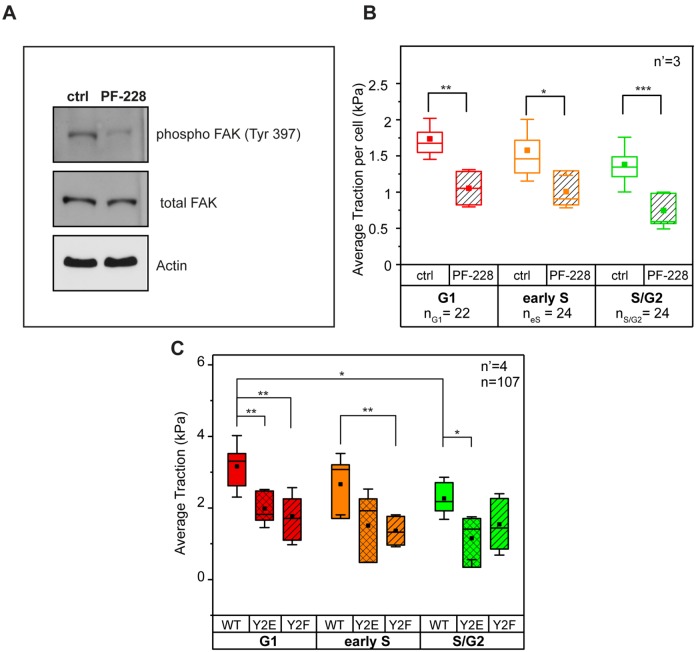
Previous work has shown that the density of stresses is dependent on the shape of the cell and the location of focal adhesions (Han et al., 2012; Oakes et al., 2014). Here, the specific biochemical inhibition of FAK activity (Figure 4A) greatly reduced the levels of paxillin phosphorylation detected at individual FAs and in all cell cycle phases (Supplemental Figure S7). However, it did not perturb the overall size and number of integrin contacts (Supplemental Figure S8). Consistently, the average tractions transmitted to the substrate dropped significantly in all analyzed cells (Figure 4B). To further demonstrate that paxillin phosphorylation is necessary for the modulation of force transmission to the substrate during the cell cycle, the force analysis was extended to HeLa Fucci2 cells transfected with wild type (WT) and with nonphosphorylatable (Y2F) or phosphomimetic (Y2E) paxillin mutants (Zaidel-Bar et al., 2007). Cells expressing either paxillin mutant displayed decreased average traction (as compared with the WT control) in the G1 phase. Moreover, Y2F mutants exhibited a reduction of average traction in the early S phase, while Y2E mutants exhibited a reduction in the S/G2 (Figure 4C). Altogether, these data indicate that the mechanical maturation of integrin contacts is supported by FAK-mediated paxillin phosphorylation regulating FA turnover. This process culminates in the G1 phase, thus colocalizing in space and time with higher transmission of actomyosin-mediated contractility.
FIGURE 4:
FAK activity and regulation of tractions. (A) Biochemical inhibition of FAK activity and (B) resulting phase-dependent reduction of traction. (C) Average traction in phosphomimetic (Y2E) and nonphosphorylatable (Y2F) paxillin mutants. n = number of cells and n’ = number of independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
To assess whether the cell cycle–dependent variation of force transmission to the substrate is a feature of epithelial cancer cells beyond the type already examined, we extended our analysis to highly metastatic human breast cancer cells MCF-7 and their tamoxifen-resistant (TamR) offspring (MCF-7 TamR; Figure 5). tamoxifen is a widely used drug in chemotherapies against hormone-dependent breast carcinoma (Chang, 2012). The molecule interacts competitively with the estrogen receptors on cancer cells, thereby blocking the estrogen activity (Harvey et al., 1999). Lack of estrogen stimulation halts cell cycle progression and directs cells toward apoptosis. Interestingly, tamoxifen treatment arrests WT MCF-7 cells in G1 (Otto et al., 1996). TamR MCF-7 cells are insensitive to the above-mentioned effects of the drug. In addition, they are characterized by dysregulated cell cycle progression and apoptotic pathway, which contribute to their survival under long-term tamoxifen treatment (Huber-Keener et al., 2012).
FIGURE 5:
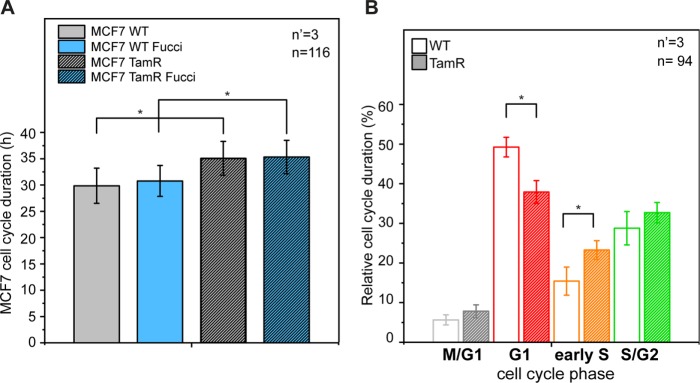
Proliferation dynamics in WT and tamoxifen-resistant MCF-7 cells. (A) Average duration of cell cycle in WT and tamoxifen-resistant (TamR) MCF-7 cells. (B) Relative duration of each cell cycle phase. *p < 0.05. n = number of cells and n’ = number of independent experiments.
