Significance
SMC–kleisin protein complexes contribute to the structural maintenance of chromosomes and are essential for the functioning of cells across all domains of life. In particular, condensin is a ring-shaped motor complex responsible for chromosome segregation; however, key structural features of this complex remain controversial. Leveraging coevolutionary information, we are able to determine an atomically detailed structure of the whole condensin complex. Converging evidence indicates that the complex is composed of a single ring and undergoes large structural rearrangements to fulfill its function. Our findings constitute the first step toward studying the structure–function relationship of the various molecular motors operating on DNA.
Keywords: bacterial condensin, SMC–kleisin complexes, coevolutionary information, direct coupling analysis, DNA translocation
Abstract
Protein assemblies consisting of structural maintenance of chromosomes (SMC) and kleisin subunits are essential for the process of chromosome segregation across all domains of life. Prokaryotic condensin belonging to this class of protein complexes is composed of a homodimer of SMC that associates with a kleisin protein subunit called ScpA. While limited structural data exist for the proteins that comprise the (SMC)–kleisin complex, the complete structure of the entire complex remains unknown. Using an integrative approach combining both crystallographic data and coevolutionary information, we predict an atomic-scale structure of the whole condensin complex, which our results indicate being composed of a single ring. Coupling coevolutionary information with molecular-dynamics simulations, we study the interaction surfaces between the subunits and examine the plausibility of alternative stoichiometries of the complex. Our analysis also reveals several additional configurational states of the condensin hinge domain and the SMC–kleisin interaction domains, which are likely involved with the functional opening and closing of the condensin ring. This study provides the foundation for future investigations of the structure–function relationship of the various SMC–kleisin protein complexes at atomic resolution.
Members of the structural maintenance of chromosomes (SMC) and kleisin families of proteins are conserved in all domains of life and have key roles in the maintenance of chromosomes (1–3). In eukaryotes, several SMC–kleisin complexes operate during and after DNA replication to promote chromosome segregation in mitosis and meiosis. One such protein complex is cohesin, which has several critical biological roles such as mediating sister-chromatid cohesin, promoting DNA double-strand break repair, and regulating gene expression (2). Defects in the cohesin complex have been related to genetic disorders (4, 5), as well as to several types of human cancer (6–10). In addition, it has been suggested that cohesin organizes DNAs into chromatids by capturing small loops of DNA and then extruding them in a progressive manner (10–15). Together with compartmentalization of chromatin (16–18), DNA loop extrusion has been used to explain intrachromosomal interactions during interphase, as measured by Hi-C. In interphase, the extrusion seems to be primarily regulated by the activity of the insulator factor CTCF (10, 12, 13) and to be involved in establishing essential distal interactions between enhancers and promoters.
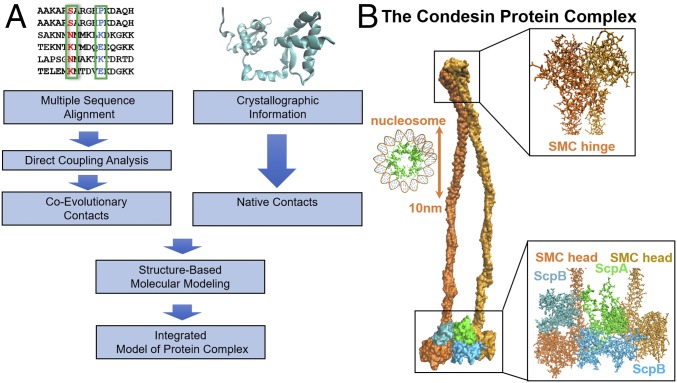
In prokaryotes, the SMC–kleisin complex condensin segregates and condenses bacterial genomes during the process of cell division (19). Condensin is formed by SMC–kleisin proteins SMC and ScpA, respectively, and a third subunit, ScpB (Fig. 1B) (7–9, 11). This family of proteins shares a unique architecture comprising two globular domains: a nucleotide-binding domain (NBD) of the ABC-type ATPase fold (SMC head) and a central hinge domain, connected by a long antiparallel intramolecular coiled coil. The two SMC monomers held together at the hinge are additionally bridged by kleisin, creating stable tripartite rings that associate with and entrap chromosomes (18–21). A recent study pointed out that the hinge domain can also adopt an asymmetrical configuration (22), potentially functioning as a flexible joint that can accommodate the opening and closing movement of the coiled-coil region. However, in what manner SMC and kleisin assemble to form condensin, as well as the homologous cohesin, and the molecular details of their biological function remain unclear.
Fig. 1.
Schematic of our integrative computational approach together with the resulting proposed structure for the prokaryotic condensin complex. (A) We obtain MSAs for the protein families of SMC, ScpA, and ScpB from Pfam (41). Using these sequence data as input, DCA is used to infer intraprotein and interprotein coevolving contacts. Known crystal structures of the condensin subunits are obtained from the PDB (27, 28). To obtain the complete structure of the condensin complex, we use both coevolutionary and crystallographic residue contacts as constraints in MD simulations. (B) The proposed structure for whole condensin complex, composed of 2,924 residues at atomic resolution. A single-ring structure is consistent with all of the available information, both structural and coevolutionary. A nucleosome (PDB ID code 5T5K) is shown on the right-hand side to illustrate the scale of the figure.
