Abstract
Long non-coding (lnc)RNAs have been demonstrated to be involved in the development of various types of cancers, such as osteosarcoma (OS). Long non-coding (lnc) RNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) expression was reported to be highly expressed in OS and promoted the development of this disease; however, the underlying molecular mechanism by which MALAT1 promotes the progression of OS requires further investigation. In the present study, the expression of MALAT1 and miR-34a was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The abundance of cyclin D1 (CCND1) was detected by RT-qPCR and western blotting. Cell viability, migration and invasion were examined by MTT and Transwell assays. The interaction between miR-34a and MALAT1 or CCND1 was probed by a dual luciferase reporter assay and RNA immunoprecipitation. Xenograft tumor assay was performed to verify the roles of MALAT1 and miR-34a in tumor growth in vivo. The results demonstrated that MALAT1 and CCND1 mRNA expression levels were upregulated and miR-34a was downregulated in OS tissues and cells. Additionally, MALAT1 expression was correlated with tumor size, clinical stage and distant metastasis in patients with OS. In addition, MALAT1 promoted OS cell viability, invasion and migration, while MALAT1 silencing exhibited opposing effects. Moreover, MALAT1 functioned as a ceRNA to suppress miR-34a expression and in turn upregulate CCND1 in OS cells. Rescue experiments further demonstrated that MALAT1 knockdown partially reversed anti-miR-34a-mediated promotion on OS cell viability, migration and invasion; overexpression of CCND1 partially reversed the effects of MALAT1 silencing on OS progression. Furthermore, in vivo experiments also revealed that MALAT1 promoted OS tumor growth via miR-34a inhibition and upregulating the expression of CCND1. In conclusion, the present study suggested that MALAT1 exerted its oncogenic function in OS by regulating the miR-34a/CCND1 axis in OS, which may provide novel insight into the diagnosis and therapy for OS.
Keywords: metastasis associated lung adenocarcinoma transcript 1, microRNA‐34a, cyclin D1, viability, osteosarcoma
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor among children and adolescents between 10-25-years-old, and the second leading cause of cancer-mortality in pediatric age between 10-14-years-old (1). Despite great advances in therapeutic strategies for OS, including surgical resection combined with adjuvant chemotherapy or radiotherapy, the clinical prognosis and 5-year survival rate of patients with OS (65%) remains unsatisfactory due to high metastasis and recurrence (2). The pathogenesis of OS is an extremely compli-cated process, which has been associated with multifarious molecular changes (3). Therefore, an improved understanding of the pathogenesis of OS may aid the identification of novel diagnostic biomarkers and the development of effective therapeutic strategies for the treatment of OS.
Long non-coding RNAs (lncRNAs) are defined as evolutionarily conserved non-coding RNAs (ncRNAs) that are >200 nucleotides in length with no or little protein-coding capacity (4). An increasing number of studies have revealed that lncRNAs serve important regulatory roles in a variety of pathophysiological processes, including cell proliferation, cell cycle progression, apoptosis and carcinogenesis (5-7). It is well reported that lncRNAs are frequently dysregulated in numerous types of tumors, and act as oncogenes or tumor suppressors in the development of various cancers, such as OS (8,9). As one of the first reported cancer-associated lncRNAs (10), metastasis associated lung adenocarcinoma transcript 1 (MALAT1), located on chromosome 11q13, was highly expressed in metastasizing non-small cell lung cancer (11). Subsequently, research has revealed that MALAT1 was widely expressed in human tissues, including the heart, kidney, spleen and brain (12). Previously, accumulating evidence has demonstrated that MALAT1 serves diverse roles in the carcinogenesis of numerous types of tumors, including breast cancer, hepatocellular carcinoma and cervical cancer (13-15). In addition, several studies have reported that MALAT1 expression was increased in OS tissues and cells, and correlated with the poor prognosis of patients with OS. Furthermore, enhanced expression of MALAT1 promoted the progression of OS (16-18); however, the underlying molecular mechanism by which MALAT1 promotes OS progression requires further investigation.
MicroRNAs (miRNAs/miRs) are another class of highly conserved ncRNAs with a length of 18-25 nucleotides, and regulate gene expression by inducing the degradation or post-transcriptional inhibition of mRNAs (19). Specifically, dysregulated miRNAs have been closely associated with the development and progression of human cancers by affecting biological processes, including differentiation, proliferation, invasion and migration (20). Recently, increasing evidence has demonstrated that lncRNAs could function as competing endogenous RNAs (ceRNAs) to suppress the expression and activities of miRNAs, thus regulating the expression of target mRNA (21-23). As a well-studied miRNA, miR-34a, a member of the miR-34 family, has been reported as downregulated in OS cells and functioned as a tumor suppressor in the pathogenesis of OS (24,25). Considering the importance of MALAT1 and miR-34a in the pathogenesis of OS, the present study investigated whether MALAT1 functions as a molecular sponge of miR-34a in OS.
In the present study, it was reported that MALAT1 was upregulated in OS tissues and cells. Additionally, the expression of MALAT1 was associated with tumor size, clinical stage and distant metastasis in patients with OS; however, MALAT1 silencing suppressed the viability, invasion and migration in OS cells. Mechanistically, it was demonstrated that MALAT1 depletion-mediated suppression on OS progression was attributed to its function as a ceRNA of miR-34a and thus the regulation of cyclin D1 (CCND1) expression. The findings of the present study may contribute to the development of lncRNA-directed diagnosis and treatment of patients with OS.
Materials and methods
Human OS tissue collection
A total of 30 pairs of OS and adjacent normal tissue samples were surgically resected from patients at the Shangqiu First People’s Hospital (Shangqiu, China) between 2016 and 2017. All specimens were confirmed as OS samples via histological analysis following surgery. None of the patients with OS had received any preoperative treatments, including radiotherapy or chemotherapy prior to surgery. All tissues were immediately snap-frozen in liquid nitrogen following surgical removal and stored at -80°C until RNA extraction. The present study was approved by the Research Ethics Committee of Shangqiu First People’s Hospital and written informed consent was obtained from each patient. Patients with OS were classified into high (n=16) and low (n=14) MALAT1 expression groups according to the median expression levels of MALAT1 and the details of clinical characteristics of patients are presented in Table I.
Table I.
Association of MALAT1 expression with clinicopathological factors in osteosarcoma.
| Clinicopathological feature | n | MALAT1 expression
|
P-value | |
|---|---|---|---|---|
| Low [(n) %)] | High [(n) %] | |||
| Age | ||||
| <20 years | 16 | 7 (43.8) | 9 (56.2) | >0.05 |
| ≥20 years | 14 | 7 (50.0) | 7 (50.0) | |
| Sex | ||||
| Female | 13 | 5 (38.5) | 8 (61.5) | >0.05 |
| Male | 17 | 9 (52.9) | 8 (47.1) | |
| Tumor size | ||||
| >8 cm | 18 | 5 (27.8) | 13 (72.2) | <0.05 |
| ≤8 cm | 12 | 9 (75.0) | 3 (25.0) | |
| Clinical stage | ||||
| IIA | 8 | 6 (75.0) | 2 (25.0) | <0.05 |
| IIB/III | 22 | 8 (36.4) | 14 (63.6) | |
| Distant metastasis | ||||
| Absent | 20 | 12 (60.0) | 8 (40.0) | <0.05 |
| Present | 10 | 2 (20.0) | 8 (80.0) | |
MALAT1, metastasis associated lung adenocarcinoma transcript 1.
