Significance
We overcame the difficulty of disentangling biotic and abiotic effects on decomposition by using the largest field-based reciprocal transplant experiment to date. We showed that decomposition responses to climate depend on the composition of microbial communities, which is not considered in terrestrial carbon models. Microbial communities varied in their effects on both mass loss and types of carbon decomposed in an interactive manner not predicted by current theory. Contrary to the traditional paradigm, bacterial communities appeared to have a stronger impact on grassland litter decomposition rates than fungi. Furthermore, bacterial communities shifted more rapidly in response to changing climates than fungi. This information is critical to improving global terrestrial carbon models and predicting ecosystem responses to climate change.
Keywords: leaf litter decomposition, reciprocal transplant, bacteria, fungi, elevation gradient
Abstract
Bacteria and fungi drive decomposition, a fundamental process in the carbon cycle, yet the importance of microbial community composition for decomposition remains elusive. Here, we used an 18-month reciprocal transplant experiment along a climate gradient in Southern California to disentangle the effects of the microbial community versus the environment on decomposition. Specifically, we tested whether the decomposition response to climate change depends on the microbial community. We inoculated microbial decomposers from each site onto a common, irradiated leaf litter within “microbial cages” that prevent microbial exchange with the environment. We characterized fungal and bacterial composition and abundance over time and investigated the functional consequences through litter mass loss and chemistry. After 12 months, microbial communities altered both decomposition rate and litter chemistry. Further, the functional measurements depended on an interaction between the community and its climate in a manner not predicted by current theory. Moreover, microbial ecologists have traditionally considered fungi to be the primary agents of decomposition and for bacteria to play a minor role. Our results indicate that not only does climate change and transplantation have differential legacy effects among bacteria and fungi, but also that bacterial communities might be less functionally redundant than fungi with regards to decomposition. Thus, it may be time to reevaluate both the role of microbial community composition in its decomposition response to climate and the relative roles of bacterial and fungal communities in decomposition.
Microbial communities are the engines of decomposition (1), a fundamental process regulating the carbon cycle. In ecosystems, microbial decomposition converts detritus into CO2 and releases nutrients for plant growth. While much is understood about how changes in abiotic conditions (2, 3) and substrate quality (4) affect decomposition rates, the role of microbial community composition remains elusive (5, 6).
This knowledge gap may be key for predicting how ecosystems will respond to climate change (7). Most terrestrial ecosystem models assume that biogeochemical rates are invariant with changes in the size and composition of microbial communities (8). However, recent work from laboratory manipulations of microbial communities (9, 10) and common garden field experiments (11, 12) demonstrate that bacterial and fungal community composition affects decomposition rates. These studies find that, under the same environmental conditions, decomposer communities are not functionally redundant. What is not yet known, however, is the importance of community-by-environment interactions on microbially driven functioning. Even if communities are functionally distinct, if they respond proportionally to changes in climate, then one could still ignore community differences and predict changes in functioning. Recent evidence suggests that such interactions are likely. For instance, microbial community-by-environment interactions influence respiration rates in laboratory microcosms (13–15), and historical precipitation altered the relationship between soil moisture and extracellular enzyme production across a natural climate gradient (16).
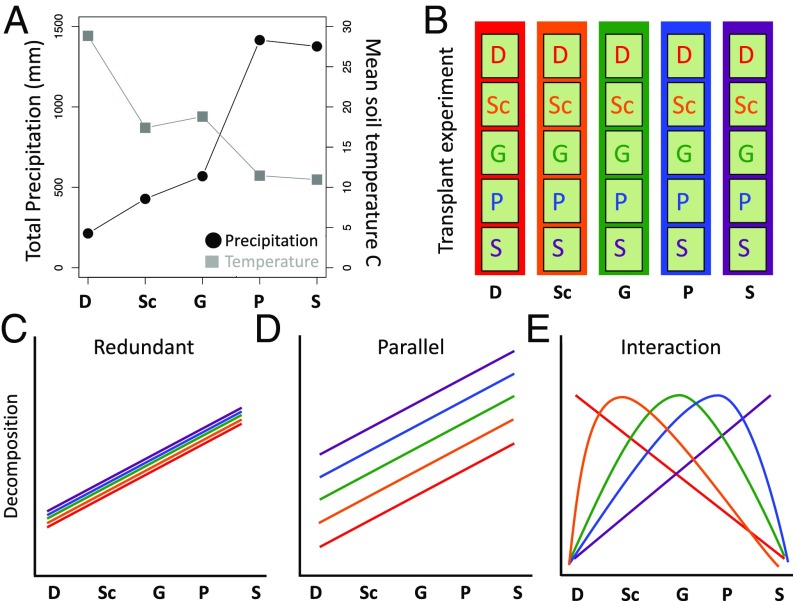
Given that the factors regulating decomposition are often context dependent and can vary in their influence across a range of spatial and temporal scales (17, 18), we hypothesized that decomposition responses to changing climatic conditions would depend on microbial community composition. To test this hypothesis, we conducted the largest microbial community transplant experiment to date. Such transplant experiments are necessary to disentangle the confounding effects of microbial community composition and abiotic environment on functional processes (18). We reciprocally transplanted five leaf litter microbial communities into five sites across an elevation gradient in Southern California that varies in temperature and precipitation (Fig. 1 A and B) (19). Moving the communities from colder, wetter sites at higher elevations to hotter, drier sites at lower elevations mimics the expected shift with climate change to more arid conditions in the southwest United States (20). While elevation gradients have long been used as “space for time” substitutions to predict how plant and animal communities will respond to climate change (21), the approach has only recently been applied to microbial communities (22, 23).
Fig. 1.
