Abstract
A series of novel 2-phenylaminopyrimidine (PAP) derivatives structurally related to STI-571 were designed and synthesized. The abilities of these compounds to inhibit proliferation were tested in human chronic myeloid leukemia K562 cells. (E)-3-(2-bromophenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acryla- mide(12d) was the most effective cell growth inhibitor and was 3-fold more potent than STI-571.
Keywords: 2-Phenylaminopyrimidine (PAP) derivative, STI-571, antiproliferative activity
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic disorder characterized by the uncontrolled expansion of myeloid progenitors in the bone marrow [1,2,3]. Bcr-Abl tyrosine kinase, a gene product of the reciprocal translocation between chromosomes 9 and 22 has been found to be a main cause of CML and has been used as a successful target for the treatment of CML [4,5,6].
The 2-phenylaminopyrimidine (PAP) derivative STI-571 (4-(4-methylpiperazin-1-ylmethyl)-N-[4- methyl-3-(4-pyridine-3-yl-pyrimidin-2-ylamino)phenyl]benzamide mesylate, imatinib mesylate, Gleevec) is a potent and relative selective Bcr-Abl tyrosine kinase inhibitor that has shown potent efficacy in CML patients in chronic phase [7,8]. Although newly diagnosed patients with chronic phase disease achieve durable responses to STI-571 therapy, relapse and resistance are frequently observed in patients with advanced disease [9,10]. Mutations in the kinase domain of Bcr-Abl have been found to be one of the main reasons of resistance to STI-571 [11]. Crystallographic studies have revealed a basic structure of Abl in which mutations in the kinase domain confer STI-571 resistance [12]. STI-571 binds to an inactive conformation of the active loop of Abl. Mutations in the kinase domain influence the residues to which the drug binds or change the conformation of Abl making STI-571 unable to bind [12,13]. Potent Bcr-Abl activity inhibition by 2-phenylaminopyrimidine (PAP) derivatives which bind to Abl in a different way suggests that novel PAP derivatives would be more potent CML cell growth inhibitors than STI-571 [14,15,16,17,18]. Recently, we synthesized a series of novel PAP cinnamamide derivatives structurally related to STI-571 and compared their antiproliferative activity in CML K562 cells. Several derivatives with more potent antiproliferative activity were found.
Results and Discussion
Design and synthesis of PAP cinnamamide derivatives
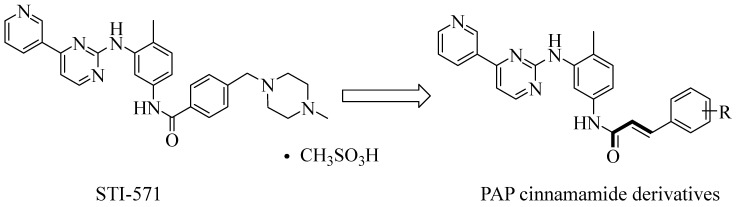
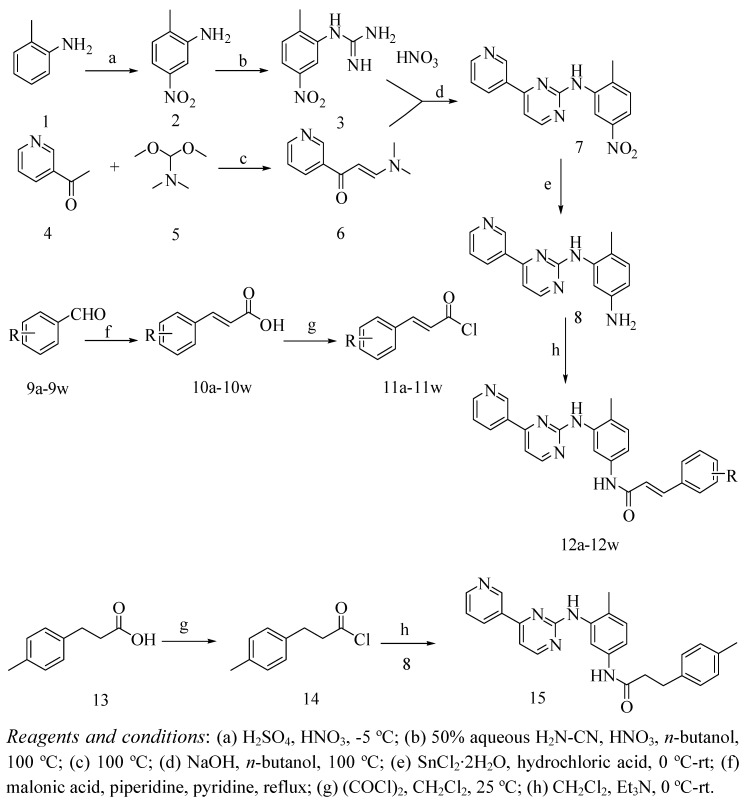
The design of PAP derivatives was based on the vinylogy rule. A double bond was introduced into the benzamide moiety to construct an α,β-unsaturated acylamide (Figure 1), with the reasoning that α,β-unsaturated enones could react with the nuclephilic groups of residues in the Abl kinase domain. Therefore, a series of PAP cinnamamide derivatives with various substituents on the right hand benzene ring were synthesized. In order to investigate the effect of the conjugate moiety, compound 15 was also synthesized. The synthetic routes of the target compounds are shown in Scheme 1.
Figure 1.
The structures of the designed compounds related to STI-571.
Scheme 1.
Synthetic routes of the target compounds.