Cell culture and transfection
The OS cell lines (Saos-2, MG63 and SOSP-9607) and the normal human osteoblastic cell line, hFOB were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), and incubated at 37°C in a humidified atmosphere containing 5% CO2.
PcDNA-MALAT1 (MALAT1) with sites of restriction enzymes NheI and BamHI, pcDNA vector (Vector), small interfering (si)RNAs against MALAT1 (si-MALAT1) and pcDNA-CCND1 (CCND1) with sites of restriction enzymes NheI and BamHI, CCND1 (si-CCND1), an siRNA scrambled control (si-NC), miR-34a mimics (miR-34a), miRNA scram-bled control (miR-NC), miR-34a inhibitor (anti-miR-34a) and inhibitor control (anti-miR-NC) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: si-MALAT1, sense 5′-GATCCATAATCGGTTTCAAGG-3′, antisense, 5′-TTGAAACCGATTATGGATCAT-3′; si-CCND1, sense 5′-GGAGCAUUUUGAUACCAGATT-3′, antisense, 5′-UCUGGUAUCAAAAUGCUCCGG-3′; si-NC, sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; miR-34a mimics, sense 5′-UGGCAGUGUCUUAGCUGGUUGU-3′, antisense, 5′-AACCAGCUAAGACACUGCCAUU-3′; miR-NC, sense 5′-UUCUCCGAACGUGUCACGTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′; and anti-miR-34a 5′-ACAACCAGCUAAGACACUGCCA-3′. SOSP-9607 and Saos-2 cells exhibited greater variations in MALAT1 expression due to transfection, hence subsequent experiments were conducted in SOSP-9607 and Saos-2 cells. Transient transfection with the 40 nM aforementioned oligonucleotides or 1 µg plasmids into SOSP-9607 and Saos-2 cells was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were collected for further analyses at 48 h post-transfection and transfection efficacy was investigated by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) as described below.
Construction of MALAT1 lentiviral expression vector
The MALAT1 lentiviral vector was produced by Shanghai GenePharma Co., Ltd. The full-length of MALAT1 or miR-34a cDNA was synthesized and subcloned into pGLV/H1/green fluorescent protein lentiviral frame plasmids using the restriction enzymes XhoI and KpnI (Addgene, Inc., Cambridge, MA, USA). Following DNA sequencing, lentivirus containing MALAT1 or miR-34a and the packaging vectors (pCMV-VSVG, pMDLg/pRRE and pRSV-REV) were transduced into 293T cells (ATCC, multiplicity of infection=50) to generate the recombinant lentiviral vectors MALAT1 (lenti-MALAT1) and miR-34a (lenti-miR-34a). At 48 h following cotransduction, the lentivirus-containing supernatant was collected and purified with a 0.45-µm filter. Lentivirus was collected via ultracentrifugation for 60 min at 50,000 × g (4°C) and stored at −80°C until further use. Empty lentiviral vectors (lenti-NC) were used as the control. SOSP-9607 cells were inoculated in a 24-well plate and infected with the aforementioned recombinant vector-containing lentiviruses at a multiplicity of infection of 20 until 60% confluence was attained. The stably transfected cells were used for in vivo analysis.
RT-qPCR analysis
Total RNA from the tissue samples and cells was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The first strand cDNA was synthesized from 1 µg of total RNA using a PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer’s protocols. The expression levels of MALAT1 were determined using SYBR Green Real-Time PCR Master Mix (Roche Diagnostics, Basel, Switzerland) on an ABI 7500 PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), with GAPDH as an internal control. qPCR was performed to detect miR-34a expression using the TaqMan microRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) on an ABI 7500 PCR system and U6 small nuclear RNA was used as an endogenous control. Three replicates were involved in this experiment. RT-qPCR was conducted with the following amplification protocol: 95°C for 1 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. The relative gene expression was calculated using the 2−ΔΔCq quantification method (26). The primer sequences used were as follows: MALAT1 forward, 5′-AAAGCAAGGTCTCCCCACAAG-3′, reverse, 5′-GGTCTGTGCTAGATCAAAAGGCA-3′; GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′; miR-34a forward, 5′-GTGCAGGGTCCGAGGT-3′, reverse, 5′-GCCGCTGGCAGTGTCTTAGCTG-3′ and U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
MTT assay
Cell viability was assessed using an MTT assay. Following transfection, SOSP-9607 and Saos-2 cells were seeded into 96-well plates at a density of 3,000 cells/well and incubated at 37°C for 0, 24, 48 and 72 h. Then, 20 µl of 0.5 mg/ml MTT (Beyotime Institute of Biotechnology, Haimen, China) was added into each well and incubated for another 4 h at 37°C. Following removal of the supernatant, 150 µl dimethyl sulfoxide was administrated to cells to dissolve formazan. The relative absorbance at wavelength of 490 nm was measured with a microplate reader (Model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell invasion and migration assays
Cell invasion assay was performed using a Transwell chamber (8-µm; Corning Incorporated, Corning, NY, USA) containing a Matrigel-coated membrane (BD Bioscience, San Jose, CA, USA) and cell migration assay was conducted in a similar fashion without Matrigel. The transfected SOSP-9607 and Saos-2 cells (5×105 cells) in serum-free DMEM medium were seeded in the upper chambers, and the DMEM containing 10% FBS was added to the lower chambers as a chemoat-tractant. Following incubation at 37°C for 24h, the cells that did not migrate or invade on the upper chamber were removed with a cotton swab, while cells that invaded or migrated to the lower chamber were fixed with 100% methanol for 20 min and stained with 0.5% crystal violet at room temperature for 5 min. The number of invaded or migrated cells was counted under a light microscope (Olympus Corporation, Tokyo, Japan).
Luciferase reporter assay
The potential binding sites of miR-34a and 3′-untranslated region (UTR) of MALAT1 or CCND1 were predicted by bioinformatics software miRcode 11 (http://www.mircode.org/) or TargetScan Human Release 7.1 (http://www.targetscan.org/vert_71/). For the luciferase reporter assay, SOSP-9607 cells were seeded into 24-well plates at 3×104 cells/well and cotransfected with 50 nM miR-34a or miR-NC, 50 ng of recombinant luciferase pGL3 vectors (Promega Corporation, Madison, WI, USA) with sites of restriction enzymes NheI and XhoI containing the wild-type or mutant MALAT1 or 3′-UTR of CCND1 fragments and 2 ng pRL-TK (Promega Corporation) using Lipofectamine 2000. Cells were harvested at 48 h post-transfection and then the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation). Renilla lucif-erase activity was used for normalization.
