Significance
To investigate the epigenetic mechanism of pregnancy failure in mammals, we exploited the high rate of fetal loss in pig induced pluripotent stem cell (iPSC) nuclear transfer. We generated methylomes of pig iPSCs and associated nuclear transfer embryos from reciprocal crosses between two distinct pig breeds. Our methylome analysis revealed that misregulation of RTL1 as the principal basis of pregnancy failure using pig iPSCs. Remarkably, RTL1 has broad fertility implications across mouse, rat, pig, cattle, and human from nuclear transfer cloning, tetraploid complementation, and artificial insemination, to natural fertilization. In all of these procedures, low RTL1 expression consistently corresponds to pregnancy failures.
Keywords: RTL1, pregnancy failure, DNA methylation, genomic imprinting, nuclear transfer
Abstract
Substantial rates of fetal loss plague all in vitro procedures involving embryo manipulations, including human-assisted reproduction, and are especially problematic for mammalian cloning where over 90% of reconstructed nuclear transfer embryos are typically lost during pregnancy. However, the epigenetic mechanism of these pregnancy failures has not been well described. Here we performed methylome and transcriptome analyses of pig induced pluripotent stem cells and associated cloned embryos, and revealed that aberrant silencing of imprinted genes, in particular the retrotransposon-derived RTL1 gene, is the principal epigenetic cause of pregnancy failure. Remarkably, restoration of RTL1 expression in pig induced pluripotent stem cells rescued fetal loss. Furthermore, in other mammals, including humans, low RTL1 levels appear to be the main epigenetic cause of pregnancy failure.
Decades since the cloning of Dolly the sheep, nuclear transfer (NT) technology has been successfully used in more than 20 mammalian species, but cloning efficiency (defined as the survival rate to birth per reconstructed oocyte) remains perplexingly low: for example, 1–2% for mice (1), 1.5% for monkeys (2), 0.3% for pigs, 0.3% for sheep, 0.8% for horses, 1.7% for cattle, and 6% for goats (3). That is, over 90% of cloned embryos fail to survive to term and are aborted at different gestational stages. The process of NT involves epigenetic reprogramming, and epigenetic errors are presumably responsible for NT-related miscarriages, but the underlying mechanism is not clear.
NT-derived epigenetic abnormalities can arise at either of the two stages hypothesized to occur during nuclear reprogramming (4). The first stage involves the erasure of epigenetic patterns in the terminally differentiated somatic cells and the reestablishment of a totipotent embryonic epigenetic state. The second stage involves redifferentiation of the totipotent embryonic status to various differentiated somatic cell types during postimplantation development (4). Analysis of the preimplantation cloned embryos revealed that DNA methylation, histone modification, and X chromosome inactivation (XCI) are common types of critical epigenetic errors in the first stage of nuclear reprogramming (5–7). For example, histone H3 lysine 9 trimethylation (H3K9me3) and histone H3 lysine 4 trimethylation (H3K4me3) were found to account for the arrest of two-cell and four-cell stage cloned embryos (7, 8). Modifying H3K9me3 and H3K4me3 in cloned mouse embryos improved development to the blastocyst stage from about 30% to over 95%, comparable to embryos produced in vivo.
For abnormal XCI found in cloned mouse blastocysts, by blocking expression of Xist, a gene responsible for XCI in NT embryos, the development of cloned embryos were improved 10-fold as early as embryonic day 5.5 and the high development rate persisted until term (9). Modifying other epigenetic errors in preimplantation NT embryos, such as histone acetylation and DNA methylation by inhibitors, was also reported to improve mammalian cloning efficiency (1).
Although the preimplantation development of NT embryos can be greatly improved by modifying epigenetic errors, vast cloned embryos are still lost during the postimplantation stage. For example, the maximum survival rate to birth for cloned blastocysts is improved to only about 10% after modifying histone modification or XCI, compared with a birth rate of ∼50% for in vitro fertilization blastocysts in mice. In a recent report of monkey NT, 86% (24 of 28) of postimplantation stage embryos were aborted even after modification of H3K9me3 and histone acetylation (2). Studies in the mouse and large-animal models, as well as epidemiologic studies in humans, suggest that aberrant genomic imprinting may account for postimplantation pregnancy failures (10). However, direct evidence for a common epigenetic mechanism for postimplantation fetal loss has been missing due to the lack of suitable animal models.
The pig has similar genetics, anatomy, and physiology to humans, and is increasingly employed as a model for studies of human disease and organ donation. This is exemplified by pig models of human cystic fibrosis and human Huntington’s disease models (11–13), and safer pig-to-human organ donors (14). Because there are no functional pig embryonic stem cells (ESCs) for use in the generation of gene-modified pig models, researchers have tried to utilize pig induced pluripotent stem cells (iPSCs) as an alternative (15–17). Compared with mouse and human iPSCs, pig iPSCs derived to date have not been shown to maintain pluripotency without continuing expression of reprogramming factors. The endogenous pluripotency genes of these pig iPSCs are typically not expressed, and when the reprogramming factors are withdrawn, these cells differentiate or undergo apoptosis (18). No pig iPSCs have been shown to reliably contribute to chimera formation or germline transmission (19–22). This is a much broader and puzzling problem in the SC field. In fact, the failure to derive true iPSCs extends to all large animals also including goat, sheep, and cattle.
Previously, it has been reported that when pig iPSCs were used as donor cells for NT cloning, ∼99.99% of cloned embryos failed to survive to term, a 100-fold greater loss than has been reported with fibroblast NT cloning (19–21). These iPSC NT embryos can develop to blastocyst stage with an efficiency of 10–30%, similar to fibroblast cloning, suggesting that the pregnancy failure mostly occurs postimplantation. On the one hand, the prominent epigenetic abnormalities in these pig iPSCs is an opportunity to investigate the decade-long puzzle as to why large-animal iPSCs have failed to achieve the same reprogramming potential as mouse iPSCs. On the other hand, the strikingly high rate of fetal loss prompted us to use this opportunity to explore the underlying epigenetic mechanisms of postimplantation abortions.
Using pig iPSC NT cloning as a model of fetal loss in this study, we performed a comprehensive genome-wide, allele-specific methylome analysis of the pig. Our methylome analysis narrowed the abortion problem down to the DLK1-DIO3 locus. We report here that misregulation of the imprinted RTL1 gene in the DLK1-DIO3 locus appears to be the principal epigenetic basis of pregnancy failure in mammals.
Results
Methylation Abnormalities in Pig iPSCs and Demise of Cloned Fetuses.
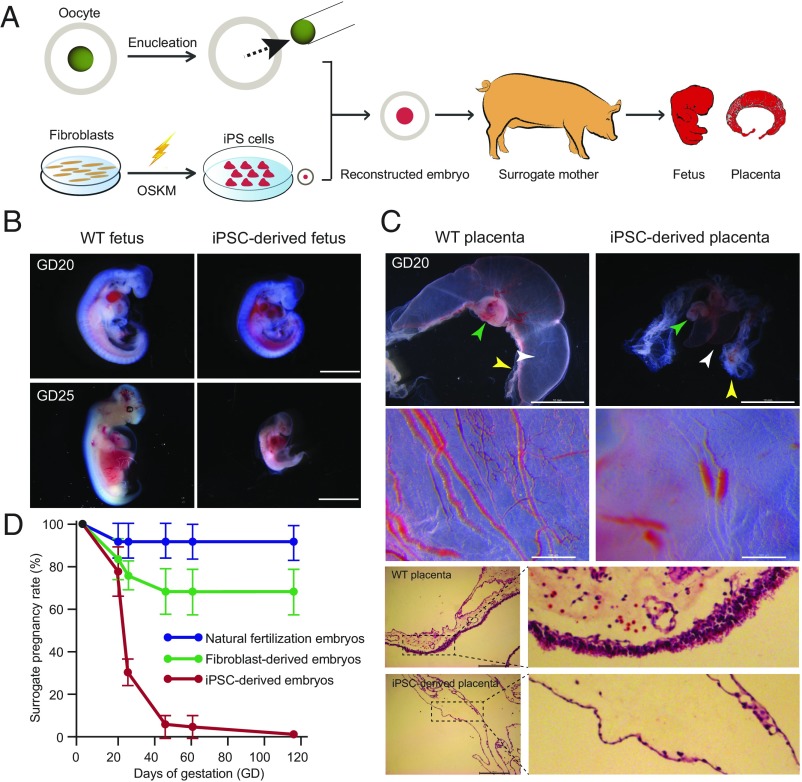
To explore the underlying epigenetic mechanism of abortions in the pig iPSC NT model, we first performed a detailed examination of the early development of iPSC-derived pigs (Fig. 1A and SI Appendix, Fig. S1). At 20 d of gestation (GD20), iPSC-derived fetuses of normal size with beating hearts were detected in most surrogates (77.78%, 7 of 9) (Fig. 1B), a pregnancy rate similar to that for fibroblast cloning (83.33%, 10 of 12). However, severe defects could already be detected in fetal-placental capillaries in iPSC-derived conceptuses at this stage (Fig. 1C), and by GD25 most of the iPSC-derived fetuses were being resorbed (68.75%) (Fig. 1 B and C). By GD45, <10% surrogates remained pregnant (6.25%), in contrast to 70% for fibroblast cloning (Fig. 1D). By the end of gestation, all iPSC-derived fetuses had aborted: no live piglets survived to term (Fig. 1D and SI Appendix, Table S1).