The general method of Bredereck [14,19] was used to synthesize the pyrimidine ring system, which involved reacting 3-acetylpyridine (4) with N,N-dimethylformamide dimethyl acetal (5) to give the enaminone 6 in 92.6% yield. The phenylaminopyrimidine derivative ring system 7 was constructed by reacting enaminone 6 with guanidinium 3, which was prepared in 66% yield via the reaction of o-toluidine 1 with 50% aqueous cyanamide in refluxing n-butanol. Reduction of 7 with SnCl2·2H2O afforded 6-methyl-N1-(4-pyridin-3-yl-pyrimidin-2-yl)benzene-1,3-diamine (8) in 75% yield, which reacted with trans-cinnamyl chloride to afford the target compounds 12a-12w in yields ranging from 33.9% to 78.8%. Compound 15 was synthesized via the same method with 3-p-tolylpropanoic acid as the starting material.
Antiproliferative activities of PAP cinnamamide derivatives in CML cells and proposed binding mode to c-Abl
The antiproliferative activities of these PAP cinnamamide derivatives were tested in human leukemia K562 cells (Table 1), and the structure-activity relationship was analyzed. The results revealed that compounds with sterical small substituents at position 2 or at position 4 on the right hand benzene ring were more potent than the corresponding unsubstituted compound 12a. Compounds with a substituent at position 2 were more potent than compounds with the same substituent at position 4. Among them, compound 12c and 12d were more effective than STI-571 in inhibiting K562 cell growth. Comparison of activities of the compounds with substitution at position 2 revealed that electron-drawing groups were better than alkoxy groups for cell growth inhibition. The order of activity of the compounds with a substituent at position 2 was Br > Cl > NO2 > F> OCH3 > OCH2CH3. Introduction of an additional substituent into compound 12f with a -OCH3 at position 3 (i.e. compound 12o) improved the antiproliferative activity, while other compounds with double substituents were less active than the compounds with a single substituent. Compound 12l was 7-fold more potent than compound 15, suggesting that the vinylogy modification might be important for the antiproliferative activity of these PAP cinnamamide derivatives.
Table 1.
The substituents of target compounds and their antiproliferative effects in K562 cells.
| Compound | R | GI50 (μM) in K562a |
|---|---|---|
| 12a | H | 2.01 ± 0.10 |
| 12b | 2-F | 0.67 ± 0.07 |
| 12c | 2-Cl | 0.10 ± 0.01 |
| 12d | 2-Br | 0.07 ± 0.01 |
| 12e | 2-NO2 | 0.26 ± 0.02 |
| 12f | 2-OCH3 | 0.83 ± 0.05 |
| 12g | 2-OCH2CH3 | 1.55 ± 0.21 |
| 12h | 4-F | 1.83 ± 0.09 |
| 12i | 4-Cl | 1.26 ± 0.04 |
| 12j | 4-Br | 0.55 ± 0.05 |
| 12k | 4-NO2 | 0.39 ± 0.04 |
| 12l | 4-CH3 | 0.51 ± 0.02 |
| 12m | 4-OCH3 | 1.05 ± 0.06 |
| 12n | 4-OCH2Ph | > 5 |
| 12o | 2,3-di-OCH3 | 0.14 ± 0.01 |
| 12p | 2,4-di-OCH3 | 1.26 ± 0.05 |
| 12q | 2,3,4-tri-OCH3 | 0.78 ± 0.06 |
| 12r | 3,4-di-OCH3 | 0.99 ± 0.17 |
| 12s | 3-OCH3,4-OCH2CH3 | > 5 |
| 12t | 3-OCH2CH3,4-OCH3 | 0.48 ± 0.04 |
| 12u | 3-OCH3,4-OCH2Ph | > 5 |
| 12v | 3-OCH2Ph,4-OCH3 | > 5 |
| 12w | 3,4(-OCH2O-) | 0.57 ± 0.02 |
| 15 | 3.51 ± 0.33 | |
| STI-571 | 0.23 ± 0.01 |
a GI50 is the concentration that inhibits 50% of cell growth. Cells were seeded at 4.0 × 104 cells/mL and incubated with various concentrations of tested compounds for 72 h. The data shown are the mean ± SE of three independent experiments.
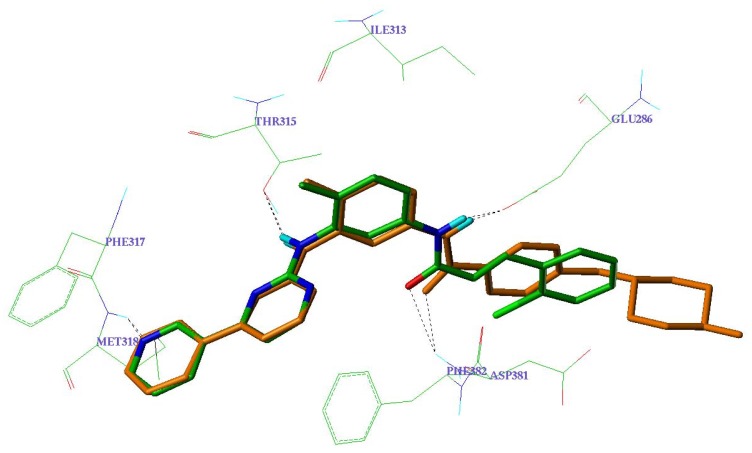
The proposed binding mode may partially rationalize above pharmacological results. The most potent compound 12d was found to share the same binding mode as that of STI-571 observed in the crystal structure of c-Abl kinase. Both STI-571 and compound 12d form hydrogen bonds with Met318, Thr315, Glu286 and Phe382. Furthermore, the olefinic bond of compound 12d matches the stereo conformation of the phenyl group of STI-571, which might form π-π interaction with c-Abl residues.
Figure 2.
Binding modes of STI-571 (orange; X-ray crystal structure) and compound 12d (green; docked pose) to the c-Abl kinase domain.