Despite major progress in recent years, many questions related to the structure and function of SMC–kleisin complexes remain open. These open questions include the precise mechanisms that lead to loading, entrapment, release, and stable cohesin, as well as the exact role of the NBD and ATPase domains (23). A recent study demonstrated the ability of both eukaryotic condensin (24) and cohesin (10) to serve as a mechanochemical motor that can translocate along DNA. Importantly, the complete structure of SMC–kleisin complexes has yet to be established. The two main models of the pictorial structure of these complexes are (i) a single ring that embraces two double-stranded DNAs (3, 23) or (ii) two rings acting as a pair of molecular “handcuffs” where each ring can embrace its own double-stranded DNA (25). Although structural data exist for each subunit of the condensin protein complex (26–28), such data are limited for interprotein interactions between the subunits. Furthermore, existing crystallographic data cannot capture the dynamics of interaction surfaces and therefore are limited in their ability to disentangle the various conformations of the complex.
Here, we set out to determine the molecular architecture of bacterial SMC–ScpAB protein complex by using the limited available structural data together with the information encoded in the abundant protein sequence data (26–28). We take advantage of residue coevolution to predict residue contacts in a 3D structure. Because of the evolutionary constraint to maintain protein fold and function, correlations arise between the amino acid identities at different residues sites of the proteins ScpA, ScpB, and SMC, which form the complex. According to this phenomenon, highly coevolving residue pairs are likely to be spatially proximate in a 3D structure or complex. To disentangle the complex network of cross-correlations between amino acids, we construct a global statistical model of the sequence data using direct coupling analysis (DCA). This methodology, which uses a maximum entropy approach (29), has already been used to successfully identify both intraprotein (30–35) and interprotein (30, 33, 34, 36, 37) contacts.
Using DCA, we first identify the highly coevolving residue pairs as a proxy for intraprotein and interprotein residue pairs within the SMC–kleisin complex that form contacts in a 3D complex (30–34, 38). We use the residue contacts obtained from publicly available crystal structures to validate our approach. Then, we combine the coevolutionary information and the limited structural data with molecular-dynamics (MD) simulations to determine the complete structures of the subunits of the condensin complex, including the disordered region of ScpA. We use a structure-based model (SBM) enhanced by the newly obtained coevolutionary information to study the formation of the condensin complex. Using this approach, we also test the plausibility of alternative stoichiometries; all of the evidence discussed below seems to suggest a single-ring structure for prokaryotic condensin. Additionally, we find evidence pointing to alternative functional conformations encoded in the sequence data, particularly within the SMC hinge domain and SMC–kleisin interaction domains. Finally, we report an atomic-scale structure for the whole condensin complex that is consistent with all of the structural and coevolutionary information currently available (Fig. 1B).
Results
Establishing the Full Structures of the Condensin Subunits.
In establishing the full structure of the three subunits, it should be noted that the structures of several fragments of the SMC–kleisin complex have already been obtained separately, in multiple experimental studies. Available experimental data include several fragments of SMC: the hinge domain [Protein Data Bank (PDB) ID code 1GXL], the head domain along with fragments of the coiled coil region (PDB ID code 3ZGX). The ScpA polypeptide chain is composed of an N-terminal winged-helix domain (PDB ID code 4I98) and a C-terminal winged-helix domain (PDB ID code 4I99) connected by a winding segment (amino acids 86–150). Crystallographically, it was demonstrated that the C-domain of ScpA is disordered in the absence of an intrinsic binding partner, the SMC head domain. ScpB is also composed of N- and C-terminal domains, connected by an interprotein contact region.
Using DCA, we predict the structure of the whole condensin complex, including all of the parts mentioned above that have known crystal structures. The crystallographically determined contacts are then used to assess the validity of the coevolutionary approach. We begin by obtaining coevolutionary contacts for each subunit separately. The top coevolving intraprotein residue pairs (see Materials and Methods for the ranking score) are compared with the available, experimentally determined physical contacts for the following systems: the SMC hinge domain (27) (see Fig. 4A; PDB ID code 1GXL), the ScpB subunit (28) (see Fig. 3B; PDB ID code 4I98), and the ScpA subunit (28) (see Fig. 3A; PDB ID codes 4I98 and 4I99). In addition, for the overall 1,175-residue SMC protein, including N- and C- head domains, we introduce the top 1,075 DCA contacts (28) (see Fig. 5A; PDB ID code 3ZGX). One can easily see in the contact maps that highly coevolving residue pairs largely agree with physical contacts in the native structures indicating that the DCA methodology can successfully predict secondary and tertiary structural contacts. After validating our approach, we proceed in studying the condensin complex using both crystallographic (wherever available) and coevolutionary information (entire complex). Additional maps comparing the DCA-derived contacts with crystallographic results taken from PDB can be found in SI Appendix.
