Abstract
Background/Aims
Helicobacter pylori eradication rates are decreasing because of increases in clarithromycin resistance. Thus, finding an easy and accurate method of detecting clarithromycin resistance is important.
Methods
We evaluated 70 H. pylori isolates from Korean patients. Dual-labeled peptide nucleic acid (PNA) probes were designed to detect resistance associated with point mutations in 23S ribosomal ribonucleic acid gene domain V (A2142G, A2143G, and T2182C). Data were analyzed by probe-based fluorescence melting curve analysis based on probe-target dissociation temperatures and compared with Sanger sequencing.
Results
Among 70 H. pylori isolates, 0, 16, and 58 isolates contained A2142G, A2143G, and T2182C mutations, respectively. PNA probe-based analysis exhibited 100.0% positive predictive values for A2142G and A2143G and a 98.3% positive predictive value for T2182C. PNA probe-based analysis results correlated with 98.6% of Sanger sequencing results (κ-value=0.990; standard error, 0.010).
Conclusions
H. pylori clarithromycin resistance can be easily and accurately assessed by dual-labeled PNA probe-based melting curve analysis if probes are used based on the appropriate resistance-related mutations. This method is fast, simple, accurate, and adaptable for clinical samples. It may help clinicians choose a precise eradication regimen.
Keywords: Helicobacter pylori, Clarithromycin, Melting array, Peptide nucleic acids
INTRODUCTION
Helicobacter pylori infection is involved in the pathogenesis of chronic gastritis, peptic ulcer disease, and mucosa-associated lymphoid tissue lymphoma and is a major risk factor for the development of gastric cancer.1 The rate of H. pylori infection is higher in Korea than in other countries.2 Traditionally, triple therapy including a proton pump inhibitor, amoxicillin, and clarithromycin has been recommended as the primary regimen for H. pylori eradication.3,4 However, clarithromycin resistance is prevalent and exceeds more than 20% in Korea.5 Therefore, increased resistance to clarithromycin has become the most important factor of treatment failure. In recent years, eradication rates with first-line therapy in South Korea have ranged from 74.6% to 75.8%.6–8 Clarithromycin resistance is due to point mutations in the peptidyl transferase encoding region in domain V of the H. pylori 23S ribosomal ribonucleic acid (rRNA) gene.9,10
Techniques to evaluate clarithromycin resistance can be divided into two categories: standard susceptibility tests and molecular methods.11–14 However, phenotypical tests such as the agar dilution method and Epsilometer (E)-test require complicated conditions for H. pylori growth and serial subcultivations for several days to determine clarithromycin resistance. Furthermore, the agar dilution method and E-test fail to give any information about point mutations.15–17 Molecular methods such as polymerase chain reaction (PCR), real-time PCR, and fluorescence in situ hybridization with deoxyribonucleic acid (DNA) probes have been developed to overcome these shortcomings.9,13,18,19
Peptide nucleic acid (PNA) molecules are synthetic DNA mimics, with a negatively-charged sugar-phosphate backbone that confers high affinity for DNA or RNA complementary sequences.20,21 PNA molecules are stable at higher melting temperatures and more resistant to nucleases and proteases than DNA molecules.21 PNA probes are also usually smaller (13–18 bp) than DNA probes (≥18 bp); therefore, they penetrate the bacterial cell wall more easily.21,22
In this study, we developed and validated a new genotypic method to detect clarithromycin resistance in H. pylori, which involved probe-based fluorescence melting curve analysis (Sea-Sun Biomaterials, Daejeon, Korea) using a set of PNA probes. Sanger sequencing was used as the reference method.
MATERIALS AND METHODS
1. Study settings
We used 70 H. pylori strains from clinical isolates consecutively which were collected from patients who underwent gastric biopsy at Gangnam Severance Hospital, Seoul, Korea, between March 2015 and February 2017. We performed PNA probe-based fluorescence melting curve analysis compared with the standard susceptibility culturing test (E-test; BioMerieux SA, Marcy I’Etoile, France) and PCR sequencing to assess resistance status of 70 H. pylori strains.
