Abstract
Background and aim
Tripartite motif containing (TRIM) family protein has been involved in multiple pathogenesis of cancers. TRIM2 is a member of the family, and its role in clear cell renal cell carcinoma (ccRCC) remains to be unclarifid. Here, we showed the clinical value and biological role of TRIM2 in ccRCC.
Methods
ROC curves analyzed the clinicopathological parameters, Kaplan-Meier survival analysis determined the correlation of OS and DFS time, multivariate analysis demonstrated the prognostic indicator in overall survival and disease-free survival of ccRCC with TRIM2 expression in The Cancer Genome Atlas Kidney Clear Cell Carcinoma (TCGA-KIRC) database. Western blotting and immunohistochemistry were used to check the level of TRIM2 expression. Gain-of-function assay by exogenous overexpression of TRIM2 studied the biological role of TRIM2 in renal cell carcinoma cells.
Results
TRIM2 expression was associated with various clinicopathologicalfactors and lower TRIM2 expression was interrelated to a poor prognosis. The levels of TRIM2 expression were also scanty in ccRCC tissues and renal cancer cell lines than in normal control. The biological role of TRIM2 in ccRCC was identifid by bioinformatics analysis and functional analysis. Exogenous overexpression of TRIM2 with the gain-of-function assay in renal cell carcinoma cells showed that the cell proliferation, migration, and invasion were signifiantly suppressed.
Conclusion
These results showed that TRIM2 acted as an antitumor gene and a specifi prognostic indicator for patients with ccRCC, which indicated that positive modulation of TRIM2 might be a novel treatment strategy for ccRCC.
Keywords: TRIM2, ccRCC, tumor suppressor, prognostic indicator
Introduction
Renal cell carcinoma (RCC), which accounts for 3.79% in all adult malignancies, is the most common and lethal adult kidney malignancy.1 There are 65,340 new cases and 14,970 deaths estimated for 2018 in USA.2 Clear cell renal cell carcinoma (ccRCC) is the highest mortality, invasion, and metastasis subtype of RCC and develops easily into resistance to traditional radiotherapy and chemotherapy.3 Recently, 90.7% of RCC had been ccRCC, which was reported in a Chinese study.4 Progression has been made to manage the patients for different diagnoses and treatments of ccRCC. Cancer metastasis and invasion lead to 90% of patient deaths, and ~30% of patients have been diagnosed with local invasion or distant metastasis, resulting in a poor prognosis.5,6 Standard surgical resection is normally used for localized tumor treatment, but the outcome varies greatly for different progressions.7 Similarly, patients with local-regional tumor still have a high risk for tumor recurrence after nephrectomy.8 Patients with metastatic ccRCC benefit from chemotherapy drugs based on the tyrosine kinase inhibitor (TKI), which is a specific molecular target drugs.9,10 Unfortunately, because of intrinsic resistance or acquired resistance, TKI-treated ccRCC patients still develop poor clinical progression.11 It is essential to explore more effective prognostic molecular biomarkers and its molecular mechanisms underlying tumor progression and metastasis to evaluate and manage patients with ccRCC.
In recent years, the novel tumor promoters or tumor suppressors’ role of tripartite motif containing (TRIM) proteins in various cancers has attracted much attention to different tumor researchers.12,13 TRIM protein has been widely reported to have an extensive range role in biological processes of autophagy, inflammation, immunity, and cancer.14 NHL is the subgroup of TRIM protein, which contains four mammalian paralogs (TRIM2, TRIM3, TRIM32, and TRIM71), and TRIM3 acts as tumor suppressor, can block tumor growth by preventing cyclin D1-cdk4 accumulation through sequestering p21 in glioblastomas, and also can regulate stem cell factors and epithelial–mesenchymal transition (EMT) regulators in gastric cancer.15,16 TRIM2 has been verified with weird pathogenesis in multiple cancers. As a binding partner protein of TRIM2, TRIM8 is downregulated in glioma and affects cancer cell proliferation and associated with patients’ survival.17 Recently, TRIM8, which can stabilize p53, has been considered as a new p53 modulator and has an antip-roliferative role in chemo-resistant RCC, but the expression and the role of TRIM2 in ccRCC still remain to be unknown.18
In this study, we investigated the expression of TRIM2 with clinicopathological features and patient’s survival in an integrate investigation system including The Cancer Genome Atlas (TCGA) database, clinical ccRCC tissues, gene set enrichment analysis (GSEA), and the biological function in cellular models. Our study demonstrated that TRIM2 acted as an antitumor gene and low TRIM2 expression predicts poor progression.
Patients and methods
Patient samples
Surgical specimens (paired human ccRCC tissues and adjacent noncancerous tissues) were obtained from 48 patients between 2017 and 2018 in the Department of Urology, The First Affiliated Hospital of Xiamen University. Resected tissues were first frozen into liquid nitrogen and then stored at –80°C for further experiment. Informed consent was obtained from patients, the procedures of experiment and study were approved by the Institutional Review board of Xiamen University, and all patients provided written informed consent, in accordance with the Declaration of Helsinki.
RNA extraction and qRT-PCR
Tissue RNA was extracted according to the instructions of manufacturer with the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). The purity and concentration of the RNA solution were tested by NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). qPCR analysis was performed according to the instructions of manufacturer (LightCycler 480II; Hoffman-La Roche Ltd., Basel, Switzerland) with SYBR Green mix (Thermo Fisher Scientific). Relative expression of TRIM2 was calculated by 2−ΔCt (ΔCt = CtTRIM2 – CtGAPDH). Primers were purchased from GENEWIZ (Suzhou, China): TRIM2, (forward) 5′-TGGAGAAGGAAATGGGCATG-3′ and (reverse) 5′-CTGCAACCACAACATGCACCA-3′; GAPDH, (forward) 5′-GAGTCAACGGATTTGGTCGT-3′ and (reverse) 5′-GACAAGCTTCCCGTTCTCAG-3′.
Immunohistochemistry (IHC)
Adjacent normal tissues and ccRCC tissues were fixed in formalin, dehydrated, embedded, and then incubated with primary rabbit TRIM2 polyclonal antibody (1:100; A14394; ABclonal Biotech Co., Ltd., Wuhan, China) at 4°C overnight. The sections were washed three times with PBS and, then, incubated with goat antirabbit secondary antibody (1:200; GB23303; Servicebio, Inc., Woburn, MA, USA) for 2 hours at room temperature.
