Abstract
Paclitaxel is a first-line chemotherapeutic agent for gastric cancer; however, resistance limits its effectiveness. Investigation into the underlying mechanisms of paclitaxel resistance is urgently required. In the present study, a paclitaxel-resistant gastric cancer cell line (MGC-803R) was generated with a morphological phenotype of epithelial-to-mesenchymal transition (EMT) and increased expression levels of microRNA (miR)-155-5p. MGC-803R cell-derived exosomes were effectively taken up by paclitaxel-sensitive MGC-803S cells, which exhibited EMT and chemoresistance phenotypes. miR-155-5p was enriched in MGC-803R-exosomes and could be delivered into MGC-803S cells. miR-155-5p overexpression in MGC-803S cells via transfection with mimics resulted in similar phenotypic effects as treatment with MGC-803R exosome and increased miR-155-5p content in MGC-803S exosomes, which then capable of inducing the malignant phenotype in the sensitive cells. GATA binding protein 3 (GATA3) and tumor protein p53-inducible nuclear protein 1 (TP53INP1) were identified as targets of miR-155-5p. Exosomal miR-155-5p inhibited these targets by directly targeting their 3′ untranslated regions. Knockdown of miR-155-5p was observed to reverse the EMT and chemoresistant phenotypes of MGC-803R cells, potentially via GATA3 and TP53INP1 upregulation, which inhibited MGC-803R-exosomes from inducing the malignant phenotype. These results demonstrated that exosomal delivery of miR-155-5p may induce EMT and chemoresistant phenotypes from paclitaxel-resistant gastric cancer cells to the sensitive cells, which may be mediated by GATA3 and TP53INP1 suppression. Targeting miR-155-5p may thus be a promising strategy to overcome paclitaxel resistance in gastric cancer.
Keywords: exosome, microRNA-155-5p, paclitaxel, epithelial-to- mesenchymal transition, chemoresistance, gastric cancer
Introduction
Gastric cancer ranks as the second most common malignant disease and the third leading cause of cancer-associated mortality in developing countries (1). Chemotherapy is one of the principal therapeutic approaches used in the treatment of cancer. Chemoresistance has been acknowledged as a major obstacle in successful cancer treatment. Paclitaxel is widely used as a front-line chemotherapeutic agent for gastric cancer; however, resistance limits the effectiveness of chemotherapy and results in treatment failure in the majority of cases (2,3). An improved understanding of the mechanism of chemoresistance to paclitaxel may provide novel therapeutic approaches for gastric cancer therapy; however, the underlying mechanisms of paclitaxel resistance in gastric cancer are not well understood.
Exosomes are a subset of extracellular microvesicles with a size ranging from 40-100 nm and are released by all types of cells (4). Increasing evidence has demonstrated that exosomes are intercellular mediators that contribute to the hallmarks of cancer by influencing tumor growth, metastasis, angiogenesis, immune regulation (5-7). Recently, increasing attention to the role of exosomes in chemoresistance has been gained (8-10). It is well known that exosomes are nanosized vesicles with a lipid bilayer membrane and can carry a variety of cell-of-origin cargo, including DNA, RNA and proteins (11). Chemotherapy may alter the exosomal composition of tumor cells (12-14); the potential role of exosomes in chemoresistance regulation may be determined by analyzing exosomal cargo.
Among the molecular components present as exosomal cargo, microRNAs (miRNAs/miRs), as a type of noncoding small RNA, have been extensively studied in the regulation of chemoresistance (15,16); however, the role and mechanism of exosomal miRNAs from chemoresistant cancer cells in the variations of phenotypic chemoresistance remain unclear. miRNA expression profile analysis of chemoresistant cancer cell-derived exosomes has been conducted in recent years. Bioinformatics analysis has revealed that target genes regulated by these dysregulated exosomal miRNAs were associated with drug resistance (17-19). Experimental evidence has demonstrated that exosomal miRNAs could be shuttled from chemoresistant to chemosensitive cancer cells to transmit chemoresistance (19-21).
Recently, researchers have revealed that exosomal miR-155-5p served an important role in the regulation of chemoresistance. Challagundla et al (22) firstly reported that exosomal miR-155-5p mediated cross-talk between monocyte and neuroblastoma cells to promote cancer cell chemoresistance. In addition, Patel et al (23) and Mikamori et al (24) revealed that miR-155-5p expression levels were upregulated in cancer cells and their exosomes following exposure to gemcitabine. Exosomes derived from gemcitabine-treated pancreatic cancer cells mediated the acquisition of chemo-resistance via the delivery of miR-155-5p into the sensitive cells (23,24). Additionally, Santos et al (25) reported that doxorubicin (DOX)- and paclitaxel-resistant breast cancer cells transmitted chemoresistance to neighboring cancer cells by exosomal delivery of miR-155-5p. These findings suggested that exosomal miR-155-5p may be a very important signaling molecule to transmit chemoresistance from drug-resistant to drug-sensitive cancer cells; however, the role and mechanism of chemoresistant cancer cell-derived exosomal miR-155-5p in this process require further investigation. Whether exosomal miR-155-5p mediates the transmission of paclitaxel resistance in gastric cancer cells remains unknown.
In the present study, a paclitaxel-resistant gastric cancer cell line MGC-803 (MGC-803R) was established, and the cellular morphological characteristics and miR-155-5p expression levels between MGC-803R cells and sensitive (MGC-803S) cells were compared. Cancer cell-derived exosomes were then isolated and characterized, followed by analysis of the role and mechanism of exosomal miR-155-5p in transmitting a chemoresistance phenotype from paclitaxel-resistant to paclitaxel-sensitive gastric cancer cells.
Materials and methods
Establishment of a paclitaxel-resistant MGC-803 cell line
The human gastric cancer cell line MGC-803 was obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientifics, Inc.) and incubated at 37°C in a humidified incubator with 5% CO2. Paclitaxel-resistant MGC-803R cells were established by continuous exposure to stepwise-increasing concentrations of paclitaxel (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). MGC-803 cells were initially cultured in DMEM containing a low concentration of paclitaxel (1 µg/l). The cells were then sub-cultured every 2 weeks in DMEM with increasing concentrations of the drug, a 25% increase each time. Finally, cells that were viable in the cell culture medium with a high concentration of paclitaxel (100 µg/l) were designated as paclitaxel-resistant MGC-803R cells. These cells were maintained in the drug-containing medium following induction, but were cultured in drug-free medium for 1 week at 37°C prior to subsequent experimentation. Parental cells, denoted as MGC-803S, were cultured under the same conditions without treatment.
Cell proliferation analysis
MGC-803R and MGC-803S cells were seeded in 96-well plates (5,000 cells/well) and exposed to increasing concentrations of paclitaxel for 48 h at 37°C. The concentrations of paclitaxel used for the drug dose-response curve analysis of MGC-803R cells were 0, 100, 200, 300, 400, 500, 600 and 700 µg/l, while the concentration of drug used for MGC-803S cells was 0, 2, 6, 8, 10, 12 and 14 µg/l. The proliferative ability of cells was determined with a Cell Counting Kit-8 (CCK-8) kit (MedChemExpress, Monmouth Junction, NJ, USA) according to the manufacturer’s protocols and presented as cell viability (%). The absorbance was measured at 450 nm using a Cytation 5 cell imaging multi-mode reader (BioTek Instruments, Inc., Winooski, VT, USA). The half-maximal inhibitory concentration (IC50) of drugs was calculated using GraphPad Prism version 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The resistant index (RI) was calculated utilizing the following formula: RI = IC50 of resistant cells/IC50 of sensitive cells. Exosomes were isolated from MGC-803S cells, MGC-803R cells and cancer cells following transfection with oligonucleotides (described below). For exosome treatment analysis, each well in a 96-well plate was seeded with 1,000 cells and loaded with exosomes at 100 µg/ml for 48 h for functional analysis at 37°C; untreated cells served as the control. The cell culture medium was then removed and fresh medium containing the IC50 concentration of paclitaxel was added to each well for 48 h. At the end of treatment, cell proliferation was measured. The inhibition rate (%) of cells by drug was calculated as 1-cell viability (%).