2. Isolation of Helicobacter pylori strains and growth conditions
One antral and one corporal biopsy specimen were placed in a transport medium. The gastric tissue specimens were seeded in egg yolk emulsion (EYE) agar plates (Yuhan LabTech, Seoul, Korea). EYE agar contained Columbia agar 43.82 μg/mL, EYE 112.36 μL/mL, IsoVitaleX 11.23 μL/mL, and 2,3,5-triphenyltetrazolium chloride 45.0 μg/mL (for colony staining). The plates were subsequently stored in a multigas incubator (microaerophilic atmosphere: 10% carbon dioxide, 5% oxygen, and 85% nitrogen at 37°C) for 3 to 7 days. If H. pylori was not isolated after 7 days of incubation, the plates were incubated for 3 more days. Isolation of H. pylori was identified on the basis of colony morphology and confirmed by matrix-assisted laser desorption/ionization-time-of-fight mass spectrometry (MALDI-TOF) using the Microflex LT system (Bruker Daltonics, Bremen, Germany), and the measured profiles were compared to a database with MALDI Biotyper 3.1 software (Bruker Daltonics). The colonies were re-identified by subculture analysis. In our institution, isolation of H. pylori is routinely performed from gastric biopsy specimens using the above methodology. In the present study, we used isolates of H. pylori that had been in storage.
3. Design of peptide nucleic acid oligonucleotide probes for the detection of clarithromycin resistance
PNA probes were designed by adapting existing DNA probes targeting three point mutations (A2142G, A2143G, and T2182C) of 23S rRNA in H. pylori. The three point mutations were chosen based on prior reports. 23S rRNA point mutations typically involve A2142G or A2143G; in particular, the A2143G mutation has been previously identified in approximately 70% to 90% of patients with clarithromycin-resistant H. pylori.23,24 T2182C has also been regarded as an important point mutation in clarithromycin-resistant isolates.24 We used the U-TOP™ HPy ClaR Detection Kit (SeaSun Biomaterials) to construct the PNA probes.
4. Validation of the real-time PCR-based MeltingArray analysis
Using the three PNA probes for probe-based fluorescence melting curve analysis (SeaSun Biomaterials), real-time PCR was performed with the CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s manual. Real-time PCR reactions were performed in 20-μL reaction mixtures containing 10 μL ×2 qPCR PreMix (SeaSun Bio-materials), 5 μL primer and detection PNA probe mixture, and 3 μL DNA template (15 ng/μL). The reaction conditions for amplification and melting point analysis were 95°C for 10 minutes; 42 cycles of 95°C for 30 seconds, 58°C for 45 seconds, and 72°C for 45 seconds; followed by melting point analysis. Melting point analysis was performed using a denaturation step of 95°C for 5 minutes; 1-minute hybridization steps of 75°C, 55°C, and 45°C; and a stepwise temperature increase from 20°C to 85°C at 1°C per step, with a 5-second interval between each step. The data were analyzed using Bio-Rad CFX manager v1.6 software (Bio-Rad). Mutations were distinguished by the fluorescence signal of detection probes and corresponding melting temperatures (Tm).
5. Sanger sequencing
Sanger sequencing was performed as follows. A total of five sets of primers were used to amplify genomic DNA (gDNA) regions containing the mutations. PCR was performed using 20-μL reaction mixtures containing 3 μL purified gDNA (15 ng/μL), 10 μL ×2 qPCR PreMix (SeaSun Biomaterials), and 0.5 μL each of a forward and reverse primer (10 mM). And, thermocycling was done using 40 cycles at 95°C for 30 seconds, 58°C for 45 seconds, and 72°C for 45 seconds, with a final elongation step at 72°C for 5 minutes. The PCR products were electrophoresed on a 2% agarose gel and showed with ethidium bromide staining under ultraviolet light to check their sizes and evaluate their quantities. PCR products were sequenced using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA). Sequencing reaction products were electrophoresed on the ABI 3500XL genetic analyzer (Applied Bio-systems). Sequence data were analyzed using the ABI 3500XL DNA Analyzer (Applied Biosystems). All gDNA samples were tested individually with each of the eight sets of primers in both directions.
6. Antimicrobial susceptibility test
The in vitro minimum inhibitory concentration (MIC) of clarithromycin against the H. pylori clinical isolates was evaluated two times using the E-test. The MIC was determined in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations, with the clinical breakpoint for clarithromycin defined as >0.5 mg/L.25 The standard strain, H. pylori ATCC 43504 (NCTC 11637, Australia), was used as a quality control organism.