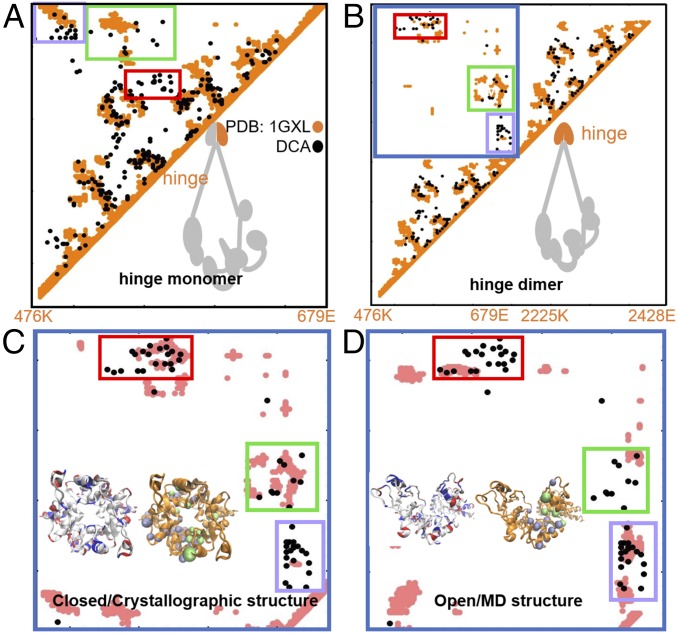
Fig. 4.
DCA predicts two alternative configurations for the SMC hinge domain. Contact maps show the comparison between DCA-derived residue contacts and crystallographic contacts (A and B) as well as between DCA contacts and simulation-derived contacts (C and D). DCA contacts are shown in black, crystallographic contacts are shown in orange, and MD simulation-derived contacts are shown in pink. (A) DCA recapitulates available crystallographic data for a single hinge domain as well as (B) for two hinge domains (27). DCA identifies three clusters of contacts, enclosed in red, green, and purple rectangles, as the main interfacial residue contacts. These DCA-derived contacts are inconsistent with the structure of a single SMC monomer (A), while consistent with the interprotein region of the contact maps, which is marked by the blue border in B and magnified in C and D. Contact maps for two alternative structures for hinge interface region obtained from MD shown in the following: (C) The “closed” configuration, consistent with the available crystallographic data about the interface region. The red and green DCA-derived clusters of interfacial coevolutionary interactions are satisfied by this structure. (D) The “open” configuration, which does not resemble any known crystallographic data. This simulated structure satisfies the coevolutionary constraints represented by the contacts mainly in the purple rectangle. Neutral, positively charged, and negatively charged amino acids are represented in white, blue, and red, respectively. MD configurations are shown as orange cartoons with top DCA contacts from the green and purple rectangles shown as green and purple beads. These two alternative configurations of the complex may be involved with the opening and closing of the ring and, therefore, shed light on the hinge interaction with DNA.
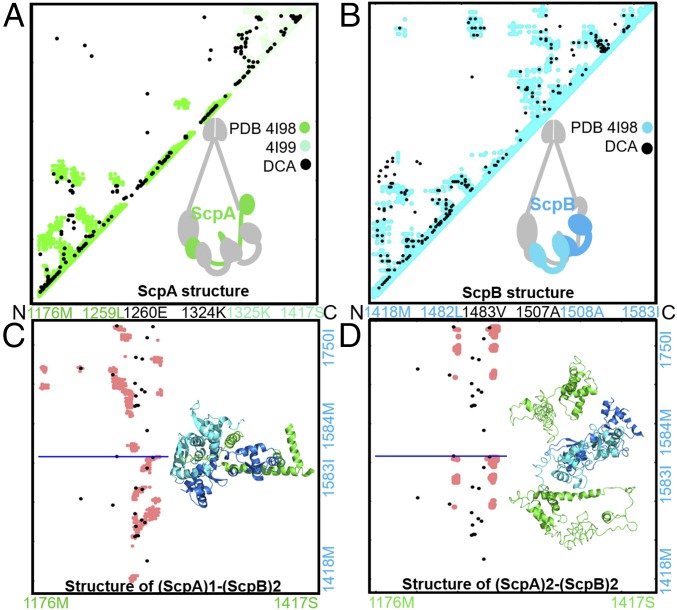
Fig. 3.
The ScpAB system favors a single ScpA subunit accommodating two ScpB proteins. Contact maps show a comparison between DCA-derived contacts and crystallographic data (A and B) as well as between DCA contacts and simulation-derived contacts (C and D). DCA contacts are shown in black. Crystallographic contacts are shown in green and cyan for the ScpA and ScpB subunits, respectively. Contacts from MD simulation are shown in pink (C and D). The residue indices and corresponding amino acid identities are labeled along the axes. (A) DCA recapitulates available crystallographic data for ScpA subunit (28). (B) DCA recapitulates available crystallographic data for ScpB subunit (28). (C) Comparison between simulated data from MD and DCA contacts for the stoichiometry (ScpA)1–(ScpB)2. Residue indices are shown on the axis, with the horizontal blue line separating the contact maps of the two ScpB proteins. It is evident that DCA-derived contacts are consistent with the simulated structure of either the two copies of ScpB. (D) Comparison between simulated data from MD and DCA contacts for the stoichiometry (ScpA)2–(ScpB)2. Residue indices are shown on the axis, with the horizontal blue line separating the contact maps of the two ScpB proteins. This stoichiometry results in a simulated structure inconsistent with the DCA-derived coevolutionary contacts. The comparison between C and D strongly favors the (ScpA)1–(ScpB)2 stoichiometry vs. the (ScpA)2–(ScpB)2.