Exosome isolation and characterization
Gastric cancer cells were cultured in freshly prepared DMEM containing exosome-free FBS for 48 h until cells had reached 90% confluence. Cell culture supernatants were collected and filtered using 0.22-µm pore filters (Merck KGaA), followed by differential centrifugation at 4°C: 300 × g for 10 min to remove cells, 2,000 × g for 15 min to remove cell debris and 10,000 × g for 30 min to remove large particles. The cell culture supernatants were then concentrated by ultrafiltration with 30 kDa Amicon filter membranes (Merck KGaA) to remove a large amount of small soluble proteins. Finally, the exosomes were isolated using ExoQuick exosome precipitation solution (SBI System Biosciences, Palo Alto, CA, USA) as previously described (26). Exosomes were fixed in 1% glutaraldehyde for 5 min at room temperature. The morphology of exosomes was examined via transmission electron microscopy (×30,000) in a total of 6 fields. The exosomal protein concentration was measured by a Bicinchoninic Acid colorimetric method and exosome-associated protein cluster of differentiation 9 (CD9) expression was analyzed by western blotting (described below). The number and size distribution of exosomes were detected by a Nanosight NS300 system (Nanosight™ Technology; Malvern Panalytical Ltd., Malvern, UK) and analyzed using Nanoparticle Tracking Analysis (NTA) software (version 3.1, Malvern Panalytical Ltd.).
Exosome labeling, internalization and immunofluorescence staining
Exosomes (25 µg/µl) from MGC-803S cells and MGC-803R cells were pre-labeled with the red fluorescent dye CM-Dil (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 1 h, washed with PBS and centrifuged at 110,000 × g at 4°C for 70 min to remove excess dye; 8×104 of MGC-803S cells were seeded in 12-well plates and incubated at 37°C with the labeled exosomes for 4 h. Unlabeled exosomes were used as a negative control. The cells were fixed with 4% paraformaldehyde at room temperature for 1 h, permeabilized with 0.1% Triton X-100 and blocked with 5% bovine serum albumin (BSA) (Wuhan Boster Biological Technology, Ltd., Wuhan, China) at room temperature for 20 min. The cells were then incubated with rabbit monoclonal anti-α-smooth muscle actin (α-SMA) antibody (cat. no. A03744, dilution: 1:100, Wuhan Boster Biological Technology Co. Ltd.) at 4°C overnight, followed by incubation with Alexa Fluor® 488-conjugated anti-rabbit secondary antibody (cat. no. A-11034, dilution: 1:1,000, Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 1 h; the nuclei were stained with Hoechst 33342 dye at room temperature for 10 min. Incorporation of exosomes into target cells was visualized ≥6 field by fluorescence microscopy (magnification, ×400, Zeiss AG, Oberkochen, Germany).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from cells and exosomes was extracted using an miRNeasy Mini Kit (Qiagen, Inc., Hilden, Germany). miRNAs and mRNAs were reverse transcribed using the miScript II RT Kit (Qiagen, Inc.) and detected by miScript SYBR Green PCR Kit (Qiagen, Inc.) according to the manufacturer’s protocols. The amplification of fluorescence signals was detected by a fluorescence thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). RNU6B and β-actin were set as internal controls for cellular miRNAs and mRNAs, respectively. The relative expression levels of exosomal miRNA were normalized to miR-16-5p. The 2−ΔΔCq method was used for miRNAs and mRNAs quantification (27); miScript primers for miRNAs and RNU6B (cat. no. 218300) were obtained from Qiagen, Inc. The primers sequences for mRNAs detection and the thermocycling conditions applied are listed in Table I; experiments were conducted ≥3 times.
Table I.
Primers sequences and amplification conditions for reverse transcription-quantitative polymerase chain reaction of mRNAs.
| Genes | Primers sequences (5′-3′) | Annealing temperatures (°C) | Product length (bp) |
|---|---|---|---|
| β-actin | For: CACGAAACTACCTTCAACTCC | 56 | 265 |
| Rev: CATACTCCTGCTTGCTGATC | |||
| E-cadherin | For: CGCATTGCCACATACACTCT | ||
| Rev: TTGGCTGAGGATGGTGTAAG | 60 | 252 | |
| Vimentin | For: GAGCTGCAGGAGCTGAATG | ||
| Rev: AGGTCAAGACGTGCCAGAG | 60 | 344 | |
| GATA3 | For: ATCTGACCGAGCAGGTCGTA | ||
| Rev: GGCGACGACTCTGCAATTCT | 63 | 145 | |
| TP53INP1 | For: CTTCCTCCAACCAAGAAC | ||
| Rev: CTCCTCCATTGGACATGAC | 55 | 249 |
For, forward; Rev, reverse.
Western blot analysis
Cellular proteins were extracted with radioimmunoprecipitation buffer (Vazyme, Piscataway, NJ, USA), supplemented with protease inhibitors. Protein concentration was determined by Bicinchoninic Acid protein quantification kit (Vazyme). A total of 40 µg of protein for each group was separated on 12% SDS-PAGE gels and transferred to 0.45 µm polyvinylidene difluoride membranes (Merck KGaA). Membranes were then blocked with 5% BSA and incubated with primary antibodies against E-cadherin (CDH1 polyclonal antibody, dilution: 1:1,000, cat. no. A3044, ABclonal Biotechnology Co., Ltd., Wuhan, China), Vimentin (vimentin mouse monoclonal antibody, dilution: 1:800, cat. no. MB9006, Bioworld Technology, Inc., St. Louis Park, MN, USA) and CD9 (CD9 polyclonal antibody, dilution: 1:1,000, cat. no. BS60359, Bioworld Technology, Inc.) followed by incubation with peroxidase-conjugated secondary antibodies [goat anti-rabbit IgG (H+L)-horseradish peroxidase (HRP), dilution: 1:2,500, cat. no. BS13278; goat anti-mouse IgG (H+L)-HRP, dilution: 1:2,000, cat. no. BS50350, Bioworld Technology, Inc.) at 37°C for 1 h. β-actin [β-actin (I102) poly-clonal antibody, dilution: 1:5,000, cat. no. AP0060, Bioworld Technology, Inc.] was used as the loading control for cellular protein. Bands were visualized by chemiluminescence using immobilon ECL Ultra Western HRP Substrate (Merck KGaA) according to the manufacturer’s protocols.
Cell transfection
Oligonucleotides, including miR-155-5p mimics, mimics negative control (MNC), miR-155-5p inhibitor (inhibitor) and inhibitor negative control (INC) were synthesized and purified by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences of the oligonucleotides were described as previously (28). Cells were seeded at a density of 1.0×105 cells/well in 6-well plates and incubated overnight, followed by transfection with 5 nM mimics or 100 nM inhibitor using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.); correspondingly, MNC and INC were used as controls for mimics and inhibitor, respectively. Following transfection for 24 h, the cells were collected, seeded into 96-well plates at a density of 5,000 cells per well and treated with IC50 level of paclitaxel at 37°C for 48 h. Cell proliferation was then measured as aforementioned. Successful transfection was determined via RT-qPCR. Following transfection for 48 h, additional cells were collected for RNA and protein extraction. For exosome isolation, cells were refreshed with DMEM containing exosome-free FBS after transfection for 4-6 h. Following culture for 48 h, the cell culture medium was collected for exosome isolation.
Luciferase reporter construction and analysis
The potential binding sites of miR-155-5p to the 3′ untranslated region (UTR) of GATA binding protein 3 (GATA3) and tumor protein p53-inducible nuclear protein 1 (TP53INP1) mRNAs were predicted by TargetScan analysis (http://www.targetscan.org, release 7.2). The 3′UTR fragments of the two mRNAs containing the binding sites were synthesized and purified by Invitrogen (Thermo Fisher Scientific, Inc.); corresponding mutant fragments were also designed and synthesized. These oligonucleotides were annealed and inserted between SacI and SalI restriction sites of the pmirGLO dual-luciferase miRNA target expression vectors (Promega Corporation, Madison, WI, USA). Sequences of these oligonucleotides are listed in Table II. For miRNA functional analysis, MGC-803S cells were seeded into 24-well plates overnight and then co-transfected with 5 nM of mimics, MNC and 250 ng of reporter plasmids using Lipofectamine 2000. For exosome analysis, MGC-803S cells were transfected with luciferase reporter vectors for 4-6 h and then incubated with exosomes at 37°C for 48 h. Luciferase activities were quantified using the Dual Glo Luciferase Assay System (Promega Corporation) and normalized to activity of Renilla luciferase (Promega Corporation).