RNA immunoprecipitation (RIP)
RIP was conducted using a Magna RNA-binding protein immunoprecipitation kit (EMD Millipore, Billerica, MA, USA). Briefly, after the centrifugation at 2,000 × g for 10 min at 4°C, the cell pellets of SOSP-9607 transfected with miR-34a mimics were collected and resuspended in NP-40 lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 1 mM PMSF (Sigma-Aldrich; Merck KGaA), 1 mM dithiothreitol (Invitrogen; Thermo Fisher Scientific., Inc.), 1% protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA), as well as 200 U/ml RNase inhibitor (Invitrogen; Thermo Fisher Scientific., Inc.). The supernatant from whole cell lysate was incubated with RIP buffer containing 0.5 M EDTA, 0.1% RNase inhibitor, A + G magnetic beads conjugated with human anti-Argonaute2 (Ago2) antibody (ab32381, 1:200; Abcam, Cambridge, MA, USA) and IgG (PP6421-K, 1:200; EMD Millipore), as well as a positive control (input). Following incubation overnight at 4°C, the magnetic beads were rinsed with cold NT2 buffer to remove the non-specific binding and then incubated with 10 mg/ml proteinase K (Sigma-Aldrich; Merck KGaA) at 55°C for 30 min. The co-precipitated RNAs were isolated and detected by RT-qPCR as aforementioned to demonstrate MALAT1 and miR-34a in the precipitates.
Western blotting
Total cell lysates were obtained from tissues and cells with radioimmunoprecipitation assay buffer solution (Beyotime institute of Biotechnology) containing 1% proteinase and phosphatase inhibitors (Sangon Biotech Co., Ltd., Shanghai, China). The protein concentration was determined using a Bicinchoninic Acid protein assay kit (Sigma-Aldrich; Merck KGaA). The supernatant (20 µg) from cell lysates was separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.). Following blocking with 5% non-fat milk for 1 h in Tris-buffer saline with 0.05% Tween-20 buffer at room temperature, the membranes were incubated overnight with primary antibody against CCND1 (ab134175, 1:10,000; Abcam) and β-actin (ab20272, 1:5,000; Abcam) at 4°C. Following washing, the membranes were incubated for 2 h with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The protein bands were detected and visualized using an electrochemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.) and analyzed by Image Lab software 5.2 (Bio-Rad Laboratories, Inc.). Three replicates were involved in this experiment.
Xenograft tumor growth assay
The animal study was approved by the Institutional Animal Care Committee of Shangqiu First People’s Hospital. A total of 24 female nude BALB/c mice (5-weeks-old, 15-18 g) were purchased from Henan Experimental Animals Centre (Zhengzhou, China) and maintained in an air content (10-15 air change/h), temperature (20-25°C) and humidity (50-60%) controlled environment with 12-h light/dark cycle and free access to food and water under specific pathogen-free conditions. SOSP-9607 cells (5×106) stably transduced with lenti-NC, lenti-MALAT1 or lenti-MALAT1 + lenti-miR-34a were subcutaneously inocu-lated into the posterior flank of nude mice. Tumor size was measured every 5 days starting from 5 days post-inoculation; tumor volumes were calculated by using the equation: Volume (mm3) = Length × width2/2. After 25 days, the mice were sacrificed, and the tumors were imaged and weighted. The xenograft tumors were excised and subjected to RT-qPCR analysis of MALAT1 and miR-34a expression and western blot analysis of CCND1 expression as aforementioned.
Statistical analysis
Data are presented as the mean ± standard deviation from three independent experiments. All statistical analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). The differences between two groups were evaluated by using a Student’s t-test or one-way analysis of variance followed by a Bonferroni post hoc test. Spearman’s correlation analysis was conducted on the abundances of lncRNA and miRNA or mRNA. P<0.05 was considered to indicate a statistically significant difference.
Results
MALAT1 is highly expressed in OS and correlates with clinical features
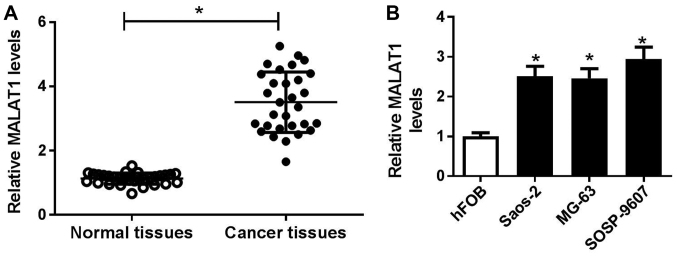
To address the function of MALAT1 in OS, the expression of MALAT1 in 30 pairs of OS and adjacent normal tissues were detected by RT-qPCR. As presented in Fig. 1A, MALAT1 expression was significantly higher in OS tissues than in adjacent normal tissues. To determine the association of MALAT1 expression with clinicopathologic features, including age, sex, tumor size, clinical stage and distant metastasis were analyzed in high (n=16) and low (n=14) MALAT1 expression groups. The results revealed that high MALAT1 expression levels were associated with tumor size, clinical stage and distant metastasis, respectively (P<0.05; Table I); however, the expression of MALAT1 was independent of age and sex (P>0.05). Additionally, the expression levels of MALAT1 were detected in OS cell lines (Saos-2, MG63 and SOSP-9607) and the normal human osteoblastic cell line hFOB. RT-qPCR analysis demonstrated that MALAT1 expression was significantly upregulated in OS cell lines compared with in hFOB cells (Fig. 1B), particularly in SOSP-9607 and Saos-2 cells. Therefore, SOSP-9607 and Saos-2 cells were selected for further experiments.
Figure 1.
Expression of MALAT1 in OS tissues and cells. (A) Expression of MALAT1 in OS tissues (n=30) and adjacent normal tissues (n=30) were analyzed via RT-qPCR. (B) Expression of MALAT1 in OS cell lines (Saos-2, MG63 and SOSP-9607) and the normal human osteoblastic cell line, hFOB, by RT-qPCR. *P<0.05 vs. normal tissues or hFOB cells. MALAT1, metastasis associated lung adenocarcinoma transcript 1; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
MALAT1 knockdown suppresses cell viability, migration and invasion in OS cells
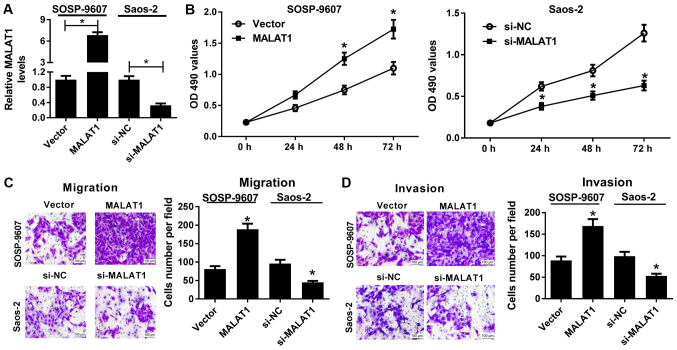
To confirm the association between MALAT1 and the development of OS, gain-of-function and loss-of-function approaches were performed in the present study. MALAT1 was stably overexpressed in SOSP-9607 cells via transfection of MALAT1 and downregulated in Saos-2 cells via transfection of si-MALAT1, as demonstrated by RT-qPCR (Fig. 2A). An MTT assay revealed that, compared with in the control, ectopic expression of MALAT1 significantly promoted the viability of SOSP-9607 cells, whereas MALAT1 downregulation significantly reduced the viability of Saos-2 cells (Fig. 2B). In addition, Transwell migration and invasion assays demonstrated that MALAT1 overexpression led to significant enhancements of the migration and invasive abilities of SOSP-9607 cells compared with the control. On the contrary, downregulation of MALAT1 resulted in a significant reduction in the migration and invasive abilities of Saos-2 cells compared with the control (Fig. 2C and D). Collectively, these findings indicated that MALAT1 knockdown delayed the progression of OS.