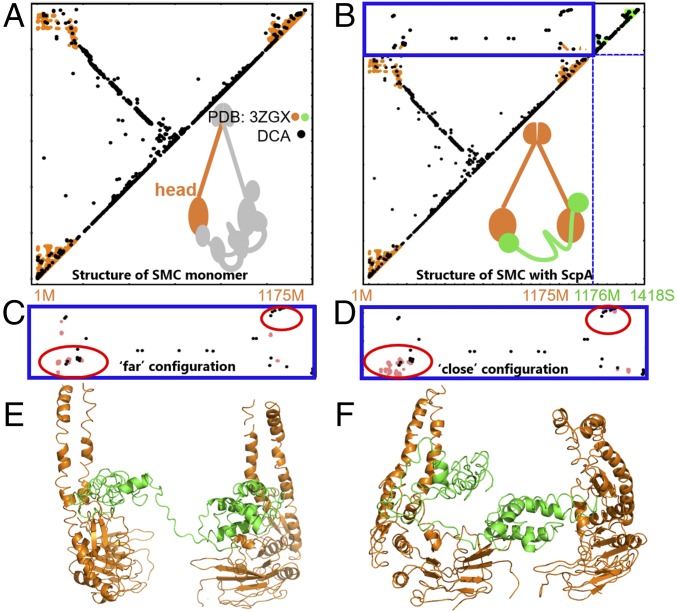
Fig. 5.
The SMC–kleisin system has several possible configurations. Contact maps show the comparison between the DCA contacts with crystallographic contacts (A and B) as well as the DCA contacts with simulation-derived contacts (C and D). SMC–ScpA interprotein contact domain is shown in blue border (B–D). DCA contacts are shown in black, crystallographic contacts are shown in orange and green, and MD simulations-derived contacts are shown in pink. (A) DCA recapitulates available crystallographic data for a single SMC head domain as well as for (B) a single SMC head domain (orange) with crystallographically determined contacts from a single ScpA (green) protein complex. (C–F) MD simulations reveal two alternative interfacial configurations between SMC heads and kleisin. One configuration in C and E shows the two heads relatively far from each other, while the other in D and F shows the two SMC heads closer together. (C) The interprotein contact map shows the comparison between simulated data from MD and DCA contacts for the “far” configuration. DCA contacts are satisfied in the bottom red circle. (D) The interprotein contact map shows the comparison between simulated data from MD and DCA contacts for the “close” configuration. Here, DCA contacts are satisfied in both top and bottom red circles. (E) A representative structure of the far configuration. SMC and ScpA are shown in orange and green, respectively. (F) A representative structure of the close configuration. SMC and ScpA are shown in orange and green, respectively. Our results demonstrate several alternative configurations for the SMC–kleisin system, suggesting overall dynamics of the condensin ring.
Coevolutionary Information Suggests Condensin as a Single Ring Structure.
To resolve the stoichiometry of the condensin protein complex, we begin with the simpler single SMC–ScpAB single-ring structure (Fig. 2). It has been suggested that, for the Streptococcus pneumoniae bacteria, a single ScpA subunit binds to ScpB with 1:2 stoichiometry, resulting in an overall trimetric ScpAB subcomplex. This subcomplex connects with two SMC head domains, one on each side of ScpA, thus forming an overall 2:1:2 SMC–ScpA–ScpB stoichiometry.
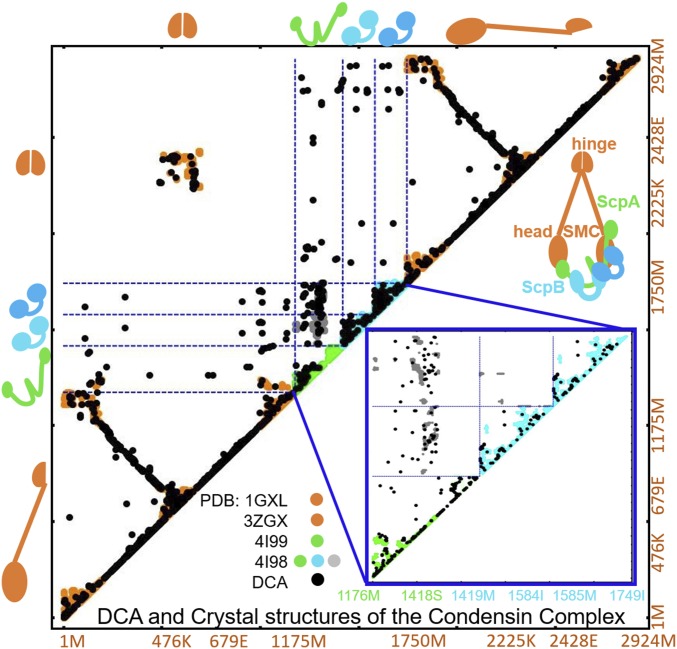
Fig. 2.