Fig. 1.
Most iPSC-derived nuclear transfer fetuses result in miscarriage during postimplantation development. (A) A schematic showing the construction of iPSC-derived embryos. OKSM indicates OCT4, KLF4, SOX2, and C-MYC. (B) WT fetuses and iPSC-derived fetuses. Whereas iPSC-derived fetuses appear alive and similar to WT fetuses at GD20, most are resorbing by GD25. [Scale bars, 2 mm (Upper), 5 mm (Lower).] (C) Chorioallantoic fusion defects were observed on day 20 iPSC-derived placentas. The white arrowhead indicates allantois. The yellow arrowhead indicates chorion. The green arrowhead indicates fetus. iPSC-derived allantois exhibited impaired blood flow within the vessels. Histological abnormalities of chorion observed in day 20 iPSC-derived placentas manifest by a thin trophoblast layer. WT placentas at day 20 showing normal morphology of chorion with primary chorionic villi. [Scale bars, 10 mm (Upper), 500 μm (Middle), 200 μm (Lower).] (D) Pregnancy rates for recipients carrying pig natural fertilization embryos, pig fibroblast-derived embryos, and pig iPSC-derived embryos. Values are pregnancy rate means ± SEM and were obtained from three independent experiments. The iPSC-derived embryos show significant lower pregnancy rate compared with the fibroblast-derived embryos at GD25 (P = 0.009, two-tailed Student’s t test), GD45 (P = 0.006, two-tailed Student’s t test), GD60 (P = 0.006, two-tailed Student’s t test), and full term (0.003, two-tailed Student’s t test).
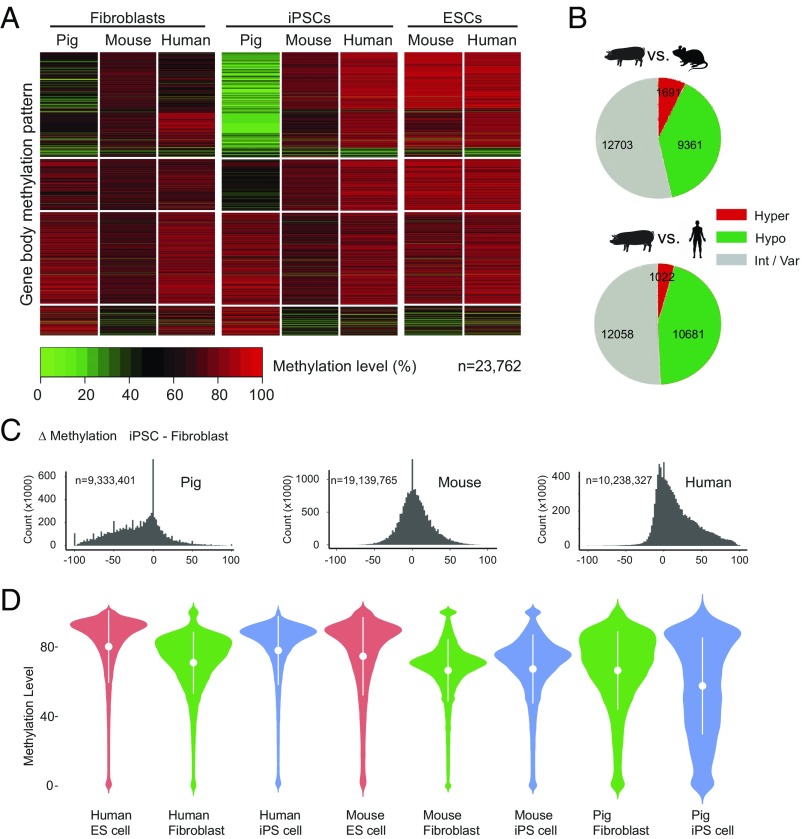
We next performed genome-wide methylome analyses of pig iPSCs and their derived fetuses and placentas. We compared the methylation state of orthologous gene bodies during the induction process from fibroblast to iPSC in pig, mouse, and human (23–27) (Fig. 2 and Datasets S1 and S2). At the cellular level, the DNA methylation patterns of pig fibroblasts mirror those of mouse and human while the pig iPSC has a strikingly different DNA methylation pattern (Fig. 2A). Methylation patterns in mouse iPSCs and human iPSCs are similar, and both resemble methylation patterns in ESCs. Pig iPSCs, however, showed widespread hypomethylation (9,361 genes vs. mouse, 10,681 genes vs. human) and some hypermethylation (1,691 genes vs. mouse, 1,022 genes vs. human) (Fig. 2B). Collectively, pig iPSCs did not appear to be properly reprogrammed compared with mouse or human iPSCs (Fig. 2 C and D).
Fig. 2.
Pig iPSCs are reprogrammed differently and show genome-wide hypomethylation compared with mouse and human. (A) Clustered heat map to visualize the methylation pattern of the gene body for orthologous genes. Each line represents one orthologous gene; n indicates number of orthologous genes. (B) Pie charts showing the number of the gene body methylation types of orthologous genes in pig, mouse, and human iPSCs. The gene body methylation types are partitioned according to their status in pig iPSC: hypomethylated (pig iPSC–mouse iPSC ≤ −0.25), intermediate or variable (−0.25 < pig iPSC–mouse iPSC > 0.25), or hypermethylated (pig iPSC–mouse iPSC ≥ 0.25). (C) Histograms of methylation changes (Δ methylation = iPSC-fibroblast) for 100-bp tiles across pig, mouse, and human genomes. The genome was split into tiles of 100-bp length; n indicates number of tiles. (D) Violin plots for methylation level of all Ensembl genes in different cell types. The plots show the distribution for gene body methylation level of all genes in pig tissues. The methylation distribution difference is dramatically evened out after NT. The x axis represents the probability density of the data at each methylation level. The mean and SD are indicated by the white dots and error bars.
NT Can Reprogram Most Epigenetic Abnormalities.
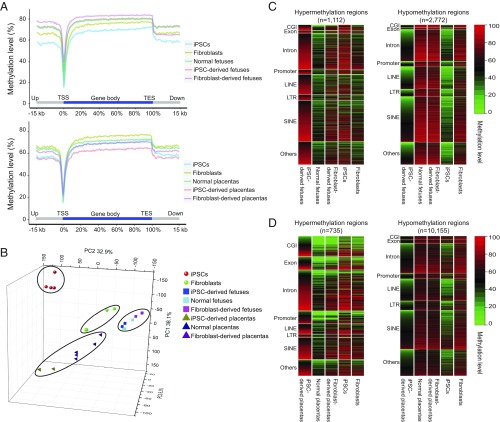
NT resulted in similar methylation levels between iPSC-derived fetuses and normal fetuses, with the highest methylation levels found in fibroblast-derived fetuses (Fig. 3A and SI Appendix, Fig. S2 A and B). In contrast to fetal methylation patterns, methylation levels in iPSC placentas were lower than in either normal or fibroblast-derived placentas, implying that hypomethylation persists in the NT-derived iPSC placentas (Fig. 3A and SI Appendix, Fig. S2 A and B). Principal component analysis (PCA) based on gene methylation or RNA-seq expression data showed similar trends (Fig. 3B and SI Appendix, Fig. S2 C and D). In short, NT markedly normalizes methylation and gene-expression differences in fetuses and placentas derived from different types of donor cells, although iPSC-derived placentas appear the least reprogrammed. In addition, in comparison with mouse and human (28–31), the methylation patterns of pig fetus and placenta are more similar to the human counterparts (SI Appendix, Fig. S3).
