Significance
MicroRNAs repress genes controlling important decisions in animal development, but miRNA production and their interaction with larger genetic programs of development are not well understood. We found that the initial transcript of an important neuronal miRNA, miR-124, is expressed much earlier in development than the mature functional miRNA. Conversion of this primary RNA into its active form is blocked by the RNA binding protein PTBP1, whose down-regulation upon neuronal differentiation allows mature miR-124 expression. To understand the need for this regulation, we developed a mathematical model of the PTBP1/miR-124 regulatory circuit, finding that early expression of unprocessed miR-124 creates a sharp transition in gene expression during neuronal differentiation, making this developmental decision less affected by random variation and noise.
Keywords: genetic regulatory circuit, miRNA-124, miRNA biogenesis, neuronal differentiation, embryonic stem cells
Abstract
MicroRNA (miRNA)-124 is expressed in neurons, where it represses genes inhibitory for neuronal differentiation, including the RNA binding protein PTBP1. PTBP1 maintains nonneuronal splicing patterns of mRNAs that switch to neuronal isoforms upon neuronal differentiation. We find that primary (pri)-miR-124-1 is expressed in mouse embryonic stem cells where mature miR-124 is absent. PTBP1 binds to this precursor RNA upstream of the miRNA stem–loop to inhibit mature miR-124 expression in vivo and DROSHA cleavage of pri-miR-124-1 in vitro. This function for PTBP1 in repressing miR-124 biogenesis defines an additional regulatory loop in the already intricate interplay between these two molecules. Applying mathematical modeling to examine the dynamics of this regulation, we find that the pool of pri-miR-124 whose maturation is blocked by PTBP1 creates a robust and self-reinforcing transition in gene expression as PTBP1 is depleted during early neuronal differentiation. While interlocking regulatory loops are often found between miRNAs and transcriptional regulators, our results indicate that miRNA targeting of posttranscriptional regulators also reinforces developmental decisions. Notably, induction of neuronal differentiation observed upon PTBP1 knockdown likely results from direct derepression of miR-124, in addition to indirect effects previously described.
MicroRNAs (miRNAs) are an abundant class of molecules that regulate many important developmental events (1). Assembled into an RNA-induced silencing complex (RISC), miRNAs base-pair with the 3′ UTRs of their target messenger RNAs to inhibit translation and induce mRNA decay (2–5). MiRNA biogenesis starts with transcription of a primary miRNA transcript (pri-miRNA) containing a hairpin structure that is cleaved by the DROSHA–DGCR8 (Microprocessor) complex to generate a ∼70-nt stem–loop intermediate, the precursor miRNA (pre-miRNA) (6). This pre-miRNA is exported to cytoplasm and further processed by the DICER–TRBP complex to produce a mature ∼22-nt miRNA that is loaded onto an Argonaute protein within the RISC (7, 8). Through base pairing to its seed region, each miRNA mediates binding of RISC to miRNA-responsive elements within a target mRNA to inhibit its expression (9). Besides the initial transcription of the pri-miRNA, miRNA expression can be regulated at later stages of biogenesis including the DROSHA and DICER processing steps (10–12).
The differentiation of embryonic cells into neurons is mediated by complex regulatory networks involving all of the steps of the gene expression pathway. Numerous molecules control this process, including transcription factors, chromatin modifiers, RNA binding proteins (RBPs), and miRNAs (13–19). The miRNA miR-124 has been described as a master regulator of neuronal differentiation (17, 20, 21). MiR-124 is up-regulated as neuronal progenitors exit mitosis and begin to differentiate and acts to repress many genes that maintain the nonneuronal state. Expression of miR-124 has been shown to be sufficient to drive cells into the neuronal pathway (14, 22, 23). Known targets of miR-124 include the RE1-silencing transcriptional factor (REST) that represses neuronal transcription programs and the polypyrimidine tract-binding protein (PTB, PTBP1, or hnRNP I) that represses neuronal alternative splicing patterns (20, 21, 24).