Overview of all crystallographic structural data for the condensin complex together with coevolutionary information. Residue–residue contacts map is shown for the DCA-derived contacts (represented by black dots) together with the contacts from available crystal structures obtained from PDB for the SMC–ScpA–ScpB–ScpB–SMC system. The residue indices for the whole system and their respective amino acid identities of each protein subunit are shown on the x and y axes. Orange, green, and cyan represent crystallographic results of SMC, ScpA, and ScpB subunits, respectively (27, 28). In the intraprotein regions, wherever data are available, the agreement between crystallography and coevolutionary contacts is evident. A magnified view of the ScpA–ScpB–ScpB crystallographic (ScpA, ScpB, and ScpAB interprotein region shown in green, cyan, and gray, respectively) and predicted DCA contacts (represented in black) are shown in the bottom right corner in a blue box.
Fig. 2 shows the top coevolving contacts for the whole single-ring structure, composed of five parts: SMC (amino acids 1–1175)–ScpA (amino acids 1176–1418)–ScpB (amino acids 1419–1584)–ScpB (amino acids 1585–1749)–SMC (amino acids 1750–2924) (black dots mark the DCA-derived contacts). In the following, we will refer to the N- and C-terminal domains of SMC, which form the SMC heads, as SMC Ndomain and SMC Cdomain, respectively.
Using DCA, we establish the top 15 coevolving residue pairs for the following systems: (i) SMC Ndomain–ScpA, (ii) SMC Cdomain–ScpA, (iii) ScpB–ScpA, (iv) SMC Ndomain–ScpB, and (v) SMC Cdomain–ScpB; these residue pairs were assumed to form a structural contact. For the ScpB–ScpB dimer, instead, 10 contacts were obtained by ranking the PDB experimental dimer contacts (28) by the strength of coevolutionary couplings between those residues inferred from DCA. This number of contacts was chosen as a result of predictive positive values (PPV) analysis, compared between our predicted DCA contacts and crystallographic contacts. For all systems, PPV = 0.8 and higher (for more details, see SI Appendix). All of these contacts were then used as constraints in MD simulations (see Materials and Methods for further MD parameters) to obtain the complete structure of the condensin protein complex. This structure, beyond satisfying all of the known intraprotein contacts, also satisfies all of the DCA-derived interprotein contacts between all of the subunits (see SI Appendix, Fig. S2 for discussion on DCA-derived intraprotein and interprotein datasets). This suggests that of the cohesin protein complex might indeed exhibit a single-ring structure, as this structure recapitulates all of the available information, both structural and coevolutionary.
Alternative Higher-Order Stoichiometries.
Leveraging the information contained in the full DCA contact map, we now turn to investigate the plausibility of higher-order stoichiometries for condensin by examining several interprotein contacts between ScpA with two ScpB monomers and with two SMC head domains.
We first focus on the interaction of the ScpAB subcomplex. In the single ring obtained above, a single ScpA subunit can interact with two ScpB proteins. The region composed of residues 66–90 of each ScpB protein forms an interface with the winding segment region of ScpA. It is currently unclear what sequence of event forms the complex. Does ScpA form its interaction surfaces with both ScpB proteins simultaneously, or does it first bind to one protein followed by the second protein? In Fig. 3, we show the predicted intraprotein contacts from DCA of ScpA (Fig. 3A), ScpB (Fig. 3B), respectively.
Using the top 15 DCA contacts between ScpA–ScpB as constraints in our SBM, we performed simulations where one ScpA monomer was capable of forming the top contacts with either ScpB proteins. A resultant interprotein contact map between ScpA and two monomers of ScpB generated from MD simulation is shown in Fig. 3C (represented in pink). The top DCA contacts between ScpA and ScpB are shown in Fig. 3C (marked by black dots). The ScpAB subcomplex was stabilized only when performing simulations in which first a single ScpB forms a complex with ScpA, and only successively a second ScpB monomer binds. This suggests that there exists a precise order of events in the association of the ScpAB subunits.
As was mentioned earlier, the unresolved structure of SMC–kleisin protein complexes has created a controversy regarding the stoichiometry of its subunits. The two dominant models for the SMC–kleisin complex are (i) a single-ring structure, for which a single ScpA protein would interact simultaneously with an ScpB dimer in a 1:2 stoichiometry, that is, the (ScpA)1–(ScpB)2 system; or (ii) a double-ring structure for which the ScpB dimer would simultaneously interact with two ScpA subunits in a 2:2 stoichiometry, that is, the (ScpA)2–(ScpB)2 system. To further investigate the single- vs. double-ring question, we repeated simulations for a (ScpA)2–(ScpB)2 system again using coevolutionary information, that is, providing the same top DCA interprotein ScpA–ScpB contacts between each of the two ScpA and two ScpB proteins. In Fig. 3D, we show the contact map for the (ScpA)2–(ScpB)2 system obtained using MD simulations. As can be seen, in trying to satisfy the top DCA constraints, the system results in a sandwich structure in which the ScpB protein is located in the middle of the two ScpA proteins. In this configuration, which is characterized by identical interactions between ScpA and the ScpB subunits, coevolutionary constraints appear to diverge from physical contacts (Fig. 3D). Once again, our analysis favors the (ScpA)1–(ScpB)2 stoichiometry, corresponding with a single-ring structure.