Fig. 3.
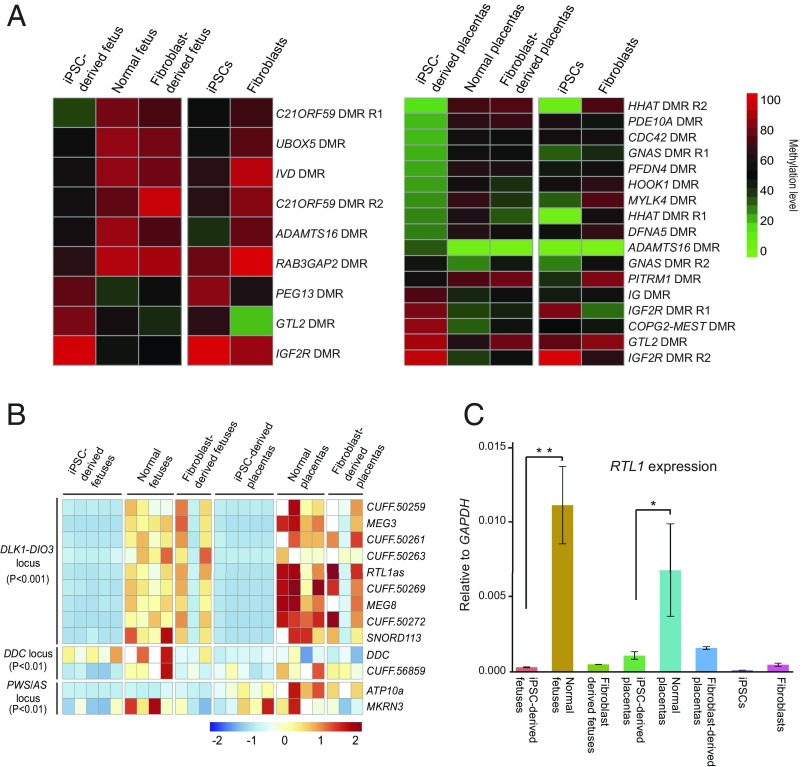
Key features of DMRs in iPSC-derived fetus/placenta vs. normal. (A, Upper) Averaged DNA methylation levels along the gene bodies, 15 kb upstream of the TSS, and 15 kb downstream of the transcription end sites (TES) of all Ensembl genes in the normal pig fetus, iPSC-derived pig fetus, and fibroblast-derived pig fetus. (Lower) Averaged DNA methylation levels along the gene bodies and 15 kb upstream of TSS and 15 kb downstream of TES of all Ensembl genes in normal, iPSC-derived placenta, and fibroblast-derived placentas. (B) PCA based on the gene methylation level of all WGBS samples. PCA 3D scatter plots were generated based on the gene body methylation of all genes in all porcine WGBS samples (four iPSCs, three fibroblasts, two iPSC-derived fetuses, two fibroblast-derived derived fetuses, three normal fetuses, two iPSC-derived placentas, two fibroblast-derived placentas, and three normal placentas). Of the total variance, 77.3% was explained by the first three PCs (PC1, 38.1%; PC2, 32.9%; PC3 6.3%). (C) Heat maps showing methylation levels of pig iPSC-derived fetus hypermethylation (Left) and hypomethylation (Right) DMRs across different genomic regions in the iPSC-derived fetus vs. normal fetus. Hypermethylated regions were strongly enriched in CG islands (P < 0.001, hypergeometric enrichment test). (D) Heat map of the methylation level of iPSC-derived placenta hypermethylation/hypomethylation DMRs among different genomic regions in iPSC-derived placentas vs. normal placentas; n indicates the number of DMRs. Hypermethylated regions were strongly enriched in CGIs (P < 0.001, hypergeometric enrichment test).
To better define the genomic regions bearing methylation abnormalities causing the demise of iPSC-derived piglets, we next compared differentially methylated regions (DMRs) between iPSC-derived fetus/placenta and normal fetus/placenta. We found that DMRs were widely distributed across the whole genome (SI Appendix, Fig. S4 and Dataset S3). In the iPSC-derived fetuses, we identified 3,884 DMRs compared with normal fetuses: 1,112 were hypermethylated and 2,772 were hypomethylated (Fig. 3C). In iPSC-derived placentas, we identified 735 hypermethylated DMRs and 10,155 hypomethylated DMRs (Fig. 3D). This broad distribution of persistent DMRs in iPSC-derived conceptuses suggests hypomethylation memory of the patterns seen in pig iPSC donor cells. We also noticed that of the hypermethylation aberrations, NT is particularly ineffective at reprogramming CG islands (CGIs). In summary, NT reprograms most but not all methylation discrepancies in pig iPSCs, and is more successful in the fetus than placenta.
Disruption of DNA Methylation and Expression of Imprinted Genes.
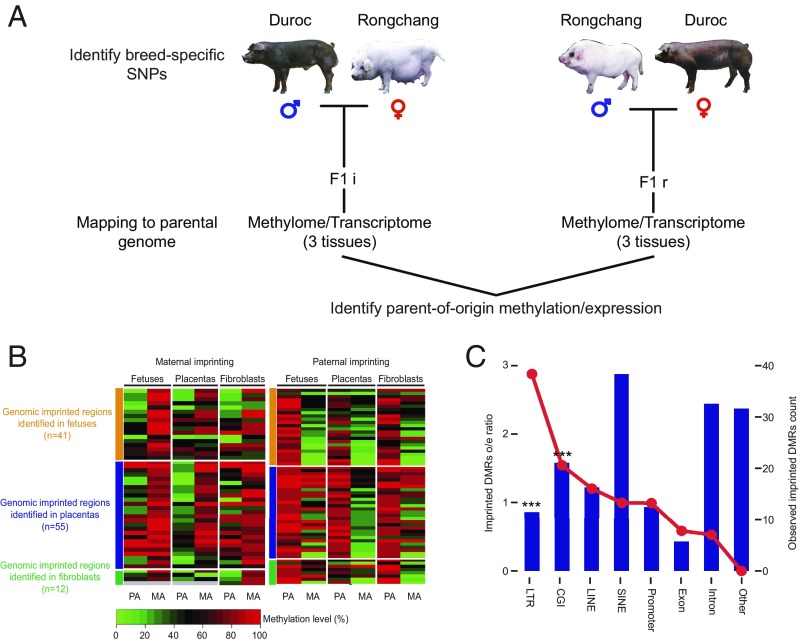
Genomic imprinting plays an important role in embryogenesis and cell pluripotency (32–34), and previous studies show that aberrant genomic imprints present in donor cells persist in cloned animals after NT (35, 36). Compared with human and mouse (30, 32, 37, 38), considerably fewer imprinted loci have been reported in the pig (39). To systematically investigate the pig imprinting map, we performed allele-specific methylation analysis on individuals from reciprocal crosses between two distinct breeds, Duroc and Rongchang (a local Chinese pig breed). Identification of 5.4 million SNPs in the genome that differ between the two breeds allowed us to distinguish parental origin of alleles in the F1 progeny (Fig. 4A). In total, we identified 41, 55, and 12 imprinted DMRs in fetus, placenta, and fibroblasts, respectively (Fig. 4B, SI Appendix, Fig. S5, and Datasets S4 and S5). Comparing these regions with the known imprinted region in mouse and human, we examined the distribution preference in the pig. The imprinted DMRs tend to congregate in the CGI and long terminal repeat (LTR) regions (Fig. 4C), suggesting intriguing origins and complex regulatory mechanisms of the imprinted DMRs.
Fig. 4.