Table II.
Sequences of the synthesized oligonucleotides employed for the dual-luciferase assay.
| Oligonucleotides | Sequences
|
|
|---|---|---|
| Sense (5′-3′) | Antisense (5′-3′) | |
| GATA3 mRNA | CTAGCGGCCGCTAGTCAGTTGTT | TCGACTTTATTTTCTTTTAATGCATCAA |
| 3′UTR containing the binding sites (wild-type) | TGATGCATTAAAAGAAAATAAAG | ACAACTGACTAGCGGCCGCTAGAGCT |
| GATA3 mRNA | CTAGCGGCCGCTAGTCAGTTGTTT | TCGACTTTATTTTCTTCGCCGATATCAA |
| 3′UTR containing the mutant sites (mutant type) | GATATCGGCGAAGAAAATAAAG | ACAACTGACTAGCGGCCGCTAGAGCT |
| TP53INP1 mRNA | CTAGCGGCCGCTAGTCACACTAA | TCGACCTATCACCTGTTAATGCTAATGT |
| 3′UTR containing the binding sites (wild-type) | CATTAGCATTAACAGGTGATAGG | TAGTGTGACTAGCGGCCGCTAGAGCT |
| TP53INP1 mRNA | CTAGCGGCCGCTAGTCACACTAA | TCGACCTATCACCTGGCCGCTAGAATGT |
| 3′UTR containing the mutant sites (mutant type) | CATTCTAGCGGCCAGGTGATAGG | TAGTGTGACTAGCGGCCGCTAGAGCT |
GATA3, GATA binding protein 3; TP53INP1, tumor protein p53-inducible nuclear protein 1; UTR, untranslated region.
Statistical analysis
All data are presented as the mean ± standard error of the mean from at least three independent experiments. A Student’s t-test, one-way analysis of variance and a Tukey’s post hoc test were performed using GraphPad Prism 5 software. P<0.05 was considered to indicate a statistically significant difference.
Results
Paclitaxel-resistant gastric cancer cell line MGC-803R exhibits epithelial-mesenchymal transition (EMT) and upregulated levels of miR-155-5p
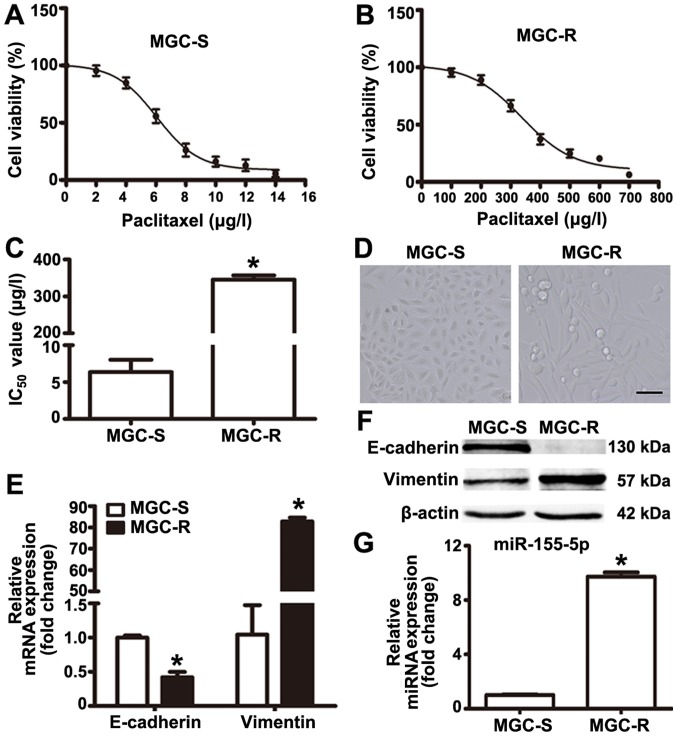
Paclitaxel-resistant MGC-803R cells were generated via the treatment of paclitaxel-sensitive parental MGC-803S cells with gradually increasing doses of paclitaxel in successive passages. MGC-803R and MGC-803S cells were exposed to various concentrations of paclitaxel for 48 h to determine paclitaxel-induced-cytotoxicity. The drug dose-response curves for the two cell lines are presented based on cell proliferation as determined by CCK-8 assays (Fig. 1A and B). The IC50 values for MGC-803R and MGC-803S cells were 345.4±11.67 and 6.06±1.68 µg/l, respectively (Fig. 1C). Compared with MGC-803S cells, the RI of MGC-803R cells was determined to be 57.5 (Fig. 1C).
Figure 1.
Comparison of IC50 values, epithelial-mesenchymal transition markers and miR-155-5p levels between MGC-803R and MGC-803S cells. MGC-R cells and MGC-S cells were exposed to various concentrations of paclitaxel to determine paclitaxel-induced-cytotoxicity. Cell proliferation was determined by a Cell Counting Kit-8 assay. (A and B) Drug dose-response curves for MGC-803S and MGC-803R cells; (C) IC50 values for MGC-803R and MGC-803S cells. (D) Morphology of MGC-803S and MGC-803R cells. Magnification, ×100, Scale bar, 50 µm. (E) E-cadherin and Vimentin mRNA expression levels were detected by RT-qPCR. (F) Western blotting of E-cadherin and Vimentin. (G) RT-qPCR analysis of miR-155-5p. *P<0.05 vs. MGC-S cells. IC50, half-maximal inhibitory concentration; MGC-R, paclitaxel-resistant; MGC-S, paclitaxel-sensitive; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
In addition, it was observed that MGC-803R cells became elongated with a mesenchymal-like morphology, which was markedly different from the epithelial-like morphology of MGC-803S cells (Fig. 1D). To confirm that these morphological changes were indicative of EMT in MGC-803R cells, the mRNA and protein expression levels of E-cadherin and Vimentin were determined by RT-qPCR and western blotting. The results revealed notably increased levels of Vimentin and reduced levels of E-cadherin protein in MGC-803R cells compared with in MGC-803S cells (Fig. 1E); however, a significant increase and decrease in the expression levels of E-cadherin and Vimentin were respectively reported in MGC-803R cells compared with MGC-803S cells (Fig. 1F). miRNA detection analysis demonstrated that the expression levels of miR-155-5p were significantly higher in MGC-803R cells than in MGC-803S cells (Fig. 1G). These results indicate that MGC-803 cells exhibit EMT phenotype and increased levels of miR-155-5p following induction by paclitaxel treatment.