Experimental
General
All reagents and solvents (analytical grade) were commercially available and used without further purification. Melting points were determined with a Yanaco apparatus and are uncorrected. 1H-NMR spectra were recorded in DMSO-d6 (unless otherwise specified) on a Bruker AV-500 spectrometer. The coupling constants were recorded in hertz (Hz) and chemical shifts were reported in parts per million (δ, ppm) downfield from tetramethylsilane (TMS). High-resolution mass spectra (HRMS) were recorded on a high-resonance electrospray time-of-flight mass spectrometer LC/MSD QTOF 6520 (Agilent).
2-Methyl-5-nitroaniline (2)
To sulfuric acid (225 g) cooled at -5 °C, o-toluidine (1, 15 g, 140 mmol) was added dropwise with vigorous stirring. Mixed acid (14 g of 65% nitric acid and 50 g of sulfuric acid) was then added dropwise at -5 °C during 2 h. Then the mixture was poured onto crushed ice and made alkaline with aqueous sodium hydroxide. The precipitate formed was collected by filtration and air-dried. Subsequent recrystallization from 50% ethanol provided 17.28 g (81%), of product as maroon crystals, mp: 105-107 °C.
N-(2-Methyl-5-nitrophenyl)guanidinium nitrate (3)
To a solution of 2-methyl-5-nitroaniline (2, 20.46 g, 135 mmol ) in n-butanol (120 mL), 65% nitric acid (10.5 mL) was added dropwise followed by a 50% aqueous solution of cyanamide (22.7 mL, 22.81 g, 270 mmol). The reaction mixture was refluxed for 12 h and then cooled to 0 °C. The precipitate was collected by filtration and washed with chilled solution of methanol and diethyl ether (1:1, 20 mL) and air-dried to afford 18.48 g (53%) of the title product as a yellow solid, mp: 216-218 °C.
3-Dimethylamino-1-(pyridin-3-yl)propenone (6)
A mixture of 3-acetylpyridine (4, 24.21 g, 200 mmol) and N,N-dimethylformamide dimethyl acetal (5, 30.96 g, 260 mmol) was refluxed for 16 h under nitrogen and then concentrated under reduced pressure. To the residue, cyclohexane (100 mL) was added, and the mixture was cooled to 0 °C. The precipitate was collected by filtration to afford 32.61 g (92.6%) of product as yellow crystals, mp: 78-80 °C. 1H-NMR (CDCl3) δ: 9.05 (d, 1H, J = 1.6, Py-2-H), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.21 (dt, 1H, J1 = 7.9, J2 = 1.9, Py-4-H), 7.86 (d, 1H, J = 12.2, COCH=CH), 7.37 (dd, 1H, J1 = 7.3, J2 = 4.9, Py-5-H), 5.69 (d, 1H, J = 12.2, COCH=CH ), 3.20 (s, 3H, CH3), 2.97 (s, 3H, CH3).
N-(2-Methyl-5-nitrophenyl)-4-pyridin-3-yl-pyrimidin-2-ylamine (7)
To a suspension of 3-dimethylamino-1-(pyridin-3-yl)propenone (6, 26.96 g, 153 mmol) and N-(2-methyl-5-nitrophenyl)guanidinium nitrate (3, 51.40 g, 200 mmol) in n-butanol (200 mL), solid sodium hydroxide (8.63 g, 216 mmol) was added. The mixture was refluxed for 16 h and then cooled to 0 °C. The precipitate was collected by filtration and washed with methanol and diethyl ether, and air-dried to afford 43.62 g (92.4 %) of product as a yellow solid, mp: 196-197 °C.
6-Methyl-N1-(4-pyridin-3-yl-pyrimidin-2-yl)benzene-1,3-diamine (8)
To a solution of stannous chloride dihydrate (11.29 g, 50 mmol) in hydrochloric acid (30 mL) cooled at 0 °C, N-(2-methyl-5-nitrophenyl)-4-pyridin-3-yl-pyrimidin-2-ylamine (7, 3.69 g, 12 mmol) was added in portions while the suspension was vigorously stirred for 6 h. The mixture was then poured onto crushed ice, made alkaline with solid sodium hydroxide, and extracted three times with ethyl acetate (100 mL). The combined organic phase was dried over anhydrous sodium sulfate and the filtrate was evaporated to dryness in vacuo. The residue was recrystallized from methylene chloride to afford 2.52 g (75%) of product as a yellow solid, mp: 142-144 °C. 1H-NMR δ: 9.24 (d, 1H, J = 2.2, Py-2-H ), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.63 (s, 1H, NH), 8.46 (d, 1H, J = 5.1, pyrimidyl-6-H), 8.39 (dt, 1H, J1 = 8.1, J2 = 2.0, Py-4-H), 7.53 (dd, 1H, J1 = 7.9, J2 = 5.1, Py-5-H), 7.35 (d, 1H, J = 5.1, pyrimidyl-5-H ), 6.87 (d, 1H, J = 8.2), 6.80 (d, 1H, J = 2.3), 6.35 (dd, 1H, J1 = 8.0, J2 = 2.3), 4.83 (br, 2H, NH2), 2.07 (s, 3H, CH3).
General procedure for the synthesis of substituted cinnamic acids 10a-10w
Malonic acid (1.92 g 18.33 mmol) and the appropriate substituted benzaldehyde 9a-9w (12.23 mmol) were dissolved in pyridine (2.5 mL) and a catalytic amount of piperidine was added. The reaction mixture was refluxed for 1-3 h, and then cooled to room temperature and acidified with diluted hydrochloric acid. The precipitate was collected by filtration, air-dried and recrystallized from ethanol to yield corresponding cinnamic acid.