Figure 2.
Effects of MALAT1 on the viability, migration and invasion of osteosarcoma cells. SOSP-9607 cells were transfected with MALAT1 or Vector, and Saos-2 cells were transfected with si-MALAT1 or si-NC. (A) Reverse transcription-quantitative polymerase chain reaction was conducted to detect the expression of MALAT1 in the transfected SOSP-9607 and Saos-2 cells. (B) MTT assay was employed to assess cell viability at 0, 24, 48 and 72 h of the transfected SOSP-9607 and Saos-2 cells. (C and D) Transwell migration and invasion assays were conducted to evaluate the migration and invasive abilities in the transfected SOSP-9607 and Saos-2 cells. Scale bar, 100 µm. *P<0.05 vs. Vector or si-NC. MALAT1, metastasis associated lung adenocarcinoma transcript 1; NC, negative control; NS, not significant; si, small interfering RNA; Vector, empty vector control.
MALAT1 suppresses miR-34a expression via functioning as a ceRNA
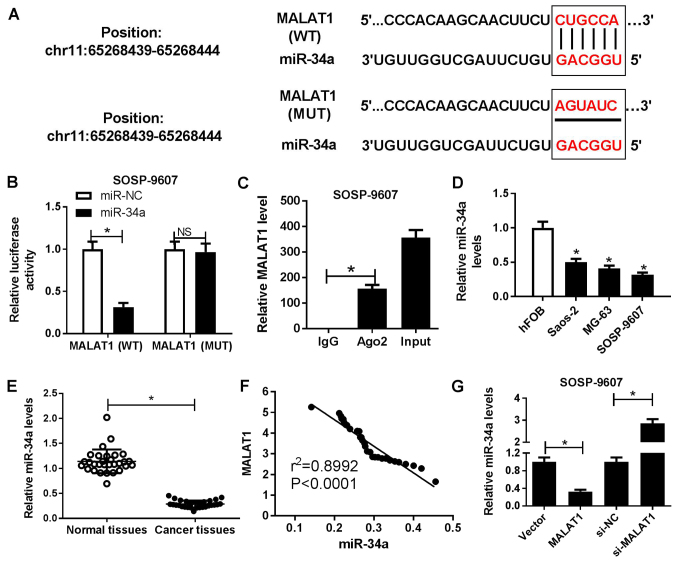
Bioinformatics software miRcode 11 was employed to predict the potential miRNA candidates and the results revealed that MALAT1 contained the binding sites of miR-34a (Fig. 3A). To confirm the interaction between MALAT1 and miR-34a, luciferase reporter and RIP assays were performed. The results of the luciferase reporter assay demonstrated that miR-34a mimics significantly reduced the luciferase activity of cells transfected with wild-type MALAT1 compared with in the control, but not within cells containing mutant MALAT1 (Fig. 3B). The RIP assay demonstrated that MALAT1 and miR-34a were preferentially enriched with Ago2-targeting beads compared with the IgG-treated group (Fig. 3C). miR-34a was observed to be significantly downregulated in OS cell lines (Saos-2, MG63 and SOSP-9607) compared with in hFOB cells (Fig. 3D). In addition, a significant decrease in miR-34a expression levels within OS tissues was observed compared with in corresponding adjacent normal tissues (Fig. 3E). Furthermore, Spearman’s correlation analysis suggested a negative correlation between MALAT1 and miR-34a in OS tissues (Fig. 3F). To further assess the potential association between MALAT1 and miR-34a, SOSP-9607 and Saos-2 cells were transfected with MALAT1, si-MALAT1 or corresponding controls, respectively. RT-qPCR analysis demonstrated that miR-34a expression levels were significantly decreased in MALAT1-overexpressed SOSP-9607 cells and significantly increased in si-MALAT1-transfected Saos-2 cells compared with in the corresponding controls (Fig. 3G). The results of the present study suggested that MALAT1 served as a molecular sponge of miR-34a to inhibit miR-34a expression in OS cells.
Figure 3.
Regulatory association between MALAT1 and miR-34a in OS cells. (A) Predicted binding sites between miR-34a and MALAT1, and the mutations in the MALAT1 sequence that disrupts the interaction between miR-34a and MALAT1. (B) Luciferase reporter assay was performed to detect the luciferase activity in SOSP-9607 cells following cotransfection with miR-34a or miR-NC, and the luciferase reporter plasmids containing the WT or MUT MALAT1 sequence. (C) Association between MALAT1 and miR-34a with Ago2 was determined by an RNA immunoprecipitation assay. MALAT1 and miR-34a expression levels in the immunoprecipitates were examined by RT-qPCR. (D) miR-34a expression OS cell lines (Saos-2, MG63 and SOSP-9607) and hFOB cells was estimated by RT-qPCR. (E) miR-34a expression levels in OS tissues and adjacent normal tissues were detected by RT-qPCR. (F) Correlation between MALAT1 and miR-34a expression in OS tissues. (G) Expression of miR-34a in SOSP-9607 or saos-2 cells transfected with MALAT1, si-MALAT1 or corresponding controls was measured by RT-qPCR. *P<0.05 vs. miR-NC, IgG, hFOB, normal tissues, Vector or si-NC. Ago2, Argonaute2; IgG, immunoglobulin G, MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; MUT, mutant; NC, negative control; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering RNA; Vector, empty vector control; WT, wild-type.