Identification of imprinted genes and imprinted DMRs. (A) Identification of parent-of-origin methylation. Tissue samples were collected from F1 offspring of reciprocal crosses between genetically distinct pig breeds. F1i: Duroc (PA) × Ronghcang (MA); F1r: Rongchang (PA) × Duroc (MA). DNA samples were subject to bisulfite sequencing, and the methylation level of parental alleles was distinguished by breed-specific SNPs. Parent-of-origin methylation was identified by identification of a reciprocal methylation bias on a parental allele. (B) Heatmap for the imprinted DMR in three tissues. MA, maternal allele; PA, paternal allele. Each tile indicates the average methylation level of the parental allele in the imprinted regions. Gray tiles indicate missing values. (C) Plot showing the distribution of all imprinted DMRs across different genomic features. The red line plot represents the imprinted DMRs observed/expected ratio (corresponding to the left y axis). The bar plot represents the counts of the observed imprinted DMRs in each genomic feature (corresponding to the right y axis). The imprinted DMRs include both candidate-imprinted DMRs identified in this study and homologous imprinted DMRs inferred from the mouse. The imprinted regions were strongly enriched in CGIs (P < 0.001, hypergeometric enrichment test). ***P < 0.001.
Next, we examined the aberrant methylation of the imprinted regions in the iPSC-derived fetuses or placentas. Most of them appeared to be inherited from iPSCs, indicating a reprogramming-resistant nature in these regions (Fig. 5A and Datasets S6 and S7). By detailed analysis of coding and long-noncoding RNAs (lncRNAs) across all of the known and candidate imprinted regions, we identified 13 differentially expressed imprinted genes (iDEGs) in iPSC-derived tissues compared with normal fetuses and placentas. Among these, nine localized to the DLK1-DIO3 cluster (P < 0.001), two localized to the DDC cluster (P < 0.01), and two localized to the PWS/AS cluster (P < 0.01) (Fig. 5B). By comparison with the fibroblast-derived tissues, six abnormally imprinted genes were found, all of which reside in the DLK1-DIO3 cluster (P < 0.001) (Fig. 5B and SI Appendix, Fig. S6).
Fig. 5.
Aberrant methylation and gene expression of imprinted regions in iPSC-derived fetus and placenta. (A) Heat map of DMRs in iPSC-derived fetus/placenta compared with normal fetus/placenta in imprinted loci. The imprinted loci include the candidate imprinted regions and homologous imprinted regions inferred from mouse. (B) Heat map of iDEGs (known plus candidate) in different tissues. The list of iDEGs includes iDEGs between iPSC-derived fetus and normal fetus, and iDEGs between iPSC-derived placenta and normal placenta. DEGseq adjusted P value is used for detecting DEGs. (C) RTL1 expression measured by strand-specific quantitative PCR. The iPSC-derived fetus/placenta show significant lower RTL1 expression than the normal fetus/placenta (n = 3, *P < 0.05; **P < 0.01, two-tailed Student’s t test).
Aberrant Silencing of RTL1 Is Key to Resorption of iPSC-Derived Fetuses.
In the DLK1-DIO3 imprinted domain of eutherian mammals, three protein-coding genes, Delta-like homolog 1 (DLK1), Retrotransposon Gag like 1 (RTL1), and Type 3 deiodinase (DIO3) are expressed from the paternal allele, while multiple long- and short-noncoding RNA genes are expressed from the maternal allele (40). We confirmed that RTL1 was completely silenced in GD25 conceptuses by real-time quantitative PCR (Fig. 5C). It was reported that RTL1 plays a critical role in capillary endothelial cells for the maintenance of fetomaternal interface and development of the placenta (41). Consistent with this role, differential expression of genes between iPSC-derived fetuses and placentas (vs. normal) at GD20 suggested that failure to establish fetomaternal contact profoundly disrupted fetal metabolism and led to resorption at this stage (SI Appendix, Figs. S7 and S8 and Datasets S8 and S9).
We reasoned that silencing of the DLK1-DIO3 locus, specifically RTL1, could result from hypermethylation of imprinting control regions IG DMR and GTL2 DMR (SI Appendix, Fig. S9). In contrast to previous mouse studies (36, 42, 43) showing that the disruption of regulation-associated genes (TET1, ZFP57, DNMT3A, and DNMT3L) could lead to the aberrant methylation of the DLK1-DIO3 locus, no disruption of these genes were detected in the iPSC NT tissues (SI Appendix, Fig. S10). We examined the expression of these genes in iPSCs as shown in SI Appendix, Fig. S10. Tet1 and DNMT3L showed significantly lower expression in the iPSCs. Furthermore, neither valproic acid (36) or ascorbic acid (42) (included in our culture conditions) was unable to rescue the aberrant methylation of the DLK1-DIO3 locus.
The previous study in the mouse reported a transhomolog interaction between the Rtl1 and its maternal antisense transcript Rtl1as (44, 45). The maternal antisense transcript Rtl1as is an essential regulator of Rtl1, maintaining Rtl1 expression at a proper level. However, in the pig iPSC-derived placentas/fetuses, the abnormal hypermethylation in the DMRs of DLK1-DIO3 locus leads to the complete silencing of the locus, including both the paternal (RTL1) and maternal (RTL1as) alleles (SI Appendix, Fig. S11).
Rescue of Pregnancy Failure by Forced RTL1 Expression.
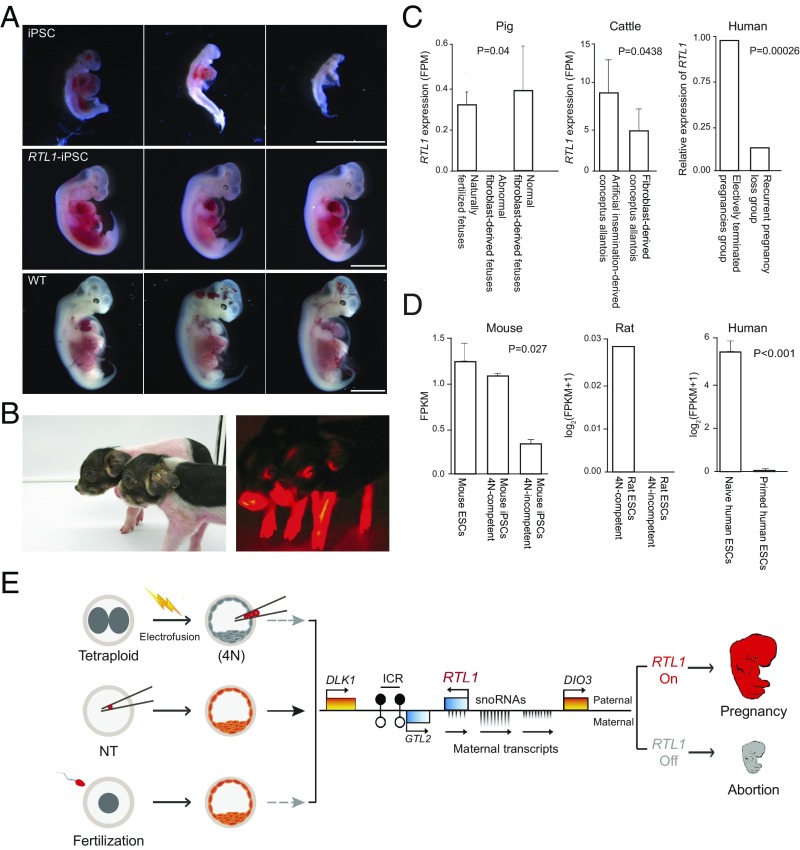
To confirm that aberrant silencing of RTL1 contributes to the high failure rate in pig iPSC cloning, we used plasmid for RTL1 overexpression. In picked and expanded iPSC clones, we used quantitative PCR to quantify RTL1 expression level. We chose those iPS cells (RTL1-iPSCs) that have comparable RTL1 expression level with normal fibroblasts for NT cloning experiments. RTL1-iPSC embryos were transferred into 10 recipients and, remarkably, 9 were pregnant, as detected by ultrasonography at GD25 after embryo transfer. Fetuses from two of these pregnant surrogates were examined at GD25 to evaluate their early development in comparison with fetuses derived from unmodified pig iPSC. Of seven fetuses retrieved from one RTL1-iPSC surrogate, four were morphologically normal and three were resorbing. In the other RTL1-iPSC surrogate, of six fetuses retrieved, three were morphologically normal and three were resorbing. In contrast, in the control iPSC surrogate, four fetuses were retrieved and all of them were resorbing (Fig. 6A and Table 1). Because IGF1 and LYVE1 are reportedly important for the maintenance of fetal capillaries, and may function downstream of RTL1 during development (46, 47), we performed quantitative PCR of their expression. The correlation of IGF1 and LYVE1 with RTL1 in fetuses was confirmed (SI Appendix, Fig. S12), as restoration of RTL1 in RTL1-iPSC fetuses increased expression of both genes.