(A) Total precipitation (in millimeters) and mean annual soil temperature (in degrees Celsius) at the five sites along the elevation gradient. Sites are represented in increasing precipitation order in the same color scheme: desert (D) = red, scrubland (Sc) = orange, grassland (G) = green, pine–oak (P) = blue, and subalpine (S) = purple. (B) Schematic of microbial transplant experiment. Microbial communities from all sites were placed in a common garden experiment in all sites using a common substrate (irradiated grassland litter represented with light green box; n = 5 inocula × 5 sites × 4 plots × 3 time points = 300 litterbags). Three possibilities for decomposition responses are: (C) redundant, in which all microbes function similarly in every site and are only affected by abiotic conditions; (D) parallel, in which microbes differentially affect decomposition, but respond to climate in a proportional manner; and (E) and interaction, in which decomposition is a result of an interaction between microbial communities and their environment. While any interaction is possible, we illustrate an example in which a community decomposes most in its home site (home-field advantage).
Our experimental design allowed us to quantify the decomposition response of five microbial communities across a climatic gradient in the field. We disentangled the effects of abiotic environment versus microbial community on carbon cycling functioning by inoculating microbial communities onto common, gamma-irradiated leaf litter in nylon mesh litterbags that allow for transport of water and nutrients but prevent immigration of microbial cells (12). We then tracked the microbial community (bacterial and fungal biomass and community composition) and its functioning (litter chemistry and mass loss) after 6, 12, and 18 mo. While many biotic and abiotic conditions vary along the gradient, including vegetation and soil nutrients (19, 24), the litterbags allowed us to control for changes in vegetation by using a common litter substrate. They also physically separate the litter communities from the soil. Thus, we presume that the main site differences that the litterbag communities experience are differences in temperature and precipitation (Fig. 1A).
Although there are many potential outcomes, the possibilities for decomposition responses to the climate gradient fall under three general theoretical models, depending on whether the communities are functionally redundant, parallel, or interact with their local climate. Functional redundancy predicts that all microbial communities decompose leaf litter similarly when transplanted to the same abiotic conditions (Fig. 1C) (9). Thus, decomposition may differ as the climate varies across sites (for instance, increasing with increasing precipitation), but it is indifferent to the microbial community within a site. Under the functionally parallel model (Fig. 1D), different communities are not functionally redundant. They decompose differently even when exposed to the same climate conditions at a site. However, the responses of the communities to climate change are predictable based on observations from any one site; decomposition rates of the communities change in proportion to one another across sites. Finally, under the interaction model, the manner in which the communities are functionally distinct depends on the environment (Fig. 1E). One such example of microbial interaction with its environment is home-field advantage, where a community decomposes litter quickest in its native environment (25). This outcome (or any outcome where the microbial community and climate interact to influence decomposition) would indicate that decomposition responses to climate depend on the microbial community.
Results
Across the five sites, soil temperature ranged from an average of 11–26 °C and total precipitation from 214 to 1,416 mm over the duration of the experiment (Fig. 1A and SI Appendix, Fig. S1 and Table S1). Microbial community composition of the initial inocula (n = 20), transplanted litterbags (n = 300), and in situ communities (n = 80) were assayed by amplicon sequencing [16S ribosomal RNA for bacteria and Internal Transcribed Spacer 2 (ITS2) for fungi], yielding 18.7 M and 24.6 M quality reads for bacteria and fungi, respectively. Bacterial diversity in the leaf litter inocula consisted of at least 135 families belonging to 26 phyla with the vast majority of reads belonging to four phyla: Acidobacteria, Actinobacteria, Bacteroidetes, and Proteobacteria (SI Appendix, Fig. S2A). Fungal diversity in the leaf litter inocula consisted of at least 145 families belonging to 6 phyla with the vast majority in the phylum Ascomycota (SI Appendix, Fig. S2B). Operational taxonomic unit (OTU) richness was higher for bacteria than fungi in all sites (SI Appendix, Fig. S3). Bacterial biomass dominated in both the in situ leaf litter and in the transplanted litterbags, with fungal:bacterial ratios, estimated as grams carbon per gram dry leaf litter converted from bacterial flow cytometry counts and fungal hyphal abundance counts, ranging from 0.15 to 0.72 across the sites (SI Appendix, Table S2), similar to previous measurements (22).
As expected, given the differences in temperature and precipitation across the sites (Fig. 1A), the initial communities used as inocula differed in their composition (SI Appendix, Fig. S4). The inocula were also representative of the in situ leaf litter communities adjacent to the experimental plots during the experimental period (SI Appendix, Fig. S4), with microbial community composition varying strongly by site (PERMANOVA: bacteria R2 = 0.66; fungi R2 = 0.58; P < 0.001) but much less over time (bacteria R2 = 0.05; fungi R2 = 0.04; P < 0.001; SI Appendix, Table S3). Fungi and bacteria displayed similar patterns in community similarity along the gradient; communities from the highest elevation sites (pine–oak, subalpine) and the lowest elevation sites (desert, scrubland) were relatively similar to one another and distinct from the midelevation site (grassland) (SI Appendix, Fig. S4).
Composition of the Transplanted Communities.
We transplanted the microbial communities from each site along the elevation gradient to disentangle the effect of abiotic environment (site) and microbial community (inoculum) on decomposition. Within the transplanted litterbags, both fungal and bacterial richness were reduced compared with the original inoculum (SI Appendix, Fig. S5), suggesting that the litterbags prevented new immigration while nonlitter specialists were outcompeted (11).