Cell culture and transient transfection
The renal cancer cell lines of human, such as HK2, 786-O, ACHN, and OSRC-2, were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Cancer cells were cultivated in high-glucose DMEM (Wuhan Boster Biological Technology, Ltd., Wuhan, China), which contained 10% FBS (Thermo Fisher Scientific) at 37°C in a 5% CO2 incubator, as described in the previous study.19 The plasmid vectors expressing TRIM2 (TRIM2) or negative control (NC) were constructed by GeneChem (Shanghai, China). A total of 3 µg of TRIM2 or NC plasmid were transfected in each six-well plates with the Lipofectamine® 2000 reagent (Thermo Fisher Scientific) according to the recommendations of manufacturer.
Cell proliferation analysis
786-O and ACHN cells were first transfected with TRIM2 or NC, and then, the cells were added to a 96-well plate at the density of 3×103/well. Cell proliferation rate (OD value) was detected by cell counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc, Rockville, MD, USA) according to the protocol of manufacturer. After adding each well with 10 µL of CCK-8 solution 2 hours, the OD value was measured at 450 nm. Three independent experiments were conducted in each experiment.
Migratory and invasion assays
Migratory assays were performed as previously described.20 The cells were homogenizated withdrawing of plasma for 24 hours and, then, cultured 1×105 cells/well in 24-well transwell plates with polycarbonate membrane inserts (Corning Incorporated, Corning, NY, USA). In the invasion assays, the membrane was coated with matrigel (Thermo Fisher Scientific) first and cultured with 2×105 cells/well. Incubating for 24 hours, the cells were fixed with 100% methanol, then stained with 0.05% crystal violet, and counted five random fields. Three independent experiments were conducted in each experiment.
Western blotting
Tissues and cells were pyrolysised in protein lysis system containing RIPA (Wuhan Boster Biological Technology, Ltd.), protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA), and phenylmethylsulfonyl fluoride (PMSF) (Wuhan Boster Biological Technology, Ltd.). Concentrations of protein were measured using the bicinchoninic acid kit (Beyotime Institute of Biotechnology, Haimen, China) at 562 nm. A total of 30 µg of proteins were subjected to SDS-PAGE and, then, separated and transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA) for 90 minutes. After transferred to PVDF membranes, the proteins were blocked in PBS with 5% nonfat milk for 1 hour and then incubated with antibodies against TRIM2 (1:1,000; A14394; ABclonal Biotech Co., Ltd.) and GAPDH (1:2,000; BM3876; Wuhan Boster Biological Technology, Ltd.) at 4°C overnight. The membranes were washed and incubated with secondary antibodies (1:5,000; BA1020; Wuhan Boster Biological Technology, Ltd.) on the next day at room temperature for 2 hours. Finally, the membranes were washed and, then, detected by ChemiDoc-XRS+ (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Bioinformatics analysis
The RNA-seq data of ccRCC patients (TCGA-KIRC) and the clinical information of patients were obtained from the Xena Functional Genomics Explorer (https://xenabrowser.net/heatmap/) of the University of California Santa Cruz.21 GSEA was used to understand the TRIM2 pathway involved in the ccRCC pathogenesis of TCGA-KIRC (http://www.broadinstitute.org/gsea).22 Nominal P<0.05 and false discovery rate <25%, which was following the performance of 1,000 permutations, had considered to be significantly enriched for enriched gene sets analysis.
Statistical analyses
The RNA result data of paired samples were analyzed by paired sample t-test, and unpaired samples were analyzed by one-way ANOVA or t-test. Area under the curve (AUC) and receiver operator characteristic (ROC) curve were used to distinguish the clinical classifications, overall survival (OS) good (≥5 years, alive) and OS poor (≤2 years, dead) and disease-free survival (DFS) good (≥5 years, disease free) and DFS poor (≤2 years, recurred/progressed). The Kaplan–Meier (KM) curve evaluated survival rate and expression level of TRIM2 with log-rank test. Prognostic significance of TRIM2 was analyzed by univariate and multivariate Cox proportional hazard regressions in ccRCC. P<0.05 was considered to be statistically significant. All statistical analyses were performed using the SPSS Statistics 22.0 (IBM Corporation, Armonk, NY, USA).
Results
TRIM2 is significantly downregulated and associated with various clinicopathological parameters in TCGA-KIRC
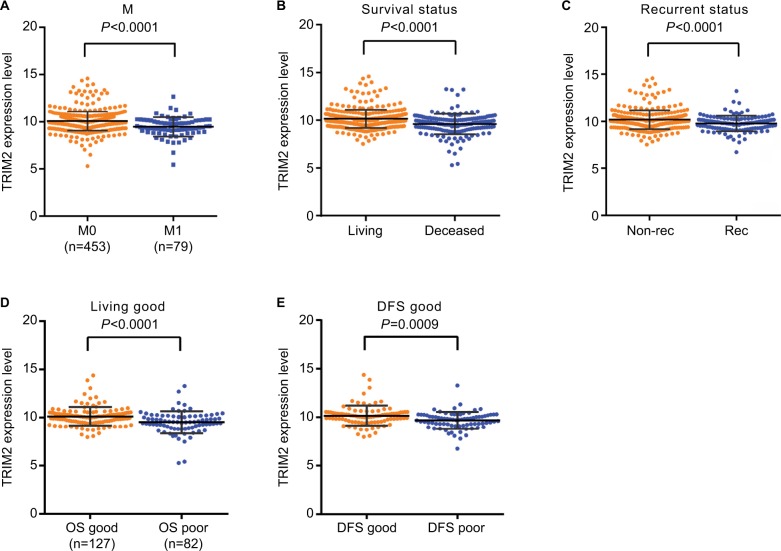
We investigated TRIM2 mRNA expression levels in ccRCC cancer tissues and corresponding normal tissues of TCGA-KIRC. Detailed clinicopathological information of these ccRCC patients is presented in Table 1. TRIM2 relative expression ranged from 9.7489 to 13.3865 in normal tissues and 5.2964 to 14.5911 in tumor tissues. The expression of TRIM2 was significantly lower in ccRCC tissues than in corresponding noncancerous normal tissues (Figure 1A). Similar result was showed in 72 paired ccRCC tissues and corresponding noncancerous normal tissues (Figure 1B). Significantly lower TRIM2 expression was found in T stage IV and III, when contrasted to T stage I and II (Figure 1C); furthermore, lower TRIM2 expression was found in T stage IV or III compared with T stage I or II, and the same result was showed in T stage IV compared with T stage III (Figure 1D). Meanwhile, the lower level of TRIM2 was associated with grade stage and pathological TNM stage in ccRCC patients (Figure 1E–H), but the expression of TRIM2 was not significantly associated with gender, age, or lymph node metastasis (data not shown). These data indicated that the TRIM2 expression is downregulated and its expression was significantly associated with various clinicopathological parameters in ccRCC.