WT and TamR MCF-7 stably transfected with the Fucci2 sensor were generated by viral infection (see Materials and Methods). The introduction of the Fucci sensor did not affect the cell cycle duration of either cell type (Figure 5A). When compared, the two cancer strains displayed a significantly different cell cycle dynamic (Figure 5, A and B). In the absence of tamoxifen treatment, WT MCF-7 were characterized by faster proliferation (average cell cycle duration 27.3 and 32 h for MCF-7 WT and TamR, respectively; Figure 5A). In particular, WT cells lingered relatively longer in G1 and showed reduced permanence in all other cell cycle phases (Figure 5B). This observation indicates a delayed progression from the G1 to the S phase, consistent with the previously reported proliferative anomalies (Ward et al., 2012).
We aimed at investigating the dosage-dependent effect of tamoxifen (4-hydroxytamoxifen on ethanol vehicle) treatment on the Fucci transfected MCF7 strains, by means of live microscopy for a total period of 84 h. As expected (Sutherland et al., 1983; Ward et al., 2012), the two strains had different responses to the drug (Supplemental Figure S9). Treatment with a 5-μM dosage of 4-hydroxytamoxifen induced the apoptosis of WT MCF7, while the TamR population remained stable. In both strains, upon treatment with the drug, surviving cells accumulated in the G1 phase of the cell cycle. A dosage increase to 10 μM caused faster apoptosis in the WT population, while no significant change was detected in TamR cells. Again, in both strains, the remaining cells mostly populated the G1 phase. Finally, a dose of 20 μM of 4-hydroxytamoxifen was cytotoxic for both strains, with rapid cell death starting after 1 d of exposure.
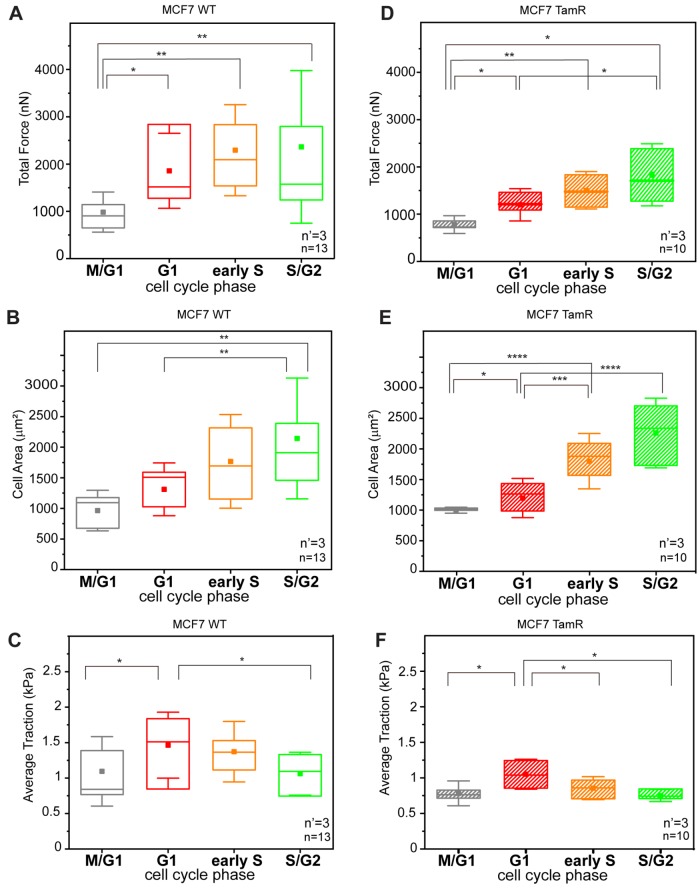
In both strains, the phase-resolved profiles of forces transmitted to the cTFM were obtained from complete cell cycle analyses of individual cells (Figure 6). The resulting trends reproduced the phase-dependent dynamics measured for HeLa Fucci2 cells (Figure 1). In particular, the absolute total force peaked in the G1 and early S phases for the WT (Figure 6, A and D). A significant drop in force transmitted in the late S/G2 phase was detected both in WT MCF-7 and in TamR cells. Effects of traction upon cell cycle progression were obtained by integrating the corresponding cell spreading dynamics (Figure 6, C and F) and confirmed the presence of a peak in G1 for both cell strains. Altogether, these data contribute to establishing the presence of a periodic variation of traction forces exerted by cancer cells on the substrate during their proliferative cycles. In addition, they individuate the G1 phase as the most mechanically intense, during which the cells are able to impart the highest levels of force at the interface with the surrounding extracellular environment.
FIGURE 6:
Cell cycle–dependent force transmission in MCF-7 cells. (A, D) Phase-resolved dynamics of absolute forces during cell cycle progression for WT (A) and TamR (D) MCF-7 cells, respectively. (B, E) Corresponding dynamics of cell spreading over the cell cycle. (C, F) Tractions transmitted to the substrate during individual cell cycles for WT (left) and TamR cells (right), respectively. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n = number of cells from four independent experiments per cell line variant and n’ = number of independent experiments.