MGC-803R-exosomes induce EMT and a paclitaxel-resistant phenotype in MGC-803S cells
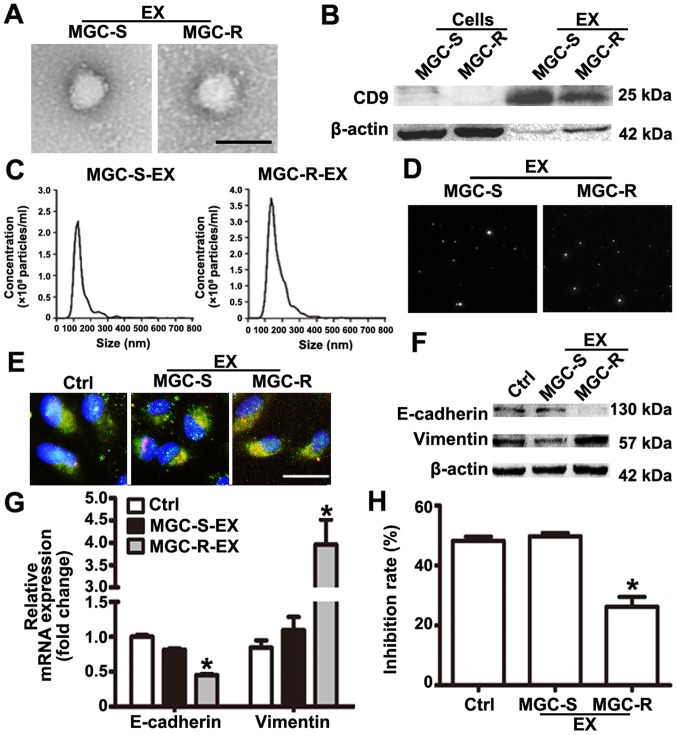
Exosomes act as an important mediator of intercellular communication (11). Studies have indicated that chemoresistant tumor cells can release exosomes that transmit drug resistance during tumorigenesis (12-14). To analyze whether MGC-803R cell-derived exosomes may confer a malignant phenotype on paclitaxel-sensitive cancer cells, exosomes from the culture medium of MGC-803R and MGC-803S cells were isolated. Purified exosomes from the cultures exhibited typical cup-shaped morphology with positive expression of exosomal marker CD9 (Fig. 2A and B). The mean size distribution and concentration of exosomes were determined with a Nanosight™ system. The mean size of MGC-803S-exosomes was 150±12.7 nm, while that of MGC-803R-exosomes was 168±13.2 nm (Fig. 2C). According to the size distribution of tumor exosomes detected by NTA, these results indicated that the detected size corresponded with the expected size of exosomes (29). Typical images of two cell-derived exosomes demonstrated the presence of particles with a concentration of more than 2.0×108 particles/ml (Fig. 2D). There was no notable difference in exosome quantities extracted from MGC-803R and MGC-803S cells. To demonstrate that exosomes could be taken up by the recipient cells, MGC-803S cells were incubated with CM-Dil-labeled exosomes. As presented in Fig. 2E, CM-Dil red fluorescence signals were visible around the nuclei and were also in the cytoplasm of MGC-803S cells following exposure to MGC-803R-derived exosomes or MGC-803S-exosomes; however, the negative control did not exhibit any red fluorescence. This suggested the uptake of CM-Dil-labeled exosomes by MGC-803S cells.
Figure 2.
MGC-803R-exosomes induce epithelial-mesenchymal transition and a paclitaxel-resistant phenotype in MGC-803S cells. (A) Exosome isolation from the cell culture medium of MGC-803S and MGC-803R cells were analyzed by transmission electron microscopy. Scale bar, 100 nm. (B) Exosomal marker CD9 was detected by western blotting. Nanoparticle tracking analysis of the size and number of exosomes was conducted. (C) Size distribution graph. (D) Representative images with Nanoparticle Tracking Analysis were presented. (E) Exosome internalization analysis; exosomes labeled with CM-Dil (red fluorescence) were visible around the nuclei and located in the cytoplasm (green fluorescence) of MGC-803S cells (nuclei were blue). Magnification, ×400. Scale bar, 25 µm. (F) Analysis of E-cadherin and Vimentin mRNA levels by reverse transcription-quantitative polymerase chain reaction. (G) E-cadherin and Vimentin protein expression as detected by western blotting; (H) The paclitaxel chemosensitivity of MGC-803S cells was detected following incubation with exosomes. *P<0.05 vs. Ctrl and MGC-S-EX. CD, cluster of differentiation; CD9, cluster of differentiation 9; Ctrl, control; MGC-S-EX, MGC-803S cell-derived exosomes; MGC-R-EX, MGC-803R cell-derived exosomes.
To investigate whether MGC-803R-exosomes were responsible for the spread of chemoresistance, MGC-803S cells were incubated with MGC-803R-exosomes or MGC-803S-exosomes for 48 h. The treated MGC-803S cells were then collected for the analysis of E-cadherin and Vimentin mRNA and protein expression. The protein expression levels of E-cadherin in MGC-803R-exosome-treated cells were notably lower and those of Vimentin were markedly increased compared with the control (Fig. 2F). Compared with MGC-803S-exosome treatment and the untreated control groups, MGC-803R-exosomes significantly reduced the mRNA expression levels of E-cadherin and increased that of Vimentin in MGC-803S cells (Fig. 2G). Furthermore, MGC-803S cells were also collected and treated with paclitaxel for chemosensitivity analysis following incubation with exosomes. CCK-8 assays revealed that MGC-803R-exosome treatment significantly reduced the chemosensitivity of MGC-803S cells, while MGC-803S-exosomes had no such effect compared with the control (Fig. 2H). These data suggested that the induction of EMT and a paclitaxel-resistant phenotype in MGC-803S cells may be ascribed to MGC-803R-exosomes.
Overexpression of miR-155-5p in MGC-803S cells exhibits similar effects to treatment with MGC-803R-exosomes
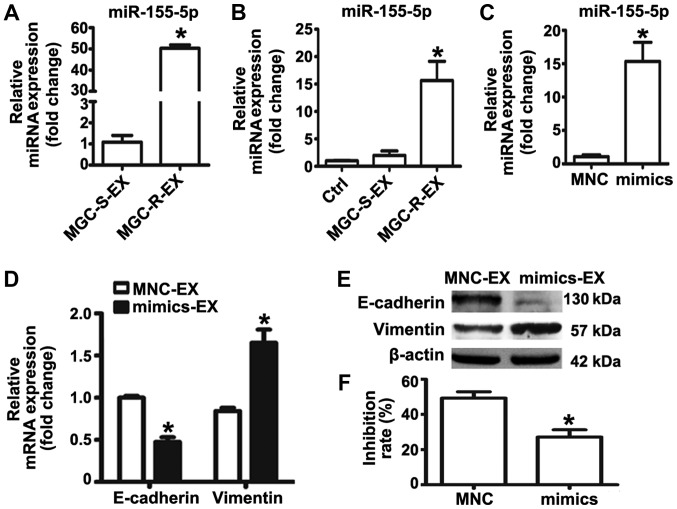
Previous studies revealed that exosomal miR-155-5p may serve an important role in chemoresistance (22-24). The present study reported that miR-155-5p expression levels were significantly elevated in MGC-803R cells. MGC-803R-exosome treatment was proposed to confer EMT and chemoresistant phenotypes to MGC-803S cells in the present study. To investigate whether miR-155-5p was selectively compartmen-talized into MGC-803R-exosomes, RT-qPCR was performed to detect miR-155-5p expression levels in MGC-803R- and MGC-803S-exosomes. The results revealed that miR-155-5p was significantly enriched in MGC-803R-exosomes compared with MGC-803S-exosomes (Fig. 3A). In addition, miR-155-5p expression levels in MGC-803S cells following treatment with exosomes were determined. The expression levels of miR-155-5p were significantly increased in MGC-803S cells following incubation with MGC-803R-exosomes compared with the control (Fig. 3B). These results indicated that miR-155-5p was enriched in MGC-803R-exosomes and could be delivered to MGC-803S cells via exosomal transfer.
Figure 3.
miR-155-5p overexpression confers epithelial-mesenchymal transition and paclitaxel-resistant phenotypes on MGC-803S cells. (A) miR-155-5p expression levels in MGC-S-EX and MGC-R-EX were detected by RT-qPCR; (B) Analysis of miR-155-5p expression levels in MGC-803S cells following incubation with MGC-S-EX or MGC-R-EX. (C) MGC-803S cells were transfected with miR-155-5p mimics or MNC. Following transfection for 48 h, miR-155-5p expression levels were measured by RT-qPCR. (D) Alterations in the mRNA expression levels of E-cadherin and Vimentin in MGC-803S cells after transfection; (E) E-cadherin and Vimentin protein expression levels as analyzed by western blotting. (F) Analysis of the proliferation of MGC-803S cells following transfection. *P<0.05 vs. MGC-S-EX, Ctrl, MNC-EX or MNC. Ctrl, control; EX, exosome; miR, microRNA; MNC, mimics negative control; MGC-R, paclitaxel-resistant; MGC-S, paclitaxel-sensitive; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
To further investigate the role of miR-155-5p in EMT and the regulation of paclitaxel chemoresistance, miRNA mimics were used to overexpress miR-155-5p in MGC-803S cells. miR-155-5p expression was observed to be significantly upregulated in the mimics group compared with the control (Fig. 3C); the miR-155-5p expression levels were similar to that of MGC-803S cells following MGC-803R-exosome treatment (Fig. 3B). Detection of mRNA and protein expression levels of EMT markers revealed that transfection of miR-155-5p mimics resulted in significantly reduced E-cadherin and increased Vimentin expression compared with the control (Fig. 3D and E). MGC-803S cells transfected with miRNA mimics were then exposed to paclitaxel. Compared with MNC-transfected cells, those transfected with miR-155-5p mimics exhibited significantly lower proliferative abilities in response to paclitaxel treatment (Fig. 3F). The results of the present study indicated that MGC-803R-exosomes may induce EMT and a paclitaxel-resistant phenotype in MGC-803S cells via the transfer of miR-155-5p.