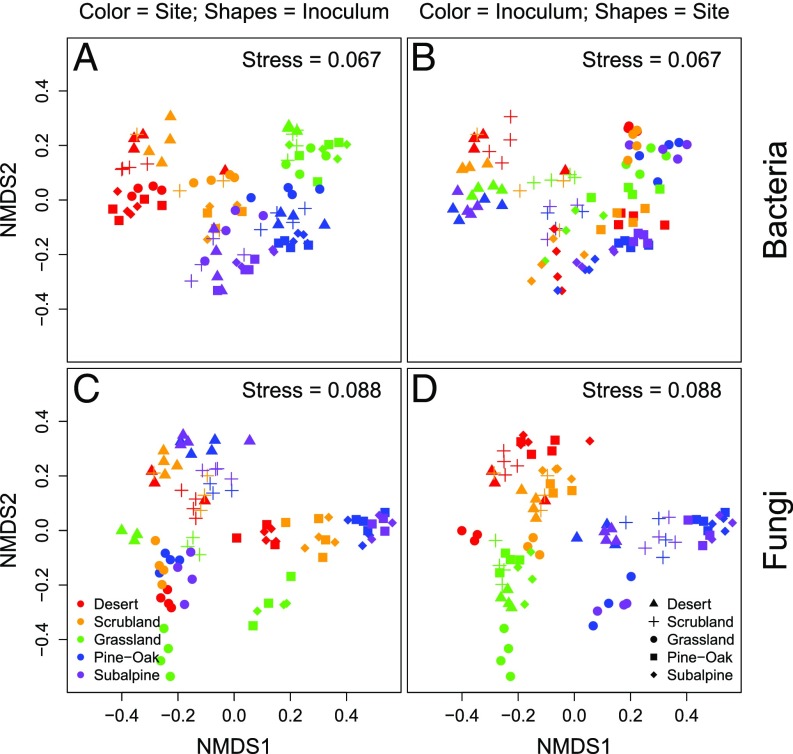
After 18 mo, the transplanted bacterial communities shifted to reflect the surrounding abiotic environment, clustering largely by site (Fig. 2A and SI Appendix, Fig. S6A) rather than inoculum (Fig. 2B and SI Appendix, Fig. S6B). In total, site explained 47% of variation in bacterial composition, yet community composition still displayed significant legacy effects, with inoculum and site-by-inoculum interactions together accounting for 24% of the variation in bacterial composition (SI Appendix, Table S4).
Fig. 2.
NMDS of Bray–Curtis microbial community composition at 18 mo for (A) bacteria colored by site and shapes by inoculum and (B) bacteria colored by inoculum and shapes by site. The Bottom two panels are both fungal community composition with either (C) colored by site and shapes by inoculum or (D) colored by inoculum and shapes by site.
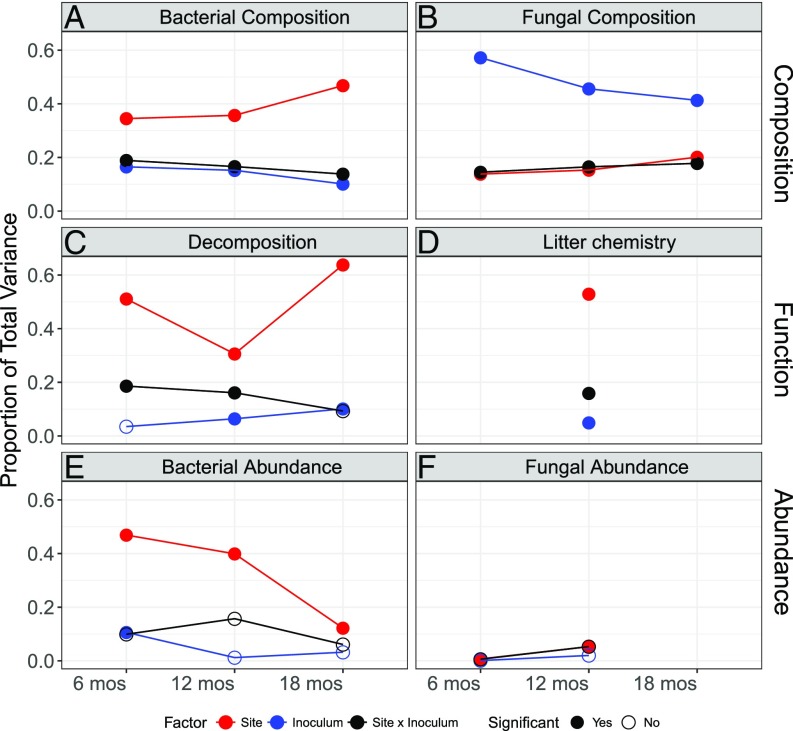
Meanwhile, fungal communities retained much stronger legacy effects, with site explaining only 20% of the variation, and inoculum and site-by-inoculum interactions together accounting for 59% of the variation in fungal composition (SI Appendix, Table S5), even after 18 mo. Thus, although there were significant effects of site on fungal community composition, visual clustering of communities by site was only visible within an inoculum type (Fig. 2C and SI Appendix, Fig. S7A) because of the overriding effect of the inoculum treatment (Fig. 2D and SI Appendix, Fig. S7B). As expected, for both bacteria and fungi, the effect of inoculum on composition was highest at the first sample collection and decreased over time (blue points in Fig. 3 A and B), while conversely, the effect of site on composition increased over time (red points in Fig. 3 A and B).
Fig. 3.
Proportion of variance explained by the treatments (site, inoculum, site x inoculum) on (A) bacterial community composition, (B) fungal community composition, (C) decomposition, (D) litter chemistry, (E) bacterial abundance, and (F) fungal abundance. The proportions for bacterial and fungal community composition and litter chemistry are calculated based on variance estimates from PERMANOVA (SI Appendix, Tables S4, S5, and S7), whereas those for microbial abundance and decomposition are calculated from the total variance explained by two-way ANOVA multiplied by the partial eta-squares for each explanatory variable (SI Appendix, Tables S6 and S9).
Functioning of the Transplanted Communities.