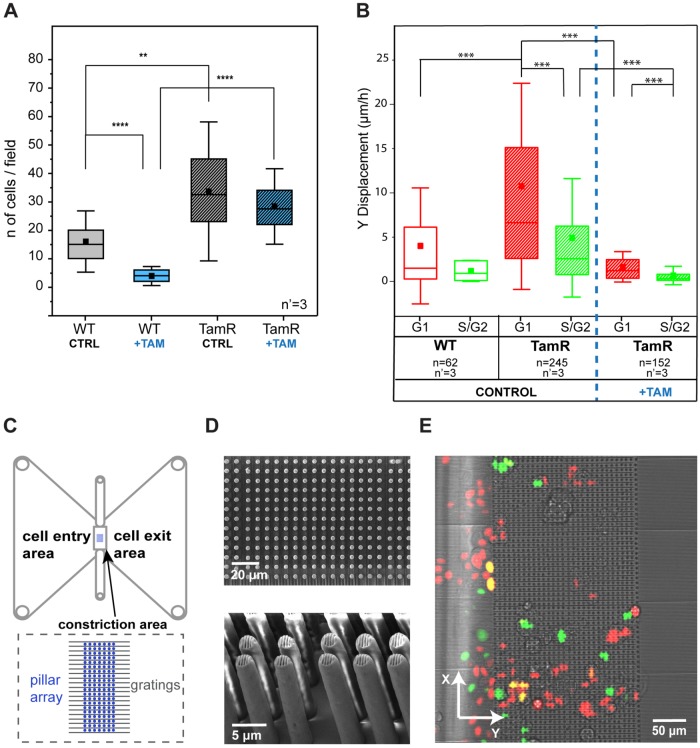
To assess whether this finding is linked to a specific phenotype in MCF-7 cells, we performed two independent evaluations of cell invasiveness. The Boyden chamber assay (Figure 7A; Kleinman and Jacob, 2001) revealed a more efficient pervasive invasion of TamR MCF-7 cells than of the WT population. This difference was further increased by the addition of 10 μM of tamoxifen. The drug treatment almost completely ablated the pervasive invasion of WT cells while showing a minor effect on the invasion of the resistant counterpart. In addition, a 2D invasion assay was performed to evaluate cell migration within a dense array of vertical obstacles generating narrow constrictions (Corallino et al., 2018). The patterned area was planar and conducive to live-cell microscopy, thus allowing the detection of pervasive migration and the concomitant resolution of the cell cycle phase (Figure 7, B–E). Directional movements were pronounced in the G1 phase in both WT and TamR MCF-7 cells, yielding more efficient advance in the dense array of vertical obstacles in this phase of the cell cycle. Enhanced invasiveness in G1 was also measured upon treatment with tamoxifen, which induced, however, an overall reduction of cell motility.
FIGURE 7:
Migratory and invasive phenotypes of MCF7-WT and MCF7-TamR cells. (A) Comparison of invasion capacity of WT and TamR MCF-7 cells, with and without tamoxifen, using a Boyden chamber–based invasion assay. The results come from three independent experiments, each performed in triplicate. **p < 0.01, ****p < 0.0001. (B) Migratory properties of MCF7-WT and MCF7-TamR cells in a 2D invasion assay. ***p < 0.001. n = number of cells and n’ = number of independent experiments per condition. (C) Schematic of the device as reported in Corallino et al. (2018). (D) Scanning electron microscope pictures (top panel, top view; bottom panel, side view) of the pillar array. (E) Merged transmission and fluorescent pictures from the live microscopy experiments.
In summary, the results presented here demonstrate a significant, periodic variation of tractions transmitted to the substrate by proliferating cancer cells. Force transmission in correspondence with integrin contacts is low immediately after division, a phase in which focal adhesions are not yet fully reassembled. A rapid increase in transmitted forces is detected in the G1 and early S phases. The mechanical activity of cells changes trend in the S/G2 phase, during which transmitted forces are reduced. These measurements obtained on different strains of human epithelial cancer are in line with what reported for nontransformed epithelial cells from the human retina (RPE-1; Vianay et al., 2018), indicating that this pattern of mechanical activity may be a conserved feature beyond the analyzed cell types.
It is generally accepted that cell-generated tractions increase with cell spreading. Therefore, cells with a larger surface should transmit more force to the substrate. This assumption is confirmed by the correlative analysis of individual cells over the entire cell cycle. However, the correlation is not linear, as the two cellular activities, force transmission and spreading, follow different dynamics. In fact, the density of transmitted forces is maximal in G1. During this phase, cancer cells are able to exert tractions concentrated on a limited surface, a behavior that may be conducive to higher efficiency in the penetration of complex environments requiring a major deformation of the cell nucleus (Denais et al., 2016; Panagiotakopoulou et al., 2016; Paul et al., 2017; Green et al., 2018).