Table 1.
Correlation between TRIM2 mRNA expression and clinicopathological parameters of ccRCC patients
| Parameter | TRIM2 mRNA expression
|
P-value | |||
|---|---|---|---|---|---|
| Number | Low (n=266) | High (n=265) | |||
|
| |||||
| Age (years) | ≤60 | 263 | 130 | 133 | |
| >60 | 268 | 136 | 132 | 0.795 | |
| Gender | Male | 343 | 172 | 171 | |
| Female | 188 | 94 | 94 | 1.000 | |
| T stage | T1 + T2 | 340 | 140 | 200 | |
| T3 + T4 | 191 | 126 | 65 | 0.000 | |
| N stage | N0 + NX | 515 | 256 | 259 | |
| N1 | 16 | 10 | 6 | 0.447 | |
| M stage | M0 + MX | 452 | 210 | 242 | |
| M1 | 79 | 56 | 23 | 0.000 | |
| G stage | G1 + G2 | 249 | 96 | 153 | |
| G3 + G4 | 282 | 170 | 112 | 0.000 | |
| TNM stage | I + II | 325 | 130 | 195 | |
| III + IV | 206 | 136 | 70 | 0.000 | |
Abbreviations: ccRCC, clear cell renal cell carcinoma; TRIM2, tripartite motif 2.
Figure 1.
The level of TRIM2 is downregulated and correlated with various clinicopathological parameters in ccRCC tissues.
Notes: The mRNA level of TRIM2 in ccRCC was downloaded from the Cancer Genome Atlas Kidney Clear Cell Carcinoma dataset containing 72 normal tissues and 533 ccRCC tissues. The mRNA levels of TRIM2 were compared in different clinicopathological parameters: (A) cancer vs paracancer, (B) cancer vs paired paracancer, (C and D) T stage, (E and F) TNM stage, and (G and H) G stage. **P<0.01, ***P<0.001.
Abbreviations: ccRCC, clear cell renal cell carcinoma; TCGA-KIRC, The Cancer Genome Atlas Kidney Clear Cell Carcinoma; TRIM2, tripartite motif 2, N, normal; T, tumor.
Low TRIM2 predicts poor prognosis of ccRCC
We first confirmed that the TRIM2 expression was down-regulated in cancer tissues and associated with various clinicopathological parameters. Then, we identified whether expression of TRIM2 in ccRCC contributed to disease progression. Statistical analysis of TCGA-KIRC showed that patients with metastasis had a lower level of TRIM2 expression (Figure 2A). The database revealed that significantly lower TRIM2 expression in dead ccRCC patients than in alive ccRCC patients (Figure 2B). To explore the prognostic significance of TRIM2, we compared TRIM2 mRNA level between recurrence and disease free of ccRCC patients and the TRIM2 mRNA expression was significantly higher in disease-free ccRCC (Figure 2C). Moreover, the TRIM2 mRNA level was able to classify good or poor prognosis in OS and DFS time of ccRCC patients (Figure 2D and E), indicating that lower TRIM2 predicts poor prognosis of ccRCC and it may be a potential prognostic indicator for ccRCC.
Figure 2.
Low level of TRIM2 mRNA predicts poor prognosis of ccRCC patients.
Note: The TRIM2 mRNA level was decreased and negatively correlated with (A) nonmetastasis and metastasis, (B) living status, (C) nonrecurrent and recurrent, (D) OS good and OS poor, and (E) DFS good and DFS poor.
Abbreviations: ccRCC, clear cell renal cell carcinoma; DFS, disease-free survival; OS, overall survival.
The association between TRIM2 and diagnostic value in ccRCC
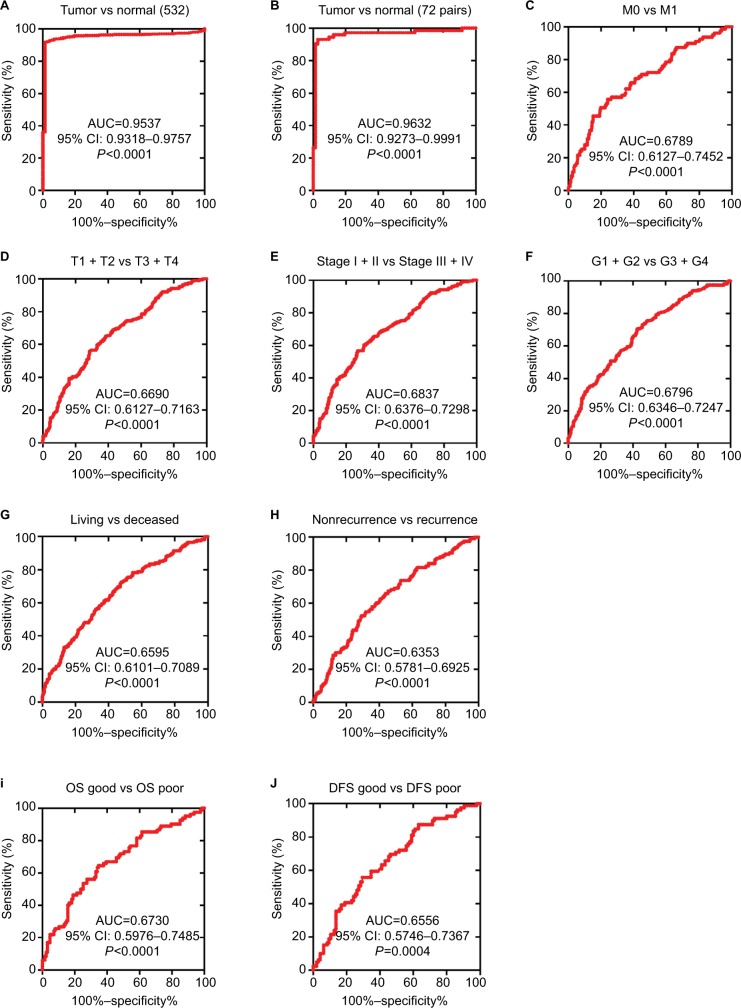