Exosomal transfer of miR-155-5p promotes EMT and paclitaxel chemoresistance in MGC-803S cells
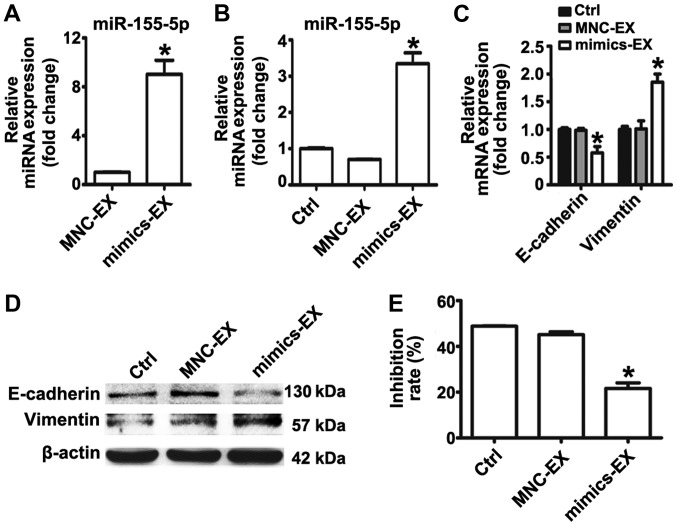
To further validate that exosomal miR-155-5p is involved in EMT and the induction of a paclitaxel-resistant phenotype, exosomes from MGC-803S cells following transfection with miRNA mimics or MNC were isolated. RT-qPCR analysis demonstrated that miR-155-5p was significantly enriched in exosomes isolated from mimic-transfected MGC-803S cells (Fig. 4A). MGC-803S cells were then treated with mimics or MNC-transfected-MGC-803S cell-derived exosomes (mimics-exosomes or MNC-exosomes, respectively). RT-qPCR analysis revealed that miR-155-5p expression levels were consistently elevated in MGC-803S cells following incubation with mimics-exosomes (Fig. 4B). This suggested that miR-155-5p may be delivered into MGC-803S cells via exosomal transfer.
Figure 4.
Exosomal miR-155-5p promotes epithelial-mesenchymal transition and chemoresistance in MGC-803S cells. (A) Exosomes were isolated from MGC-803S cells following transfection with mimics or MNC. miR-155-5p levels in these exosomes were detected by reverse transcription-quantitative polymerase chain reaction. (B) miR-155-5p expression levels were measured in MGC-803S cells following treatment with mimics-EX or MNC-EX. (C) mRNA expression levels of E-cadherin and Vimentin were analyzed in MGC-803S cells following treatment with mimics-EX or MNC-EX. (D) Western blotting of E-cadherin and Vimentin protein expression levels. (E) Analysis of the MGC-803S cell proliferation following treatment with mimics-EX or MNC-EX. *P<0.05 vs. MNC-EX or Ctrl. Ctrl, control; EX, exosome; miR, microRNA MNC, mimics negative control.
To evaluate whether mimics-exosomes treatment could replicate the effect of MGC-803R-exosomes on MGC-803S cells, alterations in the expression of EMT markers and the chemosensitivity of MGC-803S cells following incubation with mimics-exosomes were determined. The results revealed that mimics-exosome treatment significantly inhibited E-cadherin expression and enhanced Vimentin expression at the mRNA level; notable differences at the protein level were observed (Fig. 4C and D). Additionally, the proliferation of MGC-803S cells was significantly reduced by mimics-exosomes treatment following incubation with paclitaxel compared with MNC-exosomes and the control (Fig. 4E). These results indicated that exosomal transfer of miR-155-5p may be involved in EMT and the induction of a paclitaxel-resistant phenotype.
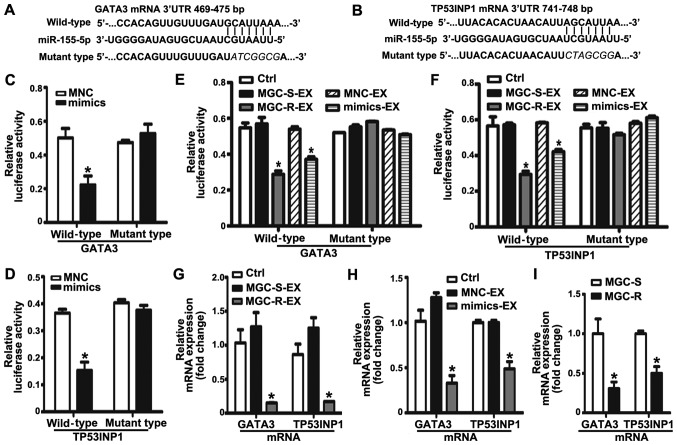
Exosomal miR-155-5p suppresses GATA3 and TP53INP1 by directly targeting their 3′ UTRs
To determine the potential mechanism of exosomal miR-155-5p in acquiring a malignant phenotype, the potential targets of miR-155-5p were predicted by TargetScan. Among the list of predicted targets, GATA3 and TP53INP1 have been reported to be associated with the regulation of chemoresistance (30,31). Therefore, GATA3 and TP53INP1 were selected as candidate targets for further validation in the present study. According to the predicted binding sites in the 3′UTR of GATA3 and TP53INP1 mRNAs, the corresponding wild-type and mutant-type luciferase reporter vectors were constructed, which were then co-transfected with mimics or MNC into MGC-803S cells (Fig. 5A and B). Luciferase activity analysis demonstrated that miR-155-5p mimics significantly suppressed the relative luciferase activity of the wild-type luciferase reporter vector groups compared with the MNC and mutant groups (Fig. 5C and D). Conversely, the relative luciferase activity was not affected by mimics in the mutant-type luciferase reporter vector groups. The results suggested that GATA3 and TP53INP1 were targets of miR-155-5p. To further investigate the effect of MGC-803R-exosomes and mimics-exosomes on relative luciferase activity, MGC-803S cells were transfected with luciferase reporter vectors and then incubated with MGC-803R-exosomes or mimics-exosomes. Compared with the control, MGC-803R-exosomes and mimics-exosome treatment significantly reduced the relative luciferase activity in wild-type luciferase reporter vector groups; significant alterations in luciferase activity in the mutant-type luciferase reporter vector groups were not observed (Fig. 5E and F). RT-qPCR analysis revealed that MGC-803R-exosome and mimics-exosome treatments significantly downregulated GATA3 and TP53INP1 mRNA levels compared with in the control group (Fig. 5G and H). Furthermore, the mRNA expression levels of the two target genes were also compared between MGC-803S and MGC-803R cells. As expected, TP53INP1 and GATA3 mRNA expression levels were significantly suppressed in MGC-803R cells compared with MGC-803S cells (Fig. 5I). The results suggested that exosomal transfer of miR-155-5p may induce chemoresistance via the suppression of GATA3 and TP53INP1 expression.
Figure 5.
Exosomal miR-155-5p suppresses GATA3 and TP53INP1 by directly targeting their 3′-UTRs. The position and sequences of the binding sites in the 3′UTR of target gene mRNAs, (A) GATA3 and (B) TP53INP1 were predicted by TargetScan. DNA fragments containing the wild-type or mutant binding sites were designed and inserted into pmirGLO dual-luciferase miRNA target expression vectors. (C and D) MGC-803S cells were co-transfected with reporter vectors, and mimics or MNC. Cells were then collected for luciferase activity assay. (E and F) The effects of exosomes on the luciferase activity in MGC-803S cells. (G) GATA3 and TP53INP1 mRNA expression levels in MGC-803S cells following treatment with MGC-R-EX and MGC-S-EX. (H) GATA3 and TP53INP1 mRNA expression levels in MGC-803S cells after co-culturing with MNC-EX and mimics-EX. (I) Comparison of GATA3 and TP53INP1 mRNA expression levels between MGC-803S and MGC-803R cells. *P<0.05 vs. Ctrl, MNC, MNC-EX, MGC-S-EX or MGC-S. EX, exosome; GATA3, GATA binding protein 3; MGC-R, paclitaxel-resistant; MGC-S, paclitaxel-sensitive; TP53INP1, tumor protein p53-inducible nuclear protein 1; 3′UTR, 3′-untranslated region.