Finally, the use of a recently developed reference-free approach (Bergert et al., 2016) to monitor the mechanical activity of cells during their progression in the cell cycle offered the possibility of investigating the role of focal adhesion maturation. The involvement of FAK-mediated paxillin phosphorylation is supported by the direct spatial and temporal covariance between the protein activity and the local variation of traction forces transmitted to the substrate. Using this experimental framework, future analysis may reveal the full mechanism linking cell migration, proliferation, and force transmission.
MATERIALS AND METHODS
Confocal traction force microscopy
Substrate traction was measured using cTFM substrates with red-emitting QDs with an interdisc spacing of 2 μm (Bergert et al., 2016). After being coated with fibronectin, adhering cells generated deformations of the substrate, which were captured as distortions of the fluorescent nanodisc array. The subpixel detection of the QD nanodisc center was followed by computational reconstruction of the triangular mesh. From the displacement field, the surface tractions were reconstructed using FEA, as previously reported (Bergert et al., 2016).
Absolute force per adhesion calculation
The absolute force per adhesion was determined based on three elements: the focal adhesion area (pixels in an image belonging to an individual adhesion), the overall traction forces, and the conversion from pixels to micrometers. To determine the area of the adhesions, a threshold was applied to the paxillin signal as previously reported in (Bergert et al., 2016). This yielded the positions and sizes of individual adhesions, as well as a list of pixels belonging to each of them. For the calculation of the forces per adhesion, traction forces acquired from the FEA were interpolated onto a quadratic grid corresponding to the pixels in the paxillin image. For each pixel, the Euclidean norm of the traction was calculated. Based on the adopted magnification and the pixel size of the microscope camera, the actual pixel’s size (in micrometers) was defined. Finally, traction magnitudes multiplied by the area of one pixel were summed for all the pixels within the adhesion area to compute the absolute force per adhesion.
Cell lines and culture conditions
MCF-7 WT and tamoxifen-resistant cells, a kind gift of Stefan Wiemann (German Cancer Research Center, Heidelberg, Germany), have been extensively characterized previously (Jurmeister et al., 2012; Ward et al., 2012; Bonnans et al., 2014). HeLa Fucci2 cells stably expressing a Fucci construct (Sakaue-Sawano et al., 2008) were purchased from Riken Bioresource Center (Japan) and previously characterized (Panagiotakopoulou et al., 2016).
MCF7 and MCF7-TamR cells expressing the Fucci reporter system were obtained by lentiviral infection of mKO2-hCdt1(30/120) and mAG-hGem (1/110) probes (Sakaue-Sawano et al., 2008). HeLa cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS) at 37°C under a 5% CO2 humidified atmosphere. WT and TamR MCF-7 cells were maintained in DMEM phenol red free medium (Sigma-Aldrich, St. Louis, MO) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% FBS at 37°C under a 5% CO2 humidified atmosphere. Cells were routinely analyzed by reverse transcriptase-PCR for mycoplasm contamination.
Lentiviral production and infection
HEK293T packaging cells (1 × 106) in a 10-cm (diameter) dish were transfected with equimolar ratios of pMD2, psPAX2 (Invitrogen), and Fucci expression vectors. Transfection was performed with LipofectAMINE 3000 (Invitrogen) in OPTIMEM in accordance with the manufacturer’s instructions. After transfection, cells were incubated for 18 h, and then the medium containing the transfection reagent has been removed and replaced with fresh culture medium. After 24 h, culture medium containing lentiviral particles was collected and passed through a 0.45-μm filter, 8 μg/ml polybrene was added, and the culture was immediately used to infect the target cells.
Live microscopy
Cell cycle movies were acquired using an inverted Nikon-Ti wide-field microscope (Nikon, Japan) equipped with an Orca R-2 CCD camera (Hamamatsu Photonics, Japan) or a Nikon-Ti spinning disk confocal microscope (Nikon, Japan) equipped with an Andor DU-888 camera (Oxford Instruments, United Kingdom), both with an incubation chamber (Life Imaging Services, Switzerland) to control temperature, CO2, and humidity. Images were collected using a 40× objective (Plan Fluor 40× Oil DIC H N2). Multiple nonoverlapping fields capturing all positions with quantum dots were recorded in parallel (ΔT = 1 h, total duration ∼30 h for HeLa Fucci and ∼48 h for MCF7).
At each time of measurement, a transmission and two fluorescent images of the nuclei of the cells were acquired using differential interference contrast (DIC), an FITC (fluorescein isothiocyanate) filter set, and a TRITC (tetramethylrhodamine isothiocyanate) filter set. Focal drift during the experiments was avoided using the autofocus system of the microscope.
For the analysis of the effect of tamoxifen on the phase partition and viability of the cells, cells were seeded in the presence of 4-hydroxytamoxifen in ethanol solvent (Sigma-Aldrich, St Louis, MO) or ethanol solvent control.
Image analysis
Cell cycle duration was quantified as the time between birth and division of the same randomly chosen cell. The cell cycle phase of Fucci2 transfected cells was determined from the ratio of the absolute intensities of red and green fluorescence of the nucleus. Absolute intensities were calculated by subtracting the intensity of the background from the relative intensity of the nucleus, both measured in ImageJ. Transparent cells (M/G1) were distinguished from the rest of the population because their relative intensity was equal to that of the background.