We now discuss the interactions between the ScpA subunit with the SMC head domains. SMC head domain is composed of the protein’s N and C terminals interacting with each other to form V-shaped heterodimers. Each SMC head, which contains a conserved ATPase domain, associates with the ScpA kleisin subunit (20, 21). The N-terminal region of ScpA forms a helical bundle with the coiled coil emerging from the ATPase of one SMC monomer (23), while its winged helical C terminal binds to the base of the ATPase of the second SMC monomer. This general structure is formed in both prokaryotic and eukaryotic SMC–kleisin complexes, suggesting that asymmetric ring formation is a universal feature (3, 39). A recent study suggested evidence that the ATP-dependent head–head engagement induces a lever movement of the SMC neck region, which might help to separate the coiled-coil arms (22, 26).
We investigate whether a single ScpA subunit interacts with two SMC head domain, that is, the (ScpA)1–(SMC)2 system, or whether the system contains more than one ScpA subunit, that is, the (ScpA)2–(SMC)2 system. As before, we repeated the MD simulations for the (ScpA)2–(SMC)2 system, providing the same top DCA interprotein ScpA–SMC contacts for each of the two ScpA proteins. The MD simulations could not establish stable interaction surfaces between two ScpA subunits with two SMC head domains. Steric hindrance within the (ScpA)2–(SMC)2 system prevents both ScpA interfaces from existing at the same time, resulting in the loss of one of the ScpA subunits. This result suggests that only one ScpA is bound to SMC at any one time, with a single ScpA bridging two SMC complexes. This last evidence, along with the analysis in Fig. 3, further supports the condensin complex being composed of a single ScpA subunit forming a single-ring structure.
DCA Reveals Several Hinge Configurations for the Condensin Protein Complex.
It is widely believed that SMC has two major interaction surfaces: the SMC hinge domain and the SMC–kleisin interaction surface, both of which have been suggested to serve as entry and exit gates for DNA, respectively (3). In this section, we analyze the coevolutionary information obtained from DCA for the SMC hinge domain, which is composed of an interaction surface between two SMC monomers. In Fig. 4, we show contacts obtained from the crystal structure of a single hinge domain (Fig. 4A) and from a hinge dimer (Fig. 4B) as was crystallographically obtained for Thermotoga maritima (PDB ID code 1GXL). The intraprotein contacts of each monomer and their interfacial contacts are represented in orange. The interprotein domain is shown in the blue rectangle. The top 210 DCA contacts are also shown, represented by black circles. In this case, DCA cannot distinguish between intraprotein and interprotein contacts, because bacterial SMC is composed of two identical monomers. However, there exist three clusters of coevolving pairs that do not agree with the contacts from the crystallographic structure of the monomer (see orange in Fig. 4A); as a result, these contacts are hypothesized to belong to the SMC dimer interfaces at the hinge (see red, green, and purple rectangles in Fig. 4B).
While two clusters (see red and green rectangles) agree well with the crystallographic data of the hinge interface, the third (in purple) has poor agreement with the crystallographically determined contacts. We used the top 15 DCA contacts of both green and purple clusters as spatial constraints in MD simulations. The resulting trajectories show two distinct SMC hinge configurations (Fig. 4 C and D): the first appears as a closed configuration (Fig. 4C) and agrees with crystallography, while the additional one produced from DCA data appears as an open configuration (Fig. 4D). As a result, simulations form one of either two clusters of contacts (enclosed by green and purple rectangles in Fig. 4), while satisfying different regions of the cluster of contacts enclosed by the red rectangle.
It is important to note that several studies have suggested that SMC–kleisin ring structures can open up in the presence DNA strands. One hypothesis is that this opening occurs via the SMC hinge domain (22, 28). When considering the arrangement of charged amino acids in both of our obtained configurations: closed (Fig. 4C, Inset) and open (Fig. 4D, Inset). The negatively charged amino acids Asp and Glu are represented in red, the positively charged amino acid Lys and Arg are represented in blue, while neutral amino acids are represented in white. While the closed configuration of the SMC dimer hinge domain produces a symmetrical charge arrangement (Fig. 4C), the open configuration presents a nonsymmetric arrangement, with two Lys groups now facing a mutual direction. The possibility of several hinge configurations are of major functional significance since it suggests the potential rearrangement of the hinge region in the vicinity of DNA. The positively charge arrangement in this “open” SMC hinge configuration could potentially bind to the negatively charged phosphate groups of DNA. It was previously reported that DNA binding-mediated ATPase activity in SMC heads is regulated by the acetylation of several positively charged Lys residues located both in the SMC head and in the coiled coil (29, 40), further demonstrating the importance of charge arrangement in the tripartite SMC–kleisin complex structure.
In Fig. 5B, we show the crystallographic data regarding the interactions between a single SMC monomer (shown in orange) and the ScpA subunit (shown in green) for the Bacillus subtilis bacteria (PDB ID code 3ZGX). For comparison, the top 325 coevolving contacts from DCA are shown in black. The dashed blue lines separate the intraprotein from the interprotein contacts. The interaction between the helical N domain and ScpA is already captured in the top 50 contacts from DCA.