As lower TRIM2 predicts poor prognosis of ccRCC, then we investigated the diagnostic value of TRIM2. ROC curves were analyzed the clinicopathological parameters with TRIM2. TRIM2 could sufficiently discriminate ccRCC with the AUC of 0.9537 (95% CI: 0.9318–0.9757; P<0.0001) from normal tissues (Figure 3A), and it could effectively distinguish ccRCC with the AUC of 0.9632 (95% CI: 0.9273–0.9991; P<0.0001) from paired normal tissues (Figure 3B). In addition, we found the significant difference of TRIM2 mRNA expression in ccRCC subgroups, including T stage, N stage, M stage, G stage, TNM stage, living status, recurrent status, good or poor OS, good or poor DFS, and M0/M1 (Figure 3C, AUC =0.6789, P<0.0001), (T1 + T2)/(T3 + T4) stage (Figure 3D, AUC =0.66690, P<0.0001), TNM (I + II)/(III + IV) stage (Figure 3E, AUC =0.6837, P<0.0001), (G1 + G2)/(G3 + G4) stage (Figure 3F, AUC =0.6796, P<0.0001), alive/dead status (Figure 3G, AUC =0.6595, P<0.0001), non-recurrent/recurrent (Figure 3H, AUC =0.6353, P<0.0001), OS good/poor prognosis (Figure 3I, AUC =0.6730, P<0.0001), and DFS good/poor prognosis (Figure 3J, AUC =0.6556, P<0.0001). However, low TRIM2 level could not differentiate ccRCC patients with lymph node metastasis or without (data not shown). These results illustrated that TRIM2 expression could be a valid and a potential diagnostic indicator for ccRCC.
Figure 3.
Low TRIM2 expression serves as a diagnostic indicator in ccRCC patients.
Notes: (A) ROC curve showed that TRIM2 could effectively distinguish ccRCC from paracancer tissues, AUC 0.9537 (P<0.0001), and paired-cancer tissues, AUC 0.9632 (P<0.0001). (B-F) ROC curve analysis toward the expression of TRIM2 mRNA in subgroups of ccRCC patients against (C) metastasis status, (D) T stage, (E) TNM stage, (F) G stage, (G) living status, (H) recurrent status, (I) OS good vs poor, and (J) DFS good vs poor.
Abbreviations: AUC, area under the curve; ccRCC, clear cell renal cell carcinoma; DFS, disease-free survival; OS, overall survival; ROC, receiver operator characteristic; TRIM2, tripartite motif 2.
Low TRIM2 expression correlated with poor OS in ccRCC patients
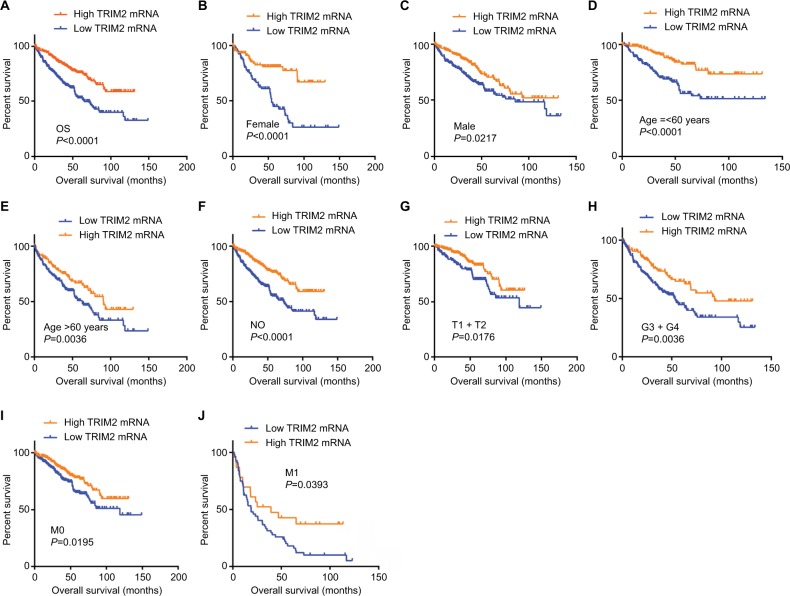
KM survival analysis determined the correlation of OS time and TRIM2 expression of ccRCC patients. Based on the median expression value of TRIM2 mRNA, 532 patients from TCGA-KIRC were divided into two groups. Patients with low TRIM2 expression exhibited shorter OS time (Figure 4A, P<0.0001). Furthermore, we carried out OS analysis between the TRIM2 mRNA expression in various subgroups of ccRCC patients. The results illustrated that TRIM2 expression could be a potential prognostic factor for ccRCC patients in subgroups, including female (Figure 4B, P<0.0001), male (Figure 4C, P=0.0217), age ≤60 years (Figure 4D, P<0.0001), age >60 years (Figure 4E, P=0.0036), N0 stage (Figure 4F, P<0.0001), T1 + T2 stage (Figure 4G, P=0.0176), G3 + G4 stage (Figure 4H, P=0.0036), M0 (Figure 4I, P=0.0195), and M1 (Figure 4J, P=0.0393). However, there was no significant correlation between the TRIM2 mRNA expression and OS of ccRCC patients with N1 stage and TNM stage (data not shown). The prognostic value of TRIM2 expression and each clinicopathological factor evaluated for OS are shown in Table 2. Univariate Cox proportion HR analysis showed the following the results: age >60 years (HR, 1.803; P<0.001), T stage (T III + VI) (HR, 3.120; P<0.001), N stage (N1) (HR, 3.823; P<0.001), M stage (M1) (HR, 4.346; P<0.001), G grade (G3 + G4) (HR, 2.639; P<0.001), and low TRIM2 expression status (HR, 1.951; P<0.001). Multivariate analysis showed the following results: age >60 years (HR, 1.631; P=0.002), T stage (T3 + T4) (HR, 1.66; P=0.005), M stage (M1) (HR, 2.562; P<0.001), G grade (G3 + G4) (HR, 1.681; P=0.006), and low TRIM2 expression (HR, 1.388; P=0.049); these results indicated that TRIM2 could be an independent prognostic indicator of OS.