Fig. 6.
RTL1 rescues excessive loss of iPSC-derived fetuses and has broad influence in mammals. (A) Cloned fetuses at GD25 derived from pig iPSCs, RTL1-iPSCs, and natural fertilization (WT). The RTL1-iPSC–derived fetuses show normal morphology similar with WT fetuses. (Scale bars, 5 mm.) (B) One-week-old cloned piglets derived from RTL1 overexpressed iPSCs. (C) RTL1 has wide-ranging influence in pig, cattle, and human for embryo development in postimplantation. (Left) RTL1 shows a lower expression in abnormal SCNT fetuses compared with the naturally fertilized fetuses (P = 0.04, two-tailed Student’s t test) in pigs. The bar graph (Right) represents the average of RTL1 expression in the three groups. (Center) RTL1 shows a lower expression in the allantois of NT-derived conceptuses in cattle. The bar graph (Right) is the average of RTL1 expression in the AI and NT groups. RTL1 shows a lower expression in SCNT fetuses compared with the AI fetuses (P = 0.0438, DEGseq P value). The Right shows the relative expression of RTL1 in human first trimester placental chorionic villi derived from eight electively terminated pregnancy (ETP) individuals and one recurrent pregnancy loss individual. RTL1 expression is scaled according to gene expression in ETP, which is considered normal. Bars indicate the relative expression; DEGseq P values are shown. (D) RTL1 expression is negatively correlated with the developmental potential of SCs in mammal. (Left) The RTL1 expression in different mouse SC lines. The 4N-competent groups indicate the iPSC cell lines (MEF as starting cell) that support the development of all-iPSC mice using tetraploid (4N) embryo complementation. The 4N-incompetent groups indicate the iPSC lines which fail to support the development of all-iPSC mice. The 4N-incompetent iPSCs show significantly lower expression than 4N-competent iPSCs (P = 0.027, one-tailed Student’s t test). (Center) The 4N-competent groups indicate the ES cell lines which support the development of all-ESC rat using tetraploid complementation. The 4N-incompetent groups indicate the ESC lines, which fail support the development of all-ESC rat using tetraploid complementation. (Right) The RTL1 expression in different human stem cell lines. The human naïve ESC showed significantly higher expression than human primed ESC and human iPSC (P < 0.001, two-tailed Student’s t test). (E) Model for RTL1 as an important marker for the imprinting status of donor cells for NT, ESCs/iPSCs for tetraploid complementation/chimera formation. Lack of RTL1 in these processes all results in fetal demise and abortion.
Table 1.
In vivo development of NT embryos derived from pig iPSCs and RTL1 overexpressed iPSCs
| Donor cells | No. of transferred embryos | No. of recipients | No. of pregnancies GD25, GD90 | No. of live cloned piglets |
| iPSCs | 3,564 | 15 | 3, 1 | 0 |
| RTL1- iPSCs | 2,316 | 10 | 9, 6* | 16 |
| Fibroblasts | 5,307 | 20 | 15, 14 | 69 |
Note: Two of the nine pregnant surrogates were used at 25 d of gestation to detect the early development.
All of the remaining seven RTL1-iPSC recipients were pregnant at GD60 by ultrasonography, and six carried to term and gave birth to a total of 16 live cloned piglets (Fig. 6B and Table 1). Thus, restoration of RTL1 expression decreased fetal losses significantly, greatly improving our capability to generate iPSC-derived piglets. Most RTL1-iPSC piglets were morphologically identical to normal fertilized piglets and overall cloning efficiency rivaled that achieved with fibroblasts.
RTL1 Has Broad Influence in Eutherian Mammals.
We compared RTL1 expression levels in different pig fibroblast lines, and found that the lines with high RTL1 expression performed better in terms of fetal survival after NT (SI Appendix, Fig. S13). Similarly, our analysis of a recent report on pig cloning (48) with fibroblasts shows that RTL1 expression was consistently decreased in abnormal fibroblast-derived NT fetuses (Fig. 6C). To validate the essential role of RTL1 in cloning, we created homozygous RTL1 mutant pig fibroblasts via CRISPR technology and performed NT with these cells. Among four surrogates transferred to date, no RTL1-knockout embryos could be detected beyond GD30. Aberrant silencing of RTL1 is not unique to iPSC cloned pigs. Examination of cattle allantois RNA-Seq data from four artificial insemination (AI) conceptuses and five NT conceptuses (49) revealed that the NT groups exhibited variable RTL1 expression at GD34 compared with the AI groups, with silencing of RTL1 in three of five NT conceptuses. In short, RTL1 is the most frequent abnormally expressed gene among all aberrant imprinted genes across different species (SI Appendix, Fig. S14 A–D).
By reanalysis of human fertility clinic data (50) comparing gene expression in spontaneously aborted conceptuses vs. induced abortions, RTL1 was found to be significantly lower in the former (Fig. 6C). In addition, it has been described (51, 52) that >10% of infertile patients have sperm with abnormal DNA methylation patterns in the imprinting control region of RTL1 (SI Appendix, Fig. S15). Furthermore, the expression of RTL1 decreases with increased maternal age. All of these human data support our hypothesis that RTL1 may be a critical epigenetic determinant of human spontaneous abortions.
Importantly, when reanalyzing the SC datasets from previous studies (53–55) we noticed that the expression of RTL1 could be a marker to evaluate developmental potential of SCs in mammals. We found that 4N “on” mouse iPSCs (competent for tetraploid complementation) showed higher RTL1 expression than 4N “off” mouse iPSCs (incompetent for tetraploid complementation) (Fig. 6D). A similar result was observed for rat ESCs. Furthermore, the expression of RTL1 differs in human SC lines with different pluripotency. These analyses suggest that RTL1 is associated with the developmental potential of SCs.
Discussion
We have presented genome-wide, base-pair resolution, imprinted allele-specific DNA methylation maps for pig iPSCs and associated NT embryos from reciprocal crosses between two distinct pig breeds. These datasets greatly expand our understanding of genomic imprinting in mammals, and provides a good resource for comprehensive investigation of the abnormalities of DNA methylation during animal cloning. Compared with previous methylation landscape analyses in mouse and human embryos, our pig methylome study focused on the relationship between aberrant imprinting and abortion during postimplantation development. We found that the retrotransposon-derived RTL1 gene within the DLK1-DIO3 locus is essential for the generation of live piglets via NT. More importantly, the aberrant silencing of RTL1 appears to be the common epigenetic reason for pregnancy failure in mammalian cloning and could be a major cause of the failure of fetal survival in general (Fig. 6E). The RTL1 is also known as MART1, PEG11, SIRH2, Mar1, HUR1 among others, and it is implicated in various processes. In addition to implantation, it also serves as a pathogenic gene for postnatal growth failure or facial abnormality (56), an antigen expressed in melanoma (57), and a novel driver of hepatocarcinogenesis (58). These intriguing characteristics of RTL1 suggest a similarity of implantation and carcinogenesis.
In our RTL1 overexpression rescue experiments, the birth rate of iPSC NT has increased from ∼0–0.89%. Besides RTL1, our methylation analysis found that other imprinted genes, such as IGF2R and PEG13, are also aberrantly methylated. Therefore, silencing of RTL1 appears to represent the main, but not all causes of pregnancy failure of pig iPSC NT. Our RTL1 rescue experiment is constitutive overexpression, lacking precise spatiotemporal and dosage control. It has been reported that too high or too low expression all affect the normal development of the fetus (41).