In the human and mouse genomes, there are three genes encoding miR-124 precursors: miR-124-1 (also called Rncr3), miR-124-2, and miR-124-3 (25–27). In the mouse, miR-124-1 (chr14:65209494-65209578; mm9) and miR-124-2 (chr3:17695662-17695770) are highly expressed in neurons, while expression of miR-124-3 (chr2:180628745-180628812) is more limited. During neuronal development, the three loci show different levels and timing of induction (26, 28, 29).
miR-124 has also been described as a suppressor of the tumor phenotype in glioma and colorectal cancers (30–32). In a colon cancer cell line, this suppressor of tumor growth has been linked to miR-124 repression of DDX6, c-Myc, and PTBP1 (33).
PTBP1 represses neuronal patterns of alternative splicing in a variety of nonneuronal cell types including embryonic stem cells (ESCs) and neural progenitor cells (NPCs) (34–38). During neuronal differentiation, PTBP1 expression is turned off, in part through the action of miR-124 (21). This allows the induction of PTBP2, a paralogous protein which has different regulatory properties and allows the production of spliced isoforms specific to differentiating neurons (19, 21, 36, 38–44). In addition to its role in splicing, PTBP1 is also found in the cytoplasm, where it can antagonize the action of miR-124 by binding in the 3′ UTRs of transcripts such as CoREST and SCP1 (37). Like the ectopic expression of miR-124, the depletion of PTBP1 can induce neuronal differentiation through effects on both splicing and translation (37). Although these two posttranscriptional regulatory programs, miR-124 induction and PTBP1 depletion, are each sufficient to drive differentiation, a systems-level understanding of how these regulatory circuits interact in controlling the commitment decision is lacking.
Here we show that PTBP1 directly represses miR-124 expression at the level of pri-miRNA processing. We find that pri-miR-124-1 RNA is expressed in mouse ESCs (mESCs) without production of mature miRNA. We show that PTBP1 binds pri-miR-124-1 and blocks DROSHA cleavage in the nucleus. Through mathematical modeling, we find that this regulatory loop connecting miR-124 levels with those of PTBP1 enforces a sharp regulatory transition during neuronal development as PTBP1 is depleted and miR-124 expression is increased.
Results
Primary miR-124-1 Is Expressed but Not Processed into Mature miRNA in mESCs.
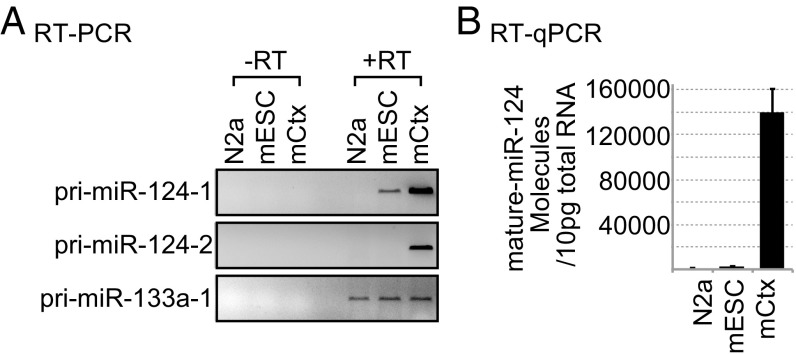
Examining the expression of pri-miR-124 by RT-PCR in mESCs, N2a neuroblastoma cells, and cultured mouse cortical neurons (mCtx), we detected strong expression of pri-miR-124-1 and pri-miR-124-2 (Fig. 1A) but not of pri-miR-124-3 in cortical neurons. Unexpectedly, we also observed expression of pri-miR-124-1 in mESCs (Fig. 1A and Table 1). Similar expression was observed by RNA sequencing (discussed below). In contrast, mature miR-124 was abundant in neurons as expected but only present at the limit of RT-qPCR detection in E14 mESCs (Fig. 1B and Table 1).
Fig. 1.
Pri-miR-124-1, but not mature miR-124, is expressed in mESCs. (A) RT-PCR of pri-miR-124-1, pri-miR-124-2, and pri-miR-133a-1 from N2a, mESC and, mCtx. Representative gel image is shown from three biological replicate cultures. PCR products generated in the absence of reverse transcription (−RT) controlled for the presence of contaminating genomic DNA. RT-PCR analysis of pri-miRNA was done using primers spanning of the stem–loop (SI Appendix, Fig. S2B). A primer pair detecting the upstream region of pri-miR-124-1 gave equivalent results (SI Appendix, Fig. S2A). RT-qPCR measurement of these molecules is reported in Table 1. N2a, mouse neuroblastoma cells; mESC, E14 mouse embryonic stem cells; mCtx, mouse cortical neurons, days in vitro 5. (B) Quantification of mature miR-124 molecules in 10 pg of total RNA from the same cells as in A. Mean molecule numbers were determined by RT-qPCR from three cultures; error bars are SEM.