Figure 4.
Low level of TRIM2 mRNA predicts poor overall survival rate in ccRCC patients.
Notes: (A) The ccRCC patients from TCGA-KIRC database were divided into low TRIM2 expression group and high TRIM2 expression group according to the median expression value of TRIM2 mRNA level. The correlation between TRIM2 expression and overall survival time of total ccRCC patients was analyzed by Kaplan–Meier. Overall survival analysis toward the expression of TRIM2 mRNA was performed in subgroups of ccRCC patients: (B) female, (C) male, (D) age ≤60 years, metastasis status, (E) age >60 years, (F) N0 stage, (G) T1+ T2, (H) G3+ G4 stage, (I) nonmetastasis status, and (J) metastasis status.
Abbreviations: ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas; TRIM2, tripartite motif 2.
Table 2.
Univariate and multivariate analyses of TRIM2 mRNA level and patient’s overall survival
| Variable | Univariate analysis | Multivariate analysisc | |||||
|---|---|---|---|---|---|---|---|
| HRa | 95% CIb | P-value | HR | 95% CI | P-value | ||
| Overall survival (n=526) | |||||||
| Age (years) | |||||||
| >60 (n=264) | 1.803 | 1.318–2.468 | 0.000 | 1.631 | 1.198–2.219 | 0.002 | |
| ≤60 (n=262) | |||||||
| Gender | |||||||
| Male (n=336) | 0.948 | 0.697–1.290 | 0.825 | ||||
| Female (n=181) | |||||||
| T stage | |||||||
| T3 or T4 (n=188) | 3.120 | 2.306–4.220 | 0.000 | 1.669 | 1.171–2.379 | 0.005 | |
| T1 or T2 (n=338) | |||||||
| N stage | |||||||
| N1 (n=15) | 3.832 | 2.070–7.061 | 0.000 | ||||
| N0 or NX (n=511) | |||||||
| M stage | |||||||
| M1 (n=78) | 4.346 | 3.192–5.918 | 0.000 | 2.562 | 1.796–3.654 | 0.000 | |
| M0 or MX (n=448) | |||||||
| G grade | |||||||
| G3 or G4 (n=281) | 2.639 | 1.885–3.697 | 0.000 | 1.681 | 1.164–2.428 | 0.006 | |
| G1 or G2 (n=245) | |||||||
| TRIM2 | |||||||
| Low (n=263) | 1.951 | 1.428–2.664 | 0.000* | 1.388 | 1.002–1.923 | 0.049* | |
| High (n=263) | |||||||
Notes:
HR, estimated from Cox proportional hazard regression model.
CI of the estimated HR.
Multivariate models were adjusted for T, N, M classification, age, and gender
P<0.05 was considered statistically significant.
Abbreviation: TRIM2, tripartite motif 2.
Low TRIM2 expression correlated with poor DFS in ccRCC
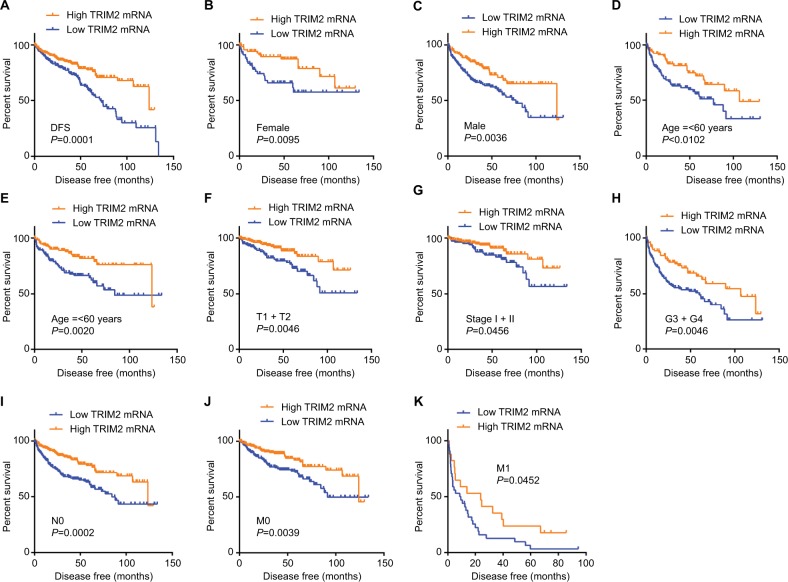
As the results showed, TRIM2 could be an independent prognostic indicator of OS and, then, we tested the prognostic value of TRIM2 expression and DFS of patients with ccRCC. Based on the median expression value of TRIM2 mRNA, 421 patients from TCGA-KIRC were divided into two groups. The poor DFS patients had a low TRIM2 expression (Figure 5A, P<0.0001), and TRIM2 expression analysis indicated that TRIM2 could be a potential prognostic factor for ccRCC patients with female (Figure 5B, P=0.0095), male (Figure 5C, P=0.0036), age >60 years (Figure 5D, P=0.0102), age ≤60 years (Figure 5E, P=0.0020), T1 + T2 stage (Figure 5F, P=0.0046), TNM stage I + II (Figure 5G, P=0.0456), G3 + G4 stage (Figure 5H, P=0.0046), N0 stage (Figure 5I, P=0.0002), M0 (Figure 5J, P=0.0039), and M1 (Figure 5K, P=0.0452), but DFS of the ccRCC patients in N1 stage and G1 + G2 stage had no significant correlation with different TRIM2 expressions (data not shown). The prognostic value of TRIM2 expression and each clinicopathological factor evaluated for DFS are shown in Table 3. Univariate Cox proportion HR analysis showed the following results: T stage (T3 + T4) (HR, 4.519; P<0.0001), N (N1) stage (HR, 5.937; P<0.0001), M (M1) stage (HR, 8.520; P<0.0001), G (G3 + G4) grade (HR, 3.350; P<0.0001), and low TRIM2 expression status (HR, 2.060; P<0.0001). Multivariate analysis showed the following results: T stage (T3 + T4) (HR, 2.014; P=0.001), N stage (N1) (HR, 3.273; P=0.002), M stage (M1) (HR, 5.166; P<0.0001), G grade (G3 + G4) (HR, 2.173; P<0.0001), and low TRIM2 expression (HR, 1.502; P=0.040); these results indicated that TRIM2 could be an independent prognostic indicator of DFS.