We and others have tried blastocyst injection and early embryo aggregation experiments with pig iPSCs (21, 22). In our previously reported aggregation experiment (21), pig iPSC-derived NT embryos were aggregated with normal NT embryos, providing the iPSC embryos with a normal trophectoderm and placenta. Still, no chimera animals were ever born. All survived newborns contained no contributions of the iPSC-derived cells. Expression of RTL1 is required both in the fetus and the placenta for normal fetal development.
A recent paper published by Matoba et al. (59) showed complete loss of H3K27me3 imprinting in mouse preimplantation SC NT (SCNT) embryos, and suggested it likely accounts for the postnatal developmental defects observed in SCNT embryos. We examined the expression of these H3K27me3 genes in pig and cattle postimplantation NT embryos. No significantly differential expression of these genes was found in postimplantation NT embryos compared with normal embryos (SI Appendix, Fig. S14 E and F). This indicated that the H3K27me3-imprinting system, as well as its loss-of-imprinting in mouse SCNT embryos, may not be conserved in large-animal species. In fact, Sfmbt2, one of the H3K27me3-imprinted genes, loss-of-imprinting of which was found to contribute to the placenta overgrowth phenotype of SCNT embryos in mice, is not an imprinted gene in pig, bovine, or human (60). Future studies should identify H3K27me3-dependent imprinted genes in large-animal species and address whether the imprinting status of these genes contributes to the developmental defects observed in SCNT embryos of large-animal species, including primates.
Although 12 y have passed since iPSC technology was first published in 2006, true iPSCs have yet to be generated for large animals, including pigs, sheep, and cattle (15, 61, 62). Germline-competent iPSCs are only recognized for one species, the mouse. Previous studies have pointed out that gene expression within the DLK1-DIO3 region correlates with pluripotency in mouse SCs (32, 36), but the specific gene/mechanism underlying this potential remained undetermined. Our current study indicates that RTL1 may be the core element in determining the developmental potential of mammalian SCs. Inducing RTL1 expression in pig, sheep, and cattle iPSCs may have implications in the derivation of functional iPSCs that are free of continuous expression of Yamanaka factors. Transient expression of RTL1 along with the other factors may help achieve real pluripotency. This is important to the exploration of using artificial gametes from iPSCs in efforts to rescue endangered species.
Functional RTL1 is specific to eutherians, as no homologous sequences of RTL1 can be identified in the platypus or chicken genome (40, 63). A truncated form with very few remaining regions of homology can be found in the marsupial genome, but it shows no sign of expression in fetal and pouch tissue (SI Appendix, Fig. S16A) (40, 63). Interestingly, the premature birth of marsupial pups corresponds to the peak phase of spontaneous abortion in mammals (SI Appendix, Fig. S16B). All of these coincidences together imply that the insertion of RTL1 may have been a driving force in the evolutionary acquisition of the placenta.
In this study, we found that restoration of RTL1 expression in pig iPSCs rescues fetal loss of cloned animals. Silencing of RTL1 was also found in porcine NT embryos and bovine NT embryos. Therefore, RTL1 appears to be a good marker for the imprinting status of donor cells in animal cloning. In the future, selecting starting donor cells with proper RTL1 expression for NT may help increase the overall cloning efficiency. RTL1 is also a promising biomarker for prediction and diagnosis of pregnancy complications in mammals, including humans. Retrotransposon-derived genes play important roles during different developmental stages, such as totipotency of SCs and synaptic transmission (64–66). It would be interesting to elucidate whether RTL1 exploits similar mechanisms as other retrotransposon-derived genes in the development of placenta.
Materials and Methods
Ethics Statements.
This study was approved by the Animal Welfare Committee of China Agricultural University (SKLAB-2012-11). All pigs used in this study were taken care of and operated according to the relevant regulations.
Cell Culture and Derivation of Pig iPSCs.
Porcine fetal fibroblasts (PFFs) were isolated from day 20–30 porcine embryos of laboratory minipigs and reciprocal hybrids of Duroc and Rongchang pigs. PFFs were cultured in serum-based EF medium [DMEM containing 10% FBS, 1% nonessential amino acids (Invitrogen), 1% penicillin-streptomycin (Gibco)]. Doxycycline (DOX)-inducible vectors with human OCT4, KLF4, SOX2, C-MYC (OKSM) were electroporated into PFFs with a Nucleofector 2b Device (Lonza). The transfected cells were replated onto 100-mm dishes covered with a γ-ray–treated mouse embryonic fibroblast (MEF) feeder layer and cultured for 5 d in serum-based ESC medium [DMEM containing 10% FBS, 1× NEAA (Gibco), 1% penicillin-streptomycin (Gibco), 0.1 mM b-mercaptoethanol (Sigma), 106 unit/L mouse leukemia inhibitory factor (LIF; Gibco), supplemented with 600 mg/mL G418 (Calbiochem)] and 2 mg/mL DOX. The transfected cells were cultured in 2i/LIF medium [500 mL neurobasal medium (Gibco), 500 mL DMEM-F12 medium (Gibco), 5 mL N2 supplement (Gibco), 10 mL B27 supplement (Gibco), 3 μM CHIR99021 (Selleck), 1 μM PD0325901 (Selleck), 0.1 mM b-mercaptoethanol (Sigma), 1% penicillin-streptomycin (Invitrogen), and 106 unit/L human LIF (Gibco)] for 2 wk. Pig iPSC colonies similar to mouse or rat ESCs were selected for further cultivation and evaluation.
Nuclear Transfer.
To exclude the potential effect of residual expression of reprogramming factors (OKSM) on the development of cloned embryos, the iPSCs were cultured in the absence of DOX for more than 1 wk and then used for NT. Matured oocytes were enucleated by micromanipulation. Then, a single donor cell was injected into the perivitelline space, and fusion was accomplished using a BTX Electro-cell Manipulator 200 (BTX) with two direct current pulses (1-s interval) of 1.2 kV/cm for 30 μs in fusion medium [0.3 M mannitol, 1.0 mM CaCl2, 0.1 mM MgCl2, and 0.5 mM Hepes (pH adjusted to 7.0–7.4)]. Oocytes were then incubated for 30 min in PZM3, and the fusion percentage was calculated under a stereomicroscope. Fifty fused embryos were placed into a four-well dish (Nunc) containing 500 μL of PZM3 at 38.5 °C and 5% CO2 with maximum humidity.
Embryo Transfer.
Day 1 NT zygotes were transferred surgically into surrogate mothers (250–300 zygotes per surrogate). About 25 d later, the pregnancy status of the surrogates was diagnosed by ultrasonography.
Sample Collection.
On GD20, all pregnant surrogates were killed, and the reproductive tract was removed. In the laboratory, the uterus was opened, each conceptus was isolated from the uterine wall, and a stereomicroscope was used to separate fetal and placental tissues, which were then used for RNA or DNA extraction.
RNA and DNA Extraction.
Fetuses and placentas were first frozen in liquid N2 and pounded in a mortar. Half of the tissue was used for RNA extraction, and the remaining half was used for DNA extraction. RNA samples were extracted using the RNeasy mini kit (Qiagen) following the manufacturer’s protocol. To eliminate contamination of RNA with DNA, removal of genomic DNA was performed with DNase I. Samples were eluted in RNase free water and stored at −80 °C. DNA samples were extracted using QIAamp DNA Mini Kit (Qiagen) following the manufacturer’s protocol. Samples were eluted in sterile MilliQ water and stored at −20 °C ready for use.
RNA Reverse-Transcription and Quantitative PCR Analysis.
Two micrograms of total RNA were converted to cDNA in a 25-μL final volume using MMLV reverse transcriptase (Promega) according to the manufacturer’s protocol. Quantitative PCR was performed using a LightCycler 480 with SYBR Green Master Mix (Roche). The procedure included preincubation (95 °C, 5 min), amplification (95 °C, 10 s; 60 °C, 10 s; 72 °C, 10 s) for 30 cycles, melting curve (95 °C, 5 s; 65 °C, 1 min), and cooling (40 °C, 10 s). Expression of the housekeeping gene GAPDH was used as a control. For strand-specific quantitative PCR, we used gene-specific primers to replace oligo dT in the RT-PCR. Statistical analyses and graphs were made using Excel and GraphPad Prism 5 with statistical significance set at 0.05. The primers used for strand-specific RT-PCR were shown in Dataset S10.
