Significance
A chemoenzymatic method was developed that permits site-selective Fab and Fc glycan remodeling of intact antibodies. Thus, homogeneous glycoforms of cetuximab were generated in which the immunogenic Fab N-glycans and the less active core-fucosylated Fc N-glycans were precisely remodeled to a desired sialylated N-glycan and an optimal nonfucosylated complex N-glycan, respectively. The glycoengineered cetuximab showed increased affinity for the FcγIIIa receptor and significantly enhanced antibody-dependent cell-mediated cytotoxicity activity. This approach opens a new avenue to manipulating antibody glycosylations to produce specific homogeneous glycoforms useful for functional studies and therapeutic applications.
Keywords: Fc receptor, antibody glycosylation, antibody therapy, N-glycan, glycoengineering
Abstract
The N-glycans attached to the Fab and Fc domains play distinct roles in modulating the functions of antibodies. However, posttranslational site-selective modifications of glycans in antibodies and other multiply glycosylated proteins remain a challenging task. Here, we report a chemoenzymatic method that permits independent manipulation of the Fab and Fc N-glycans, using cetuximab as a model therapeutic monoclonal antibody. Taking advantage of the substrate specificity of three endoglycosidases (Endo-S, Endo-S2, and Endo-F3) and their glycosynthase mutants, together with an unexpected substrate site-selectivity of a bacterial α1,6-fucosidase from Lactobacillus casei (AlfC), we were able to synthesize an optimal homogeneous glycoform of cetuximab in which the heterogeneous and immunogenic Fab N-glycans were replaced with a single sialylated N-glycan, and the core-fucosylated Fc N-glycans were remodeled with a nonfucosylated and fully galactosylated N-glycan. The glycoengineered cetuximab demonstrated increased affinity for the FcγIIIa receptor and significantly enhanced antibody-dependent cell-mediated cytotoxicity (ADCC) activity.
Monoclonal antibodies are an important class of therapeutic proteins widely used for the treatment of cancer and autoimmune and infectious diseases (1, 2). IgG antibodies contain two heavy chains and two light chains that form three major domains: two identical Fab domains responsible for antigen binding and an Fc domain that is engaged in Fc receptor-mediated effector functions. All IgG1 antibodies contain N-glycans at the highly conserved N297 glycosylation sites (3). It has been shown that the fine structures of the Fc N-glycans (e.g., the status of core fucosylation or terminal sialylation) are critical in defining an antibody’s effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) and antiinflammatory activities (2, 4–8). On the other hand, Fab glycosylation, which is present in some monoclonal antibodies and about 20% of circulating i.v. immunoglobulins, can play an important role for immunity, antigen recognition, and serum half-life of antibodies (9, 10). Thus, a method to selectively manipulate Fc and Fab glycosylation is highly desirable for better understanding the roles of antibody glycosylation and for developing more efficient antibody-based therapeutics. Unfortunately, the conventional genetic method has been unable to selectively alter the nature of glycans at different sites of a multiply glycosylated protein.
We and others have previously described a chemoenzymatic method for glycan remodeling of glycoproteins (including antibodies) that uses an endoglycosidase and endoglycosynthase pair for deglycosylation and subsequent glycosylation with defined N-glycans (11–19). In particular, we have shown that the Endo-S and Endo-S2 enzymes and their corresponding glycosynthase mutants, including Endo-S D233Q and Endo-S2 D184M, are efficient and highly specific for antibody Fc glycosylation (13–15), whereas the Endo-F3 and its glycosynthase mutant (such as Endo-F3 D165A) demonstrated more relaxed substrate specificity but required core-fucosylated GlcNAc as an acceptor (16, 20). Recently, we have shown that Endo-F3 mutant could achieve certain site-selectivity in enzymatic glycan remodeling of human erythropoietin that carries N-glycans at three distinct sites (20). Despite these progresses, discriminative and site-selective glycan modification of antibodies carrying both Fc and Fab glycosylation remains to be achieved.