General procedure for the synthesis of the target compounds 12a-12w and 15
To a solution of cinnamic acid 10a-10w or 13 (0.84 mmol) in methylene chloride (15 mL), oxalyl chloride (1 mmol) was added. The reaction mixture was stirred for 2-3 h at room temperature, and then evaporated to dryness in vacuo. The cinnamoyl chloride or 14 yielded was dissolved in methylene chloride (5 mL) and used in the subsequent reaction. To a solution of 6-methyl-N1-(4-pyridin-3-yl- pyrimidin-2-yl)benzene-1,3-diamine (8, 0.21 g, 0.76 mmol) and triethylamine (0.25g, 2.48 mmol) in methylene chloride (15 mL) cooled at 0 °C, the above methylene chloride solution of the cinnamoyl chloride or 14 was added dropwise and the reaction mixture was then stirred for 2-8 h at room temperature. The precipitate was collected by filtration, washed with methylene chloride and water, and air-dried to afford the corresponding cinnamamide derivatives.
(E)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-phenylacrylamide (12a): yield: 74.5%; mp:181-182 °C. 1H-NMR δ: 10.12 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.2, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.8, Py-4-H), 7.99 (d, 1H, J = 1.6), 7.61 (d, 2H, J = 7.1), 7.57 (d, 1H, J = 15.8, COCH=CH), 7.54 (dd, 1H, J1 = 7.9, J2 = 4.8, Py-5-H), 7.46-7.39 (m, 5H), 7.19 (d, 1H, J = 8.3), 6.84 (d, 1H, J = 15.8, COCH=CH), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H21N5ONa [M+Na]+ 430.16438, found: 430.21130.
(E)-3-(2-fluoropheny)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12b): yield: 76.8%; mp: 243-245 °C. 1H-NMR δ: 10.26 (s, 1H, NHCO), 9.28 (d, 1H, J = 1.6, Py-2-H), 8.95 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.52 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.48 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 8.04 (d, 1H, J = 1.9), 7.84 (d, 1H, J = 15.6, COCH=CH ), 7.76-7.71 (m, 2H), 7.54 (dd, 1H, J1 = 7.9, J2 = 4.8, Py-5-H), 7.50 (t, 1H, J = 8.4), 7.44 (d, 2H, J = 5.2), 7.37-7.34 (m, 1H), 7.21 (d, 1H, J = 8.3), 6.87 (d, 1H, J = 15.6, COCH=CH), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H21FN5O [M+H] + 426.17301, found: 426.20733.
(E)-3-(2-chlorophenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12c): yield: 77.8%; mp: 270-272 °C. 1H-NMR δ: 10.24 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.52 (d, 1H, J = 5.2, pyrimidinyl-6-H), 8.48 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 8.01 (d, 1H, J = 1.6), 7.87 (d, 1H, J = 15.5, COCH=CH), 7.78-7.76 (m, 1H), 7.56-7.51 (m, 2H), 7.45-7.42 (m, 4H), 7.20 (d, 1H, J = 8.3), 6.90 (d, 1H, J = 15.5, COCH=CH), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H21ClN5O [M+H] + 442.14346, found: 442.14943.
(E)-3-(2-bromophenyl)-N-[4-methyl-3-(4-(pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12d): yield: 80.8%; mp: 268-269 °C. 1H-NMR δ: 10.26 (s, 1H, NHCO), 9.28 (d, 1H, J = 1.7, Py-2-H), 8.95 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.52 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.49 (dd, 1H, J1 = 8.0, J2 = 1.8, Py-4-H), 8.04 (d, 1H, J = 1.9), 7.84 (d, 1H, J = 15.6, COCH=CH ), 7.76-7.71 (m, 2H), 7.54 (dd, 1H, J1 = 7.9, J2 = 4.8, Py-5-H), 7.50 (t, 1H, J = 5.2), 7.44 (d, 2H, J = 5.2), 7.37-7.34 (m, 1H), 7.21 (d, 1H, J = 8.3), 6.87 (d, 1H, J = 15.6, COCH=CH), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H21BrN5O [M+H] + 486.09295, found: 486.09298.
(E)-N-[4-methyl-3-(4-(pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-(2-nitrophenyl)acrylamide (12e): yield: 78.8%; mp: 270-272 °C. 1H-NMR δ: 10.26 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.52 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.49 (dt, 1H, J1 = 8.0, J2 = 1.8, Py-4-H), 8.08 (d, 1H, J = 7.9), 8.01 (d, 1H, J = 1.8), 7.84 (d, 1H, J = 15.8, COCH=CH), 7.82-7.80 (m, 2H), 7.68-7.65 (m, 1H), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.44-7.40 (m, 2H), 7.21 (d, 1H, J = 8.4), 6.83 (d, 1H, J = 15.5, COCH=CH), 2.22 (s, 3H, CH3); HRMS m/z calcd. for C25H20N6O3Na [M+Na]+ 475.14946, found: 475.16374.
(E)-3-(2-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12f): yield: 66.3%; mp: 246-247 °C. 1H-NMR δ: 10.08 ( s, 1H, NHCO), 9.26 (d, 1H, J = 1.7, Py-2-H), 8.92 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.2, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 7.99 (d, 1H, J = 1.8), 7.79 (d, 1H, J = 15.8, COCH=CH), 7.56 (dd, 1H, J1 = 7.8, J2 = 1.5), 7.53(dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.43-7.37 (m, 3H), 7.18 (d, 1H, J = 8.3), 7.10 (d, 1H, J = 8.2), 7.01 (t, 1H, J = 7.5), 6.87 (d, 1H, J = 15.8, COCH=CH), 3.89 (s, 3H, OCH3) 2.21(s, 3H, CH3); HRMS m/z calcd. for C26H23N5O2Na [M+Na]+ 460.17494, found: 460.16676.