Figure 5.
Low level of TRIM2 mRNA predicts poor DFS rate in ccRCC patients.
Notes: (A) The ccRCC patients from TCGA-KIRC database were divided into low TRIM2 expression group and high TRIM2 expression group according to the median expression value of TRIM2 mRNA level. The correlation between TRIM2 expression and DFS time of total ccRCC patients was analyzed by Kaplan–Meier. DFS analysis toward the expression of TRIM2 mRNA was performed in subgroups of ccRCC patients: (B) female, (C) male, (D) age ≤60 years, (E) age >60 years, (F) T1 + T2, (G) TNM (I + II), (H) G3 + G4 stage, (I) N0 stage, (J) nonmetastasis status, and (K) metastasis status.
Abbreviations: ccRCC, clear cell renal cell carcinoma; DFS, disease-free survival; TCGA, The Cancer Genome Atlas; TRIM2, tripartite motif 2.
Table 3.
Univariate and multivariate analyses of TRIM2 mRNA level and patient’s disease-free survival
| Variable | Univariate analysis | Multivariate analysisc | ||||
|---|---|---|---|---|---|---|
| HRa | 95% CIb | P-value | HR | 95% CI | P-value | |
| Disease-free survival (n=421) | ||||||
| Age (years) | ||||||
| ≤60 (n=231) | 1.369 | 0.961–1.950 | 0.082 | |||
| >60 (n=193) | ||||||
| Gender | ||||||
| Female (n=145) | 1.431 | 0.963–2.127 | 0.076 | |||
| Male (n=280) | ||||||
| T stage | ||||||
| T3 or T4 (n=141) | 4.519 | 3.129–6.527 | 0.000 | 2.014 | 1.323–3.066 | 0.001 |
| T1 or T2 (n=284) | ||||||
| N stage | ||||||
| N1 (n=12) | 5.937 | 2.981–11.826 | 0.000 | 3.273 | 1.573–6.814 | 0.002 |
| N0 or NX (n=414) | ||||||
| M stage | ||||||
| M1 (n=53) | 8.520 | 5.870–12.365 | 0.000 | 5.166 | 3.401–7.848 | 0.000 |
| M0 or MX (n=372) | ||||||
| G grade | ||||||
| G3 or G4 (n=217) | 3.350 | 2.218–5.058 | 0.000 | 2.173 | 1.406–3.357 | 0.000 |
| G1 or G2 (n=208) | ||||||
| TRIM2 | ||||||
| Low (n=212) | 2.060 | 1.422–2.985 | 0.000 | 1.502 | 1.018–2.214 | 0.040* |
| High (n=213) | ||||||
Notes:
HR, estimated from Cox proportional hazard regression model.
CI of the estimated HR.
Multivariate models were adjusted for T, N, M classification, age, and gender,
P<0.05 was considered to have statistical significance.
Abbreviation: TRIM2, tripartite motif 2.
TRIM2 was downregulated in RCC cells and involved in biological pathogenesis of ccRCC
As TRIM2 was downregulated and significantly associated with various clinicopathological parameters in TCGA-KIRC database, we also confirmed this observation in RCC cells (Figure 6A) and Western blotting confirmed that the protein level of TRIM2 was downregulated in RCC cells (Figure 6B). Then, we tested TRIM2 expression in ccRCC patient samples by qRT-PCR and IHC (Figure 6C and D). These results revealed that TRIM2 mRNA and protein levels from RCC cells and cancer tissues were significantly lower than immortalized normal renal epithelial cells and corresponding noncancerous renal tissues.
Figure 6.
TRIM2 is downregulated in ccRCC cells and tissues.
Notes: (A and B) Gene expression levels of TRIM2 in renal cancer cell lines. (C and D) qRT-PCR and IHC analyses of TRIM2 expression in ccRCC tissues and paracancer tissues. (E) Gene set enrichment analysis compared low TRIM2 and high TRIM2 expression groups in TCGA database. Enrichment curves are shown for activated gene sets related to G2M checkpoint, EMT, hypoxia pathway, and Myc signaling.
Abbreviations: ccRCC, clear cell renal cell carcinoma; EMT, epithelial–mesenchymal transition; FDR, false discovery rate; IHC, immunohistochemistry; TCGA, The Cancer Genome Atlas; TRIM2, tripartite motif 2; NES,normalized enrichment score.
GSEA, which was a computational tool to determine whether a gene set had statistically significant between two biological statuses, was used to elucidate whether TRIM2 involved in ccRCC pathogenesis. The results demonstrated that the expression of TRIM2 expression was associated with the gene signatures of G2M checkpoint, EMT, hypoxia pathway, and Myc signaling (Figure 6E).