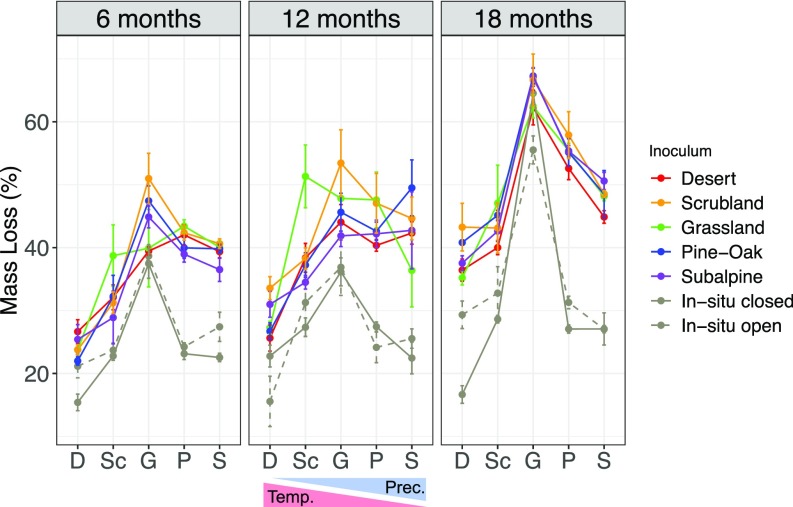
The initial compositional differences among the transplanted communities allowed us to test whether decomposition responses to climate differed by community. Decomposition rate (percent mass loss of litter) was primarily influenced by site, explaining 30–64% of the total variation in mass loss at all three collection times (Fig. 3C and SI Appendix, Table S6). Generally, mass loss was lowest at the two lowest elevation sites and peaked at the midelevation grassland site with intermediate temperature and precipitation (Fig. 4). However, in agreement with our main hypothesis, decomposition rates also depended on the microbial community and in particular, an interactive effect of the community with environment. Litterbags inoculated with communities from different sites differed in mass loss at both the 12- and 18-mo collections (ANOVA: P < 0.05; SI Appendix, Table S6). While the main effect of inoculum was small (explaining only 3–10% of mass loss variation; Fig. 3C and SI Appendix, Table S6), it indicates that communities were not functionally redundant. Moreover, decomposition rate was also influenced by a significant inoculum-by-site interaction (Fig. 3C). This interaction did not result in a home-field advantage for the microbial communities; in particular, scrubland microbial communities decomposed more litter in the grassland site and vice versa. In addition, communities from the lowest (desert) and highest (pine–oak, subalpine) sites appeared to respond more similarly to one another that the midelevation sites (Fig. 4). We also plotted decomposition against total precipitation (SI Appendix, Fig. S8A) and average soil temperature (in degrees Celsius) (SI Appendix, Fig. S8B), yielding similarly shaped curves. This inoculum-by-site effect explained 19% and 16% of mass loss variation at the 6- and 12-mo collections, respectively (Fig. 3C). The functional differences between communities peaked at 12 mo, when the grassland community decomposed on average 39% more than any other community in the scrubland site, and scrubland microbes decomposed 20% more than any other community in the grassland site. By 18 mo, however, these differences disappeared as the microbial communities in the litterbags, and particularly bacterial composition, converged to reflect the abiotic environment (Fig. 2A).
Fig. 4.
Variation in leaf litter decomposition (mean ± SE percent mass loss) for the full factorial transplant experiment (5 inoculum treatments × 5 sites) along the gradient across the three time points. Sites ordered in order of increasing precipitation: D, desert; G, grassland; P, pine–oak; S, subalpine; and Sc, scrubland. In addition to transplant litterbags, we included open or closed in situ litterbags for comparison.
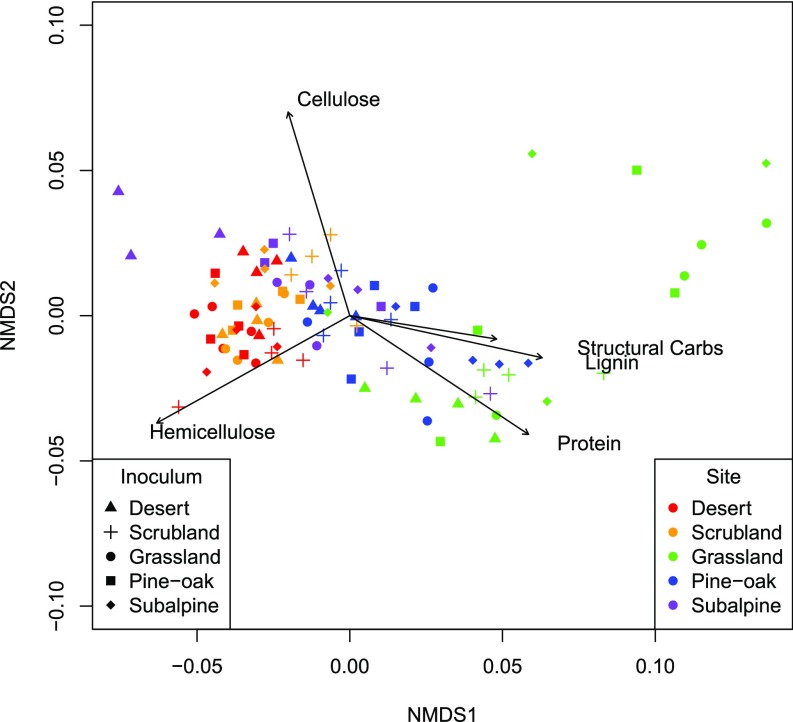
We next tested whether these functional differences between the microbial communities also affected the residual chemistry of the leaf litter. As with mass loss, litter chemistry depended primarily on the abiotic environment (main effect of site: R2 = 0.53), but also on the initial microbial community; together the effect of inoculum and the inoculum-by-site interaction accounted for 21% of the variance observed in litter chemistry (Fig. 3D and SI Appendix, Table S7). Cellulose and hemicellulose were significantly more degraded in grassland than in any other site, whereas other litter components (lignin, crude protein, structural carbohydrates) were less degraded in the grassland (Fig. 5 and SI Appendix, Fig. S9). An ad hoc analysis revealed that the site effects were most strongly driven by differences in the proportion of hemicellulose degraded, although effects were fairly evenly spread across carbon components (SI Appendix, Table S8). For the inoculum effects, the ad hoc comparisons were not significant, but the trends were similar to the site effects; the difference between the grassland and scrubland communities appeared to be driven by differences in the resulting proportions of hemicellulose, followed by protein and cellulose. Thus, not only do inoculum source and site-by-inoculum interactions affect the ecosystem process of decomposition, but also which specific carbon compounds are degraded, indicating that the five microbial communities had unique impacts on carbon cycling along the gradient.
Fig. 5.