The top 15 coevolutionary contacts between both the N (amino acids 1–200) and C domains (amino acids 975–1175) of ScpA were used as constraints for the MD simulations of the (ScpA)1–(SMC)2 system; the same contacts are shown in the amplification of the interprotein domain in Fig. 5 C and D. MD simulations resulted in two main configurations, in which the contacts were satisfied, especially in the N-domain region (see blue circles): (i) the two SMC heads remain separated, while the winding segment region of the ScpA demonstrates flexibility to connect the two SMC monomers (Fig. 5 C and E); and (ii) the two SMC heads both make contacts with each other and with ScpA. We note that not all DCA contacts could be simultaneously satisfied in our MD simulations and that the winding segment region of ScpA appears to facilitate engagement between the two SMC head domains (Fig. 5 D and F). These results further demonstrate that, using DCA evolutionary information, we were able to find several plausible SMC–ScpA configurations, which demonstrate the possible dynamics in the overall condensin system as well.
Discussion
In this study, we propose a structure for the whole condensin protein complex. Using an integrative approach, we combined fragmentary crystallographic data, which were previously known, with coevolutionary information to resolve a series of controversial issues regarding the structure of SMC–kleisin complexes. Condensin is composed of three subunits: two SMC monomers interacting with each other to form a dimer, the ScpA subunit bridging the two monomers of SMC, and the ScpB dimer. Using DCA, we were able to predict all of the known tertiary structural contacts for each of those subunits; this validates our contact predictions for the specific protein families based on sequence data alone. The single-protein structures resulting from our integrative approach are complete, regardless of usual limitations of crystallography, such as the large size of the SMC protein or the partially disordered structure of ScpA. We further exploited coevolutionary information to obtain the interfaces between the subunits of the complex. Here, with only limited experimental data, the contacts obtained from DCA are essential to generate a complete complex structure.
Currently, one of the significant controversies in the context of SMC–kleisin protein complexes is the question of the complex stoichiometry, namely, whether it is a single-ring or double-ring structure. To address this controversy, we used structure-based MD simulations informed by the coevolutionary contacts from DCA. In this approach, we examined the plausibility of the single-ring model, to see whether this simple model could explain all of the available information. We found that a single ring could account for all contacts, both crystallographically determined and coevolutionary, leaving behind no unexplained evidence. We also analyzed the feasibility of higher-order ring models. The results of our simulations for both the ScpAB and the SMC–ScpA subcomplexes suggest that coevolutionary constraints tend to be more consistent with the single-ring model. Overall, all of our findings support the one-ring model more than the two-ring model.
The precise biological mechanism of operation of the SMC–kleisin complexes remains unclear. While it has been recently proved that the human condensin (24) and cohesin (10) extrudes DNA by using ATP, it has also been suggested that there might be an active molecular mechanism driving the ring opening. Binding and hydrolysis of ATP by SMC proteins is indeed essential for DNA loading and has been suggested to provide the energy for transient reconfiguration of the hinge (24). In eukaryotic cohesin, the SMC hinge domain and the SMC–kleisin are suggested to serve as entry and exit gates for DNA, respectively (3). In prokaryotic condensin, the binding of ScpA to SMC dimers presumably forces the head domains into different configurations, owing to their association with opposite ends of the kleisin subunit. ATP binding or hydrolysis might trigger distinct conformational changes into the two head domains that could act synergistically to open the tripartite ring for DNA entry (26). Despite all of this fragmentary evidence, how the opening of the ring is achieved is not yet understood. Our integrative approach enables us to investigate the coevolutionary signature of the function of the complex, exploring all of its evolutionarily preserved configurations. Our analysis suggests the existence of an unknown configuration of the hinge domain; such rearrangement might be involved in the opening of the ring in the presence of DNA. Moreover, the flexibility of the ScpA might also favor the bistability of the condensin complex, allowing possible interaction with the DNA strand and its release from the ring.
Our study, by providing the structure for the whole prokaryotic condensin complex, constitutes a step toward investigating the structure–function relationship of many similar SMC–kleisin complexes. Upon obtaining the full structure of the condensin protein complex, we now are able to gain further insight into the mechanism of nucleosome interaction with SMC–kleisin complexes, including their potential motor capabilities.
Materials and Methods
Full details are provided in SI Appendix.
Obtaining Protein Sequence Data for Condensin Subunits.
To predict coevolved contacts between the various subunits, the databases of multiple sequence alignments (MSAs) for each protein were extracted from Pfam (41).
Experimental Protein Structural Data.
All experimental crystal structures in this study were retrieved from PDB (42).
DCA.
To quantify the amino acid coevolution between residue sites in a MSA, we construct a probabilistic model of a sequence, that is most consistent with the MSA data. The probability distribution resulting from our model, , must reproduce the single-site and pairwise frequencies of the dataset. We adopted the pseudolikelihood maximization approach of Ekeberg et al. (43) to estimate the parameters of a probabilistic model that is most consistent with the sequence data. The coevolution between a pair of residue sites i and j is calculated using an average product corrected Frobenius norm of the couplings matrix (43).