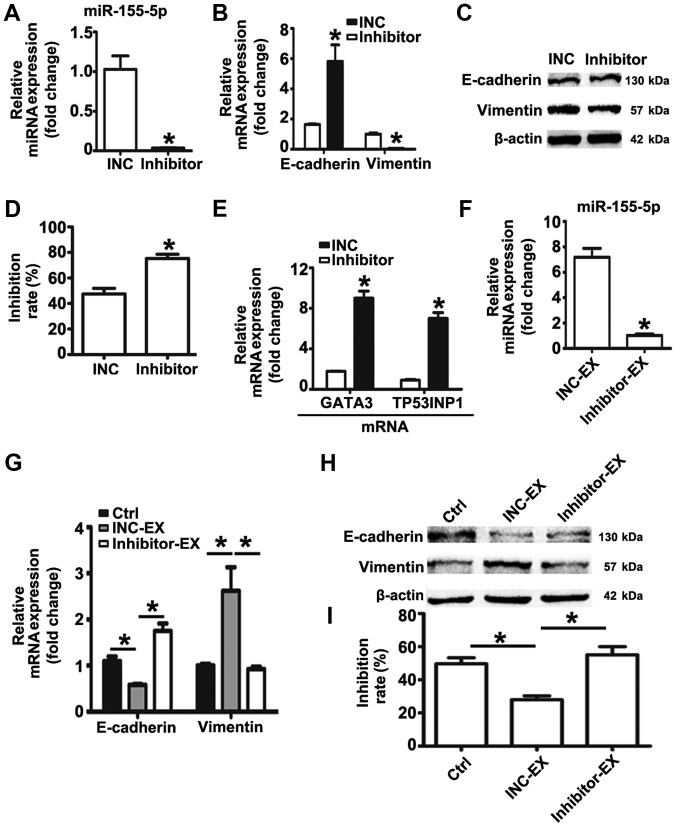
miR-155-5p inhibition reverses EMT and the chemoresistant phenotype of MGC-803R cells, and suppresses the induction of malignant phenotypes by MGC-803R-exosomes
The expression levels of miR-155-5p in MGC-803R cells were significantly higher than in MGC-803S cells. MGC-803R-exosomes was proposed to deliver miR-155-5p into chemosensitive cancer cells, inducing EMT and paclitaxel-resistant phenotypes in the present study. Based on these data, it was suggested that knockdown of miR-155-5p may dysregulate the malignant phenotypes of gastric cancer cells. Therefore, an miRNA inhibitor was employed to down-regulate the expression levels of miR-155-5p in MGC-803R cells. RT-qPCR detection revealed that miR-155-5p expression levels were significantly suppressed by miRNA inhibitor compared with the control (Fig. 6A). Compared with the INC group, significantly increased mRNA expression levels of E-cadherin, and reduced mRNA and protein levels of Vimentin were observed in MGC-803R cells following transfection with miRNA inhibitor (Fig. 6B and C); the protein expression levels of E-cadherin were not significantly altered. MGC-803R cells transfected with miRNA inhibitor or INC were then subjected to paclitaxel treatment. Knockdown of miR-155-5p significantly increased the chemosensitivity of MGC-803R cells to paclitaxel compared with the INC group (Fig. 6D), which was accompanied with significantly increased expression levels of GATA3 and TP53INP1 mRNA (Fig. 6E).
Figure 6.
miR-155-5p inhibition reverses EMT and the chemoresistant phenotype of MGC-803R cells and suppresses the induction of malignant phenotypes by MGC-803R-exosomes. (A) MGC-803R cells were transfected with inhibitor or INC. Following transfection for 48 h, miR-155-5p expression levels were measured by RT-qPCR. (B) mRNA expression levels of E-cadherin and Vimentin in MGC-803R cells following transfection. (C) E-cadherin and Vimentin protein expression levels were analyzed by western blotting. (D) Analysis of the chemosensitivity of MGC-803R cells following transfection. (E) GATA3 and TP53INP1 mRNA levels in MGC-803R cells following transfection. (F) Exosomes were isolated from MGC-803R cells following transfection with inhibitor or INC. Exosomal miR-155-5p levels were detected by RT-qPCR. (G) mRNAs expression levels of E-cadherin and Vimentin in MGC-803S cells following INC-EX and inhibitor-EX treatment. (H) Western blotting of E-cadherin and Vimentin protein expression levels. (I) Analysis of the chemosensitivity of MGC-803S cells following treatment with inhibitor-EX or INC-EX. *P<0.05 vs. INC, INC-EX or Ctrl. EX, exosome; inhibitor, miR-155-5p inhibitor; INC, inhibitor negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Furthermore, exosomes from MGC-803R cells transfected with INC or inhibitor were isolated. RT-qPCR analysis revealed that miR-155-5p levels were significantly reduced in exosomes derived from inhibitor-transfected MGC-803R cells compared with the exosomes secreted by INC-transfected MGC-803R cells (Fig. 6F). To analyze whether miR-155-5p suppression influenced MGC-803R-exosomes spreading the malignant phenotypes to sensitive cells, MGC-803S cells were incubated with inhibitor-exosomes or INC-exosomes. The results revealed that INC-exosomes treatment induced the EMT phenotype with markedly reduced levels of E-cadherin and increased levels of Vimentin, and significantly increased the paclitaxel resistance of MGC-803S cells compared with the untreated control group, while inhibitor-exosomes exhibited no such effect (Fig. 6G-I).
Discussion
Paclitaxel has been reported to possess notable antitumor activity; the development of chemoresistance is commonly observed in patients with gastric cancer (2,3). At present, paclitaxel resistance in tumors has been reported to be associated with underlying mechanisms, including increased drug efflux, alteration of intercellular signaling, tubulin mutation and over-expression of β-tubulin isotype composition (32,33). Recently, it has been demonstrated that drug-resistant cancer cells could transmit chemoresistant phenotypes to chemosensitive cancer cells via exosomes (8). Whether the acquisition of paclitaxel resistance by gastric cancer cells occurs in this way remains unknown. In the present study, the paclitaxel-resistant gastric cancer cell line MGC-803R was generated; it was proposed that MGC-803R cells could confer EMT characteristics and a paclitaxel-resistant phenotype on parental cells by exosomal transfer of miR-155-5p.
Increasing evidence suggests that tumor cell-derived exosomes provide critical signals in the development of chemoresistance (8-10). Several studies have revealed that delivery of key molecules by exosomes to tumor cells could lead to chemoresistance (34,35). Colon cancer cell exosomes highly enriched in N-terminal truncated isoforms of p73 mRNA were able to confer drug resistance on recipient cells (34). Exosomes from glioblastoma cancer cells harboring a protein tyrosine phosphatase receptor type Z1-MET fusion conferred temozolomide resistance on parental cells (35). It has been demonstrated that chemotherapeutic agents can also alter exosomal cargo components of cancer cells (12,14,19). Kreger et al (14) reported that paclitaxel treatment stimulated the secretion of specific exosomes from breast cancer cells, which were highly enriched with survivin protein. Bandari et al (12) observed that chemotherapy notably promoted exosome secretion in myeloma and resulted in a distinct exosomal proteome profile. miRNA microarray analysis revealed that a total of 11 miRNAs were upregulated in cisplatin (DDP)-resistant A549 cells and in A549/DDP-exosomes compared with A549 cells and their exosomes (19). These tumor cell-exosomes could be taken up by tumor cells, altering their behavior in ways that enhanced tumor survival and progression (19). Additionally, chemotherapeutic agents also enhanced exosome release from cancer cells and were also exported into exosomes (36). This finding suggests that cancer cells may protect themselves from the cytotoxicity of therapeutic drugs by secluding them in exosomes.