NMDS of litter chemistry of transplanted litterbags at 12 mo. Each point represents the chemical composition of the litter from each litterbag for each site (color) and inoculum (shape) combination (4 plots × 4 inocula × 5 sites = 100). Each vector represents whether each of the five organic compounds (cellulose, crude proteins, hemicellulose, lignin, and structural carbohydrates) increases or decreases in abundance in that site. Stress = 0.045.
Microbial Abundance.
The functional differences among the microbial communities do not appear to be due to initial differences in abundance. Decomposition rate in the litterbags was not a function of either bacterial or fungal abundance in the inoculum leaf litter (SI Appendix, Fig. S10). Both bacterial and fungal abundances in the litterbags quickly responded to the abiotic environment (site explained 12–47% variation for bacteria and 1–5% variation for fungi), whereas the original inoculum only explained 11% of the variation in bacterial abundance at 6 mo (Fig. 3E and SI Appendix, Table S9). Further, decomposition rates were highest in the grassland, but bacterial abundance peaked at higher elevations (pine–oak, subalpine) (SI Appendix, Fig. S11A), and grassland fungal abundance did not significantly differ from the other sites (SI Appendix, Fig. S11B). At the same time, microbial abundance and decomposition rates were correlated during the experiment (Pearson r = 0.63, 0.30, and 0.30 at 6, 12, and 18 mo, respectively, for bacteria; r = 0.21 at 6 mo for fungi), presumably because higher decomposition rates lead to higher microbial biomass.
In Situ Litter Decomposition.
We used two additional sets of “in situ” litterbags (n = 120) to investigate how the experimental manipulation itself influenced decomposition rates relative to natural rates of leaf litter decay at each site. Beyond the manipulation of the microbial communities, decomposition within the litterbags might differ from local rates for two additional reasons: (i) the use of a common, ground, grassland substrate and (ii) the use of closed (0.22-µm mesh) litterbags. To quantify these effects separately, one set of litterbags contained snipped (much coarser than ground) in situ litter with their natural microbial communities in open (window screen) litterbags. The second set of litterbags contained the same snipped, in-situ litter but packaged in the closed mesh. As expected, ground litter in the inoculated transplant bags decomposed faster than the snipped in situ litter, but the mass loss patterns across the gradient were similar to that observed in the main experiment (Fig. 4). Further, litterbag material (open versus closed) had a significant effect (explaining 18% of variation versus 72% explained by site) on decomposition at 6 mo, but no effect at 12 and 18 mo (SI Appendix, Table S10). Interestingly, there was a significant site-by-material interaction at 18 mo, where higher decomposition occurred in the open than closed in situ litterbags at the desert, perhaps due to increased exposure to UV radiation (26).
Discussion
Experimental manipulations of abiotic and biotic factors are essential for disentangling the mechanisms through which climate change will affect biodiversity and ecosystem processes (27, 28). By performing a fully reciprocal microbial transplant experiment across a gradient of nearly 2,000-m elevation and 15 °C soil temperature, we were able to expand upon previous findings from laboratory manipulations that show that microbial composition can influence decomposition (9, 10, 13). We find that natural communities of microbial decomposers are not only functionally distinct, but their functioning depends on an interaction between the community and its climate, supporting a functional interaction model (Fig. 1E). Moreover, community differences accounted for a large fraction of the variation in decomposition observed. After a year, the abiotic environment was still the largest factor explaining decomposition rates (30%) and litter chemistry (53%), but the microbial community and its interactions with the environment also accounted for 22% of the variation in decomposition rate and 21% of the variation in litter chemistry. These results counter the often implicit assumption that the contribution of microbial community composition to decomposition is negligible relative to climate or litter quality (29).
While decomposition responses to climate depended on the microbial community, they were not consistent with home-field advantage. Surprisingly, microbial communities from the extreme sites along the gradient were functionally similar across the climate gradient, with desert microbes decomposing just as much litter in the subalpine environment as subalpine microbes and vice versa. At the same time, communities from the midelevation sites appeared to perform opposite from what would be predicted by home-field advantage (25), with the scrubland microbes performing best in grassland and the grassland microbes performing best in scrubland, with decomposition rates varying by as much as 40% in a single environment (Fig. 4). Notably, home-field advantage is usually considered in terms of litter quality (25), whereas in this experiment we kept litter quality constant and considered performance in terms of a home climate. Nonetheless, other studies have found inconsistent effects of home-field advantage (30) or observed that these effects were limited to recalcitrant litter types (31, 32). Ultimately, although the exact mechanisms remain unclear, our results suggest that ecosystem predictions can be improved by considering the relationship between a microbial community’s ability to degrade leaf litter and its response to new climate conditions.
In addition to having large impacts on decomposition rates, the microbial community also had smaller but significant effects on the types of carbon compounds degraded. This is important because the types of carbon compounds left behind can influence carbon storage (1, 33). These results indicate that litter communities from different environments vary in carbohydrate degradation traits, similar to previous work in the grassland site, showing that communities subjected to drought conditions shift in composition and glycoside hydrolase (GH) abundance (11).
The duration of our experiment also allowed us to tease apart the timing of the bacterial and fungal compositional shifts and their corresponding functional consequences. After 18 mo, bacterial composition was more reflective of their new environment than of their initial inoculum, while fungal composition still primarily reflected the initial inoculum. This result—that fungal communities are more resistant to change—was also found in an earlier litter transplant experiment within the grassland site (11). One potential explanation is faster rates of turnover in bacterial communities, which are known to respond more quickly to disturbances (34) than fungi (35).