7. Statistical analysis
PNA probe-based fluorescence melting curve analysis was compared with Sanger sequencing to assess resistance status. We calculated the positive predictive value (PPV), negative predictive value (NPV), and corresponding 95% confidence intervals of PNA probe-based fluorescence melting curve analysis using Sanger sequencing results as the reference. To assess the agreement between PNA probe-based fluorescence melting curve analysis and Sanger sequencing, we calculated the κ-value. All statistical analyses were conducted using SPSS software version 20.0 (IBM Corp., Armonk, NY, USA).
RESULTS
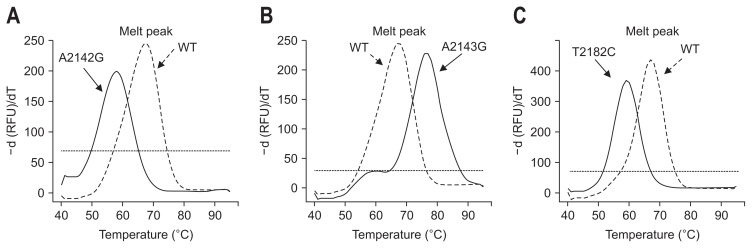
Among 70 H. pylori strains, 22 strains (31.4%) were clarithromycin resistant and 48 strains (68.6%) were clarithromycin susceptible by E-test (Table 1). Among 22 clarithromycin resistant isolates, the A2143G mutation was observed in 16 (72.7%) isolates and the A2142G mutation was not observed in any isolate. We also conducted PNA probe-based fluorescence melting curve analysis and compared the results with those obtained using Sanger sequencing. Fig. 1 shows representative melting peaks for each mutation, as well as wild-type sequences. The PNA probe-based fluorescence melting curve analysis showed distinct melting curves specific to each allele, with Tm values for each diagnostic melting peak detected exactly as predicted.
Table 1.
Minimum Inhibitory Concentrations of Clarithromycin for 70 Helicobacter pylori Strains
| Strain no. | MIC, mg/L | Strain no. | MIC, mg/L | Strain no. | MIC, mg/L |
|---|---|---|---|---|---|
| H-1 | 0.032 | H-34 | 0.032 | H-67 | 0.016 |
| H-2 | 0.032 | H-35 | 0.094 | H-68 | 0.016 |
| H-3 | 0.064 | H-36 | 0.064 | H-69 | 256* |
| H-4 | 0.047 | H-37 | 0.047 | H-70 | 0.047 |
| H-5 | 0.016 | H-38 | 0.016 | H-71 | 0.064 |
| H-6 | 192* | H-39 | 0.064 | H-72 | 0.094 |
| H-7 | 256* | H-40 | SF | H-73 | 24* |
| H-8 | 256* | H-41 | 0.064 | H-74 | 256* |
| H-9 | 0.064 | H-42 | SF | H-75 | 128* |
| H-10 | 0.064 | H-43 | 0.016 | ||
| H-11 | 0.125 | H-44 | 0.064 | ||
| H-12 | 0.016 | H-45 | 256* | ||
| H-13 | 256* | H-46 | 0.032 | ||
| H-14 | SF | H-47 | SF | ||
| H-15 | 0.19 | H-48 | 48* | ||
| H-16 | 0.047 | H-49 | 0.125 | ||
| H-17 | 24* | H-50 | 32* | ||
| H-18 | 0.032 | H-51 | 0.016 | ||
| H-19 | 0.023 | H-52 | 0.016 | ||
| H-20 | 0.016 | H-53 | 32* | ||
| H-21 | SF | H-54 | 0.032 | ||
| H-22 | 0.023 | H-55 | 64* | ||
| H-23 | 3* | H-56 | 0.023 | ||
| H-24 | 48* | H-57 | 0.023 | ||
| H-25 | 0.047 | H-58 | 64* | ||
| H-26 | 0.094 | H-59 | 0.023 | ||
| H-27 | 0.023 | H-60 | 0.023 | ||
| H-28 | 0.094 | H-61 | 0.064 | ||
| H-29 | 0.094 | H-62 | 0.032 | ||
| H-30 | 256* | H-63 | 0.016 | ||
| H-31 | 256* | H-64 | 16* | ||
| H-32 | 0.016 | H-65 | 32* | ||
| H-33 | 256* | H-66 | 0.064 |
MIC, minimum inhibitory concentrations; H, H. pylori strain; SF, subculture failure.