(E)-3-(2-ethoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12g): yield: 66.6%; mp: 255-257 °C. 1H-NMR δ: 10.09 ( s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 ( s,1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.48 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 8.01 (d, 1H, J = 1.6), 7.84 (d, 1H, J = 15.8, COCH=CH), 7.57 (dd, 1H, J1 = 7.8, J2 = 1.3), 7..53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.43-7.40 (m, 2H), 7.38-7.35 (m, 1H), 7.18 (d, 1H, J = 8.3), 7.08 (d, 1H, J = 8.3), 7.01 (t, 1H, J = 7.5), 6.85 (d, 1H, J = 15.8, COCH=CH), 4.15 (q, 2H, J = 7.0, OCH2CH3).), 2.22 (s, 3H, CH3), 1.41(t, 3H, J = 7.0, OCH2CH3); HRMS m/z calcd. for C27H26N5O2 [M+H]+ 452.20865, found: 452.24296.
(E)-3-(4-fluorophenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12h): yield: 70.6%; mp: 194-198 °C. 1H-NMR δ: 10.28 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.47 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 7.97(d, 1H, J = 1.9), 7.68 (dd, 2H, J1 = 8.5, J2 = 5.5), 7.56 (d, 1H, J = 15.8, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.43 (d, 1H, J = 5.3), 7.40 (d, 1H, J = 2.0), 7.28 (t, 2H, J1 = 8.5), 7.19 (d, 1H, J = 8.4), 6.78 (d, 1H, J = 15.8, COCH=CH), 2.22 (s, 3H, CH3); HRMS m/z calcd. for C25H20FN5ONa [M+Na]+ 448.15496, found: 448.13271.
(E)-3-(4-chlorophenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12i): yield: 65.8%; mp:268-269 °C. 1H-NMR δ: 10.14 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51 (d, 1H, J = 5.2, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 2.0, Py-4-H), 7.98 (d, 1H, J = 1.6), 7.64 (d, 2H, J = 8.6), 7.56 (d, 1H, J = 15.6, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (t, 2H, J1 = 8.5), 7.43 (d, 1H, J = 5.2), 7.41 (d, 1H, J = 1.9), 7.19 (d, 1H, J = 8.3), 6.84 (d, 1H, J = 15.6, COCH=CH ), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H21ClN5O [M+H] + 442.14346, found: 442.14943.
(E)-3-(4-bromophenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12j): yield: 49.6%; mp:124-125 °C. 1H-NMR δ: 10.14 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.52 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 7.98 (d, 1H, J = 1.5), 7.64 (d, 2H, J = 8.5), 7.59 (d, 1H, J = 15.6, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (t, 2H, J1 = 8.5), 7.43 (d, 1H, J = 5.2), 7.41 (d, 1H, J = 1.9), 7.19 (d, 1H, J = 8.3), 6.86 (d, 1H, J = 15.7, COCH=CH ), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C25H20BrN5ONa [M+Na]+ 508.07489, found: 508.05714.
(E)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-(4-nitrophenyl)acrylamide (12k): yield: 70.0%; mp: 252-254 °C. 1H-NMR δ: 10.28 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.94 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.7, Py-6-H), 8.52 (d, 1H, J = 5.2, pyrimidinyl-6-H), 8.47 (dt, 1H, J1 = 8.0 Hz, J2 = 1.9, Py-4-H), 8.29-8.27 (m, 2H), 8.00 (d, 1H, J = 1.9) 7.89 (d, 2H, J = 8.9), 7.67 (d, 1H, J = 15.8, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.44-7.42 (m, 2H), 7.21 (d,1H, J = 8.4), 7.02 (d, 1H, J = 15.8, COCH=CH), 2.22 (s, 3H, CH3); HRMS m/z calcd. for C25H20N6O3Na [M+Na]+ 475.14946, found: 475.13600.
(E)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-p-tolylacrylamide (12l): yield: 68.8%; mp: 205-208 °C. 1H-NMR δ: 10.07 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.47 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.9), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.52 (d, 1H, J = 15.6, COCH=CH), 7.50 (d, 2H, J = 8.0), 7.42 (d, 1H, J = 5.2), 7.40 (d, 1H, J = 2.0), 7.25 (d, 2H, J = 8.0), 7.18 (d, 1H, J = 8.4), 6.78 (d, 1H, J = 15.7, COCH=CH), 2.33 (s, 3H, CH3), 2.22 (s, 3H, CH3); HRMS m/z calcd. for C25H24N5O [M+H]+ 442.19809, found: 422.19907
(E)-3-(4-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12m): yield: 63.3%; mp: 115-117 °C. 1H-NMR δ: 10.03 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.92 (s,1 H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.9, 1H, Py-4-H), 7.97 (d, 1H, J = 1.7), 7.56 (d, 2H, J = 8.8), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.52 (d, 1H, J = 15.6, COCH=CH), 7.42 (d, 1H, J = 5.2), 7.40 (d, 1H, J = 2.0), 7.18 (d, 1H, J = 8.4), 7.01 (d, 2H, J = 8.8), 6.69 (d, 1H, J = 15.7, COCH=CH), 3.80 (s, 3H, OCH3), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C26H24N5O2 [M+H]+ 438.19300, found: 438.17750.