MD Simulation for Prediction of Unknown Protein Complex.
Because no complete structural data exist for the SMC–ScpA and SMC–ScpB complexes, we predict their 3D structure by combining the DCA-derived contacts with SBMs (44, 45). To reproduce docking of the selected subunits, the structure of each subunit was processed by the SMOG server (46), generating topology files that contain the SBM coarse-grained Cα potentials. DCA-derived constraints were incorporated in an SBM using a pairwise potential energy function that combines a repulsive, excluded volume interaction with an attractive Gaussian potential at long range (47–49). The obtained structures of our MD simulations (Figs. 1B, 3 C and D, and 5 E and F) were reconstructed to all-atom representations using an in-house code.
Supplementary Material
Acknowledgments
This work was supported by the Center for Theoretical Biological Physics sponsored by National Science Foundation (NSF) Grant PHY-1427654. J.N.O. was also supported by NSF Grant CHE-1614101, by Grant MCB-1241332, and by Welch Foundation Grant C-1792. D.K. acknowledges the Council for Higher Education of Israel for financial support.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates of both open and closed condensin protein complex have been deposited in the Model Archive database, https://modelarchive.org/ (Model Archive ID codes ma-abc6s and ma-ac7jp).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812770115/-/DCSupplemental.
References
- 1.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 3.Rhodes JDP, et al. Cohesin can remain associated with chromosomes during DNA replication. Cell Rep. 2017;20:2749–2755. doi: 10.1016/j.celrep.2017.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: Relevance of cohesin mutations to human disease. Curr Opin Genet Dev. 2016;37:59–66. doi: 10.1016/j.gde.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Izumi K, et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet. 2015;47:338–344. doi: 10.1038/ng.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan XW, et al. SMC1A promotes growth and migration of prostate cancer in vitro and in vivo. Int J Oncol. 2016;49:1963–1972. doi: 10.3892/ijo.2016.3697. [DOI] [PubMed] [Google Scholar]
- 7.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill VK, Kim JS, Waldman T. Cohesin mutations in human cancer. Biochim Biophys Acta. 2016;1866:1–11. doi: 10.1016/j.bbcan.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzeler KH, et al. AMLCG Study Group Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 10.Vian L, et al. The energetics and physiological impact of cohesin extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 12.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fudenberg G, et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. Transferable model for chromosome architecture. Proc Natl Acad Sci USA. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN. De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci USA. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 21.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 22.Kamada K, Su’etsugu M, Takada H, Miyata M, Hirano T. Overall shapes of the SMC-ScpAB complex are determined by balance between constraint and relaxation of its structural parts. Structure. 2017;25:603–616.e4. doi: 10.1016/j.str.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Gligoris TG, et al. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terakawa T, et al. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 2017;358:672–676. doi: 10.1126/science.aan6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brackley CA, et al. Nonequilibrium chromosome looping via molecular slip links. Phys Rev Lett. 2017;119:138101. doi: 10.1103/PhysRevLett.119.138101. [DOI] [PubMed] [Google Scholar]
- 26.Marcos-Alcalde Í, et al. Two-step ATP-driven opening of cohesin head. Sci Rep. 2017;7:3266. doi: 10.1038/s41598-017-03118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 28.Bürmann F, et al. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc Natl Acad Sci USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein–protein interaction by message passing. Proc Natl Acad Sci USA. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunt B, et al. Inference of direct residue contacts in two-component signaling. Methods Enzymol. 2010;471:17–41. doi: 10.1016/S0076-6879(10)71002-8. [DOI] [PubMed] [Google Scholar]
- 32.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng RR, Morcos F, Levine H, Onuchic JN. Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. Proc Natl Acad Sci USA. 2014;111:E563–E571. doi: 10.1073/pnas.1323734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sułkowska JI, Morcos F, Weigt M, Hwa T, Onuchic JN. Genomics-aided structure prediction. Proc Natl Acad Sci USA. 2012;109:10340–10345. doi: 10.1073/pnas.1207864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morcos F, Jana B, Hwa T, Onuchic JN. Coevolutionary signals across protein lineages help capture multiple protein conformations. Proc Natl Acad Sci USA. 2013;110:20533–20538. doi: 10.1073/pnas.1315625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaynes ET. Information theory and statistical mechanics. Phys Rev. 1957;106:620–630. [Google Scholar]
- 39.Nasmyth K. Cohesin: A catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 40.Capra EJ, et al. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 2010;6:e1001220. doi: 10.1371/journal.pgen.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 44.Punta M, et al. The Pfam protein families database. Nucleic Acid Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN. SMOG@ctbp: Simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 2010;38:W657–W661. doi: 10.1093/nar/gkq498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitford PC, et al. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins. 2009;75:430–441. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lammert H, Schug A, Onuchic JN. Robustness and generalization of structure-based models for protein folding and function. Proteins. 2009;77:881–891. doi: 10.1002/prot.22511. [DOI] [PubMed] [Google Scholar]
- 49.dos Santos RN, Morcos F, Jana B, Andricopulo AD, Onuchic JN. Dimeric interactions and complex formation using direct coevolutionary couplings. Sci Rep. 2015;5:13652. doi: 10.1038/srep13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.