The timing of these shifts relative to the functional consequences of the inoculated community suggests that bacterial decomposers may be less functionally redundant than fungal decomposers and that they might have larger effects on decomposition than previously believed. The effect of the microbial community (and particularly the community-by-site interaction) largely attenuated from 12 to 18 mo, even though the fungal communities within a site were still highly distinct at 18 mo. A review of the subject also concluded that shifts in fungal community structure do not necessarily influence decomposition rates (32), potentially due to the high overlap in metabolic activities of saprobic soil fungal species (36, 37). Indeed, both aquatic (38) and terrestrial (39) laboratory manipulations found that fungal diversity–decomposition relationships saturated rapidly after the addition of only two to six species. In contrast, a study that manipulated bacterial richness found increasing respiration function with diversity beyond 72 species (40). These results might be attributed to the larger breadth of phylogenetic diversity and thus corresponding functional traits represented by the bacterial communities. In our study, >95% of fungal taxa belonged to a single phylum (Ascomycota), whereas the vast majority of bacteria belonged to four phyla, representing hundreds of millions of years of evolution between them. In addition, a large diversity of GH genes have been found in bacteria from leaf litter (41, 42) and soil (43). Interestingly, recent work on soil microbial decomposers suggests that both fungi and bacteria are involved in complex organic matter breakdown, and that interactions between fungi and bacteria in decomposition are perhaps more lateral and less hierarchical than previously believed (44). Thus, it is becoming clear that bacteria can have strong impacts on decomposition. The larger phylogenetic diversity and breadth of metabolic capacities of bacteria may explain why shifts in bacterial composition appear to have a stronger effect on decomposition than shifts in fungal diversity.
Finally, it is notable that microbial composition, but not initial microbial biomass, predicted litter decomposition rates. Decomposition within a site was not correlated with fungal or bacterial biomass in the inoculum leaf litter. For instance, the desert community, with its low initial inoculum biomass, carried out decomposition at similar rates to communities with greater biomass. At the same time, microbial biomass and decomposition were positively correlated across all samples and sites. Many studies have observed similar correlations; for instance, a recent study found that soil microbial biomass, as measured by substrate-induced respiration, was correlated with leaf litter decomposition as much as litter quality and climate (6). However, our transplant experiment indicates that this correlation is not due to microbial biomass driving decomposition. Instead, biomass and decomposition might be correlated because they are both influenced by environment. Alternatively, faster decomposition might result in higher microbial biomass.
In conclusion, decomposition responses to changing temperature and precipitation depended on the composition of a microbial decomposer community. This study examines the decomposition response curves of different microbial communities across a range of climate conditions that could be relevant for predictions of ecosystem functioning. In fact, the dominant plant taxa along this same elevation gradient have shifted upward over the last 30 y due to climate change (24). While it is impossible to know if the microbial communities have also begun to shift their range due to a lack of historical data, our study indicates that bacterial and fungal decomposer communities take time to respond to changes in climate, and this lag has consequences for functioning. Future work should also consider invertebrate grazers (45), viruses, and fungal–bacterial interactions to obtain a more complete understanding of decomposition responses to climate change.
Materials and Methods
Field Experiment.
The five field sites (desert, scrubland, grassland, pine–oak, and subalpine, named for the vegetation present) were selected to represent a wide temperature and precipitation range within Southern California (Fig. 1A and SI Appendix, Table S1). On October 19, 2015, we deployed 300 litterbags containing irradiated grassland litter inoculated with one of the microbial communities from each of the five sites into each site (Fig. 1B). We selected litter from the midelevation grassland site as a common substrate, because the site is intermediate in temperature and precipitation and has been intensively characterized as part of the long-term Loma Ridge Global Change Experiment (11, 12). The grassland is dominated by the annual grass genera Avena, Bromus, and Lolium; the annual forb genera Erodium and Lupinus; and the native perennial grass Nassella pulchra (11).
The grassland litter was homogenized by grinding in a coffee grinder. In addition to improving litter homogenization, pregrinding (rather than clipping) aids in subsampling of the decomposed litter for downstream analyses. Homogenized litter (5 g) was placed into nylon membrane bags with 0.22-µm pores (cat. no. SPEC17970; Tisch Scientific). This pore size was selected to allow for the movement of water and nutrients but prevent the dispersal of exogenous microbial cells into the bags (12). The litterbags were then sterilized with at least 22 kGy gamma irradiation (Sterigenics). Microbial growth was not observed when the irradiated litter was plated on agar media, but we recognize that complete sterilization is unlikely. However, our goal was to knock down the existing community to such low abundance that the inocula communities could become established, which is confirmed by our results. To create the five microbial inocula, four samples of litter from each of the sites was collected on September 11, 2015, ground, homogenized within site, and 50 mg inoculum was added to each sterile litterbag to manipulate microbial community origin.
We placed 60 litterbags in each site such that four replicates of each microbial community treatment could be collected every 6 mo for 18 mo (5 sites × 5 inocula × 4 replicate plots × 3 time points = 300 litterbags). Replicates were distributed across four 1 m × 1 m plots separated by >5 m. Litterbags from each plot were collected on April 5, 2016, October 24, 2016, and on April 18, 2017. To assess the composition of the in situ microbial communities outside the litterbags, litter adjacent to the plots (the mixture of decaying plant species present) was collected at initial deployment and at each litterbag collection (5 sites × 4 replicate plots × 4 time points = 80 in situ survey samples).
To compare the decomposition rate within the transplant bags versus in situ litter decomposition, we deployed two additional sets of litterbags within each plot (2 types × 5 sites × 4 plots × 3 time points = 120 litterbags). One set of bags contained local, clipped (not ground) litter placed in litterbags made of 2-mm mesh window screening (in situ open). To test how the nylon membrane mesh contributed to these differences, the second set contained local, clipped litter placed in the nylon membrane litterbags (in situ closed).
For information on how temperature, precipitation, decomposition, litter chemistry, and bacterial and fungal abundance were assessed, and full details on sample processing, DNA extraction, genetic analysis, and bioinformatics, please see SI Appendix, Supplemental Materials and Methods. Briefly, bacterial community composition was characterized using the V4 region of the 16S ribosomal RNA gene and fungal composition was characterized using the ITS2 region with Illumina MiSeq. Sequences were submitted to the National Center for Biotechnology Information Sequence Read Archive under accession no. SRP150375. All bioinformatics processing was conducted in UPARSE (46) version 10. Analyses were conducted by defining both 97% OTUs and exact sequence variants, but since the results were nearly identical (47), we only present the analyses using 97% OTUs.
Statistical Analysis.