TRIM2 represses migration, invasion, and proliferation capabilities of renal cancer cells
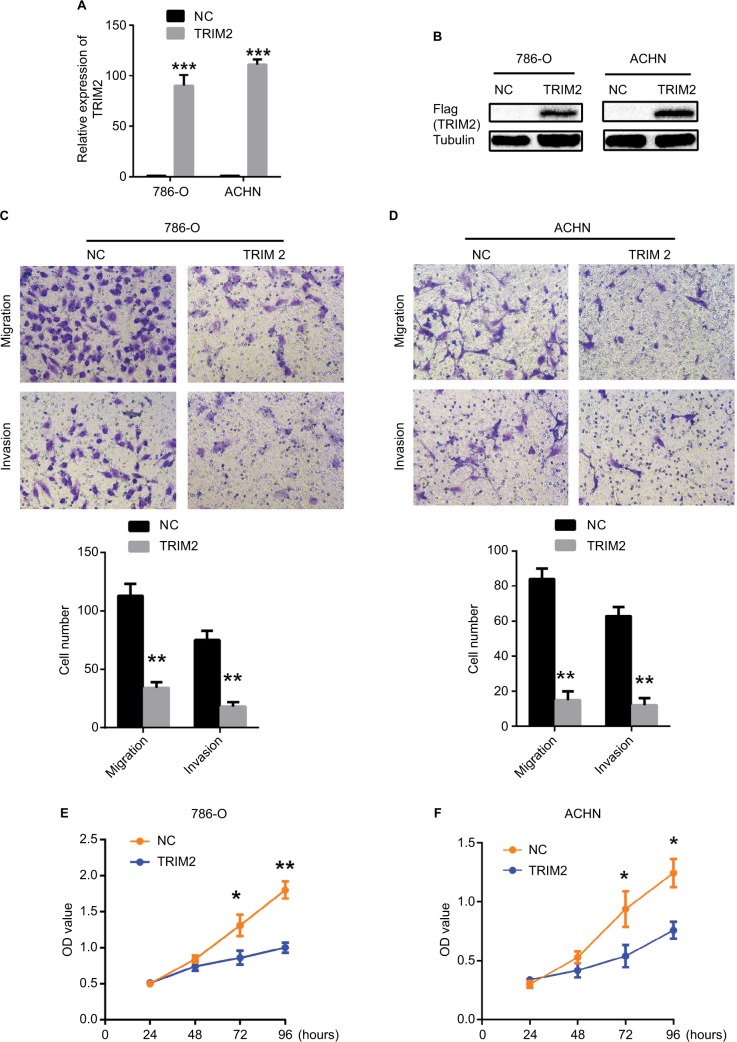
As we demonstrated that the expression of TRIM2 was associated with the gene signatures of G2M checkpoint and EMT in Figure 6, we evaluated the functional role of TRIM2 in ccRCC by the overexpression of TRIM2 in RCC cell lines. The mRNA and protein levels of TRIM2 expression were tested in 786-O and ACHN cells after transfecting cancer cells with plasmid vectors expressing TRIM2 or NC (Figure 7A and B). Overexpression of TRIM2 significantly repressed the migration and invasion abilities by transwell assays when compared with NC group (Figure 7C and D), and furthermore, the proliferation of 786-O and ACHN cells was markedly inhibited in transfection with TRIM2 compared to NC group (Figure 7E and F). These results demonstrated that TRIM2 may act as a tumor suppressor in ccRCC cells.
Figure 7.
Effects of TRIM2 overexpression on cell migration, invasion, and proliferation.
Notes: (A and B) TRIM2 mRNA and protein expressions were successfully overexpressed in 786-O and ACHN cells. (C and D) Representative images of migration and invasion assays performed using 786-O and ACHN cells. Data are presented as the mean ± SD from three independent experiments. (E and F) Cell counting kit-8 assay detected the effects of TRIM2 overexpression on the proliferation of 786-O and ACHN cells. *P<0.05, **P<0.01, and ***P<0.001.
Abbreviation: NC, negative control; TRIM2, tripartite motif 2.
Discussion
RCC is a common and lethal urological malignancy in adults. ccRCC is accounting for ~80% of all RCC histological subtype and has the highest rate of mortality, invasion, metastasis, and resistance to traditional radiotherapy and chemotherapy. The present study reports the role of TRIM2 expression in clinical significance and biological functions of ccRCC for the first time. Accumulating experimental evidence demonstrated that 1) TRIM2 expression was significantly decreased in ccRCC and was inversely associated with high T stage, TNM stage, and grades in TCGA-KIRC cohort, 2) lower TRIM2 expression level was an independent predictor of poor prognosis of OS and DFS in TCGA dataset, 3) GSEA demonstrated that lower TRIM2 expression was associated with the gene set of G2M checkpoint, EMT, hypoxia pathway, and Myc signaling, and 4) overexpression of TRIM2 suppressed cell migration, invasion, and proliferation capabilities of 786-O and ACHN RCC cells.
Recent studies confirmed that the TRIM protein was an important regulator of carcinogenesis. TRIM2 was verified as a TRIM-NHL protein, and it could regulate cell death by interacting with B-cell lymphoma 2-interacting mediator (Bim).23 Studies focused on different types of carcinoma had indicated that TRIM2 had been detected to be significantly differentially expressed in ovarian cancer, breast cancer, cervical carcinoma, buccal mucosa carcinoma, and follicular carcinoma.24–28 In a study of thyroid cancer, TRIM2 had higher expression in the nonaggressive follicular thyroid carcinomas than in the aggressive follicular thyroid carcinomas.24 TRIM2 was an important feasible gene for the observation of occurrence, development, and prognosis at all stages of buccal mucosa cancer.25 However, some certain studies showed the opposite results with the expression of TRIM2 found to be upregulated in ovarian and breast cancers.26,27 The bias between different types of cancer suggests that TRIM2 is an important gene involved in the pathogenic process of cancer by different mechanisms and microenvironments. The dysfunction of TRIM2 may be one of the factors involved in the occurrence and development of ccRCC to a certain extent, and the association between TRIM2 and renal cancer has not been reported.
Normal function of von Hippel–Lindau tumor suppressor, which could deregulate hypoxia-inducible transcription factor (HIF) loss, was one of the most pathogens in ccRCC, and HIF-2a acting as an important oncogene could promote renal tumor growth.29–32 Previous studies indicated that TRIM2 functions as a direct regulator to Bim degradation in TAM-resistant breast cancer cells and ovarian cancer,26,27 and GSEA demonstrated that low TRIM2 expression was associated with the hypoxia signaling pathways in patients with ccRCC; it may be hypothesized that TRIM2 could negatively regulated hypoxia signaling pathways, or even deregulated HIF expression in ccRCC; further experiments need to be verified this.
Cancer progression by endowing cancer cells with a more motile and invasive phenotype is critical based on EMT.33,34 GSEA demonstrated that lower TRIM2 expression was associated with the gene signatures of EMT, and it may be hypothesized that TRIM2 could negatively regulated EMT in ccRCC. Overexpressing TRIM2 represses migration, invasion, and proliferation capabilities of 786-O and ACHN renal cells. Myc protein regulates the expression of genes, which control the cell cycle progression and cell metabolism and then enhances the cell proliferation and growth.10,35 GSEA demonstrated that lower TRIM2 expression was associated with the gene signatures of G2M checkpoint and Myc signaling, and it may reveal that TRIM2 could negatively control cell cycle progression and inhibition of myc signaling in ccRCC.
The present study is the first research indicating the functional role of TRIM2 in ccRCC tumor progression, tumorigenesis, and malignant. The present results also demonstrated that TRIM2 may act as a potential novel efficacious biomarker to predict the prognosis of patients with ccRCC. However, whether TRIM2 could deregulate HIF expression and the exact mechanism of TRIM2 in ccRCC progression was not investigated.