MALAT1 knockdown partially reverses anti-miR-34a-mediated promotion of OS cell viability, migration and invasion
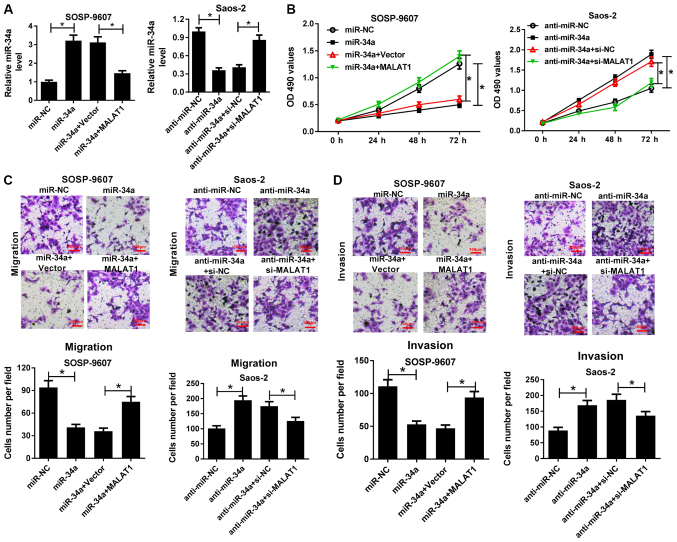
To gain insight into the mechanism by which MALAT1 regulates the progression of OS, SOSP-9607 cells were transfected with miR-34a or cotransfected with MALAT1, and Saos-2 cells transfected with anti-miR-34a or cotransfected with si-MALAT1 were employed to investigate the MALAT1-mediated effects of miR-34a on cell viability, migration and invasion. RT-qPCR demonstrated that miR-34a expression levels were significantly elevated following transfection with miR-34a mimics in SOSP-9607 cells, whereas overexpression of MALAT1 significantly suppressed the increase of miR-34a expression compared with in the corresponding control (Fig. 4A). On the contrary, anti-miR-34a significantly reduced miR-34a expression in Saos-2 cells compared with in the control, which was significantly reversed by MALAT1 knockdown (Fig. 4A). MTT assay demonstrated that miR-34a mimics significantly suppressed the viability of SOSP-9607 cells compared with in the control; overexpression of MALAT1 significantly reversed miR-34a-induced suppres-sion of cell viability. Conversely, anti-miR-34a significantly promoted the viability of Saos-2 cells compared with in the control, while MALAT1 silencing significantly abrogated the effects of anti-miR-34a on inducing cell viability (Fig. 4B). Furthermore, Transwell migration and invasion assays suggested that miR-34a overexpression significantly inhibited the migration and invasion of SOSP-9607 cells compared with in the control; however, exogenous MALAT1 significantly ameliorated the inhibitory effects of miR-34a on cell migration and invasion (Fig. 4C and D). Furthermore, MALAT1 down-regulation significantly attenuated anti-miR-34a-mediated promotion of migration and invasion of Saos-2 cells (Fig. 4C and D). These findings indicated that MALAT1 knockdown could reverse anti-miR-34a-mediated promotion of OS cell viability, migration and invasion.
Figure 4.
MALAT1 knockdown reversed anti-miR-34a-mediated promotion of OS cell viability, migration and invasion. SOSP-9607 cells were transfected with miR-34a or miR-NC, or cotransfected with MALAT1 or Vector, and Saos-2 cells were transfected with anti-miR-34a, anti-miR-NC, or cotransfected with si-MALAT1 or si-NC. (A) Expression of miR-34a in transfected SOSP-9607 and Saos-2 cells was determined by reverse transcription-quantitative polymerase chain reaction. (B) Cell viability at 0, 24, 48 or 72 h in transfected SOSP-9607 and Saos-2 cells was assessed via an MTT assay. (C and D) Migration and invasion of the transfected SOSP-9607 and Saos-2 cells were evaluated by Transwell migration and invasion assays. Scale bar, 100 µm. *P<0.05 vs. miR-NC, miR-34a + Vector, anti-miR-NC or anti-miR-34a + si-NC. Anti-miR, inhibitor; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control, si, small interfering RNA; Vector, empty vector control. anti-miR-NC, CCND1-WT, CCND1-WT + miR-34a + Vector, Vector, MALAT1 + miR-NC, si-NC or anti-miR-NC + si-MALAT1. Anti-cyclin D1; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control; si, small interfering RNA; UTR, region; Vector, empty vector control; WT, wild-type.
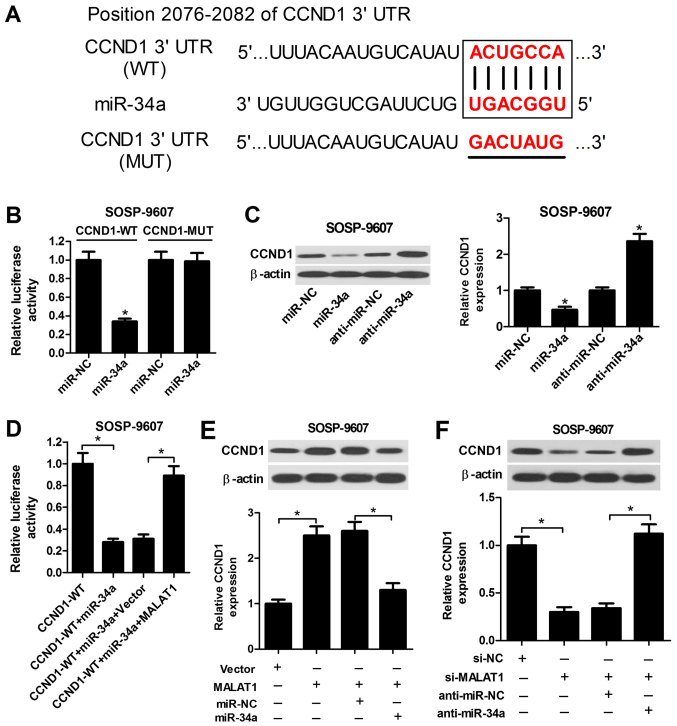
MALAT1 modulates CCND1 expression by targeting miR-34a
CCND1, a regulator of the cell cycle, which is involved in tumor progression, has been identified as a target of miR-34a in recent studies (27,28). Accordingly, to investigate whether miR-34a targets CCND1 in OS cells, the putative miR-34a recognition sequence in the 3′UTR of CCND1 was determined using TargetScan Human Release 7.1 (Fig. 5A). The subsequent luciferase reporter assay demonstrated that miR-34a significantly suppressed the luciferase activity of the wild-type reporter compared with in the control, while miR-34a did not notably affect the activity of the reporter containing the mutant sequence of the potential binding sites (Fig. 5B). miR-34a transfection led to a significant decrease in CCND1 expression levels, whereas transfection of anti-miR-34a induced a substantial increase in the expression levels of CCND1 in SOSP-9607 cells compared with in the control (Fig. 5C); MALAT1 overexpression significantly reversed the suppressive effects of miR-34a on the luciferase activity of wild-type reporter compared with in the corresponding control (Fig. 5D). Furthermore, overexpression of MALAT1 significantly enhanced the protein expression levels of CCND1 in SOSP-9607 cells, while restoration of miR-34a expression inhibited the increase in CCND1 expression levels mediated by MALAT1 overexpression (Fig. 5E). Conversely, si-MALAT1-transfected SOSP-9607 cells exhibited a significant decrease in CCND1 expression levels compared with in the control, while cotransfection with anti-miR-34a significantly abrogated this effect (Fig. 5F). Therefore, these results suggested that MALAT1 acts as an endogenous sponge of miR-34a to regulate CCND1 expression in OS cells.
Figure 5.