Here, we report an efficient chemoenzymatic method that permits highly site-selective enzymatic glycoengineering of both Fc and Fab glycans of cetuximab, a chimeric mouse-human anti-epidermal growth factor receptor (anti-EGFR) therapeutic antibody used for the treatment of colorectal cancer and squamous-cell carcinoma of the head and neck (21). Cetuximab is glycosylated in both Fab and Fc domains at the N88 and N297 sites of the heavy chain, respectively, with tremendous heterogeneity in the N-glycan structures (22, 23). The majority of the Fc N-glycans are core fucosylated, which has been shown to have much lower binding affinity to FcγIIIa receptor (FcγRIIIa) and lower ADCC activity than the corresponding afucosylated antibody. Two glycoengineered anti-EGFR antibodies with low fucose (Fuc) content, imgatuzumab (GA201) and tomuzotuximab (CetuGEX), showed enhanced ADCC activity and improved therapeutic efficacy in clinical trials for treatment of head and neck cancer (24–26). On the other hand, about 30% of the N-glycans in cetuximab are terminated with one or two α1,3Gal epitopes that turn out to be the major cause of anaphylaxis observed in some patients with cetuximab treatment (21, 27). Thus, we sought to develop a method enabling independent glycoengineering of the Fab and Fc glycans to provide a homogeneous glycoform of cetuximab with enhanced ADCC activity and without the allergic αGal epitope. By taking advantage of the substrate specificity of several endoglycosidases and their glycosynthase mutants, together with an unexpected site-selectivity of a bacterial α1,6-fucosidase, we were able to independently remodel the Fc and Fab glycans of cetuximab into a homogeneous glycoform carrying a uniformed sialylated N-glycan at the Fab domain and a nonfucosylated galactosylated N-glycan at the Fc domain that showed significantly enhanced ADCC activity.
Results and Discussion
Global Glycan Remodeling of Cetuximab Using Endo-F3 D165A and Endo-S D233Q.
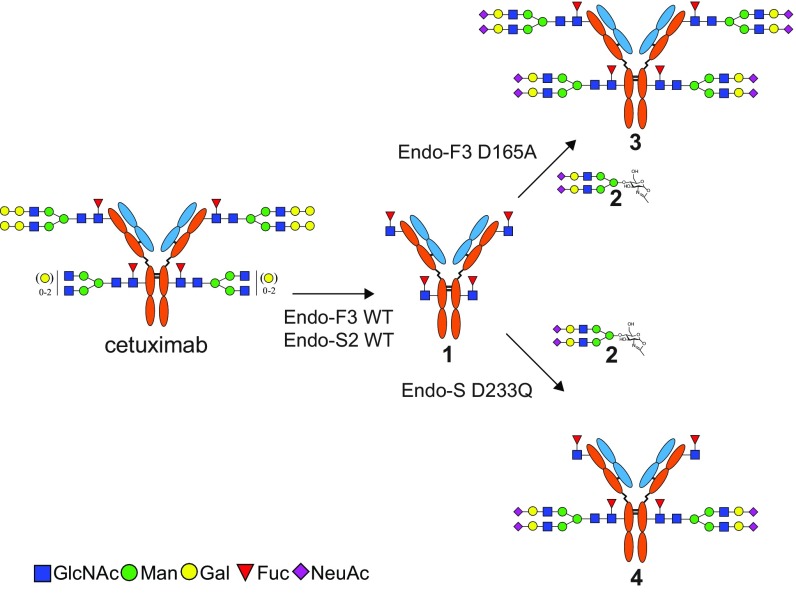
To examine the feasibility of chemoenzymatic glycan remodeling of cetuximab, we first performed a global glycan remodeling of the antibody (Fig. 1). We began our glycan remodeling by first treating cetuximab with a mixture of Endo-F3, which is efficient for cleaving core-fucosylated complex N-glycans, and Endo-S2, which is specific for hydrolyzing both complex and high-mannose glycans at the Fc region of IgG. This treatment resulted in a global deglycosylation of the Fab and Fc glycans, leaving an antibody intermediate (1) with only the core Fucα1,6GlcNAc moiety left at the original glycosylation sites. Cetuximab has two C-terminal lysine variants: the major component that has the C-terminal lysine cleaved and the minor one with the C-terminal lysine, which is uncleaved (28). To monitor this reaction and other reactions after glycan modification, we used LC-MS analysis to separate and characterize the light chain and heavy chain of cetuximab after reduction. Deconvolution of the MS data of the heavy chain of the intermediate (1) showed two distinct m/z species of 49,919 and 50,050 Da in good agreement with the calculated theoretical masses of the endoglycosidase-cleaved product (SI Appendix, Fig. S1A). The resulting homogenous glycoform of cetuximab (1) confirms previously reported hydrolyzing activity of Endo-F3 for core fucosylated glycans. In contrast to Endo-S or Endo-S2, which are specific for hydrolyzing Fc N-glycans, Endo-F3 was also efficient in removing glycans attached to the Fab domain. Next, we examined the glycosylation of the intermediate (1) with two glycosynthase mutants, Endo-S D233Q and Endo-F3 D165A, on which we previously reported (13, 16). Using a sialylated biantennary complex type N-glycan (SCT) oxazoline (2) as the donor substrate, we found that Endo-F3 D165A was able to transfer the sialylated N-glycan to all four sites when an excess of SCT oxazoline was used to give the product, 3. The result demonstrated the power of the glycosynthase mutant Endo-F3 D165A to efficiently mediate glycan transfer to both the Fab and Fc domains. The deconvoluted LC-MS data of the glycoengineered cetuximab (3) showed a major peak at 53,922 Da and a minor peak at 54,050 Da, which were consistent with the calculated masses for the heavy chain plus two SCTs (SI Appendix, Fig. S1B). On the other hand, glycosylation of 1 with the glycosynthase mutant Endo-S D233Q and the glycan oxazoline (2) led to selective glycosylation at the Fc domain, giving selectively the Fc glycosylated product (4) (Fig. 1). The deconvoluted MS of the heavy chain of 4 showed two distinct species of 51,921 and 52051 Da, which were consistent with the calculated masses for N-glycan transfer to only one of the Fucα1,6GlcNAc heavy chain sites (SI Appendix, Fig. S1C).