Resistant strains determined by the European Committee on Antimicrobial Susceptibility Testing recommendations.
Fig. 1.
Representative melting peaks of (A) A2142G, (B) A2143G, and (C) T2182C mutations. WT, wild type; RFU, relative fluorescence unit.
Table 2 summarizes the results obtained by the two methods (Sanger sequencing and PNA probe-based methods). The κ-value between PNA probe-based fluorescence melting curve analysis and Sanger sequencing was 0.990 (standard error, 0.010), indicating that PNA probe-based fluorescence melting curve analysis agreed well with Sanger sequencing. When compared with Sanger sequencing, the PPV and NPV of PNA probe-based fluorescence melting curve analysis were both 100.0% for the A2142G mutation, as well as the A2143G mutation. The PPV was 98.3% and NPV was 100% for the T2182C mutation. One isolate with a T2190C mutation was recognized as a T2182C mutation in the PNA probe-based fluorescence melting curve analysis. This is likely because the melting temperatures of T2182C and T2190C overlapped. Therefore, the concordance between PNA probe-based fluorescence melting curve analysis and Sanger sequencing in our study was 98.6% (69/70).
Table 2.
Comparison of Mutation-Detection Results Obtained with the PNA-Probe-Based Melting Curve Analysis and Sanger Sequencing
| Sanger sequencing | PNA melting curve analysis kit | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| A2142G | A2143G | T2182C | |||||
|
|
|
|
|||||
| + | − | + | − | + | − | ||
| A2142G | + | 0 | 0 | ||||
| − | 0 | 70 | |||||
| A2143G | + | 16 | 0 | ||||
| − | 0 | 54 | |||||
| T2182C | + | 58 | 0 | ||||
| − | 1 | 11 | |||||
PNA, peptide nucleic acid.
The A2142G mutation was not observed in any of the 70 H. pylori isolates. However, the T2182C mutation was found in 58 isolates (82.9%), suggesting that this mutation was not associated with clarithromycin resistance (Supplementary Table 1).
DISCUSSION
New techniques using a rapid, simple, and accurate method to detect clarithromycin resistance have become an emerging issue because of increasing eradication failure of H. pylori. In this study, we evaluated a new diagnostic method using PNA probe-based fluorescence melting curve analysis. This method has several advantages. Firstly, this method requires only a conventional real-time PCR machine, and the procedure is performed in a single step within a closed well. Therefore, sample contamination and loss are much lower than with other methods, and the reaction and analysis procedures are much faster. Accordingly, this method can provide results promptly and accurately. The commercialized DPO-PCR (Seeplex® ClaR-H. pylori ACE Detection, Seegene Institute of Life Science, Seoul, Korea) is performed in two steps. DPO-PCR is a multiplex PCR and analyzed using a semi-automated system (i.e., Screen tape®). Therefore, PNA probe-based fluorescence melting curve analysis is faster and simple. In addition, PNA probes are simple to design and modify. Therefore, we can easily modify the probes and primers to change the target mutations from relatively nonspecific mutations to mutations strongly associated with clarithromycin resistance. Furthermore, PNA probe-based fluorescence melting curve analysis can detect up to four mutations in a single well. Probe-based fluorescence melting curve analysis is also a powerful tool to detect mutations.8 PNA probes hybridize with their complementary target DNA sequences,26 and PNA–DNA hybridization is more sensitive to base mismatches than DNA–DNA hybridization. Because PNA probe-based fluorescence melting curve analysis is based on the temperature at which the probe-target hybridization dissociates,27 the results are much easier to interpret than those of Sanger sequencing. However, in this study, PNA melting curve analysis was performed on DNA isolated from cultured H. pylori, not on gastric biopsy specimen. Therefore, the performance of PNA melting curve analysis on gastric biopsy specimens need to be verified.