MALAT1 suppresses CCND1 expression by acting as a sponge of miR‐34a. (A) Predicted miR‐34a binding sites in the 3′UTR region of CCND1 and the corresponding mutant sequence. (B) Luciferase activity was measured by a luciferase reporter assay in SOSP‐9607 cells following cotransfection with CCND1 (WT) or CCND1 (MUT), and miR‐34a or miR‐NC. (C) Western blot analysis of CCND1 expression levels in SOSP‐9607 cells transfected with miR‐34a, anti‐miR‐34a or corresponding controls. (D) Luciferase activity was determined via a luciferase reporter assay in SOSP‐9607 cells following transfection with CCND1 (WT) and miR‐34a, miR‐34a + Vector, or miR‐34a + MALAT1. (E) Western blotting was performed to examine the protein expression levels of CCND1 in SOSP‐9607 cells transfected with MALAT1 or Vector, or combined with miR‐34a or miR‐NC. (F) Western blotting was conducted to detect the protein expression levels of CCND1 in SOSP‐9607 cells transfected with si‐MALAT1 or si‐NC, or with anti‐miR‐34a or anti‐miR‐NC. *P<0.05 vs. miR‐NC, anti‐miR‐NC, CCND1‐WT, CCND1‐WT + miR‐34a + Vector, Vector, MALAT1 + miR‐NC, si‐NC or anti‐miR‐NC + si‐MALAT1. Anti‐miR, inhibitor; CCND1, cyclin D1; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control; si, small interfering RNA; UTR, untranslated region; Vector, empty vector control; WT, wild‐type.
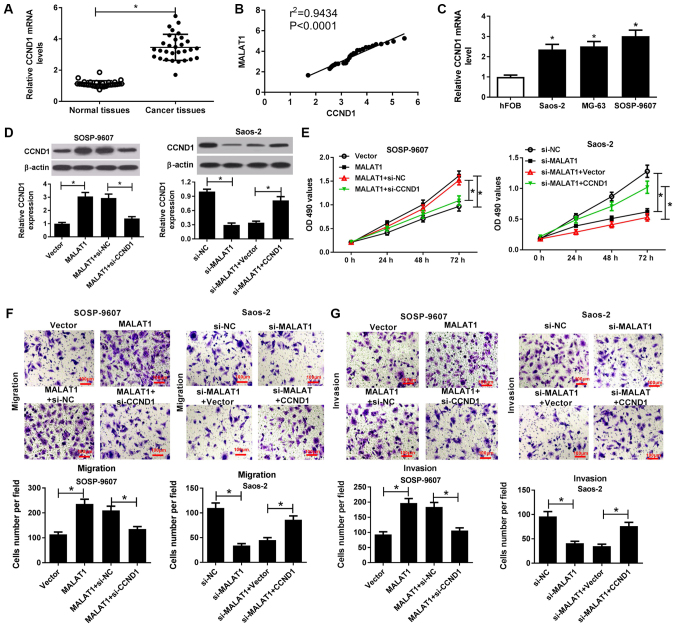
MALAT1 knockdown suppresses the viability, migration and invasion of OS cells by downregulating CCND1
RT-qPCR analysis demonstrated that CCND1 mRNA expression was significantly higher in OS tissues than in normal tissues (Fig. 6A) and positively correlated with MALAT1 expression in OS tissues (Fig. 6B). In addition, the present study reported that CCND1 mRNA expression was significantly increased within OS cells (Saos-2, MG63 and SOSP-9607) compared with in hFOB cells (Fig. 6C). To further investigate the combined effects of MALAT1 and CCND1 on the progression of OS, rescue experiments with MALAT1-transfected SOSP-9607 cells and si-MALAT1-transfected Saos-2 cells were conducted via CCND1 knockdown or overexpression, respectively. Western blotting demonstrated that overexpression of MALAT1 significantly promoted CCND1 expression in SOSP-9607 cells, while CCND1 silencing significantly reduced MALAT1 overexpression-mediated increased CCND1 expression compared with in the corresponding control (Fig. 6D). Conversely, MALAT1 silencing significantly suppressed CCND1 expression in Saos-2 cells compared with in the control, which was reversed by exogenous CCND1 expression (Fig. 6D). Subsequently, an MTT assay revealed that CCND1 downregulation significantly reduced the promotion of viability induced by MALAT1 overexpression in SOSP-9607 cells; however, increased CCND1 expression significantly reversed the inhibition of cell viability mediated by MALAT1 downregulation compared with in the corresponding control (Fig. 6E). Furthermore, Transwell migration and invasion assays indicated that MALAT1 overexpression-induced cell migration and invasion were significantly mitigated following CCND1 silencing in SOSP-9607 cells. On the contrary, MALAT1 silencing-induced inhibition of migration and invasion were significantly ameliorated by CCND1 overexpression in Saos-2 cells compared with in the control (Fig. 6F and G). Therefore, these results indicated that MALAT1 knockdown suppressed cell viability, migration and invasion in OS cells by downregulation of CCND1 expression.
Figure 6.
MALAT1 knockdown suppresses the viability, migration and invasion of OS cells by downregulation CCND1. (A) CCND1 mRNA expression in OS tissues and normal tissues was detected by RT-qPCR. (B) Correlation between MALAT1 and CCND1 expression in OS tissues. (C) CCND1 mRNA expression in OS cells (Saos-2, MG63 and SOSP-9607) and hFOB cells was evaluated by RT-qPCR. SOSP-9607 cells were transfected with MALAT1, Vector, MALAT1 + si-NC, or MALAT1 + si-CCND1; Saos-2 cells were transfected with si-MALAT1, si-NC, si-MALAT1 + CCND1 or si-MALAT1 + Vector, and subjected to analysis at 48 h post-transfection. (D) Protein expression levels of CCND1 in transfected SOSP-9607 and Saos-2 cells were detected by western blotting. (E) Viability of the transfected SOSP-9607 and Saos-2 cells was assessed by an MTT assay. (F and G) Transwell migration and invasion assays were conducted to evaluate the migration and invasive ability of transfected SOSP-9607 and Saos-2 cells. Scale bar, 100 µm. *P<0.05 vs. normal tissues, hFOB, Vector, si-NC, MALAT1 + si-NC, si-MALAT1 + Vector. CCND1, cyclin D1; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si,small interfering RNA; Vector, empty vector control.
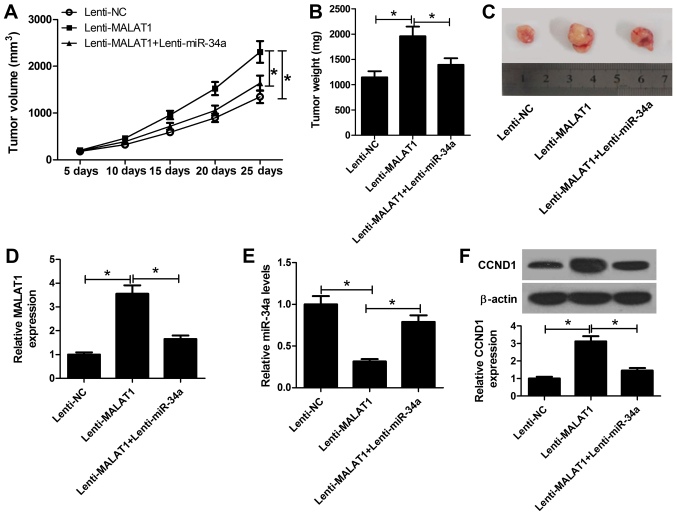
MALAT1 promotes OS tumor growth in vivo by inhibiting miR-34a and upregulating CCND1
To validate whether MALAT1 affects OS tumorigenesis in vivo, SOSP-9607 cells stably transfected with lenti-NC, lenti-MALAT1, or lenti-MALAT1 + lenti-miR-34a were subcutaneously inoculated into nude mice. As presented in Fig. 7A, tumor growth was significantly promoted via exogenous overexpression of MALAT1 compared with in the control; however, overexpression of miR-34a significantly suppressed the effects of MALAT1 on tumor growth. Additionally, tumor weight in the MALAT1-overexpressing group was significantly higher compared with in the control group, which was significantly reduced in the MALAT1 and miR-34a overexpression group (Fig. 7B and C). Then, RT-qPCR analyses of MALAT1 and miR-34a expression in the excised tumors were performed. The results indicated that MALAT1 expression was significantly upregulated in the lenti-MALAT1 group, but decreased following the transduction of lenti-miR-34a (Fig. 7D). In addition, ectopic expression of MALAT1 led to a significant decrease in miR-34a expression within xenograft tumor tissues, which was reversed via the overexpression of miR-34a (Fig. 7E). Additionally, western blot analysis revealed that the expression levels of CCND1 in the lenti-MALAT1 group were significantly increased compared with in the control; however, a significant reduction in the lenti-MALAT1 + enti-miR-34a group was also observed (Fig. 7F). Collectively, the results of the present study indicated that MALAT1 promoted OS tumor growth in vivo via the miR-34a/CCND1 axis.