Fig. 1.
Glycan remodeling of cetuximab using Endo-F3 and Endo-S and their glycosynthase mutants. Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; NeuAc, N-acetylneuraminic acid.
To further verify that the N-glycan was specifically transferred to the Fc domain, product 4 was digested with the protease IdeS and reduced, followed by LC-MS analysis. IdeS is a protease from Streptococcus pyogenes that specifically cleaves IgG antibodies below the hinge region to give a dimeric F(ab′)2 domain and two monomeric Fc domains (SI Appendix, Fig. S2) (29, 30). Treatment of product (4) with IdeS followed by reduction gave three antibody fragments corresponding to the light chain, the Fd domain, and the Fc domain. LC-MS analysis of the digested product confirmed that only the Fc domain carried a full-size sialylated N-glycan (SI Appendix, Fig. S3A), while the Fd domain still carried the Fucα1,6GlcNAc moiety (SI Appendix, Fig. S3B) and the light chain was intact without any modification (SI Appendix, Fig. S3C).
Site-Specific Defucosylation Activity of an α1,6-Fucosidase from Lactobacillus casei on the Deglycosylated Cetuximab.
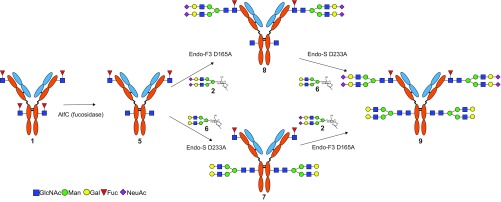
Next, we examined enzymatic defucosylation of antibody intermediate 1 aiming to produce an afucosylated Fc glycoform (Fig. 2). We have previously shown that an α1,6-fucosidase from bovine kidney could remove the Fuc from the Fucα1,6GlcNAc moiety of the deglycosylated rituximab, but with relatively low activity, requiring a long incubation time (13). Recently we have cloned and expressed an α1,6-fucosidase from L. casei (AlfC), which demonstrated a much higher activity for α1,6 defucosylation (31). The deglycosylated intermediate (1) was incubated with the AlfC overnight at 37 °C and the enzymatic reaction was monitored by LC-MS analysis. Surprisingly, even a prolonged digestion with the AlfC gave a product, compound 5, in which only one of the two core Fuc moieties on the heavy chain was removed (SI Appendix, Fig. S4A). A detailed IdeS digestion and LC-MS analysis of the generated subunits verified that the product was compound 5, in which only the Fuc moiety at the Fc domain was selectively removed by the enzyme AlfC while the Fucα1,6GlcNAc moiety, located at the Fab domain of intermediate 1, was resistant to defucosylation by the enzyme. LC-MS analysis of the IdeS-generated subunits indicated that the Fc domain carried only a GlcNAc moiety (SI Appendix, Fig. S4B), the Fd carried a Fucα1,6GlcNAc moiety (SI Appendix, Fig. S4C), and the light chain remained unchanged (SI Appendix, Fig. S4D). This result was unexpected. It is not clear whether this was due to less accessibility at the Fab glycosylation site or because of the unique substrate specificity of the α1,6-fucosidase, which may be further investigated through structural studies of the enzyme–substrate complex.
Fig. 2.
Site-specific glycoengineering of cetuximab for enhanced ADCC activity and diminished allergic reaction. Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; NeuAc, N-acetylneuraminic acid.
Site-Specific Glycoengineering of Both Fab and Fc Glycans to Generate an Optimized Glycoform of Cetuximab.