(E)-3-(4-benzyloxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12n): yield: 51.4%; mp: 221-222 °C. 1H-NMR δ: 10.02 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.91 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.49 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.45 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.7), 7.56 (d, 2H, J = 8.7), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.51 (d, 1H, J = 15.7, COCH=CH), 7.45 (d, 2H, J = 7.3), 7.43-7.39 (m, 4H), 7.35-7.32 (m, 1H), 7.18 (d, 1H, J = 8.4), 7.08 (d, 2H, J = 8.7), 6.70 (d, 1H, J = 15.6, COCH=CH), 5.20 (s, 2H, OCH2Ph), 2.20 (s, 3H, CH3); HRMS m/z calcd. for C32H28N5O2 [M+H]+ 514.22430, found: 514.23057.
(E)-3-(2,3-dimethoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12o): yield: 33.9%; mp: 146-147 °C. 1H-NMR δ: 10.13 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H ), 8.91(s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.52 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.9, Py-4-H), 8.00 (d, 1H, J = 1.5), 7.76 (d, 1H, J = 15.8, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.43 (d, 1H, J = 5.1), 7.41 (d, 1H, J = 1.9), 7.20-7.18 (m, 2H), 7.14 (t, 1H, J = 8.0), 7.10 (dd, 1H, J1 = 8.1, J2 = 1.7), 6.87 (d, 1H, J = 15.8, COCH=CH ), 3.84 (s, 3H, OCH3), 3.78 (s, 3H, OCH3), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C27H25N5O3Na [M+Na]+ 490.18551, found: 490.19200.
(E)-3-(2,4-dimethoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12p): yield: 65.1%; mp:154-156 °C. 1H-NMR δ: 9.98 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.91(s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.9), 7.70 (d, 1H, J = 15.7, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (d, 1H, J = 8.5), 7.42 (d, 1H, J = 5.1), 7.40 (d, 1H, J = 1.9), 7.17 (d, 1H, J = 8.4), 6.74 (d, 1H, J = 15.7, COCH=CH), 6.61-6.60 (m, 2H), 3.88 (s, 3H, OCH3), 3.82 (s, 3H, OCH3), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C27H25N5O3Na [M+Na]+ 490.18551, found: 490.19263.
(E)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-(2,3,4–trimethoxyphenyl) acrylamide (12q): yield: 69.0%; mp:226-227 °C. 1H-NMR δ: 10.05 (s, 1H, NHC=O), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.91(s, 1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51(d, 1H, J = 5.1, pyrimidinyl-6-H), 8.48 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.99 (d, 1H, J = 1.9), 7.65 (d, 1H, J = 15.8, COCH=CH), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.43 (d, 1H, J = 5.1), 7.40 (d, 1H, J = 1.9), 7.33 (d, J = 8.8), 7.18 (d, 1H, J = 8.4), 6.91( d, 1H, J = 8.8), 6.79 (d, 1H, J = 15.8, COCH=CH), 3.84 (s, 3H, OCH3), 3.83 (s, 3H, OCH3), 3.78 (s, 3H, OCH3), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C28H27N5O4Na [M+Na]+ 520.19607, Found: 520.19694.
(E)-3-(3,4-dimethoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12r): yield: 67.8%; mp: 234-235 °C. 1H-NMR δ: 10.03 (s, 1H, NHCO), 9.27 (d, 1H, J = 1.8, Py-2-H), 8.92 (s,1H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.50 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.98 (d, 1H, J = 1.9), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (d, 1H, J = 15.8, COCH=CH), 7.42-7.40 (m, 2H), 7.21 (d, 1H, J = 1.8), 7.18 (dd, 2H, J1 = 8.6, J2 = 3.1), 7.01 (d, 1H, J = 8.4), 6.71(d, 1H, J = 15.8, COCH=CH), 3.82 (s, 3H, OCH3), 3.80 (s, 3H, OCH3), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C27H26N5O3 [M+H]+ 468.20357, found: 468.20356.
(E)-3-(4-ethoxy-3-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino]phenyl]acryla-mide (12s): yield: 68.8%; mp: 256-258 °C. 1H-NMR δ: 10.03 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51(d, 1H, J = 5.2, pyrimidinyl-6-H), 8.47 (dt, 1H, J1 = 8.1, J2 = 1.9 Py-4-H), 7.97 (d, 1H, J = 1.9), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (d, 1H, J = 15.6,COCH=CH), 7.42 (d,1H, J = 5.1), 7.40 (d, 1H, J = 2.0), 7.20 (d, 1H, J = 1.9), 7.18 (d, 1H, J = 8.4), 7.15 (dd, 1H, J1 = 8.4, J2 = 1.9), 6.99 (d, 1H, J = 8.4), 6.70 (d, 1H, J = 15.7, COCH=CH ), 4.05 (q, 2H, J = 7.0, OCH2CH3), 3.81 (s, 3H, OCH3), 2.22 (s, 3H, CH3), 1.34 (t, 3H, J = 7.0, OCH2CH3); HRMS m/z calcd. for C28H28N5O3 [M+H]+ 482.21921, found: 482.22009.
(E)-3-(3-ethoxy-4-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acryla-mide (12t): yield: 68.5%; mp: 263-264 °C. 1H-NMR δ: 10.01 (s, 1H, NHC=O), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.92 (s, 1H, NH), 8.68 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H), 8.51 (d, 1H, J = 5.2, pyrimidinyl -6-H), 8.47 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.9), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (d, 1H, J = 15.6, COCH=CH)), 7.42 (d,1H, J = 5.1), 7.40 (d, 1H, J = 2.1), 7.19-7.16 (m, 3H), 7.00 (d,1H, J = 8.4), 6.70 (d, 1H, J = 15.7, COCH=CH), 4.06(q, 2H, J = 6.9, OCH2CH3) 3.80 (s, 3H, OCH3), 2.21 (s, 3H, CH3), 1.35 (t, 3H, J = 6.9, OCH2CH3); HRMS m/z calcd. for C28H28N5O3 [M+H]+ 482.21921, Found: 482.22036.