For the quantification of cell adhesions during the cell cycle, cells were seeded on cTFM substrates, incubated overnight (O/N), and stained for paxillin and phosphorylated paxillin (see Immunofluorescence).
Fluorescent Z-stacks of the signals emitted by the focal adhesions (Δ Z= 0.3 μm) were collected using a Nikon-Ti spinning disk confocal microscope (Nikon, Japan) equipped with an Andor DU-888 camera (Oxford Instruments, United Kingdom). Focal adhesions (total paxillin) were quantified as described in the paper by Horzum et al. (2014). After the regions of interest (ROIs) of all adhesions were registered, the phosphorylation amount per adhesion was calculated by dividing the area of the total paxillin ROI of each adhesion site by the area of the phosphorylated paxillin ROI. A designated Matlab script was used to colocalize the ROIs of total and phosphorylated paxillin that belonged to the same adhesion site by comparing their centroid coordinates.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature (RT) and permeabilized via incubation in 3% paraformaldehyde with 0.1% Triton in PBS for 5 min. After being blocked with 5% bovine serum albumin (BSA) in PBS for 2 h, samples were incubated at 4°C O/N with monoclonal primary antibodies.
The following commercial antibodies were used: mouse anti-paxillin, BD Bioscience (#610051), dilution: 1:200 (van de Water et al., 2001) and rabbit anti-paxillin Tyr118, Cell Signaling (#2541S), dilution: 1:200 (Mekhdjian et al., 2017). Secondary antibodies Alexa Fluor 647 chicken anti-rabbit (LifeTechnologies #A-21443) or DyLight 405 donkey anti-mouse (Javkson Immunofluorescence) were used at 1:200 for 1 h at RT.
FAK inhibition
HeLa cells were treated O/N with 10 μM of FAK inhibitor PF-573228 (Slack-Davis et al., 2007; SIGMA Aldrich) diluted in dimethyl sulfoxide (DMSO). The control samples were incubated with the same amount of DMSO. After the treatments, cells were immediately processed for immunofluorescence and Western blotting analyses.
Plasmids and transfection
YFP-paxillin WT and two YFP-phosphorylation mutants, in which both tyrosine 31 and 118 were replaced either by glutamic acid (Y2E, phosphomimetic) or by phenylalanine (Y2F, nonphosphorylatable), were a generous gift of Benjamin Geiger (Weizmann Institute of Science; Zaidel-Bar et al., 2007). Transfection was performed using Lipofectamine 3000 (Invitrogen) in accordance with manufacturer’s instructions and transfected cells were selected with 800 µg/ml G418.
Invasion assay
The invasive potential was tested using Transwell chambers (pore size 8 µm; Corning, Corning, NY) precoated with fibronectin (100 µg/ml; Sigma-Aldrich). Briefly, MFC7 WT and MCF7 TamR cells (5 × 104 cells/well) in 100 µl of serum-free DMEM were placed in the upper chamber, and 600 µl of culture medium containing 10% FBS was added to the lower chambers as a chemoattractant, in the presence or absence of tamoxifen (10 µM). After being incubated for 24 h, the cells remaining on the upper membrane were carefully wiped off with cotton swabs, while the cells that had invaded through the membrane were fixed in 4% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI). The stained cells were imaged and counted from five random visual fields/experiment.
For the experiments with tamoxifen,10 μM tamoxifen was added 24 h prior to starting.
Cell migration through the pillar array
The array of pillars was fabricated with two-photon polymerization on glass substrates, as previously reported in Corallino et al. (2018). The structure consisted of two components: a dense micropillar array and gratings with ridges of depth 1 μm and width 1 μm. The structure components were designed using CAD software (Autodesk Inventor, Autodesk). The interpillar distance was set to 2.5 μm after parametric testing of various dimensions to ensure the mechanical constriction of the cells during their migration through the array. The pillar thickness was set to 2.5 μm to provide the necessary mechanical stability to the structure. The structural elements of the pillars and barriers (mesh slicing and hatching) and the printing parameters of the whole structure (laser power, scan speed) were designed with the program DeScribe v.2.4.4 (Nanoscribe GmbH), which creates a file readable by the laser lithography system. The sample fabrication was completed with a 3D laser lithography system (Photonic Professional GT, Nanoscribe GmbH, Germany) used in conventional oil-immersion mode. This involved passing a two-photon laser (780-nm laser, laser power 12.5 mW, scan speed 800 μm/s) from a ×63 objective lens through a layer of oil beneath the glass substrate, polymerizing the photoresist on the top side of the glass substrate. The photoresist chosen was a biocompatible organic–inorganic hybrid polymer (Ormocomp, MicroResist Technology GmbH). The resist was drop-cast on the glass substrates and prebaked at 80°C for 2 min. After laser writing, the resist was baked at 130°C for 10 min, developed in OrmoDev developer (MicroResist Technology GmbH) for 10 min to remove the unpolymerized resist, and rinsed in isopropanol (IPA), followed by drying in a critical point dryer (Automegasamdri R 915B, Tousimis).