With the generation of the discriminately tagged glycosylation sites in intermediate 5, we sought to introduce different N-glycans at the Fab and Fc domains using the distinct substrate specificity of Endo-F3 D165A and Endo-S D233Q to generate a homogenous glycoform of cetuximab, which would be expected to have enhanced ADCC activity and diminished allergic reactions (Fig. 2). In one approach, glycosylation of intermediate 5 with SCT oxazoline (2) under the catalysis of Endo-F3 D165A gave antibody glycoform 7 in which only the fucosylated GlcNAc moiety on the Fab domain was transformed with the complex N-glycan, because Endo-F3 D165A was highly selective for glycosylating the core-fucosylated GlcNAc moiety (16). The site-selectivity of Endo-F3 D165A-catalyzed glycosylation of 5 was verified by LC-MS analysis of product 7. Deconvolution of the LC-MS data of the heavy chain of 7 indicated that only one SCT was attached to the heavy chain of 7 (SI Appendix, Fig. S5A). To identify which site the glycan was transferred to, the transglycosylation product (7) was subjected to IdeS digestion and reduction, followed by LC-MS analysis. Deconvolution of the MS data of the Fc fragment showed that the Fc domain remained in the GlcNAc-Fc form after the reaction (SI Appendix, Fig. S5B). In contrast, the Fd fragment carried a full-size sialylated N-glycan (SI Appendix, Fig. S5C), while the mass of the light chain of 7 remained the same (SI Appendix, Fig. S5D). In an alternative approach, glycosylation of the intermediate (5) with a fully galactosylated glycan oxazoline (6) under the catalysis of Endo-S D233Q yielded glycoform 8 in which only the GlcNAc moiety at the Fc domain was glycosylated, due to the fact that the Endo-S D233Q mutant was specific for transforming Fc domain glycans (13).
The site selectivity was confirmed by IdeS digestion and LC-MS analysis of the Fc and Fd fragments generated from the transglycosylation product (8) (SI Appendix, Fig. S6). With the discriminated glycosylation at the Fc and Fab domains, the intermediates (7 or 8) were further transformed at the Fab or Fc domain, respectively, with a distinct N-glycan to achieve the target glycoform of cetuximab (Fig. 2). Thus, glycosylation of 7 with glycan oxazoline 6 under the catalysis of Endo-S D233Q gave the fully glycosylated antibody (9) in which the Fab domain carries a fully sialylated N-glycan and the Fc domain carries a nonfucosylated and fully galactosylated glycan. On the other hand, starting with intermediate 8, sialylated N-glycan was successfully introduced to the Fab domain by glycosylation with glycan oxazoline (2) under the catalysis of Endo-F3 D165A mutant to afford the same final product (9) (Fig. 2). In all of the enzymatic transformations, the reaction was monitored by LC-MS analysis, and the glycosylation was pushed to completion by using an excess of glycan oxazolines. Excess glycan oxazolines were recovered in the form of free glycans and could be reused after one-step conversion to the corresponding glycan oxazolines following the previously described procedures (13, 32). All of the antibody intermediates and the final product were purified by protein A affinity chromatography.
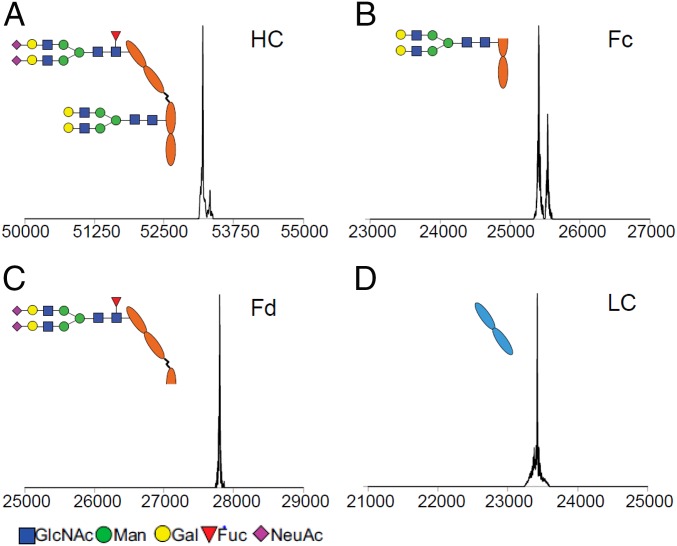
The glycosylation pattern of the final product (9) was again verified by LC-MS analysis. Deconvolution of the MS data of the reduced heavy chain of 9 indicated the presence of two N-glycans (found, 53,195 and 53,323 Da; calculated for heavy chain carrying an SCT and an asialylated complex type glycan, Mr = 53,194 and 53,322 Da, with or without the C-terminal Lys truncation, respectively) (Fig. 3A). The location of the respective N-glycan was further confirmed by using IdeS digestion and LC-MS analysis of the Fc and Fd fragments (Fig. 3 B and C); the light chain was intact without any modification (Fig. 3D), suggesting that the enzymatic transformations were highly specific and that nonenzymatic modifications did not occur in the process.
Fig. 3.
LC-electrospray ionization-MS analysis of the glycosylation pattern of the glycoengineered cetuximab (9). The deconvoluted spectra of the heavy chain and the fragments are shown. (A) Full-length heavy chain (HC). (B) Fc fragment. (C) Fd fragment. (D) Light chain (LC). Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; NeuAc, N-acetylneuraminic acid.