Figure 7.
MALAT1 promotes OS tumor growth in vivo via miR-34a inhibition and upregulating CCND1. A xenograft tumor model was established by subcutaneously injecting SOSP-9607 cells stably transfected with lenti-NC, lenti-MALAT1 or lenti-MALAT1 + lenti-miR-34a into nude mice. (A) Tumor size was measured every 5 days starting from 5 days of injection. (B and C) After 25 days, the mice were sacrificed and tumors were imaged and weighted. RT-qPCR analyses of (D) MALAT1 and (E) miR-34a in the excised tumor masses. (F) Protein expression levels of CCND1 in the excised tumor masses were detected by western blotting. *P<0.05 vs. lenti-NC, lenti-MALAT1. CCND1, cyclin D1; lenti, lentiviral vector; MALAT1, metastasis associated lung adenocarcinoma transcript 1; miR, microRNA; NC, negative control.
Discussion
Numerous studies have suggested that alterations in the expression of certain lncRNAs serve crucial roles the tumorigenesis and progression of several cancer types, such as OS (6,7,29). For instance, upregulated lncRNA Hox transcript antisense intergenic RNA promoted the viability, invasion and migration of OS cells via the activation of the protein kinase B (Akt)/mechanistic target of rapamycin pathway (30). LncRNA colorectal neoplasia differentially exerted its oncogenic role in OS cells via enhancing the activity of Notch1 signaling and promoting epithelial-mesenchymal transition (31). LncRNA forkhead box F1 adjacent non-coding developmental regulatory RNA reversed doxorubicin resistance and suppressed the progression of OS cells by downregulating ATP binding cassette subfamily B member 1 and ATP binding cassette subfamily C member 1 (32); the present study investigated the role of MALAT1.
In recent years, accumulating evidence has suggested a positive association between MALAT1 and the progres-sion of OS (33). For instance, MALAT1 silencing delayed the progression of OS by altering the expression and localization of β-catenin (34). Additionally, high serum levels of MALAT1 predicted poor survival in OS patients; overexpression of MALAT1 promoted cell metastasis and decreased E-cadherin levels by associating with enhancer of zeste homolog 2 (35). Furthermore, MALAT1 was proposed as an oncogenic lncRNA that promoted tumor growth and metastasis of OS via activation of the phosphoinositide 3-kinase/Akt signaling pathway (36). Therefore, MALAT1 may be a promising therapeutic target for the treatment of patients with OS (37). In the present study, the oncogenic role of MALAT1 was also reported in OS as MALAT1 was highly expressed in OS tissues and cells. In addition, the correlation between MALAT1 expression and the clinicopathological features of patients with OS were reported. This was in accordance with recent findings, which suggested MALAT1 as a predictor of poor survival, and was associated with tumor size, stage and metastasis in patients with OS (16). Additionally, ectopic expression of MALAT1 enhanced the progression of OS by promoting cell viability, migration and invasion, while MALAT1 silencing elicited opposing effects, which was consistent with previous studies (17).
In recent years, compelling evidence has suggested that lncRNAs act as ceRNAs or a molecular sponge to antagonize the expression and functions of miRNAs, modulating the derepression of these miRNAs targets via post-transcriptional regulation (38). The crosstalk between lncRNAs and miRNAs has been associated with the pathogenesis of human diseases, such as cancer (39). For instance, enhanced MALAT1 expression levels served as a predictor of unfavorable outcomes in OS and promoted OS progression via miR-205 suppression and activating mothers against decapentaplegic homolog 4 function (16). LncRNA X-inactive specific transcript inhibited OS cell growth and mobility by competitively binding to miR-21-5p and upregulating programmed cell death 4 (40). To determine the underlying mechanism by which MALAT1 promotes the progression of OS, a functional role of MALAT1 as a miRNA decoy for miR-34a was determined; MALAT1 suppressed miR-34a expression in OS cells in the present study. Conversely, a previous study revealed that miR-34a may be independent of MALAT1 by an RNA pulldown assay (41). This may be due to the presence of surfactant and the dilution of miR-34a by addition of cell lysate buffer, which may induce false negative results during immunoprecipitation. miR-34a has been proposed as a direct transcriptional target of the tumor suppressor p53 (42). The inactivating mutations of p53 often induce reductions in the expression of miR-34a within tumors (43). miR-34a has been frequently reported as a tumor suppressor with lost or reduced expression in various types of tumors (44). In addition, miR-34a was regarded as a novel prognostic biomarker and correlated with tumor size and stage in OS (45). The present study reported that miR-34a was downregulated in OS tissues and cells, in accordance with previous studies (24,25). Additionally, MALAT1 was negatively correlated with miR-34a expression in OS cells. Rescue experiments further revealed that MALAT1 knockdown partially reversed anti-miR-34a-mediated increases in OS cell viability, migration and invasion, suggesting that MALAT1 exerted its oncogenic role in OS by functioning as a sponge of miR-34a.
CCND1, one of the highly conserved members of the cyclin family, is well-known for its role in the response to the mitogenic signals that regulate of the G1 phase of the cell cycle (46). CCND1 has been notably associated with tumorigenesis and metastasis in clinical studies and in vivo experiments (46-48). The overexpression of CCND1 is frequently observed in a variety of tumors and is thus regarded as an oncogene in numerous human tumors, including breast, colon and prostate cancers (49,50). CCND1 was also reported to be upregulated in OS and contributed to the progression of OS (51). Interestingly, previous studies have demonstrated that miR-34a could directly target the 3′UTR of CCND3 in laryngeal carcinoma cells (27), breast cancer (28), and lung cancer (52). Similarly, it was demonstrated that CCND1 was identified as a target of miR-34a and that miR-34a suppressed CCND1 expression in OS cells in the present study. Furthermore, MALAT1 was proposed to function as a sponge of miR-34a to positively regulate CCND1 expression in OS cells. Mechanistic analyses demonstrated that overexpression of CCND1 partially reversed the effects of MALAT1 silencing on OS cell viability, invasion and migration. In addition, in vivo experiments demonstrated that MALAT1 promoted OS tumor growth via miR-34a inhibition. Therefore, the present study proposed that MALAT1 exerted its oncogenic role in OS in vitro and in vivo by regulating the miR-34a/CCND1 axis.