Table 1.
Quantification of primary and mature miR-124 and PTBP
| Molecule | N2a | mESC | mNPC | mCtx |
| RNA | ||||
| Pri-miR-124-1 | 0.3 ± 0.1 | 3.6 ± 0.9 | 0.3 ± 0.2 | 30.1 ± 9.7 |
| Pri-miR-124–2 | 0.0 ± 0.0 | 0.1 ± 0.0 | 9.7 ± 3.0 | 44.3 ± 11.7 |
| Pri-miR-124–3 | 0.4 ± 0.3 | 0.8 ± 0.2 | 0.7 ± 0.1 | 3.1 ± 1.7 |
| Mature miR-124 | 3.1 ± 1.2 × 102 | 2.2 ± 0.2 × 103 | 1.3 ± 0.4 × 103 | 1.4 ± 0.2 × 105 |
| Mature miR-294 | 4.6 ± 4.5 | 2.4 ± 0.1 × 105 | 11.6 ± 10.1 | 3.5 ± 6.0 |
| U6 | 2.5 ± 0.6 × 105 | 2.1 ± 0.8 × 105 | 2.5 ± 0.2 × 105 | 2.7 ± 0.2 × 105 |
| Protein | ||||
| PTBP1 | 1.4 ± 0.4 × 104 | 1.2 ± 0.2 × 104 | 6.7 ± 2.2 × 103 | 2.4 ± 8.8 × 102 |
| PTBP2 | 4.6 ± 1.8 × 102 | 2.3 ± 9.0 × 102 | 8.5 ± 1.3 × 102 | 3.5 ± 0.4 × 103 |
Quantification of primary and mature miR-124 and PTBP1/2 molecules per cell. Molecules of RNA per 10 pg of total cellular RNA were determined by RT-qPCR. In separate measurements, we determined that total cellular RNA varied from 10 pg to 19 pg per cell (SI Appendix, Table S1). Primer pairs for RT-qPCR were designed to detect a region upstream of the stem–loop (SI Appendix, Fig. S2B). PTBP proteins per 0.4 ng of total cellular protein were measured by fluorescent Western blot of total cell lysate and compared with a standard curve of recombinant His-PTBP protein. Total mass of protein per cell was measured to be ∼0.4 ng (SI Appendix, Table S1). N2a, mouse neuroblastoma cells; mESC, E14 mESCs; mNPC, mouse neuronal progenitor cells; mCtx, mouse cortical neurons, days in vitro 5. The mean ± SD of three cultures are given. Standard curves for RT-qPCR are presented in SI Appendix, Fig. S1.
Expression of miR-124 in mESCs was surprising given its role in driving neuronal differentiation. To better assess the amounts of mature miR-124 relative to its primary transcripts, we used RT-qPCR to quantify the absolute number of each RNA species in 10 pg of total RNA from each of the above cell types, as well as from isolated mouse NPCs (mNPCs). We determined that each of these cell types contain 10–19 pg of total RNA per cell (SI Appendix, Table S1) and that the different cell types expressed relatively constant levels of the U6 small nuclear RNA (snRNA) (Table 1). The primer pairs for RT-qPCR analysis were designed to detect a region upstream of the stem–loop and minimize changes that might derive from DROSHA processing or the strong stem–loop structure (SI Appendix, Fig. S2B). Primer pairs spanning the stem–loop were used for semiquantitative RT-PCR analyses (Figs. 1A, 2C, and 3A and SI Appendix, Fig. S2A) (45, 46). For miR-124-1, the upstream and spanning primer pairs were found to yield equivalent results (SI Appendix, Fig. S2A; pri-miR-124-1).