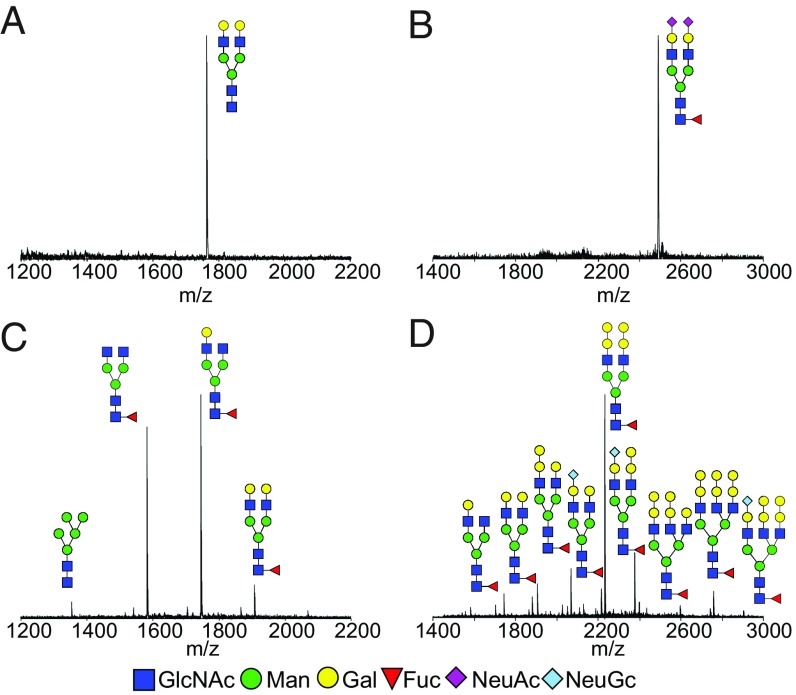
Lastly, to further confirm the homogeneity of the site-specific glycosylations, N-glycans attached at the Fc and Fab domains were released and analyzed. The final product (9) was cleaved by immobilized papain digestion to disconnect the Fab and Fc fragments, which were separated by affinity chromatography. Next, the N-glycans from the Fc and Fab fragments were released by PNGase F treatment, followed by anthranilic acid (2-AA) labeling. MALDI-TOF MS analysis indicated that the Fc of 9 carried a single fully galactosylated nonfucosylated N-glycan (Fig. 4A) and the Fab carried a single fully sialylated N-glycan (Fig. 4B). For comparison, the commercial cetuximab was also subject to the same treatment, and the released 2-AA–labeled N-glycans were analyzed by MALDI-TOF MS analysis. The results indicated that Fc glycosylation contained at least four different N-glycans (Fig. 4C) and that the Fab carried multiple N-glycans (Fig. 4D), some of which were terminated with the α1,3Gal and N-glycolylneuraminic acid epitopes. Thus, a complete site-specific glycan remodeling of cetuximab at the Fc and Fab domains was achieved through the combined enzymatic transformations. Our previous studies have demonstrated that the Endo-F3 mutant (such as D165A) was able to transfer bi- and triantennary complex type N-glycans to protein and Fc domains (16), and that the glycosynthase mutant (such as D184M) generated from Endo-S2 could transfer all three major types of N-glycans at the Fc domain (14). Therefore, it is expected that an expanded library of distinctly glycosylated antibodies could be constructed efficiently by the combined use of these enzymes.
Fig. 4.
MALDI-TOF-MS spectra (negative-ion mode) of 2-AA–labeled Fc and Fab N-glycans from commercial cetuximab and the glycoengineered cetuximab (9) released by PNGase F treatment. (A) Fc glycan from glycoengineered cetuximab (9). (B) Fab glycan from the glycoengineered cetuximab (9). (C) Fc glycan from commercial cetuximab. (D) Fab glycan from commercial cetuximab. Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; NeuAc, N-acetylneuraminic acid; NeuGc, N-glycolylneuraminic acid.
Binding of the Glycoengineered Cetuximab to FcγRIIIA.