All statistical analysis was performed in R version 3.4.0 with some additional analysis conducted in PRIMER 6+ (48). Scripts and data for R analyses are available on https://github.com/stevenallison/UCIClimateExperiment and https://github.com/sydneyg/OTUvESV. We tested the treatment main effects (site, inoculum) and interaction effect (site-by-inoculum) with a two-way ANOVA for each time point for decomposition, bacterial abundance, and fungal abundance. Bacterial and fungal abundance were square root transformed to improve normality. We estimated effect sizes–the relative importance of the two manipulated factors—on microbial abundance and decomposition with partial eta-squared as it is the preferred method for n-way ANOVA (49). We then estimated the total variance explained by multiplying the partial eta-squared by the adjusted R2 of the model.
For bacterial and fungal community composition, we calculated dissimilarity matrices with avgdist function in vegan accessed via GitHub (https://github.com/Microbiology/vegan/blob/master/R/avgdist.R). OTU tables were normalized by subsampling to the lowest common sampling depth 100× (5-17,000 sequences per sample), then the median of the Bray–Curtis dissimilarity matrices calculated from each of subsampled OTU tables was square root transformed. We then tested the treatment main effects (site, inoculum) and interaction effect on the transplanted litterbags and the main effects (site, sample date) and interaction effect on the inoculum and in situ leaf litter communities, with a two-way PERMANOVA as implemented with the Adonis function in vegan (50). We estimated the effect size of the manipulated factors on microbial composition using the estimates of the components of variation from PERMANOVA (11).
To test changes for in litter chemistry, we calculated the Euclidian distance between samples of the nonash leaf litter proportions. We tested the effect of treatments (site, inoculum) and interaction on litter chemistry with Adonis. We visualized the differences in leaf chemistry per site with nonmetric multidimensional scaling (NMDS) and used the “envfit” function to determine which organic compounds correlated well with ordination space. Figures were created in base R graphics or ggplot2.
Supplementary Material
Acknowledgments
We thank M. Goulden and N. Baker for assistance in acquiring precipitation data for the climate gradient and all members of the S.D.A., A.C.M., J.B.H.M., and K.K.T. laboratories for their assistance in setting up the reciprocal transplant experiment and data collection used for this analysis. We thank two anonymous reviewers for providing helpful tips to improve our manuscript. This work was supported by the National Science Foundation (DEB-1457160) and the Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-SC0016410).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP150375) and the data and scripts necessary to run the majority of analyses have been deposited to GitHub (available at https://github.com/stevenallison/UCIClimateExperiment and https://github.com/sydneyg/OTUvESV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811269115/-/DCSupplemental.
References
- 1.McGuire KL, Treseder KK. Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol Biochem. 2010;42:529–535. [Google Scholar]
- 2.Wang S, Ruan H, Han Y. Effects of microclimate, litter type, and mesh size on leaf litter decomposition along an elevation gradient in the Wuyi Mountains, China. Ecol Res. 2010;25:1113–1120. [Google Scholar]
- 3.Currie WS, et al. Cross-biome transplants of plant litter show decomposition models extend to a broader climatic range but lose predictability at the decadal time scale. Glob Change Biol. 2010;16:1744–1761. [Google Scholar]
- 4.Makkonen M, et al. Highly consistent effects of plant litter identity and functional traits on decomposition across a latitudinal gradient. Ecol Lett. 2012;15:1033–1041. doi: 10.1111/j.1461-0248.2012.01826.x. [DOI] [PubMed] [Google Scholar]
- 5.Handa IT, et al. Consequences of biodiversity loss for litter decomposition across biomes. Nature. 2014;509:218–221. doi: 10.1038/nature13247. [DOI] [PubMed] [Google Scholar]
- 6.Bradford MA, et al. A test of the hierarchical model of litter decomposition. Nat Ecol Evol. 2017;1:1836–1845. doi: 10.1038/s41559-017-0367-4. [DOI] [PubMed] [Google Scholar]
- 7.Graham EB, et al. Microbes as engines of ecosystem function: When does community structure enhance predictions of ecosystem processes? Front Microbiol. 2016;7:214. doi: 10.3389/fmicb.2016.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schimel J. Biogeochemical models: Implicit versus explicit microbiology. In: Schulze ED, et al., editors. Global Biogeochemical Cycles in the Climate System. Academic; New York: 2001. pp. 177–183. [Google Scholar]
- 9.Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- 10.Cleveland CC, et al. Litter quality versus soil microbial community controls over decomposition: A quantitative analysis. Oecologia. 2014;174:283–294. doi: 10.1007/s00442-013-2758-9. [DOI] [PubMed] [Google Scholar]
- 11.Martiny JBH, et al. Microbial legacies alter decomposition in response to simulated global change. ISME J. 2017;11:490–499. doi: 10.1038/ismej.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison SD, et al. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology. 2013;94:714–725. doi: 10.1890/12-1243.1. [DOI] [PubMed] [Google Scholar]
- 13.Alster CJ, Koyama A, Johnson NG, Wallenstein MD, von Fischer JC. Temperature sensitivity of soil microbial communities: An application of macromolecular rate theory to microbial respiration. J Geophys Res Biogeosci. 2016;121:1420–1433. [Google Scholar]
- 14.Matulich KL, Martiny JBH. Microbial composition alters the response of litter decomposition to environmental change. Ecology. 2015;96:154–163. doi: 10.1890/14-0357.1. [DOI] [PubMed] [Google Scholar]
- 15.Strickland MS, Keiser AD, Bradford MA. Climate history shapes contemporary leaf litter decomposition. Biogeochemistry. 2015;122:165–174. [Google Scholar]
- 16.Averill C, Waring BG, Hawkes CV. Historical precipitation predictably alters the shape and magnitude of microbial functional response to soil moisture. Glob Change Biol. 2016;22:1957–1964. doi: 10.1111/gcb.13219. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MA, Berg B, Maynard DS, Wieder WR, Wood SA. Understanding the dominant controls on litter decomposition. J Ecol. 2016;104:229–238. [Google Scholar]