In this study, we chose three specific mutations: A2142G, A2143G, and T2182C. However, the T2182C mutation, which was previously regarded as an important point mutation in clarithromycin resistance,24,28 was detected in 58 of 70 isolates (82.9%). This result indicates that T2182C is a nonspecific mutation, which does not play an important role in clarithromycin resistance. As we can change the target mutation panel by adding or removing PNA probes and primers, we can easily replace the target mutation of T2182C to up to four different resistant target mutations.
This is the first study to report on using the MeltingArray assay based on PNA probes to assess clarithromycin resistance of H. pylori. The performance of PNA melting curve analysis showed almost 100% correlation with Sanger sequencing. Clinical application of our method is easy, and it should help clinicians choose appropriate treatment strategies according to the presence or absence of clarithromycin resistance. Currently, guidelines for treating H. pylori infection in Korea recommend an algorithm of H. pylori eradication according to clarithromycin resistance status.2 Detection of clarithromycin resistance before treatment is thereby important for selecting the appropriate eradication regimen because assessing clarithromycin resistance is the first step in treating H. pylori. Therefore, rapid, easy, and accurate methods for detecting clarithromycin resistance are necessary, and the PNA probe-based fluorescence melting curve analysis is a good candidate method.
Clarithromycin resistance of H. pylori is well known. Point mutations in the 23s rRNA gene decrease the affinity between ribosomes and clarithromycin so the antibiotic is unable to inhibit protein biosynthesis.29 Mutations A2143G, A2142C/G, and A2144G are associated with clarithromycin resistance of H. pylori in Asia.30–33 In Korea, 23S rRNA gene mutations such as A2142G, A2143G, A2144G, T2182C, T2190C, A2223G, and C2195T have been reported occasionally in clarithromycin-resistant H. pylori strains.24,34–36 However, these point mutations are less prevalent, indicating that a causal link between this gene and clarithromycin resistance has not been definitively proven. In this study, among 70 H. pylori strains, 22 (31.4%) were clarithromycin resistant and 48 (68.6%) were clarithromycin susceptible, according to the E-test (Supplementary Table 1). However, among the 22 clarithromycin-resistant isolates based on the E-test, the A2143G mutation was present in only 16 isolates (72.7%), and the A2142G mutation was not observed in any isolate. Therefore, the current commercial kit targeting A2142G and A2143G cannot detect all clarithromycin-resistant strains. However, as mentioned above, probes can be easily designed and modified for PNA probe-based fluorescence melting curve analysis. As we can change the target mutations by modifying probes and primers to identify candidate mutations associated with clarithromycin resistance, we can easily and conveniently apply this kit to clinical practice for detecting H. pylori resistance.
In this study, DNA sequencing was used to detect the mechanism of clarithromycin resistance. C375T, A381C, and G382A mutations that have not been previously associated with clarithromycin resistance were detected more frequently in clarithromycin-resistant isolates than in clarithromycin-susceptible isolates (Supplementary Table 2). However, the role of these three mutations remains unclear.
In the present study, PNA melting curve analysis was performed on DNA isolated from cultured H. pylori. However, PNA melting curve analysis can be performed on gastric biopsy specimens, as well as culture-based H. pylori isolates. Considering the higher rate of H. pylori detection by PCR of gastric biopsy specimens compared with cultures,23,37 PNA melting curve analysis should likewise be more useful using gastric biopsy specimens than cultures because H. pylori is not easily isolated from cultures. Therefore, further study will be needed to validate the PNA melting curve analysis on gastric biopsy specimen.
In conclusion, although further study in a large population is necessary to confirm our results, PNA probe-based fluorescence melting curve analysis can be applied to clinical samples and may be helpful in choosing the precise H. pylori eradication regimen according to clarithromycin resistance. When more accurate information becomes available regarding resistance-related mutations, we will be able to promptly update our PNA probes and primers and thereby easily identify most clarithromycin-resistant strains.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Basic Research Project (Study No. 16-3220) of the Korea Institute of Geoscience and Mineral Resources (KIGAM), funded by the Ministry of Science, ICT, and Future Planning of Korea.