The glass substrate with the microstructures printed on top was attached on the self-adhesive underside of a commercially available microfluidic chamber (sticky-Slide Chemotaxis, Ibidi), with the micropillar array placed in the middle area. The chamber was filled with 70% ethanol for 1 h for sterilization and washed three times with 1× PBS for 5 min, and substrates were coated with 0.01% poly-l-lysine (PLL; Sigma) according to the manufacturer’s specification. Afterward, poly-l-lysine was gradually replaced with 1× PBS and finally with cell medium. Cells (104) were seeded from the inlets of the cell pool area (Figure 7C) and were gently distributed over the cell pool area, with attention that they did not float over the pillars. The chambers were incubated O/N at 37°C and once the cells were spread, cell motion was recorded using a Nikon-Ti spinning disk confocal microscope (Nikon, Japan) equipped with an Andor DU-888 camera (Oxford Instruments, United Kingdom) with a 20× objective over a 40 -h period, with pictures taken every 30 min. All the assays were performed using an environmental microscope incubator set to 37°C and 5% CO2 perfusion. For the experiments with tamoxifen,10 μM tamoxifen (4-hydroxytamoxifen on ethanol vehicle) was added 24 h prior to imaging.
The velocity of the cells was quantified using the particle tracking algorithm of Imaris (Bitplane Scientific Software, Switzerland). Time-lapse videos were uploaded into Imaris, and the voxel size and time interval were adjusted before particle tracking.
For the quantification, the component of the velocity that was parallel to the gratings and perpendicular to the pillar forest was taken.
Western blotting
Total proteins were extracted by solubilizing HeLa cells in boiling Laemmli buffer (2.5% SDS and 0.125 M Tris-HCl, pH 6.8). Lysates were incubated for 5 min at 100°C to allow protein denaturation and then spun for 5 min at 13,200 rpm to discard cell debris. Supernatants were collected and the concentration of proteins was determined using a BCA Protein Assay Kit (Pierce) according to manufacturer’s instructions. An equal amount of proteins (30 μg) was loaded on gel and separated by SDS–PAGE, transferred to a Protran nitrocellulose hybridization transfer membrane (pore size 0.2 μm; Whatman), and blocked for 1 h at RT in 1X TBST (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, and 0.05% Tween)–5% BSA. The membranes were incubated O/N at 4°C with primary antibodies diluted in 1× TBST–5% BSA.
The following primary antibodies were used: FAK (Rb polyclonal 1:1000, 3285S Cell Signaling; Cho et al., 2017) and p-FAK Y397 (Rb polyclonal 1:1000, 3283S Cell Signaling; Cho et al., 2017). Next, they were rinsed three times with 1× TBST for 5 min each and incubated for 1 h at RT with horseradish peroxidase–linked secondary antibodies (diluted in 1× TBST–5% BSA). Membranes were rinsed three times with TBST for 5 min each, and specific binding was detected by an enhanced chemiluminescence (ECL) system (Amersham Biosciences) using Hyperfilm (Amersham Biosciences). The molecular masses of proteins were estimated relative to the electrophoretic mobility of the cotransferred prestained protein marker Broad Range (Cell Signaling Technology).
Statistical analysis
Boxes in all box plots extend from the 25th to the 75th percentile, with a line at the median and a square representing the mean. Whiskers extend to the SD of the data. Data were analyzed, tested for statistical significance, fitted, and visualized using Origin or Matlab (The MathWorks). The total number of recorded events from three or more independent experiments is shown in the top or bottom right-hand corners of the presented graphs. The Shapiro–Wilk test was used to test for normality of data. For nonnormal distributed data, a nonparametric Kolmogorov–Smirnov test was performed (α = 0.05). Significant differences between slopes included in Figure 3 were calculated using the Akaike information criterion and F-test analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was part of the project “Novel Microengineered Platform for the Study of Interstitial Cell Migration” (Project 200021_146898) supported by the Swiss National Science Foundation (SNSF). C.G. is supported by the “Mäxi-Stiftung“ Foundation. T.L. and G.S. are supported by funding from ETH grant ETH-12 15–1. F.M.P. is supported by funding from ETH grant ETH-07 17-1. We thank B. Geiger for sharing the paxillin plasmids and Eleanna Papa for her help in Western blot. M.P. thanks Julian Schneider and Patrik Rohner for their help and insights on electrohydrodynamic nanoprinting and ink production.
Abbreviations used:
- cTFM
confocal traction force microscopy
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- PAX
paxillin
- QDs
quantum dots.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-12-0726) on August 16, 2018.