Construction of RTL1 Overexpression Vector.
We used Duroc liver cDNA as a template to amplify the RTL1 gene (primers: gGFP-IRES-F/IRES-pRTL1-R), while we used vector pPB-CAG-rtTA3Gipuro to amplify the IRES fragment (primers: IRES-pRTL1-F/gpRTL1-R). Then, we fused the two fragments using Q5 High-Fidelity DNA Polymerase (New England Biolabs) to get IRES-RTL1. Primers used for amplification of RTL1 and IRES were shown in Dataset S10. The backbone, pNS-EGFPpuro, was digested by SalI. IRES-RTL1 was cloned into the SalI site to create pNS-EGFPiRTL1.
Generation of RTL1 Knockout Fibroblasts for NT.
Two guide RNAs were designed to target RTL1 coding sequence. A pair of oligos for each targeting site was annealed and inserted into the BbsI site of pCRISPR-sg4 plasmid (67), resulting in sgRNAs expressing plasmids pCRISPR-sg4-RTL1-sgRNA1 and pCRISPR-sg4-RTL1-sgRNA2. Porcine embryonic fibroblasts were transfected with Cas9 expressing pCRISPR-S10 plasmid (67) and pCRISPR-sg4-RTL1-sgRNA1/2 plasmids. The oligos used were shown in Dataset S10. After 5 d of G418 selection, clones were picked for expansion. Genomic DNA was extracted for PCR screening of mutated clones. PCR products were inserted into a TA cloning vector for further sequencing identification. One homozygous mutant clone was used for NT. Day 1 NT zygotes were transferred surgically into four surrogate mothers (250–300 zygotes per surrogate).
SNP Identification and Construction of Parental Genome.
For each hybrid pig used in this study, we sequenced the parental genomes, including four Duroc pigs and four Rongchang pigs in the previous study (68). Then, we mapped the reads to SusScr3 by Bowtie2 (69), and the Genome Analysis Toolkit (GATK) 38 was utilized for SNP calling for each parent (70). Only SNPs that showed homogeneous calls were kept and those with heterogeneous calls were removed. This resulted in a total of 1.07 million SNPs between the Duroc genome and the reference genome (SusScr3), and a total of 4.3 million SNPs between the Rongchang genome and the reference genome (SusScr3). To eliminate mapping bias in the subsequent analysis, we constructed the maternal and paternal genomes (the pseudogenomes of Duroc and Rongchang pigs, respectively) by replacing the reference allele with a variant allele matching each breed (Duroc and Rongchang).
Whole-Genome Bisulfite Sequencing.
DNA was isolated using standard phenol/chloroform extraction and ethanol precipitation or using the Cell Culture DNA Midi Kit (Qiagen). Whole-genome bisulfite sequencing (WGBS) was performed as described previously (71). Paired-end DNA reads were sequenced on Illumina HiSeq 2500 or HiSeq 2000 at Novogene (Dataset S1). For the WGBS, the clean reads were aligned to the parental genomes using Bismark tools (v0.7.6) with default parameters (72). All sequencing analyses were conducted based on the average result mapped to the parental genome.
lncRNA Sequencing.
A total amount of 3 μg RNA per sample was used as input material. First, ribosomal RNA (rRNA) was removed by Epicentre Ribo-zero rRNA Removal Kit (Epicentre), and the resulting rRNA-free material was cleaned up by ethanol precipitation. Subsequently, the rRNA-depleted RNA was used to construct sequencing libraries by NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s recommendations. The libraries were sequenced on Illumina HiSeq 2500 or HiSeq 2000 at Novogene (Dataset S1). Reads from lncRNA sequencing were aligned to the parental genomes using Hisat (73). The gene annotation information used was Sus scrofa 10.2.83 from Ensembl.
Allele Assignment of Sequencing Reads.
For a hybrid individual, heterozygous SNPs were obtained by identifying the pure SNP between its maternal and paternal genome. Between 4 million and 6 million SNPs were obtained for each hybrid individual to determine the parental origin of each allele. To assign each read to its parental origin, we examined all SNPs in the read that showed high-quality base calling (Phred score ≥ 20). If the SNP included a cytosine, its bisulfite-converted form (T) was also considered. SNPs that became noninformative after bisulfite conversion were discarded. For paired-end reads, SNP information from both reads in the pair was summed. When multiple SNPs were present in a read (or a read pair) that harbors conflicting origin SNPs, the read was discarded.
Identification of Candidate Genomic Imprinting Regions.
We used WGBS data to detect parent-of-origin–dependent allele-specific methylated regions for consideration as candidate genomic imprinting regions. The parent-of-origin allele-specific methylated regions were identified by detecting imprinted DMRs between paternal and maternal allele in hybrid offspring. RADMeth (74) with a P value less than 0.05 and the absolute value of log-odds ratio greater than 5. Allele-specific methylated sites less than 100 bp away were then merged into DMR candidates. We ultimately selected candidates that contained more than five imprinted CGs (among which the mean-meth-diff is greater than 0.35) as candidate genomic imprinting regions.
Identification of Candidate Imprinted Genes.
We used lncRNA sequencing data to detect candidate imprinted genes. For each hybrid individual, only heterozygous SNPs (based on the genotypes of its parents) and SNPs identified through RNA-Seq were designated as testable. We calculated the counts of the paternal and maternal alleles for each testable locus.
Furthermore, a binomial test was performed on the allele read counts in F1 crosses to test whether the allelic expression deviated from equivalence. P values were adjusted using the Benjamini–Hochberg method in R, and an adjusted P value cut-off of 0.05 was applied to claim statistical significance. Imprinted SNPs were evaluated based on whether the parental origin of the preferred allele was the same in reciprocal crosses. A putative imprinted gene was identified if an imprinted SNP was detected in more than half of the samples in each reciprocal cross, and the parental origin of the preferred allele was the same in all samples with the detected SNP.
Identification of DEGs and DMRs.
The number of reads uniquely mapped to each gene was determined using HT-seq count (v0.5.3.p3) (75). The DEGs were identified by DESeq2 (76) with adjusted P values less than 0.001.
The DMRs were identified by RADMeth (74) with a P value less than 0.05 and the absolute value of log-odds ratio is greater than 5. Furthermore, we selected those candidates that contain more than five CGs, among which the mean-meth-diff is greater than 0.25.
Identification of Pig Homologous Genomic Imprinting Regions.
Pig homologous genomic imprinting regions were inferred from the orthologous regions of mouse through liftover (77). The regions that could not be converted were manually inspected. The genomic imprinting regions of mouse were taken from a previous study (39).
Identification of lncRNAs.
Transcripts, including mRNA and lncRNA, were assembled using Cufflinks, and those less than 200 nucleotides were filtered out. The sequences of remaining transcripts were compared with known noncoding RNAs and mRNAs, and the candidate lncRNA, intronic lncRNA, and antisense lncRNA were determined by class codes obtained from Cuffcompare (78). PLEK software was used to distinguish lncRNAs from mRNAs, and transcripts with known protein domains were excluded by Pfam Scan according to Pfam hidden Markov models (79, 80).
Genomic Features of Pigs.
For pigs, the CGI, LINE, LTR, and SINE annotations were downloaded from the University of California, Santa Cruz (UCSC) browser (SusScr3) RepeatMasker tracks. Promoters (transcription start sites, TSSs) are defined as 5 kb upstream of the TSS and are parsed from Ensembl annotation. The information for the orthologous gene between the pig, human, and mouse was downloaded from the Ensemble biomart.
PCA Based on Gene Expression/Methylation.
The PCA plot based on FPKM (fragments per kilobase of transcript per million mapped reads, or gene body methylation) values shows a nonrandom distribution of points associated with samples. The result was visualized based on the first three principal components by Origin Software.
Identification of iDEGs and Differentially Methylated Imprinted Regions Between iPSC-Derived Groups and Normal Groups.