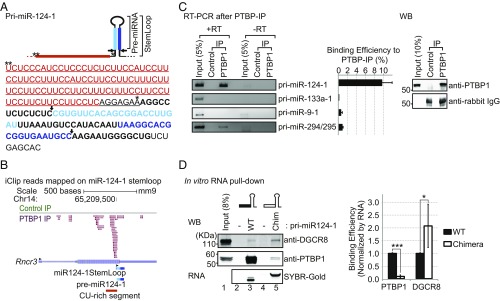
Fig. 2.
PTBP1 binds to a CU-rich segment in pri-miR-124-1. (A) The sequence of pri-miR-124-1 with its secondary structure and sites of DROSHA cleavage (arrows) are diagrammed (Top). The miR-124 duplex strands resulting from DROSHA and DICER cleavage are indicated in light and dark blue, with the final miR-124 in dark blue. Asterisks mark nucleotides 1 and 107 upstream from miR-124 stem–loop, respectively. The CU-rich segment extending from 107 nt to 8 nt upstream of the stem–loop is in red. Sequences switched with pri-miR-1a-2 to form the chimeric RNAs in D are underlined. (B) Genome browser view of PTBP1 iCLIP tags (PTBP-IP, magenta) and FLAG-rabbit (Control-IP, green) from mESCs aligned to the miR-124-1 host gene (Rncr3, AK044422). The stem–loop of miR-124-1, pre-miR-124-1, and the CU-rich segment are indicated (Bottom). Black arrowheads indicate 5′ to 3′ direction of each of RNA. (C) Immunoprecipitation of pri-miR-124-1 with PTBP1. PTBP1 was immunoprecipitated from mESC (PTBP1-IP) and bound RNA was extracted and assayed for pri-miRNAs by RT-PCR. Rabbit IgG served as a negative control (Control-IP). PCR in the absence of reverse transcription (−RT) served to control for genomic DNA contamination. Representative gel images from three biological replicates are shown (Left). RNA bound to PTBP1-IP was compared with 5% of the input RNA (Middle); error bars are SEM. Immunoprecipitated targets were checked by Western blot (Right). (D) The CU-rich segment is required for the interaction between PTBP1 and pri-miR-124-1. A chimeric miRNA had the CU-rich segment of pri-miR-124-1 (black box) replaced with the equivalent sequence from pri-miR-1a-2 (open box). WT and chimeric (Chim) pri-miR-124-1 were transcribed in vitro and hybridized to biotinylated adaptors immobilized on streptavidin beads. Beads carrying biotinylated adapters alone served as negative controls (lanes 2 and 4). Pri-miRNA–bound beads were incubated with mESC total cell extract in 2 mM EDTA to inhibit DROSHA cleavage. After washing, bound PTBP1 and DGCR8 were detected by Western blot and the bound RNA with SYBR-Gold (Left). Representative gel images are from three biological replicates. Protein bound to each RNA was quantified, normalized to the amount of bait RNA, and further normalized to WT RNA in each replicate (Right) (n = 3 biological replicates; Student’s t test; *P ≤ 0.05, ***P ≤ 0.001; error bars are SEM).
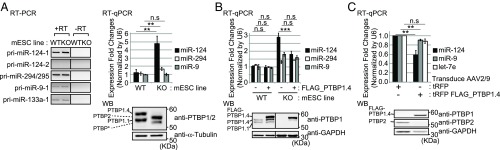
Fig. 3.