(E)-3-(4-benzyloxy-3-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl] acrylamide (12u): yield: 36.4%; mp: 178-179 °C. 1H-NMR δ: 10.08 ( s, 1H, NHCO), 9.27 (d, 1H J = 1.8, Py-2-H), 8.98 (s, 1H, NH), 8.69 (dd, 1H, J1= 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.7), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.51 (1H, J = 15.7, COCH=CH), 7.49-7.47 (m, 2H), 7.44-7.40 (m, 4H), 7.36-7.33 (m, 1H), 7.33 (d, 1H, J = 1.6), 7.22 (dd, 1H, J1 = 8.4, J2 = 1.8), 7.18 (d, 1H, J = 8.4), 7.05 (d, 1H, J = 8.7), 6.70 (d, 1H, J = 15.6, COCH=CH), 5.20 (s, 2H, OCH2Ph), 3.83 (s, 3H, OCH3), 2.20 (s, 3H, CH3);HRMS m/z calcd. for C33H30N5O3 [M+H]+ 544.23486, found: 544.23565.
(E)-3-(3-benzyloxy-4-methoxyphenyl)-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)-phenyl] acrylamide (12v) yield: 42.7%; mp: 232-234 °C. 1H-NMR δ: 10.04 ( s, 1H, NHCO), 9.27(d, 1H, J = 1.8, Py-2-H), 8.92( s,1 H, NH), 8.69 (dd, 1H, J1 = 4.8, J2 = 1.6, Py-6-H ), 8.51 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.7), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.50 (d, 1H, J = 15.8, COCH=CH), 7.46-7.44 (m, 2H), 7.42-7.38 (m, 4H), 7.35-7.33 (m, 1H), 7.23 (d, 1H, J = 1.9), 7.18 (d, 1H, J = 8.4), 7.15 (dd, 1H, J1 = 8.4, J2 = 1.8), 7.09 (d, 1H, J = 8.4), 6.70 (d, 1H, J = 15.6, COCH=CH), 5.13 (s, 2H, OCH2Ph), 3.83 (s, 3H, OCH3), 2.20 (s, 3H, CH3); HRMS m/z calcd. for C33H30N5O3 [M+H]+ 544.23486, found: 544.23565.
(E)-3-benzo[1,3]dioxol-5-yl-N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]acrylamide (12w): yield: 46.8%; mp: 244-246 °C. 1H-NMR δ: 10.02 (s, 1H, NHCO), 9.26 (d, 1H, J = 1.2, Py-2-H), 8.91 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.5, J2 = 1.1, Py-6-H), 8.51 (d, 1H, J =5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.1, J2 = 1.9, Py-4-H), 7.97 (d, 1H, J = 1.3), 7.53 (dd, 1H, J1 = 8.0, J2 = 4.8, Py-5-H), 7.49 (d, 1H, J=15.6, COCH=CH), 7.42 (d, 1H, J = 5.1), 7.40 (d, 1H, J = 1.6), 7.18 (d, 2H, J = 7.2), 7.13 (dd, 1H, J1 = 8.1, J2 = 1.4), 6.97 (d, 1H, J = 8.0), 6.67 (d, 1H, J = 15.6, COCH=CH), 6.08 (s, 2H, OCH2O), 2.21 (s, 3H, CH3); HRMS m/z calcd. for C26H21N5O3Na [M+Na]+ 474.15421, found: 474.14769.
N-[4-methyl-3-(4-pyridin-3-yl-pyrimidin-2-ylamino)phenyl]-3-p-tolylpropionamide (15): yield: 76.6%; mp: 168-169°C. 1H-NMR δ: 9.81(s, 1H, NHCO), 9.26 (d, 1H, J = 1.8, Py-2-H), 8.90 (s, 1H, NH), 8.69 (dd, 1H, J1 = 4.7, J2 = 1.6, Py-6-H ), 8.50 (d, 1H, J = 5.1, pyrimidinyl-6-H), 8.46 (dt, 1H, J1 = 8.0, J2 = 1.8, Py-4-H), 7.88 (d, 1H, J = 1.6), 7.51 (dd, 1H, J1 = 8.0 Hz, J2 = 4.8, Py-4-H), 7.41 (d, 1H, J = 5.1), 7.28 (dd, 1H, J1 = 8.0, J2 = 1.8), 7.14-7.12 (m, 3H), 7.07 (d, 2H, J = 8.0), 2.86 (t, 2H, J = 7.45, COCH2CH2), 2.58 (t, 2H, J = 7.45, COCH2CH2), 2.24 (s, 3H, CH3), 2.19 (s, 3H, CH3); HRMS m/z calcd. for C26H26N5O [M+H]+ 424.21374, found: 424.24776.
Antiproliferative activity assay
Human chronic myeloid leukemia K562 cells containing Bcl-Abl fusion kinase were cultured in RPMI 1640 medium (Gibco; New York, NY, USA) containing 10% fetal bovine serum. Cells were seeded at 4.0 × 104 cells/mL and incubated with various concentrations of test compounds for 72 h. Total cell number in each group was counted with the aid of a hemocytometer. The growth inhibition ability was calculated and expressed as the ratio of the cell number in treated group to that in the untreated group. The concentration that inhibited half of the cell growth (IG50) was calculated [20].
Molecular modeling
Molecular docking was carried out using Molegro Virtual Docker (MVD) [21]. The small molecules and the X-ray crystal structure of c-Abl in complex with STI-571 (PDB code: 1iep[12]) were imported, and STI-571 was used to define the binding cavity. Molecular docking was carried out using the Molegro Virtual Docker (MVD) [21]. The docking algorithm was set at 1,500 maximum iterations with a simplex evolution population size of 50 and a minimum of 10 runs. The schematic diagrams of interactions between c-Abl and docked poses were analyzed by Sybyl 6.91 package [22].