In conclusion, it was demonstrated that MALAT1 was upregulated in OS and associated with the clinicopathological features of OS. Knockdown of MALAT1 suppressed the progression of OS by stimulating miR-34a expression and in turn inhibiting CCND1 expression. To the best of our knowl-edge, the present study is the first to report of the crosstalk between MALAT1, miR-34a and CCND1, providing novel insight into the diagnosis and therapy of OS.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors’ contributions
GD contributed to the experimental design, most experiments and analysis. GD also drafted the manuscript and collected clinical samples. CZ, ChX and CX conducted the experiments and analyzed the data. LZ and YZ were conducted in vivo experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Shangqiu First People’s Hospital and written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cortini M, Avnet S, Baldini N. Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. doi: 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Xiao X, Wang W, Wang Z. The role of chemotherapy for metastatic, relapsed and refractory osteosarcoma. Paediatr Drugs. 2014;16:503–512. doi: 10.1007/s40272-014-0095-z. [DOI] [PubMed] [Google Scholar]
- 3.Maire G, Martin JW, Yoshimoto M, Chilton-MacNeill S, Zielenska M, Squire JA. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 2011;204:138–146. doi: 10.1016/j.cancergen.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: The dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutschner T, Hämmerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 11.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 12.Brauze D, Mikstacka R, Pelkonen O. Monoclonal antibody characterization of NADH- and NADPH-dependent hydroxylation of benzo(a)pyrene in liver microsomes from 5,6-benzoflavone-induced C57Bl/6 mice. Acta Biochim Pol. 1990;37:219–225. [PubMed] [Google Scholar]
- 13.Huang NS, Chi YY, Xue JY, Liu MY, Huang S, Mo M, Zhou SL, Wu J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7:37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016;107:149–154. doi: 10.1111/cas.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metas-tasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Pan X, Wang X, Jiao X, Zheng J, Li Z, Huo Y. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget. 2017;8:106648–106660. doi: 10.18632/oncotarget.20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, He H, Xiao W, Liu Q, Deng Z, Lu Y, Wang Q, Zheng Q, Li Y. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7:54733–54743. doi: 10.18632/oncotarget.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, Li Z, Zhang Z, Chang Y, Xia K, et al. MicroRNAs, long noncoding RNAs, and circular RNAs: Potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249–2257. doi: 10.2147/CMAR.S166308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang Y, Wei X, Xue L, Wen F, Gu J, Zheng H. Long non-coding RNA in glioma: Target miRNA and signaling pathways. Clin Lab. 2018;64:887–894. doi: 10.7754/Clin.Lab.2018.180107. [DOI] [PubMed] [Google Scholar]
- 24.Gang L, Qun L, Liu WD, Li YS, Xu YZ, Yuan DT. MicroRNA-34a promotes cell cycle arrest and apoptosis and suppresses cell adhesion by targeting DUSP1 in osteosarcoma. Am J Transl Res. 2017;9:5388–5399. [PMC free article] [PubMed] [Google Scholar]
- 25.Wen J, Zhao YK, Liu Y, Zhao JF. MicroRNA-34a inhibits tumor invasion and metastasis in osteosarcoma partly by effecting C-IAP2 and Bcl-2. Tumour Biol. 2017;39:1010428317705761. doi: 10.1177/1010428317705761. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ye J, Li L, Feng P, Wan J, Li J. Downregulation of miR-34a contributes to the proliferation and migration of laryngeal carcinoma cells by targeting cyclin D1. Oncol Rep. 2016;36:390–398. doi: 10.3892/or.2016.4823. [DOI] [PubMed] [Google Scholar]
- 28.Li ZH, Weng X, Xiong QY, Tu JH, Xiao A, Qiu W, Gong Y, Hu EW, Huang S, Cao YL. miR-34a expression in human breast cancer is associated with drug resistance. Oncotarget. 2017;8:106270–106282. doi: 10.18632/oncotarget.22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: Biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. doi: 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 30.Li E, Zhao Z, Ma B, Zhang J. Long noncoding RNA HOTAIR promotes the proliferation and metastasis of osteosarcoma cells through the AKT/mTOR signaling pathway. Exp Ther Med. 2017;14:5321–5328. doi: 10.3892/etm.2017.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Tang Y, Xing W, Dong W, Wang Z. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp Mol Pathol. 2018;104:19–25. doi: 10.1016/j.yexmp.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Kun-Peng Z, Xiao-Long M, Chun-Lin Z. LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through downregulating ABCB1 and ABCC1. Oncotarget. 2017;8:71881–71893. doi: 10.18632/oncotarget.17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20:3561–3565. [PubMed] [Google Scholar]
- 34.Zhang ZC, Tang C, Dong Y, Zhang J, Yuan T, Li XL. Targeting LncRNA-MALAT1 suppresses the progression of osteosarcoma by altering the expression and localization of β-catenin. J Cancer. 2018;9:71–80. doi: 10.7150/jca.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z, Pan X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8:46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 37.Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2016;34:932–941. doi: 10.1002/jor.23105. [DOI] [PubMed] [Google Scholar]
- 38.Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: The key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014;896206 doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 18592016:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51:1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 44.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, et al. Mir-34: A new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Jia LS, Yuan W, Wu Z, Wang HB, Xu T, Sun JC, Cheng KF, Shi JG. Low miR-34a and miR-192 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Am J Transl Res. 2015;7:111–119. [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Wang C, Prendergast GC, Pestell RG. Cyclin D1 functions in cell migration. Cell Cycle. 2006;5:2440–2442. doi: 10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 47.Ramos-García P, González-Moles MA, González-Ruiz L, Ruiz-Ávila I, Ayén Á, Gil-Montoya JA. Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2018;83:96–106. doi: 10.1016/j.oraloncology.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Luo J, Xu LN, Zhang SJ, Jiang YG, Zhuo DX, Wu LH, Jiang X, Huang Y. Downregulation of LncRNA-RP11-317J10.2 promotes cell proliferation and invasion and predicts poor prognosis in colorectal cancer. Scand J Gastroenterol. 2018;53:38–45. doi: 10.1080/00365521.2017.1392597. [DOI] [PubMed] [Google Scholar]
- 49.Inoue K, Fry EA. Aberrant expression of cyclin D1 in cancer. Signal Transduct Insights. 2015;4:1–13. doi: 10.4137/STI.S30306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju X, Casimiro MC, Gormley M, Meng H, Jiao X, Katiyar S, Crosariol M, Chen K, Wang M, Quong AA, et al. Identification of a cyclin D1 network in prostate cancer that antagonizes epithelial-mesenchymal restraint. Cancer Res. 2014;74:508–519. doi: 10.1158/0008-5472.CAN-13-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai CK, Zhao GY, Tian LY, Liu L, Yan K, Ma YL, Ji ZW, Li XX, Han K, Gao J, et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep. 2012;28:1764–1770. doi: 10.3892/or.2012.1995. [DOI] [PubMed] [Google Scholar]
- 52.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.