We extend our gratitude to the Korea Institute of Geoscience and Mineral Resources (KIGAM).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Lowe AW, Moseley RH. Helicobacter pylori eradication and gastric cancer incidence: a meta-analysis. Gastroenterology. 2016;150:1049–1051. doi: 10.1053/j.gastro.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 2.Kim SG, Jung HK, Lee HL, et al. Guidelines for the diagnosis and treatment of Helicobacter pylori infection in Korea, 2013 revised edition. J Gastroenterol Hepatol. 2014;29:1371–1386. doi: 10.1111/jgh.12607. [DOI] [PubMed] [Google Scholar]
- 3.Kim N, Kim JJ, Choe YH, et al. Diagnosis and treatment guidelines for Helicobacter pylori infection in Korea. Korean J Gastroenterol. 2009;54:269–278. doi: 10.4166/kjg.2009.54.5.269. [DOI] [PubMed] [Google Scholar]
- 4.Wong BC, Chang FY, Abid S, et al. Triple therapy with clarithromycin, omeprazole, and amoxicillin for eradication of Helicobacter pylori in duodenal ulcer patients in Asia and Africa. Aliment Pharmacol Ther. 2000;14:1529–1535. doi: 10.1046/j.1365-2036.2000.00863.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyanova L, Mitov I. Geographic map and evolution of primary Helicobacter pylori resistance to antibacterial agents. Expert Rev Anti Infect Ther. 2010;8:59–70. doi: 10.1586/eri.09.113. [DOI] [PubMed] [Google Scholar]
- 6.Kim SE, Park MI, Park SJ, et al. Trends in Helicobacter pylori eradication rates by first-line triple therapy and related factors in eradication therapy. Korean J Intern Med. 2015;30:801–807. doi: 10.3904/kjim.2015.30.6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo J, Jeon SW. Changes in the eradication rate of conventional triple therapy for Helicobacter pylori infection in Korea. Korean J Gastroenterol. 2014;63:141–145. doi: 10.4166/kjg.2014.63.3.141. [DOI] [PubMed] [Google Scholar]
- 8.Kim EM, Song MS, Hur DH, An CM, Kang JH, Park JY. Easy method for discriminating the origins of manila clam Ruditapes philippinarum with a dual-labelled PNA-probe-based melting curve analysis. Biochip J. 2015;9:247–258. doi: 10.1007/s13206-015-9402-1. [DOI] [Google Scholar]
- 9.Trebesius K, Panthel K, Strobel S, et al. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut. 2000;46:608–614. doi: 10.1136/gut.46.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Francesco V, Margiotta M, Zullo A, et al. Prevalence of primary clarithromycin resistance in Helicobacter pylori strains over a 15 year period in Italy. J Antimicrob Chemother. 2007;59:783–785. doi: 10.1093/jac/dkm005. [DOI] [PubMed] [Google Scholar]
- 11.Piccolomini R, Di Bonaventura G, Catamo G, Carbone F, Neri M. Comparative evaluation of the E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J Clin Microbiol. 1997;35:1842–1846. doi: 10.1128/jcm.35.7.1842-1846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osato MS, Reddy R, Reddy SG, Penland RL, Graham DY. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int J Antimicrob Agents. 2001;17:39–44. doi: 10.1016/S0924-8579(00)00320-4. [DOI] [PubMed] [Google Scholar]
- 13.Oleastro M, Ménard A, Santos A, et al. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41:397–402. doi: 10.1128/JCM.41.1.397-402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 15.Morris JM, Reasonover AL, Bruce MG, et al. Evaluation of sea-FAST, a rapid fluorescent in situ hybridization test, for detection of Helicobacter pylori and resistance to clarithromycin in paraffin-embedded biopsy sections. J Clin Microbiol. 2005;43:3494–3496. doi: 10.1128/JCM.43.7.3494-3496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yilmaz O, Demiray E. Clinical role and importance of fluorescence in situ hybridization method in diagnosis of H pylori infection and determination of clarithromycin resistance in H pylori eradication therapy. World J Gastroenterol. 2007;13:671–675. doi: 10.3748/wjg.v13.i5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rüssmann H, Adler K, Haas R, Gebert B, Koletzko S, Heesemann J. Rapid and accurate determination of genotypic clarithromycin resistance in cultured Helicobacter pylori by fluorescent in situ hybridization. J Clin Microbiol. 2001;39:4142–4144. doi: 10.1128/JCM.39.11.4142-4144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Doorn LJ, Glupczynski Y, Kusters JG, et al. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45:1500–1504. doi: 10.1128/AAC.45.5.1500-1504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cambau E, Allerheiligen V, Coulon C, et al. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47:3600–3607. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stender H, Williams B, Coull J. PNA fluorescent in situ hybridization (FISH) for rapid microbiology and cytogenetic analysis. Methods Mol Biol. 2014;1050:167–178. doi: 10.1007/978-1-62703-553-8_14. [DOI] [PubMed] [Google Scholar]