Conclusions
A series of novel PAP cinnamamide derivatives structurally related to STI-571 were synthesized and evaluated for their antiproliferative activity toward human leukemia K562 cells. Compounds 12c and 12d with a substitution of Cl or Br at position 2 were the most effective compounds in inhibiting K562 cell growth, being 2-3 fold more potent than STI-571. The effects of these compounds on the inhibition of Bcr-Abl activity are worthy of further study.
Acknowledgements
The authors would like to thank Molegro ApS for kindly providing us with a free evaluation copy of their software package.
Footnotes
Sample Availability: Samples of the compounds 12a-12w are available from the authors.
References
- 1.Warmuth M., Danhauser-Riedl S., Hallek M. Molecular patho-genesis of chronic myeloid leukemia: Implications for new therapeutic strategies. Ann. Hematol. 1999;78:49–64. doi: 10.1007/s002770050473. [DOI] [PubMed] [Google Scholar]
- 2.Gishizky M.L. Molecular mechanisms of Bcr-Abl-induced oncogenesis. Cytokines Mol. Ther. 1996;2:251–261. [PubMed] [Google Scholar]
- 3.Cortes J.E., Talpaz M., KantarJian H. Chronic myelogenous leukemia: A review. Am. J. Med. 1996;100:555–570. doi: 10.1016/S0002-9343(96)00061-7. [DOI] [PubMed] [Google Scholar]
- 4.Nowell P.C., Hungerford D.A. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497–1501. [Google Scholar]
- 5.Rowley J.D. A new consistent chromosmal abnormality in chronic myelgenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 6.Wong S., Witte O.N. The BCR-ABL story: Bench to beside and back. Annu. Rev. Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 7.Druker B.J., Tamura S., Buchdunger E., Ohno S., Segal G.M., Fanning S., Zimmermann J., Lydon N.B. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 8.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantariian H., Capedeville R., Ohno-Jones S., Sawyer C.L. Efficacy and safety of a specific inhibitor of Bcr-Abl tyrosine kinase in chronicmyeloid leukemia. N. Eng. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 9.Roche-Lestienne C., Soenen-Cornu V., Grardel-Duflos N., Laï J.L., Philippe N., Facon T., Fenaux P., Preudhomme C. Several types of mutations of the Abl can be found in chronic meloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.V100.3.1014. [DOI] [PubMed] [Google Scholar]
- 10.Hochhaus A., Kreil S., Corbin A.S., La Rosée P., Muller M.C., Lahaye T., Hanfstein B., Schoch C., Cross N.C.U., Berger U., Gschaidmeier H., Druker B.J., Hehlmann R. Molecular and chromosomal mechanisms of resistance to imanitib (STI-571) therapy. Leukemia. 2002;11:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 11.Gorre M.E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P.N., Sawyers C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 12.Nagar B., Bornmann W.G., Pellicena P., Schindler T., Veach D.R., Miller W.T., Clarkson., Kuriyan J. Cystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitor PD173955 and imatinib(STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 13.Schindler T., Bornmann W., Pellicena P., Miller W.T., Clarkson B., Kuriyan J. Structure Mechanism for STI-571 inhibition of Ableson Tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann J., Buchdunger E., Mett H., Meyer T., Lydon N.B. Potent and selective inhibitors of the Abl-kinase: Phenylaminopyrimidine (PAP) derivatives. Bioorg. Med. Chem. Lett. 1997;7:187–192. doi: 10.1016/S0960-894X(96)00601-4. [DOI] [Google Scholar]
- 15.Rachid Z., Katsoulas A., Brahimi F., Jean-Claude B.J. Synthesis of pyrim- idinopyridine-triazene conjugates targeted to Abl tyrosine kinase. Bioorg. Med. Chem. Lett. 2003;13:3297–3300. doi: 10.1016/S0960-894X(03)00553-5. [DOI] [PubMed] [Google Scholar]
- 16.Manley P.W., Breitenstein W., Bruggen J., Cowan-Jacob S.W., Furet P., Mesta J., Meyer T. Urea derivatives of STI-571 as inhibitors of Bcr-Abl and PDGFR kinases. Bioorg. Med. Chem. Lett. 2004;14:5793–5797. doi: 10.1016/j.bmcl.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 17.Asaki T., Sugiyama Y., Hamamoto T., Higashioka M., Umehara M., Naito H., Niwa T. Design and synthesis of 3-substituted benzamide derivatives as Bcr-Abl kinase inhibitors. Bioorg. Med. Chem. Lett. 2006;16:1421–1425. doi: 10.1016/j.bmcl.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 18.O’Hare M., Walters D.K., Deiniger M.W.N., Druker B.J. AMN 107: Tightening the grip of imatinib. Cancer Cell. 2005;7:117–119. doi: 10.1016/j.ccr.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Bredereck H., Effenberger F., Bosch H. Ber. Dtsch. Chem. Ges. 1964;97:3397–3406. [Google Scholar]
- 20.Belhacene N., Maulon L., Guerin S., Ricci J.E., Mari B., Colin Y., Cartron J.P., Auberger P. Differential expression of the Kell blood group and CD10 antigens: Two related membrane metallopeptidases during differentiation of K562 cells by phorbol ester and hemin. FASEB J. 1998;12:531–539. doi: 10.1096/fasebj.12.7.531. [DOI] [PubMed] [Google Scholar]
- 21.Thomsen R., Christensen M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 22.Sybyl Version 6.9.1. Tripos Associates; St. Louis, MO, USA: 2003. [Google Scholar]