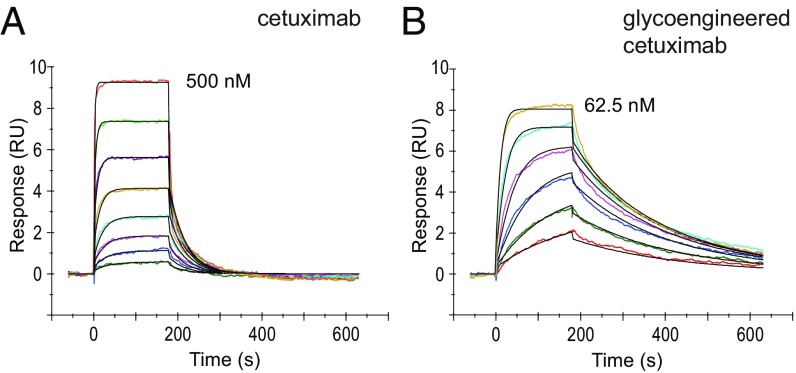
Surface plasmon resonance (SPR) was used to determine the binding affinity of commercial cetuximab and the glycoengineered cetuximab (9) to recombinant human FcγRIIIa. Protein A was immobilized to the CM5 sensor chip and used to capture each respective antibody. After capture, FcγRIIIa was injected over as the analyte at various concentrations to measure the SPR responses. Representative SPR sensorgrams for the interaction between both cetuximab products and FcγRIIIa are shown in Fig. 5. The dissociation constant (KD) between the binding of commercial cetuximab and FcγRIIIa and that between the glycoengineered cetuximab (9) and FcγRIIIa was estimated to be 42 ± 1 nM and 2.3 ± 0.5 nM, respectively, using a 1:1 binding model. The data indicated that removal of the core Fuc at the Fc glycosylation resulted in an 18-fold increase of the affinity of the glycoengineered cetuximab (9) for FcγRIIIa.
Fig. 5.
SPR analysis of the binding of FcγRIIIa with commercial cetuximab and the glycoengineered cetuximab (9). (A) Binding with commercial cetuximab with the concentration of FcγRIIIa at twofold serial dilutions starting at 500 nM. (B) Binding with glycoengineered cetuximab with the concentration of FcγRIIIa at twofold serial dilutions starting at 62.5 nM. The color curves represent the fitting lines for the analysis of the SPR data.
Binding to EGFR+ Cells and ADCC Activity of the Glycoengineered Cetuximab.
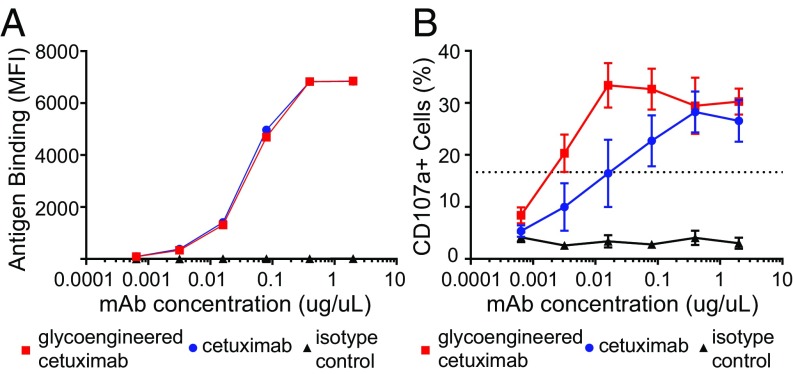
We compared the binding of the commercial cetuximab and the glycoengineered cetuximab (9) to the target antigen EGFR using the EGFR+ epidermoid carcinoma cell line A431. As shown in Fig. 6A, the glycoengineered cetuximab (9) showed essentially the same affinity as that of the commercial cetuximab, suggesting that the glycan remodeling did not affect the affinity of the antibody for the target antigen. To investigate whether the remodeled cetuximab demonstrated enhanced affinity for both CD16a polymorphisms (V/F), an ADCC assay was conducted. The EGFR+ epidermoid carcinoma cell line A431 targeted by the anti-EGFR monoclonal antibodies, glycoengineered cetuximab (9), and commercial cetuximab were used to trigger ADCC. The CD16-dependent cytotoxic function (ADCC activity) of natural killer (NK) cells was assessed by measuring surface expression of CD107a (LAMP-1) by CD56+CD3− NK cells using flow cytometry analysis of four healthy peripheral blood mononuclear cell donor populations. Percent CD107a+ NK cell population mediating ADCC activity is monitored by FACS at each antibody concentration (Fig. 6B). We found that the glycoengineered cetuximab (9) showed enhanced NK cell activation and CD107a up-regulation as compared with the commercial cetuximab control. Based on EC50 values, the glycoengineered cetuximab (9) was over eightfold more active than the commercial cetuximab.
Fig. 6.
(A) The binding of glycoengineered cetuximab (9) and commercial cetuximab with EGFR on A431 cells. (B) ADCC assays conducted using a 1:1 ratio of effector and target cells. ADCC activity was assessed by measuring the percent CD107a+ NK cell populations by FACS. Red box, Glycoengineered cetuximab (9); blue circle, commercial cetuximab; black triangle, antibody isotype control.