To improve understanding of the underlying mechanisms of chemoresistance, chemoresistant cancer cells may be an ideal cell model for investigation. The role of exosomes secreted from chemoresistant cancer cells in the induction of chemoresistance has been studied. Adriamycin (ADM/ADR)-resistant breast cancer cells (MCF7/ADM) exhibited increased expression levels of drug-resistance-associated proteins, including ubiquitin carboxyl-terminal hydrolase-L1 and P-glycoprotein (P-gp) (13). These proteins could be sorted into MCF7/ADM cell-derived exosomes, which transferred the chemoresistant phenotype into ADM-sensitive breast cancer cells (13). ADR-resistant breast cancer cells (MCF-7/ADR)-derived exosomes were reported to contain the drug-resistance-associated gene multidrug resistance-1 and P-gp. MCF-7/ADR cell-derived exosomes induced a drug resistance phenotype in MCF-7 parental cells (37). These findings demonstrated that exosomes could transfer intercellular drug resistance from drug-resistant to drug-sensitive cancer cells. To investigate the mechanism of paclitaxel resistance in gastric cancer cells, the paclitaxel-resistant gastric cancer cell line MGC-803R was established in the present study. Consistently, it was demonstrated that MGC-803R derived-exosomes conferred a paclitaxel-resistant phenotype in MGC-803S cells.
It has been observed that exosomal miRNAs shuttled from drug-resistant to drug-sensitive tumor cells were widely involved in the spread of chemoresistance (38,39). Exosomal miR-222-3p from the gemcitabine-resistant lung cancer line A549 enhanced gemcitabine resistance in parental sensitive cells (20). A549/DDP-exosomes conferred resistance to DDP in recipient cells depending on exosomal miR-100-5p (19). Thus, it was noted that miRNAs contained in chemoresistant cancer cell-derived exosomes may determine the degree of chemoresistance acquired mediated by exosomes between cancer cells. Identifying the specific miRNA cargo of exosomes may clarify their function and mechanism in the transfer of chemoresistance. Among these exosomal miRNAs, miR-155-5p has been reported to mediate transmission of a chemoresistant phenotype in breast cancer and pancreatic cancer (23-25). Notably, exosomal miR-155-5p levels were notably increased in paclitaxel-resistant breast cancer cell-derived exosomes (25). Exosomal delivery of miR-155-5p altered the response of sensitive cancer cells to treatment with chemotherapeutic agents (25). miR-155-5p as an oncogenic miRNAs is significantly elevated in gastric cancer and drives therapy resistance in numerous types of tumor (40-42). Additionally, anti-miR-155-5p increased the chemosensitivity of cancer cells to paclitaxel (43). Therefore, the present study determined whether miR-155-5p was involved in inducing the paclitaxel-resistant phenotype between gastric cancer cells. As expected, miR-155-5p was highly expressed in MGC-803R cells and was enriched in their exosomes compared with MGC-803S cells and their corresponding exosomes. MGC-803R cell-exosomes were effectively internalized by MGC-803S cells, which resulted in increased levels of miR-155-5p in the recipient cells. This suggested that miR-155-5p could be delivered into MGC-803S cells via exosomes. Increased miR-155-5p expression by miRNA mimics resulted in similar effects to those of MGC-803R cell-exosomes on MGC-803S cells. Furthermore, overexpression of miR-155-5p increased the content of miR-155-5p in MGC-803S cell exosomes, which acquired the capacity to reduce paclitaxel chemosensitivity in the parental cells. The results of the present study indicated that the paclitaxel-resistant phenotype may be transmitted from paclitaxel-resistant gastric cancer cells to sensitive cells via exosomal miR-155-5p.
EMT is characterized by loss of epithelial characteristics and the acquisition of a mesenchymal phenotype. Acquisition of an EMT phenotype has been demonstrated in numerous types of chemoresistant cancer cells (25,44,45). Consistent with previous findings, the present study reported morphological alterations in accordance with the characteristics of EMT in MGC-803R cells compared with the parental cells. EMT has been demonstrated to be associated with enhanced migration and invasion of cancer cells and has also been proposed as an important contributor to drug resistance (46). Several studies revealed that the acquisition of EMT was important for chemoresistance, but not indispensable for metastasis (47,48). Cancer cells undergoing EMT have been considered to exhibit increased resistance to chemotherapeutic agents (49). Therefore, the detection of the EMT phenotype may aid the determination of alterations in the response of cancer cells to chemotherapeutic agents. In the present study, MGC-803R exosomes induced EMT in MGC-803S cells. Overexpression of miR-155-5p and exosomal miR-155-5p also promoted EMT in MGC-803S cells. EMT induction in parental cells was an indicator of increased resistance to paclitaxel. miR-155-5p has been validated as an important regulator of EMT induction in cancer cells (50), which suggests that exosomal transfer of miR-155-5p conferred the EMT phenotype and increased paclitaxel resistance in MGC-803S cells. The mechanisms by which EMT induces chemoresistance are complex. Evidence has indicated that the contribution of EMT to chemoresis-tance may be associated with the development of cancer stem cells (51,52). miR-155-5p has been reported to target the regulation of TP53INP1 to promote cancer cell EMT and a cancer stem cell phenotype (50). In addition, morin increased the chemosensitivity of prostate cancer cells to paclitaxel by suppressing the expression of miR-155-5p and upregulating its target GATA3 (43). To determine the mechanism of exosomal miR-155-5p in the present study, the two aforementioned proteins were selected as potential targets for investigation. We reported that MGC-803R-exosomes and exosomes secreted by MGC-803S cells following transfection with miR-155-5p mimics suppressed GATA3 and TP53INP1 expression by directly targeting their 3′UTRs. Additionally, knockdown of miR-155-5p reversed EMT and the paclitaxel-resistant phenotype, which was accompanied with elevated mRNA expression levels of GATA3 and TP53INP1. Furthermore, compared with the sensitive cells, the mRNA expression levels of the two targets were significantly reduced in MGC-803R cells. These data suggested that miR-155-5p loaded in exosomes was functional. MGC-803R-derived miR-155-5p in exosomes may therefore suppress the expression of GATA3 and TP53INP1 to confer EMT and a chemoresistant phenotype on MGC-803S cells. Whether the two targets are indispensable for exosomal miR-155-5p-mediated transfer of malignant phenotypes requires further investigation in the future. In addition, EMT mechanisms are very complex. A hallmark of EMT is the functional loss of E-cadherin and the overexpression of mesenchymal markers, including N-cadherin, Vimentin and Fibronectin (53). In the present study, alterations in the expression of two classical markers, E-cadherin and Vimentin, were detected to determine whether gastric cancer cells acquire an EMT phenotype, which may be not adequate; EMT-associated transcription factors and signaling pathways require further investigation.