PTBP1 inhibits miR-124-1 maturation in vivo. (A) Mature miR-124 is enhanced in a Ptbp1 KO cell line. The levels of pri-miRNAs were measured in WT and Ptbp1 KO mESCs by RT-PCR (Left). PCR in the absence of reverse transcription (−RT) controlled for genomic DNA contamination. Spanning primer pairs as diagrammed in SI Appendix, Fig. S2B were used for RT-PCR analysis. A representative gel image is shown from three biological replicate cultures (Left). The levels of mature miR-124, miR-294, and miR-9 were measured in WT and Ptbp1 KO mESCs by RT-qPCR (Right), normalized to U6 snRNA, and further normalized to the first replicate of WT cells (n = 3 cultures; Student’s t test; **P ≤ 0.01; error bars are SEM). Ptbp1 depletion in the KO line was confirmed by Western blot using a PTB_CT antibody targeting the common C-terminal peptide of PTBP1 and PTBP2 (Bottom Right) (50). PTBP1 and PTBP2 isoforms are indicated to the left. Alpha-tubulin served as loading control. Representative gel images from three cultures. (B) Ectopic PTBP1 reverses enhancement of miR-124 expression in the KO cell line. Mature miR-124, miR-294, and miR-9 were measured by RT-qPCR in WT and Ptbp1 KO mESCs after ectopic FLAG-PTBP1.4 expression (Top). MiRNA levels were normalized to U6 snRNA, and further normalized to the first replicate of WT cells with no transfected PTBP1 (n = 3 biological replicates; Student’s t test; ***P ≤ 0.001; error bars are SEM). Expression of FLAG-PTBP1.4 was confirmed by Western blot. GAPDH served as a loading control (Bottom). Representative gel images are from three replicates. (C) Reintroduction of PTBP1 inhibits miR-124 expression in cultured cortical neurons. Cortical neurons were transduced with FLAG-PTBP1.4 expressing AAV 1 d after plating, and mature miRNA levels were measured at day 8 by RT-qPCR. Mir-9 and let-7e were assayed as controls (Top). Mature-miRNA level was normalized to U6 snRNA, and further normalized to the tRFP-only condition in each replicate (n = 3 biological replicates; Student’s t test; **P ≤ 0.01; error bars are SEM). Expression of FLAG-PTBP1.4 and endogenous PTBP2 were confirmed by Western blot. GAPDH served a loading control (Bottom). Representative gel images from three replicates. n.s, not significant.
As expected, cortical neurons expressed between 60- and 500-fold more mature miR-124 than the other cell types. Similarly, mNPCs and N2a express about 100-fold lower levels of pri-miR-124-1 than neurons, although mNPCs have begun to express some pri-miR-124-2. Notably, the level of pri-miR-124-1 in mESCs was about 10-fold higher than in N2a cells and mNPCs and only about eightfold lower than in cortical neurons (3.6 vs. 30.1 molecules of pri-miR-124-1 per 10 pg of total RNA; Table 1). Comparing the level of mature miR-124 to the total of the three pri-miR-124 RNAs in the three cell types, we find that cortical neurons appear to convert about fourfold more of the expressed pri-miR-124 into mature steady-state miR-124 than do mESCs (Table 1; not accounting for miRNA turnover). Mature miR-294 was abundant in mESCs but not the other cell types, indicating that there was not a general loss of mature miRNAs in these cells. Repeated measurements in each cell type gave similar results, indicating that the higher level of pri-miR-124-1 seen in mESCs was not due to random fluctuations or noise (Table 1). Thus, pri-miR-124-1 is expressed in mESCs, but the mature miRNA fails to accumulate due to either reduced processing or a higher decay rate.
Pri-miR-124-1 Is Bound by PTBP1.
Examining the sequences of the miR-124 genes, we noted a highly CU-rich segment upstream of the pri-miR-124-1 stem–loop that was not present in pri-miR-124-2 or pri-miR-124-3. This sequence is predicted to be bound by PTBP1, which we hypothesized may act to repress maturation of miR-124 in mESCs (Fig. 2A) (47, 48). Examining the miR-124-1 locus in an iCLIP dataset that we previously generated for PTBP1 in mESCs (38), we observed a significant peak of PTBP1 cross-linked fragments directly on this CU-rich segment of pri-miR-124-1, with a smaller peak of iCLIP tags upstream (Fig. 2B). To confirm the interaction of PTBP1 with the pri-miRNA, we immunoprecipitated PTBP1 from mESC lysates, extracted the associated RNA, and performed RT-PCR for pri-miR-124-1 (Fig. 2C). Indeed, pri-miR-124-1 was efficiently pulled down with PTBP1 and not in control immunoprecipitates (IPs). Pri-miRNAs for the muscle and neuronal miRNAs miR-133a-1 and miR-9-1, which were not predicted to be bound by PTBP1, were not pulled down with PTBP1-IP. Other pri-miRNAs observed to be bound by PTBP1 in the iCLIP data, such as miR-294/295, were also detected in the PTBP1-IP but not as strongly as pri-miR-124-1 (Fig. 2C and SI Appendix, Fig. S3).