Considering that the genomic imprinting is typically distributed in clusters and the known imprinted regions are incomplete, all of the DMRs (between iPSC-derived groups and normal groups) upstream and downstream (10 kb) of the imprinted regions were considered as differentially methylated imprinted regions. Similarly, the DEGs (between iPSC derived groups and normal groups) upstream and downstream (10 kb) of the imprinted region (including the loci with parent-of-origin expression identified in this study) were considered differentially expressed imprinted regions.
Public Data Collection and Analysis.
Different datasets across species were collected from previous studies (Dataset S2) and analyzed by the general flowchart mentioned above.
Data Availability.
The datasets generated in the current study are available in the NCBI Sequence Read Archive (accession no. SRP107099).
Supplementary Material
Acknowledgments
We thank Drs. Lara Carroll, Anne Boulet, and Anne Moon (University of Utah) for critical reading of the manuscript. This work was supported by the Transgenic Research Grants 2016ZX08010001, the National High Technology Research and Development Program (2013AA102502), the National Natural Science Foundation of China (Grant 91519316), the Transgenic Research Grant 2016ZX08009003, the Beijing Advanced Innovation Center for Food Nutrition and Human Health, Department or College of Biological Sciences, China Agricultural University, the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (Grant 2017SKLAB1-8), and the Plan 111 (B12008).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP107099).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814514115/-/DCSupplemental.
References
- 1.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, et al. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 2018;172:881–887.e7. doi: 10.1016/j.cell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Keefer CL. Artificial cloning of domestic animals. Proc Natl Acad Sci USA. 2015;112:8874–8878. doi: 10.1073/pnas.1501718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 5.Inoue K, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 6.Chan MM, Smith ZD, Egli D, Regev A, Meissner A. Mouse ooplasm confers context-specific reprogramming capacity. Nat Genet. 2012;44:978–980. doi: 10.1038/ng.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matoba S, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, et al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matoba S, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner DK, Lane M. Ex vivo early embryo development and effects on gene expression and imprinting. Reprod Fertil Dev. 2005;17:361–370. doi: 10.1071/rd04103. [DOI] [PubMed] [Google Scholar]
- 11.Rogers CS, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welsh MJ, Rogers CS, Stoltz DA, Meyerholz DK, Prather RS. Development of a porcine model of cystic fibrosis. Trans Am Clin Climatol Assoc. 2009;120:149–162. [PMC free article] [PubMed] [Google Scholar]
- 13.Yan S, et al. A huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington’s disease. Cell. 2018;173:989–1002.e13. doi: 10.1016/j.cell.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu D, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017;357:1303–1307. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, et al. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- 16.Esteban MA, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezashi T, et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sui D, et al. Fine-tuning of iPSC derivation by an inducible reprogramming system at the protein level. Stem Cell Reports. 2014;2:721–733. doi: 10.1016/j.stemcr.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, et al. Cell cycle synchronization of leukemia inhibitory factor (LIF)-dependent porcine-induced pluripotent stem cells and the generation of cloned embryos. Cell Cycle. 2014;13:1265–1276. doi: 10.4161/cc.28176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan N, et al. Piglets cloned from induced pluripotent stem cells. Cell Res. 2013;23:162–166. doi: 10.1038/cr.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du X, et al. Barriers for deriving transgene-free pig iPS cells with episomal vectors. Stem Cells. 2015;33:3228–3238. doi: 10.1002/stem.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujishiro SH, et al. Generation of naive-like porcine-induced pluripotent stem cells capable of contributing to embryonic and fetal development. Stem Cells Dev. 2013;22:473–482. doi: 10.1089/scd.2012.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DS, et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 25.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gascard P, et al. Epigenetic and transcriptional determinants of the human breast. Nat Commun. 2015;6:6351. doi: 10.1038/ncomms7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogdanović O, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet. 2016;48:417–426. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decato BE, Lopez-Tello J, Sferruzzi-Perri AN, Smith AD, Dean MD. DNA methylation divergence and tissue specialization in the developing mouse placenta. Mol Biol Evol. 2017;34:1702–1712. doi: 10.1093/molbev/msx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Court F, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kono T, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428:860–864. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- 34.Yagi M, et al. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- 35.Humpherys D, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 36.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bischoff SR, et al. Characterization of conserved and nonconserved imprinted genes in swine. Biol Reprod. 2009;81:906–920. doi: 10.1095/biolreprod.109.078139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards CA, et al. SAVOIR consortium The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6:e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekita Y, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- 42.Stadtfeld M, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405, S391–S392. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito M, et al. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development. 2015;142:2425–2430. doi: 10.1242/dev.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis E, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 46.Kitazawa M, Tamura M, Kaneko-Ishino T, Ishino F. Severe damage to the placental fetal capillary network causes mid- to late fetal lethality and reduction in placental size in Peg11/Rtl1 KO mice. Genes Cells. 2017;22:174–188. doi: 10.1111/gtc.12465. [DOI] [PubMed] [Google Scholar]
- 47.Charalambous M, da Rocha ST, Hernandez A, Ferguson-Smith AC. Perturbations to the IGF1 growth pathway and adult energy homeostasis following disruption of mouse chromosome 12 imprinting. Acta Physiol (Oxf) 2014;210:174–187. doi: 10.1111/apha.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan D, et al. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Reports. 2018;10:494–508. doi: 10.1016/j.stemcr.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biase FH, et al. Massive dysregulation of genes involved in cell signaling and placental development in cloned cattle conceptus and maternal endometrium. Proc Natl Acad Sci USA. 2016;113:14492–14501. doi: 10.1073/pnas.1520945114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sõber S, et al. RNA sequencing of chorionic villi from recurrent pregnancy loss patients reveals impaired function of basic nuclear and cellular machinery. Sci Rep. 2016;6:38439. doi: 10.1038/srep38439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi N, et al. Factors associated with aberrant imprint methylation and oligozoospermia. Sci Rep. 2017;7:42336. doi: 10.1038/srep42336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi H, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 53.Chang G, et al. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li TD, et al. Rat embryonic stem cells produce fertile offspring through tetraploid complementation. Proc Natl Acad Sci USA. 2017;114:11974–11979. doi: 10.1073/pnas.1708710114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo G, et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kagami M, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 57.Kawakami Y, Rosenberg SA. Immunobiology of human melanoma antigens MART-1 and gp100 and their use for immuno-gene therapy. Int Rev Immunol. 1997;14:173–192. doi: 10.3109/08830189709116851. [DOI] [PubMed] [Google Scholar]
- 58.Riordan JD, et al. Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet. 2013;9:e1003441. doi: 10.1371/journal.pgen.1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matoba S, et al. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell. 2018;23:343–354.e5. doi: 10.1016/j.stem.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q, et al. Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genomics. 2011;12:204. doi: 10.1186/1471-2164-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Cang M, Lee AS, Zhang K, Liu D. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS One. 2011;6:e15947. doi: 10.1371/journal.pone.0015947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang B, et al. A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under tem cell potency fluctuates with endogenous retrovirus activity. Nature. 2011;487:57–63. [Google Scholar]
- 63.Kaneko-Ishino T, Ishino F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front Microbiol. 2012;3:262. doi: 10.3389/fmicb.2012.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashley J, et al. Retrovirus-like gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018;172:262–274.e11. doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pastuzyn ED, et al. The neuronal gene Arc encodes a repurposed retrotransposon gag protein that chemically defined conditions of dual kinase inhibition. PLoS One. 2018;6:e24501. [Google Scholar]
- 66.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu C, et al. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci USA. 2017;114:722–727. doi: 10.1073/pnas.1615735114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, et al. Convergent and divergent genetic changes in the genome of Chinese and European pigs. Sci Rep. 2017;7:8662. doi: 10.1038/s41598-017-09061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 72.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dolzhenko E, Smith AD. Using beta-binomial regression for high-precision differential methylation analysis in multifactor whole-genome bisulfite sequencing experiments. BMC Bioinformatics. 2014;15:215. doi: 10.1186/1471-2105-15-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karolchik D, et al. The UCSC genome browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li A, Zhang J, Zhou Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics. 2014;15:311. doi: 10.1186/1471-2105-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the current study are available in the NCBI Sequence Read Archive (accession no. SRP107099).