In conclusion, the present study successfully generated the paclitaxel-resistant gastric cancer cell line MGC-803R and demonstrated that miR-155-5p expression levels were significantly elevated in MGC-803R cells, which was selectively sorted into MGC-803R cell-derived exosomes. Exosomal delivery of miR-155-5p may induce EMT and chemoresistance phenotypes from paclitaxel-resistant gastric cancer cells to sensitive cancer cells by suppressing the expression of GATA3 and TP53INP1. Therefore, targeting miR-155-5p may be a novel strategy to overcome paclitaxel resistance in gastric cancer.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Scientific research project of ‘six one project’ for high-level health personnel from Jiangsu Provincial Health and Family Planning Commission (grant no. LGY 2016026); The project of the Nanjing Science and Technology Commission (grant no. 201605005); Key and general program of Jiangsu Cancer Hospital (grant no. ZK201604); Project funded by China Postdoctoral Science Foundation (grant no. 2017M611740); Jiangsu province postdoctoral research funding scheme (grant no. 1701027A); Jiangsu province social development key research and development plan (grant no. BE2017694), and Science Foundation for Doctorate Research of the Affiliated Hospital of Jiangsu University (grant no. jdfyRC2016001).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
MW and BS made substantial contributions to the design of the study and wrote the paper. MW, RQ and SY performed the experiments. XX, GL and RG analyzed and interpreted the data. CT, WZ and BS revised the manuscript for critically important intellectual content and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Guo Z, Wang X, Lin R, Chen L, Fan N, Chen Y, Lin J, Yu J. Paclitaxel-based regimens as first-line treatment in advanced gastric cancer. J Chemother. 2015;27:94–98. doi: 10.1179/1973947814Y.0000000169. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Fan D. Multidrug resistance in gastric cancer: Recent research advances and ongoing therapeutic challenges. Expert Rev Anticancer Ther. 2007;7:1369–1378. doi: 10.1586/14737140.7.10.1369. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meehan K, Vella LJ. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit Rev Clin Lab Sci. 2016;53:121–131. doi: 10.3109/10408363.2015.1092496. [DOI] [PubMed] [Google Scholar]
- 6.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 7.Brinton LT, Sloane HS, Kester M, Kelly KA. Formation and role of exosomes in cancer. Cell Mol Life Sci. 2015;72:659–671. doi: 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma A. Chemoresistance in cancer cells: Exosomes as potential regulators of therapeutic tumor heterogeneity. Nanomedicine (Lond) 2017;12:2137–2148. doi: 10.2217/nnm-2017-0184. [DOI] [PubMed] [Google Scholar]
- 9.Butera G, Pacchiana R, Donadelli M. Autocrine mechanisms of cancer chemoresistance. Semin Cell Dev Biol. 2018;78:3–12. doi: 10.1016/j.semcdb.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Sousa D, Lima RT, Vasconcelos MH. Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends Mol Med. 2015;21:595–608. doi: 10.1016/j.molmed.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 12.Bandari SK, Purushothaman A, Ramani VC, Brinkley GJ, Chandrashekar DS, Varambally S, Mobley JA, Zhang Y, Brown EE, Vlodavsky I, et al. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 2018;65:104–118. doi: 10.1016/j.matbio.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning K, Wang T, Sun X, Zhang P, Chen Y, Jin J, Hua D. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surg Oncol. 2017;115:932–940. doi: 10.1002/jso.24614. [DOI] [PubMed] [Google Scholar]
- 14.Kreger BT, Johansen ER, Cerione RA, Antonyak MA. The enrichment of survivin in exosomes from breast cancer cells treated with paclitaxel promotes cell survival and chemoresistance Cancers (Basel) 2016;8:8. doi: 10.3390/cancers8120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Fu Q, Du Y, Yang Y, Cho WC. MicroRNA as regulators of cancer stem cells and chemoresistance in colorectal cancer. Curr Cancer Drug Targets. 2016;16:738–754. doi: 10.2174/1568009616666151118114759. [DOI] [PubMed] [Google Scholar]
- 16.Cui SY, Wang R, Chen LB. MicroRNAs: Key players of taxane resistance and their therapeutic potential in human cancers. J Cell Mol Med. 2013;17:1207–1217. doi: 10.1111/jcmm.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong S, Chen X, Wang D, Zhang X, Shen H, Yang S, Lv M, Tang J, Zhao J. MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget. 2016;7:19601–19609. doi: 10.18632/oncotarget.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin X, Yu S, Xu X, Shen B, Feng J. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. 2017;8:42125–42135. doi: 10.18632/oncotarget.15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, Shen B, Liu S, Yan D, Feng J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721–3733. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16:132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107:107. doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC, Singh S, Khushman M, Singh AP. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br J Cancer. 2017;116:609–619. doi: 10.1038/bjc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, et al. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:42339. doi: 10.1038/srep42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. 2018;8:829. doi: 10.1038/s41598-018-19339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Zhao C, Shi H, Zhang B, Zhang L, Zhang X, Wang S, Wu X, Yang T, Huang F, et al. Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: Novel biomarkers and a mechanism for gastric cancer. Br J Cancer. 2014;110:1199–1210. doi: 10.1038/bjc.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Zhu M, Wang M, Yang F, Tian Y, Cai J, Yang H, Fu H, Mao F, Zhu W, Qian H, et al. miR-155-5p inhibition promotes the transition of bone marrow mesenchymal stem cells to gastric cancer tissue derived MSC-like cells via NF-κB p65 activation. Oncotarget. 2016;7:16567–16580. doi: 10.18632/oncotarget.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitehead B, Wu L, Hvam ML, Aslan H, Dong M, Dyrskjøt L, Ostenfeld MS, Moghimi SM, Howard KA. Tumour exosomes display differential mechanical and complement activation properties dependent on malignant state: Implications in endothelial leakiness. J Extracell Vesicles. 2015;4:29685. doi: 10.3402/jev.v4.29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu SJ, Yang L, Hong Q, Kuang XY, Di GH, Shao ZM. MicroRNA-200a confers chemoresistance by antagonizing TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 2018;18:74. doi: 10.1186/s12885-017-3930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergara D, Tinelli A, Iannone A, Maffia M. The impact of proteomics in the understanding of the molecular basis of Paclitaxel-resistance in ovarian tumors. Curr Cancer Drug Targets. 2012;12:987–997. doi: 10.2174/156800912803251171. [DOI] [PubMed] [Google Scholar]
- 33.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 34.Soldevilla B, Rodríguez M, San Millán C, García V, Fernández-Periañez R, Gil-Calderón B, Martín P, García-Grande A, Silva J, Bonilla F, et al. Tumor-derived exosomes are enriched in ∆Np73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23:467–478. doi: 10.1093/hmg/ddt437. [DOI] [PubMed] [Google Scholar]
- 35.Zeng AL, Yan W, Liu YW, Wang Z, Hu Q, Nie E, Zhou X, Li R, Wang XF, Jiang T, et al. Tumour exosomes from cells harbouring PTPRZ1-MET fusion contribute to a malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene. 2017;36:5369–5381. doi: 10.1038/onc.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Yeung BZ, Cui M, Peer CJ, Lu Z, Figg WD, Guillaume Wientjes M, Woo S, Au JL. Exosome is a mechanism of inter-cellular drug transfer: Application of quantitative pharmacology. J Control Release. 2017;268:147–158. doi: 10.1016/j.jconrel.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Xu C, Hua Y, Sun L, Cheng K, Jia Z, Han Y, Dong J, Cui Y, Yang Z. Exosomes play an important role in the process of psoralen reverse multidrug resistance of breast cancer. J Exp Clin Cancer Res. 2016;35:186. doi: 10.1186/s13046-016-0468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220–230. doi: 10.1002/ijc.30669. [DOI] [PubMed] [Google Scholar]
- 39.Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and ‘exosomal shuttle microRNA’ in tumorigenesis and drug resistance. Cancer Lett. 2015;356B:339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 40.Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, Ying G, Ba Y. MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-β receptor 2. Cancer Sci. 2018;109:618–628. doi: 10.1111/cas.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37:33–44. doi: 10.1007/s10555-017-9724-7. [DOI] [PubMed] [Google Scholar]
- 42.Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez-Aguayo C, Fuentes-Mattei E, Xiao L, Vannini I, Redis RS, D’Abundo L, et al. Combining anti-mir-155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res. 2017;23:2891–2904. doi: 10.1158/1078-0432.CCR-16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Jin X, Meng H, Hu B, Zhang T, Yu J, Chen S, Guo X, Wang W, Jiang W, et al. Morin promotes prostate cancer cells chemosensitivity to paclitaxel through miR-155/GATA3 axis. Oncotarget. 2017;8:47849–47860. doi: 10.18632/oncotarget.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D, Lin X, Zhang C, Liu Z, Chen Z, Li Z, Wang J, Li B, Hu Y, Dong B, et al. Dual PI3K/mTOR inhibitor BEZ235 as a promising therapeutic strategy against paclitaxel-resistant gastric cancer via targeting PI3K/Akt/mTOR pathway. Cell Death Dis. 2018;9:123. doi: 10.1038/s41419-017-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, Baba H, Mori M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21:21. doi: 10.3390/molecules21070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Kong X, Lv L, Gao J. TGF-β1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015;359:288–298. doi: 10.1016/j.canlet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 51.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang ZS, Sun YZ, Wang SM, Ruan JS. Epithelial-mesenchymal transition: Potential regulator of ABC transporters in tumor progression. J Cancer. 2017;8:2319–2327. doi: 10.7150/jca.19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.