Conclusion
An efficient chemoenzymatic approach to site-specific glycoengineering of both Fab and Fc glycans of an intact monoclonal antibody is described. This was exemplified by independent Fab and Fc glycan remodeling of the monoclonal antibody cetuximab by taking advantage of the substrate specificity of three endoglycosidases (Endo-S, Endo-S2, and Endo-F3) and their glycosynthase mutants, as well as an unexpected substrate site-selectivity of the AlfC. The resulting homogeneous glycoform of cetuximab, in which the heterogeneous and immunogenic Fab N-glycans were replaced with a single sialylated N-glycan and the core-fucosylated Fc N-glycans were replaced with a nonfucosylated N-glycan, demonstrates increased affinity for FcγRIIIa and significantly enhanced ADCC activity. This method opens a new avenue to discriminately manipulate the Fc and Fab glycosylations of monoclonal antibodies to generate biobetter version of antibodies. It also provides a method to construct a library of selectively glycosylated antibody glycoforms for probing the structure–function relationship of antibody glycosylation.
Methods
Endo-S and the glycosynthase mutant Endo-S D233Q were expressed and purified as described previously (13). Endo-F3 and the glycosynthase mutant Endo-F3 D165A were expressed and purified as described previously (16). The SCT oxazoline and the asialylated complex type glycan oxazoline were synthesized following previously described procedures (33). The AlfC gene (GenBank CAQ67984.1) was codon optimized for E. coli expression and subcloned into the expression plasmid pCPD-L (34). The enzyme activity was confirmed by assaying with p-nitrophenyl-α-l-fucopyranoside following the previously reported procedures (35). The binding affinity of cetuximab and remodeled cetuximab to FcγRIIIa was determined by SPR. EGRF binding affinity and ADCC activity was measured using the EGFR+ epidermoid carcinoma cell line A431. All glycoengineered intermediates and final products were characterized using LC-MS analysis to confirm glycan removal or transfer. Detailed materials and methods are in SI Appendix, including materials, plasmid construction, enzyme expression and purification, antibody purification and glycoengineering, EGRF binding and ADCC assay, LC-MS analysis of antibodies, and MALDI-TOF MS analysis of released glycans.
Supplementary Material
Acknowledgments
We thank David Knorr (The Rockefeller University) for his assistance in preparation of the manuscript. This work was supported, in part, by the National Institutes of Health Grants R01 GM096973 (to L.-X.W.) and P01 CA190174 and R35 CA196620 (to J.V.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812833115/-/DCSupplemental.
References
- 1.Aggarwal RS. What’s fueling the biotech engine-2012 to 2013. Nat Biotechnol. 2014;32:32–39. doi: 10.1038/nbt.2794. [DOI] [PubMed] [Google Scholar]
- 2.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 3.Jefferis R. Glycosylation of recombinant antibody therapeutics. Biotechnol Prog. 2005;21:11–16. doi: 10.1021/bp040016j. [DOI] [PubMed] [Google Scholar]
- 4.Umaña P, Jean-Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17:176–180. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 6.Anthony RM, et al. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony RM, Wermeling F, Ravetch JV. Novel roles for the IgG Fc glycan. Ann N Y Acad Sci. 2012;1253:170–180. doi: 10.1111/j.1749-6632.2011.06305.x. [DOI] [PubMed] [Google Scholar]
- 8.Washburn N, et al. Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity. Proc Natl Acad Sci USA. 2015;112:E1297–E1306. doi: 10.1073/pnas.1422481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van de Bovenkamp FS, Hafkenscheid L, Rispens T, Rombouts Y. The emerging importance of IgG Fab glycosylation in immunity. J Immunol. 2016;196:1435–1441. doi: 10.4049/jimmunol.1502136. [DOI] [PubMed] [Google Scholar]
- 10.Song R, Oren DA, Franco D, Seaman MS, Ho DD. Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat Biotechnol. 2013;31:1047–1052. doi: 10.1038/nbt.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LX, Amin MN. Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem Biol. 2014;21:51–66. doi: 10.1016/j.chembiol.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairbanks AJ. The ENGases: Versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem Soc Rev. 2017;46:5128–5146. doi: 10.1039/c6cs00897f. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc. 2012;134:12308–12318. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Tong X, Yang Q, Giddens JP, Wang LX. Glycosynthase mutants of endoglycosidase S2 show potent transglycosylation activity and remarkably relaxed substrate specificity for antibody glycosylation remodeling. J Biol Chem. 2016;291:16508–16518. doi: 10.1074/jbc.M116.738765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, et al. Modulating IgG effector function by Fc glycan engineering. Proc Natl Acad Sci USA. 2017;114:3485–3490. doi: 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giddens JP, Lomino JV, Amin MN, Wang LX. Endo-F3 glycosynthase mutants enable chemoenzymatic synthesis of core-fucosylated triantennary complex type glycopeptides and glycoproteins. J Biol Chem. 2016;291:9356–9370. doi: 10.1074/jbc.M116.721597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CW, et al. A common glycan structure on immunoglobulin G for enhancement of effector functions. Proc Natl Acad Sci USA. 2015;112:10611–10616. doi: 10.1073/pnas.1513456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurogochi M, et al. Glycoengineered monoclonal antibodies with homogeneous glycan (M3, G0, G2, and A2) using a chemoenzymatic approach have different affinities for FcγRIIIa and variable antibody-dependent cellular cytotoxicity activities. PLoS One. 2015;10:e0132848. doi: 10.1371/journal.pone.0132848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons TB, et al. Optimal synthetic glycosylation of a therapeutic antibody. Angew Chem Int Ed Engl. 2016;55:2361–2367. doi: 10.1002/anie.201508723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q, et al. Glycan remodeling of human erythropoietin (EPO) through combined mammalian cell engineering and chemoenzymatic transglycosylation. ACS Chem Biol. 2017;12:1665–1673. doi: 10.1021/acschembio.7b00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung CH, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. 2008;358:1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian J, et al. Structural characterization of N-linked oligosaccharides on monoclonal antibody cetuximab by the combination of orthogonal matrix-assisted laser desorption/ionization hybrid quadrupole-quadrupole time-of-flight tandem mass spectrometry and sequential enzymatic digestion. Anal Biochem. 2007;364:8–18. doi: 10.1016/j.ab.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Janin-Bussat MC, et al. Cetuximab Fab and Fc N-glycan fast characterization using IdeS digestion and liquid chromatography coupled to electrospray ionization mass spectrometry. Methods Mol Biol. 2013;988:93–113. doi: 10.1007/978-1-62703-327-5_7. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes CA, et al. GA201 (RG7160): A novel, humanized, glycoengineered anti-EGFR antibody with enhanced ADCC and superior in vivo efficacy compared with cetuximab. Clin Cancer Res. 2013;19:1126–1138. doi: 10.1158/1078-0432.CCR-12-0989. [DOI] [PubMed] [Google Scholar]
- 25.Temam S, et al. An exploratory, open-label, randomized, multicenter study to investigate the pharmacodynamics of a glycoengineered antibody (imgatuzumab) and cetuximab in patients with operable head and neck squamous cell carcinoma. Ann Oncol. 2017;28:2827–2835. doi: 10.1093/annonc/mdx489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiedler W, et al. Phase I study of tomuzotuximab, a glycoengineered therapeutic antibody against the epidermal growth factor receptor, in patients with advanced carcinomas. ESMO Open. 2018;3:e000303. doi: 10.1136/esmoopen-2017-000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinke JW, Platts-Mills TA, Commins SP. The alpha-gal story: Lessons learned from connecting the dots. J Allergy Clin Immunol. 2015;135:589–596, quiz 597. doi: 10.1016/j.jaci.2014.12.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biacchi M, et al. Glycoform separation and characterization of cetuximab variants by middle-up off-line capillary zone electrophoresis-UV/electrospray ionization-MS. Anal Chem. 2015;87:6240–6250. doi: 10.1021/acs.analchem.5b00928. [DOI] [PubMed] [Google Scholar]
- 29.Chevreux G, Tilly N, Bihoreau N. Fast analysis of recombinant monoclonal antibodies using IdeS proteolytic digestion and electrospray mass spectrometry. Anal Biochem. 2011;415:212–214. doi: 10.1016/j.ab.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 30.An Y, Zhang Y, Mueller HM, Shameem M, Chen X. A new tool for monoclonal antibody analysis: Application of IdeS proteolysis in IgG domain-specific characterization. MAbs. 2014;6:879–893. doi: 10.4161/mabs.28762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Zhu S, Ma C, Wang LX. Designer α1,6-fucosidase mutants enable direct core fucosylation of intact N-glycopeptides and N-glycoproteins. J Am Chem Soc. 2017;139:15074–15087. doi: 10.1021/jacs.7b07906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi M, Tanaka T, Gyakushi H, Kobayashi A, Shoda S. Efficient synthesis of sugar oxazolines from unprotected N-acetyl-2-amino sugars by using chloroformamidinium reagent in water. J Org Chem. 2009;74:2210–2212. doi: 10.1021/jo8024708. [DOI] [PubMed] [Google Scholar]
- 33.Giddens JP, Wang L-X. Chemoenzymatic glyco-engineering of monoclonal antibodies. Methods Mol Biol. 2015;1321:375–387. doi: 10.1007/978-1-4939-2760-9_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen A, et al. Simplified, enhanced protein purification using an inducible, autoprocessing enzyme tag. PLoS One. 2009;4:e8119. doi: 10.1371/journal.pone.0008119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Díaz J, Carbajo RJ, Pineda-Lucena A, Monedero V, Yebra MJ. Synthesis of fucosyl-N-acetylglucosamine disaccharides by transfucosylation using α-L-fucosidases from Lactobacillus casei. Appl Environ Microbiol. 2013;79:3847–3850. doi: 10.1128/AEM.00229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.