Conclusion
We investigated the role of TRIM2 in ccRCC progression including TCGA database, GSEA, clinical specimen, and cellular model. Our study demonstrated that TRIM2 expression was significantly downregulated and associated with various clinicopathological parameters. Lower TRIM2 expression was significantly inversely correlated with disease progression, which identified TRIM2 as an independent predictor for prognosis in ccRCC patients and involved in the development of renal cancer. Further in vitro experiment verified that overexpressing TRIM2 inhibited ccRCC cells’ proliferation and malignant. All of this study provided clues to understand the potential role of TRIM2 in ccRCC and indicated that upregulating TRIM2 may play a new avenue therapeutic strategy for manipulating and managing patients with ccRCC.
Acknowledgments
We wish to thank all our colleagues in the Department of Urology, The First Affiliated Hospital of Xiamen University. This study was supported by grants from the National Natural Scientific Foundation of China (81602218).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Zhang P, Zhang G, et al. Renal cell carcinoma histological subtype distribution differs by age, gender, and tumor size in coastal Chinese patients. Oncotarget. 2017;8(42):71797–71804. doi: 10.18632/oncotarget.17894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 6.Campbell S, Uzzo RG, Allaf ME, et al. Renal Mass and Localized Renal Cancer: AUA Guideline. J Urol. 2017;198(3):520–529. doi: 10.1016/j.juro.2017.04.100. [DOI] [PubMed] [Google Scholar]
- 7.Kim SP, Alt AL, Weight CJ, et al. Independent validation of the 2010 American Joint Committee on Cancer TNM classification for renal cell carcinoma: results from a large, single institution cohort. J Urol. 2011;185(6):2035–2039. doi: 10.1016/j.juro.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 8.Ravaud A, Motzer RJ, Pandha HS, et al. Adjuvant Sunitinib in High-Risk Renal-Cell Carcinoma after Nephrectomy. N Engl J Med. 2016;375(23):2246–2254. doi: 10.1056/NEJMoa1611406. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Hill H, Christie A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539(7627):112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8(8):592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Ma X, Qiu Y, et al. TRIM50 suppressed hepatocarcinoma progression through directly targeting SNAIL for ubiquitous degradation. Cell Death Dis. 2018;9(6):608. doi: 10.1038/s41419-018-0644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Tao R, Wu SS, et al. TRIM52 up-regulation in hepatocellular carcinoma cells promotes proliferation, migration and invasion through the ubiquitination of PPM1A. J Exp Clin Cancer Res. 2018;37(1):116. doi: 10.1186/s13046-018-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Fu H, Yang H, Zhang X, et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res. 2018;37(1):162. doi: 10.1186/s13046-018-0825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Raheja R, Yeh N, et al. TRIM3, a tumor suppressor linked to regulation of p21(Waf1/Cip1.) Oncogene. 2014;33(3):308–315. doi: 10.1038/onc.2012.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Micale L, Fusco C, Fontana A, et al. TRIM8 downregulation in glioma affects cell proliferation and it is associated with patients survival. BMC Cancer. 2015;15:470. doi: 10.1186/s12885-015-1449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caratozzolo MF, Valletti A, Gigante M, et al. TRIM8 anti-proliferative action against chemo-resistant renal cell carcinoma. Oncotarget. 2014;5(17):7446–7457. doi: 10.18632/oncotarget.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao W, Lou N, Ruan H, et al. Mir-144-3p Promotes Cell Proliferation, Metastasis, Sunitinib Resistance in Clear Cell Renal Cell Carcinoma by Downregulating ARID1A. Cell Physiol Biochem. 2017;43(6):2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 20.Liu R, Qin X, Ji C, Zeng W, Yang Y, Tan W. Pygopus 2 promotes kidney cancer OS-RC-2 cells proliferation and invasion in vitro and in vivo. Asian J Urol. 2015;2(3):151–157. doi: 10.1016/j.ajur.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res. 2015;43(Database issue):D812–D817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson S, Pearson AN, Ashley MD, et al. Identification of a novel Bcl-2-interacting mediator of cell death (Bim) E3 ligase, tripartite motif-containing protein 2 (TRIM2), and its role in rapid ischemic tolerance-induced neuroprotection. J Biol Chem. 2011;286(22):19331–19339. doi: 10.1074/jbc.M110.197707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams MD, Zhang L, Elliott DD, et al. Differential gene expression profiling of aggressive and nonaggressive follicular carcinomas. Hum Pathol. 2011;42(9):1213–1220. doi: 10.1016/j.humpath.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang K, Zhang G, Mei J, Chen D, Wu M. Screening and analysis of pathogenic genes during DMBA-induced buccal mucosa carcinogenesis in golden hamsters. Oncol Rep. 2010;23(6):1619–24. doi: 10.3892/or_00000803. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Dong C, Law PT, et al. MicroRNA-145 targets TRIM2 and exerts tumor-suppressing functions in epithelial ovarian cancer. Gynecol Oncol. 2015;139(3):513–519. doi: 10.1016/j.ygyno.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Yin H, Zhu Q, Liu M, et al. GPER promotes tamoxifen-resistance in ER+ breast cancer cells by reduced Bim proteins through MAPK/Erk-TRIM2 signaling axis. Int J Oncol. 2017;51(4):1191–1198. doi: 10.3892/ijo.2017.4117. [DOI] [PubMed] [Google Scholar]
- 28.Miyatake T, Ueda Y, Nakashima R, et al. Down-regulation of insulin-like growth factor binding protein-5 (IGFBP-5): novel marker for cervical carcinogenesis. Int J Cancer. 2007;120(10):2068–2077. doi: 10.1002/ijc.22264. [DOI] [PubMed] [Google Scholar]
- 29.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14(15):4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metelo AM, Noonan H, Iliopoulos O. HIF2a inhibitors for the treatment of VHL disease. Oncotarget. 2015;6(27):23036–23037. doi: 10.18632/oncotarget.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schödel J, Grampp S, Maher ER, et al. Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer. Eur Urol. 2016;69(4):646–657. doi: 10.1016/j.eururo.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 34.Beerling E, Seinstra D, de Wit E, et al. Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Rep. 2016;14(10):2281–2288. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klatte T, Kroeger N, Rampersaud EN, et al. Gain of chromosome 8q is associated with metastases and poor survival of patients with clear cell renal cell carcinoma. Cancer. 2012;118(23):5777–5782. doi: 10.1002/cncr.27607. [DOI] [PubMed] [Google Scholar]