REFERENCES
- Alves TR, Lima FR, Kahn SA, Lobo D, Dubois LG, Soletti R, Borges H, Neto VM. (2011). Glioblastoma cells: a heterogeneous and fatal tumor interacting with the parenchyma. Life Sci , 532–539. [DOI] [PubMed] [Google Scholar]
- Beil M, Micoulet A, von Wichert G, Paschke S, Walther P, Omary MB, Van Veldhoven PP, Gern U, Wolff-Hieber E, Eggermann J, et al. (2003). Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nat Cell Biol , 803–811. [DOI] [PubMed] [Google Scholar]
- Bergert M, Lendenmann T, Zundel M, Ehret AE, Panozzo D, Richner P, Kim DK, Kress SJ, Norris DJ, Sorkine-Hornung O, et al. (2016). Confocal reference free traction force microscopy. Nat Commun , 12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol , 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. (2012). Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) , 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Sahai E. (2014). Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol , 813–824. [DOI] [PubMed] [Google Scholar]
- Cho S, Irianto J, Discher DE. (2017). Mechanosensing by the nucleus: from pathways to scaling relationships. J Cell Biol , 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. (2008). Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol , 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, Burridge K. (1994). Tyrosine phosphorylation is involved in reorganization of the actin cytoskeleton in response to serum or LPA stimulation. J Cell Sci (Pt 12), 3643–3654. [DOI] [PubMed] [Google Scholar]
- Chu TLH, Connell M, Zhou L, He Z, Won J, Chen H, Rahavi SMR, Mohan P, Nemirovsky O, Fotovati A, et al. (2018). Cell cycle-dependent tumor engraftment and migration are enabled by Aurora-A. Mol Cancer Res , 16–31. [DOI] [PubMed] [Google Scholar]
- Corallino S, Malinverno C, Neumann B, Tischer C, Palamidessi A, Frittoli E, Panagiotakopoulou M, Disanza A, Malet-Engra G, Nastaly P, et al. (2018). A RAB35-p85/PI3K axis controls oscillatory apical protrusions required for efficient chemotactic migration. Nat Commun , 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran A, Del Maestro RF. (2003). Testing the “go or grow” hypothesis in human medulloblastoma cell lines in two and three dimensions. Neurosurgery , 174–184; discussion 184–175. [DOI] [PubMed] [Google Scholar]
- Cramer LP, Mitchison TJ. (1995). Myosin is involved in postmitotic cell spreading. J Cell Biol , 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Wu A, Chen Z, Yang Y, Liu L, Shu Q. (2017). MiR-204 regulates cell proliferation and invasion by targeting EphB2 in human cervical cancer. Oncol Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame MC. (2004). Newest findings on the oldest oncogene; how activated Src does it. J Cell Sci , 989–998. [DOI] [PubMed] [Google Scholar]
- Garay T, Juhasz E, Molnar E, Eisenbauer M, Czirok A, Dekan B, Laszlo V, Hoda MA, Dome B, Timar J, et al. (2013). Cell migration or cytokinesis and proliferation?—revisiting the “go or grow” hypothesis in cancer cells in vitro. Exp Cell Res , 3094–3103. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. (2009). Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol , 21–33. [DOI] [PubMed] [Google Scholar]
- Green BJ, Panagiotakopoulou M, Pramotton FM, Stefopoulos G, Kelley SO, Poulikakos D, Ferrari A. (2018). Pore shape defines paths of metastatic cell migration. Nano Lett , 2140–2147. [DOI] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA. (2006). Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol , 211–224. [DOI] [PubMed] [Google Scholar]
- Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ. (2012). Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys J , 640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JM, Clark GM, Osborne CK, Allred DC. (1999). Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol , 1474–1481. [DOI] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. (2013). Cancer drug resistance: an evolving paradigm. Nat Rev Cancer , 714–726. [DOI] [PubMed] [Google Scholar]
- Horzum U, Ozdil B, Pesen-Okvur D. (2014). Step-by-step quantitative analysis of focal adhesions. MethodsX , 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber-Keener KJ, Liu X, Wang Z, Wang Y, Freeman W, Wu S, Planas-Silva MD, Ren X, Cheng Y, Zhang Y, et al. (2012). Differential gene expression in tamoxifen-resistant breast cancer cells revealed by a new analytical model of RNA-Seq data. PLoS One , e41333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S, Sahin O. (2012). MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol , 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. (2010). Nanoscale architecture of integrin-based cell adhesions. Nature , 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis ZI, Sahab ZJ, Sang QX. (2012). Active roles of tumor stroma in breast cancer metastasis. Int J Breast Cancer , 574025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman HK, Jacob K. (2001). Invasion assays. Curr Protoc Cell Biol Chapter 12, Unit 12.12. [DOI] [PubMed] [Google Scholar]
- Kraning-Rush CM, Reinhart-King CA. (2012). Controlling matrix stiffness and topography for the study of tumor cell migration. Cell Adh Migr , 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Wolf K. (2015). Cancer cell migration in 3D tissue: negotiating space by proteolysis and nuclear deformability. Cell Adh Migr , 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, Garre M, Parazzoli D, Shivashankar GV, Scita G, et al. (2014). ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell , 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. (1996). Cell migration: a physically integrated molecular process. Cell , 359–369. [DOI] [PubMed] [Google Scholar]