- 21.Cerqueira L, Azevedo NF, Almeida C, Jardim T, Keevil CW, Vieira MJ. DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH) Int J Mol Sci. 2008;9:1944–1960. doi: 10.3390/ijms9101944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerqueira L, Fernandes RM, Ferreira RM, et al. Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J Clin Microbiol. 2013;51:1887–1893. doi: 10.1128/JCM.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HJ, Kim JI, Cheung DY, et al. Eradication of Helicobacter pylori according to 23S ribosomal RNA point mutations associated with clarithromycin resistance. J Infect Dis. 2013;208:1123–1130. doi: 10.1093/infdis/jit287. [DOI] [PubMed] [Google Scholar]
- 24.Kim JM, Kim JS, Kim N, et al. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol. 2008;18:1584–1589. [PubMed] [Google Scholar]
- 25.European Committee on Antimicrobial Susceptibility T (EUCAST) Clinical breakpoints - bacteria version 70 [Internet] Basel: EU-CAST; [cited 2017 Mar 10]. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.0_Breakpoint_Tables.pdf. [Google Scholar]
- 26.Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr Issues Mol Biol. 1999;1(1–2):89–104. [PubMed] [Google Scholar]
- 27.Huang Q, Liu Z, Liao Y, Chen X, Zhang Y, Li Q. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One. 2011;6:e19206. doi: 10.1371/journal.pone.0019206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KS, Kang JO, Eun CS, Han DS, Choi TY. Mutations in the 23S rRNA gene of Helicobacter pylori associated with clarithromycin resistance. J Korean Med Sci. 2002;17:599–603. doi: 10.3346/jkms.2002.17.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T, Song HJ, Shin SY, et al. Clarithromycin-resistant Helicobacter pylori associated with 23S rRNA point mutations in Jeju Island. Korean J Gastroenterol. 2013;61:252–258. doi: 10.4166/kjg.2013.61.5.252. [DOI] [PubMed] [Google Scholar]
- 31.Kato S, Fujimura S, Udagawa H, et al. Antibiotic resistance of Helicobacter pylori strains in Japanese children. J Clin Microbiol. 2002;40:649–653. doi: 10.1128/JCM.40.2.649-653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xuan SH, Zhou YG, Shao B, et al. Enzymic colorimetry-based DNA chip: a rapid and accurate assay for detecting mutations for clarithromycin resistance in the 23S rRNA gene of Helicobacter pylori. J Med Microbiol. 2009;58(Pt 11):1443–1448. doi: 10.1099/jmm.0.010785-0. [DOI] [PubMed] [Google Scholar]
- 33.Maeda S, Yoshida H, Matsunaga H, et al. Detection of clarithromycin-resistant Helicobacter pylori strains by a preferential homo-duplex formation assay. J Clin Microbiol. 2000;38:210–214. doi: 10.1128/jcm.38.1.210-214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang TJ, Kim N, Kim HB, et al. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol. 2010;44:536–543. doi: 10.1097/MCG.0b013e3181d04592. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, Shin JH, Roe IH, et al. Impact of clarithromycin resistance on eradication of Helicobacter pylori in infected adults. Antimicrob Agents Chemother. 2005;49:1600–1603. doi: 10.1128/AAC.49.4.1600-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eun CS, Han DS, Park JY, et al. Changing pattern of antimicrobial resistance of Helicobacter pylori in Korean patients with peptic ulcer diseases. J Gastroenterol. 2003;38:436–441. doi: 10.1007/s00535-002-1079-2. [DOI] [PubMed] [Google Scholar]
- 37.Park CS, Lee SM, Park CH, et al. Pretreatment antimicrobial susceptibility-guided vs. clarithromycin-based triple therapy for Helicobacter pylori eradication in a region with high rates of multiple drug resistance. Am J Gastroenterol. 2014;109:1595–1602. doi: 10.1038/ajg.2014.222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.