- 18.Keiser AD, Bradford MA. Climate masks decomposer influence in a cross-site litter decomposition study. Soil Biol Biochem. 2017;107:180–187. [Google Scholar]
- 19.Baker NR, Allison SD. Extracellular enzyme kinetics and thermodynamics along a climate gradient in southern California. Soil Biol Biochem. 2017;114:82–92. [Google Scholar]
- 20.Seager R, et al. Model projections of an imminent transition to a more arid climate in southwestern North America. Science. 2007;316:1181–1184. doi: 10.1126/science.1139601. [DOI] [PubMed] [Google Scholar]
- 21.Sundqvist MK, Sanders NJ, Wardle DA. Community and ecosystem responses to elevational gradients: Processes, mechanisms, and insights for global change. Annu Rev Ecol Evol Syst. 2013;44:261–280. [Google Scholar]
- 22.Baker NR, Khalili B, Martiny JBH, Allison SD. Microbial decomposers not constrained by climate history along a Mediterranean climate gradient in southern California. Ecology. 2018;99:1441–1452. doi: 10.1002/ecy.2345. [DOI] [PubMed] [Google Scholar]
- 23.Looby CI, Treseder KK. Shifts in soil fungi and extracellular enzyme activity with simulated climate change in a tropical montane cloud forest. Soil Biol Biochem. 2018;117:87–96. [Google Scholar]
- 24.Kelly AE, Goulden ML. Rapid shifts in plant distribution with recent climate change. Proc Natl Acad Sci USA. 2008;105:11823–11826. doi: 10.1073/pnas.0802891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gholz HL, Wedin DA, Smitherman SM, Harmon ME, Parton WJ. Long-term dynamics of pine and hardwood litter in contrasting environments: Toward a global model of decomposition. Glob Change Biol. 2000;6:751–765. [Google Scholar]
- 26.Austin AT, Vivanco L. Plant litter decomposition in a semi-arid ecosystem controlled by photodegradation. Nature. 2006;442:555–558. doi: 10.1038/nature05038. [DOI] [PubMed] [Google Scholar]
- 27.Dunne JA, Saleska SR, Fischer ML, Harte J. Integrating experimental and gradient methods in ecological climate change research. Ecology. 2004;85:904–916. [Google Scholar]
- 28.Fukami T, Wardle DA. Long-term ecological dynamics: Reciprocal insights from natural and anthropogenic gradients. Proc Biol Sci. 2005;272:2105–2115. doi: 10.1098/rspb.2005.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adair EC, et al. Simple three-pool model accurately describes patterns of long-term litter decomposition in diverse climates. Glob Change Biol. 2008;14:2636–2660. [Google Scholar]
- 30.Veen GF, Keiser AD, Van der Putten WH, Wardle DA. Variation in home-field advantage and ability in leaf litter decomposition across successional gradients. Funct Ecol. 2017;32:1563–1574. [Google Scholar]
- 31.St John MG, Orwin KH, Dickie IA. No ‘home’ versus ‘away’ effects of decomposition found in a grassland-forest reciprocal litter transplant study. Soil Biol Biochem. 2011;43:1482–1489. [Google Scholar]
- 32.van der Wal A, Geydan TD, Kuyper TW, de Boer W. A thready affair: Linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiol Rev. 2013;37:477–494. doi: 10.1111/1574-6976.12001. [DOI] [PubMed] [Google Scholar]
- 33.Wickings K, Grandy AS, Reed SC, Cleveland CC. The origin of litter chemical complexity during decomposition. Ecol Lett. 2012;15:1180–1188. doi: 10.1111/j.1461-0248.2012.01837.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferrenberg S, et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013;7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glassman SI, Levine CR, DiRocco AM, Battles JJ, Bruns TD. Ectomycorrhizal fungal spore bank recovery after a severe forest fire: Some like it hot. ISME J. 2016;10:1228–1239. doi: 10.1038/ismej.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deacon LJ, et al. Diversity and function of decomposer fungi from a grassland soil. Soil Biol Biochem. 2006;38:7–20. [Google Scholar]
- 37.Talbot JM, et al. Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA. 2014;111:6341–6346. doi: 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang CK, Chauvet E, Gessner MO. Magnitude and variability of process rates in fungal diversity-litter decomposition relationships. Ecol Lett. 2005;8:1129–1137. doi: 10.1111/j.1461-0248.2005.00815.x. [DOI] [PubMed] [Google Scholar]
- 39.Setälä H, McLean MA. Decomposition rate of organic substrates in relation to the species diversity of soil saprophytic fungi. Oecologia. 2004;139:98–107. doi: 10.1007/s00442-003-1478-y. [DOI] [PubMed] [Google Scholar]
- 40.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- 41.Chase AB, Arevalo P, Polz MF, Berlemont R, Martiny JBH. Evidence for ecological flexibility in the cosmopolitan genus curtobacterium. Front Microbiol. 2016;7:1874. doi: 10.3389/fmicb.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berlemont R, et al. Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol. 2014;5:639. doi: 10.3389/fmicb.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep. 2016;6:25279. doi: 10.1038/srep25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Mondéjar R, et al. Decomposer food web in a deciduous forest shows high share of generalist microorganisms and importance of microbial biomass recycling. ISME J. 2018;12:1768–1778. doi: 10.1038/s41396-018-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cebrian J. Patterns in the fate of production in plant communities. Am Nat. 1999;154:449–468. doi: 10.1086/303244. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 47.Glassman SI, Martiny JBH. Broadscale ecological patterns are robust to use of exact sequence variants versus operational taxonomic units. MSphere. 2018;3:e00148-18. doi: 10.1128/mSphere.00148-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson MJ, Gorley RN, Clarke RK. Permanova+ for Primer: Guide to Software and Statistical Methods. Primer-E; Plymouth, UK: 2008. [Google Scholar]
- 49.Ialongo C. Understanding the effect size and its measures. Biochem Med (Zagreb) 2016;26:150–163. doi: 10.11613/BM.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oksanen J, et al. 2012. vegan: Community Ecology Package. R Package Version 2.0-10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.