- Lesman A, Notbohm J, Tirrell DA, Ravichandran G. (2014). Contractile forces regulate cell division in three-dimensional environments. J Cell Biol , 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD. (1990). Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res , 6039–6044. [PubMed] [Google Scholar]
- Marchesi S, Montani F, Deflorian G, D’Antuono R, Cuomo A, Bologna S, Mazzoccoli C, Bonaldi T, Di Fiore PP, Nicassio F. (2014). DEPDC1B coordinates de-adhesion events and cell-cycle progression at mitosis. Dev Cell , 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhdjian AH, Kai F, Rubashkin MG, Prahl LS, Przybyla LM, McGregor AL, Bell ES, Barnes JM, DuFort CC, Ou G, et al. (2017). Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol Biol Cell , 1467–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa S, Yano S, Kimura H, Yamamoto M, Toneri M, Matsumoto Y, Uehara F, Hiroshima Y, Murakami T, Hayashi K, et al. (2015). Cell-cycle fate-monitoring distinguishes individual chemosensitive and chemoresistant cancer cells in drug-treated heterogeneous populations demonstrated by real-time FUCCI imaging. Cell Cycle , 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes MJ, Bijvelt JJ, Boonstra J. (2011). Attachment of HeLa cells during early G1 phase. Histochem Cell Biol , 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw JK, Ou G, Weaver VM. (2014). Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol , 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Banerjee S, Marchetti MC, Gardel ML. (2014). Geometry regulates traction stresses in adherent cells. Biophys J , 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto AM, Paddenberg R, Schubert S, Mannherz HG. (1996). Cell-cycle arrest, micronucleus formation, and cell death in growth inhibition of MCF-7 breast cancer cells by tamoxifen and cisplatin. J Cancer Res Clin Oncol , 603–612. [DOI] [PubMed] [Google Scholar]
- Panagiotakopoulou M, Bergert M, Taubenberger A, Guck J, Poulikakos D, Ferrari A. (2016). A nanoprinted model of interstitial cancer migration reveals a link between cell deformability and proliferation. ACS Nano , 6437–6448. [DOI] [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. (2010). Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol , 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell , 241–254. [DOI] [PubMed] [Google Scholar]
- Paul CD, Mistriotis P, Konstantopoulos K. (2017). Cancer cell motility: lessons from migration in confined spaces. Nat Rev Cancer , 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penar PL, Khoshyomn S, Bhushan A, Tritton TR. (1997). Inhibition of epidermal growth factor receptor-associated tyrosine kinase blocks glioblastoma invasion of the brain. Neurosurgery , 141–151. [DOI] [PubMed] [Google Scholar]
- Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. (2012). Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell , 1513–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov SV, Sabass B, Schwarz US, Waterman CM. (2014). High-resolution traction force microscopy. Methods Cell Biol , 367–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. (2008). Collagen density promotes mammary tumor initiation and progression. BMC Med , 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retta SF, Barry ST, Critchley DR, Defilippi P, Silengo L, Tarone G. (1996). Focal adhesion and stress fiber formation is regulated by tyrosine phosphatase activity. Exp Cell Res , 307–317. [DOI] [PubMed] [Google Scholar]
- Roca-Cusachs P, Conte V, Trepat X. (2017). Quantifying forces in cell biology. Nat Cell Biol , 742–751. [DOI] [PubMed] [Google Scholar]
- Roth W, Wild-Bode C, Platten M, Grimmel C, Melkonyan HS, Dichgans J, Weller M. (2000). Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene , 4210–4220. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell , 487–498. [DOI] [PubMed] [Google Scholar]
- Schlaepfer DD, Mitra SK. (2004). Multiple connections link FAK to cell motility and invasion. Curr Opin Genet Dev , 92–101. [DOI] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, Luzzio MJ, Cooper B, Kath JC, Roberts WG, et al. (2007). Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem , 14845–14852. [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Green MD, Hall RE, Reddel RR, Taylor IW. (1983). Tamoxifen induces accumulation of MCF 7 human mammary carcinoma cells in the G0/G1 phase of the cell cycle. Eur J Cancer Clin Oncol , 615–621. [DOI] [PubMed] [Google Scholar]
- Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, et al. (2013). Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J Cell Biol , 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Water B, Houtepen F, Huigsloot M, Tijdens IB. (2001). Suppression of chemically induced apoptosis but not necrosis of renal proximal tubular epithelial (LLC-PK1) cells by focal adhesion kinase (FAK). Role of FAK in maintaining focal adhesion organization after acute renal cell injury. J Biol Chem , 36183–36193. [DOI] [PubMed] [Google Scholar]
- Vianay B, Senger F, Alamos S, Anjur-Dietrich M, Bearce E, Cheeseman B, Lee L, Thery M. (2018). Variation in traction forces during cell cycle progression. Biol Cell , 91–96. [DOI] [PubMed] [Google Scholar]
- Ward A, Balwierz A, Zhang JD, Küblbeck M, Pawitan Y, Hielscher T, Wiemann S, Sahin Ö. (2012). Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene , 1173. [DOI] [PubMed] [Google Scholar]
- Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol , 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Milo R, Kam Z, Geiger B. (2007). A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci , 137–148. [DOI] [PubMed] [Google Scholar]
- Zhang W, Law RE, Hinton DR, Couldwell WT. (1997). Inhibition of human malignant glioma cell motility and invasion in vitro by hypericin, a potent protein kinase C inhibitor. Cancer Lett , 31–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.