The CU-rich segment in pri-miR-124-1 extends from 107 nt upstream of the miR-124-1 stem–loop to 8 nt upstream (−107 to −8). A small CU-rich segment is also present in the base-paired stem immediately 5′ of the DROSHA cleavage site (Fig. 2A), and there are additional pyrimidine-rich sequences ∼200 nt upstream of the stem–loop. To examine the binding of PTBP1 to the CU-rich segment in vitro, we performed pull-down experiments using in vitro-transcribed RNAs (Fig. 2D). WT pri-miR-124-1 was compared with a chimeric RNA with nucleotides −107 to −1 upstream of the stem–loop replaced with the equivalent nucleotides from pri-miR-1a-2 (Fig. 2A; underlined). These RNAs were hybridized to complementary biotinylated DNAs immobilized on streptavidin beads, incubated in mESC lysate, and after washing the bound proteins were analyzed by Western blot. As expected, PTBP1 bound efficiently to the WT RNA and only minimally to the chimeric RNA, confirming its binding to the miR-124-1 upstream sequence (Fig. 2D). Binding of the Microprocessor component DGCR8 exhibited a contrasting pattern, with greater binding to the chimeric substrate that is not bound by PTBP1 (Fig. 2D).
To identify additional PTBP1-bound pri-miRNAs, we further analyzed the PTBP1 iCLIP data in mESCs (SI Appendix, Fig. S4) (38). Defining sequence intervals from 125 nt upstream to 125 nt downstream of each annotated miRNA stem–loop, we identified 2,020 PTBP1 iCLIP tags mapping within 295 pri-miRNA loci (SI Appendix, Fig. S4A). In addition to those described above (Fig. 2C), other pri-miRNAs showing significant PTBP1 binding included miR-5125, miR-7-1, miR-127, and others (SI Appendix, Fig. S4). Some of these exhibited extensive PTBP1 binding outside of the defined search window (>125 nt upstream or downstream of the stem–loop). Such distal PTBP1 binding may not interfere with miRNA maturation, and it will be interesting to characterize the biogenesis of these miRNAs for possible PTBP1 regulation. Within sequences immediately adjacent to the miRNA stem–loops, pri-miR-124-1 had the highest number of PTBP1 iCLIP tags (53 total tags in the clusters described above; Fig. 2B and SI Appendix, Fig. S4A).
PTBP1 Inhibits miR-124-1 Maturation.
PTBP1 binding close to the site of pri-miR-124-1 processing indicated that the protein might interfere with DROSHA–DGCR8 binding and/or cleavage. To examine this, we compared the expression of mature miR-124 in Ptbp1 WT and KO mESCs. A Ptbp1 KO line was generated from a mESC line carrying loxp sites flanking Ptbp1 exon 2, whose excision eliminates expression of the major PTBP1 isoforms PTBP1.1 and PTBP1.4. Cre recombinase was introduced into these cells and clones carrying homozygous deletions of the Ptbp1 exon were selected (Fig. 3A, Bottom Right). As observed previously, PTBP1 depletion induced the expression of PTBP2, which is encoded on a separate gene and is posttranscriptionally repressed by PTBP1 (21, 39, 49). A smaller, less-abundant protein was also observed in the KO cells using an antibody targeting the C-terminal peptide common to both PTBP1 and PTBP2 (50). This could be another isoform of PTBP2 or could result from translation initiation downstream from the deleted Ptbp1 exon. Importantly, the loss of the major PTBP1 isoforms resulted in a nearly fivefold increase in mature miR-124 over the control cells (KO vs. WT; Fig. 3A, Right). The KO cells did not show significant changes in the level of pri-miR-124-1, indicating that the increase in mature miRNA did not result from increased transcription from the locus (Fig. 3A, Left). Mature mir-294, whose precursor also exhibited some PTBP1 binding, showed only a small increase in the KO cells, while miR-9, which is not predicted to be targeted by PTBP1, but which is also induced with neuronal differentiation, showed no change.
We then introduced ectopic FLAG-PTBP1.4 into the WT and Ptbp1 KO mESCs (Fig. 3B). Ectopic PTBP1.4 had little effect on miR-124 or other miRNA levels in the WT cells where PTBP1 expression is already high. Under transfection conditions the KO cells expressed threefold more miR-124 than WT cells (Fig. 3B), somewhat less than seen in normal culture (Fig. 3A). There were also modest increases in miR-294 and miR-9 in the mock- and PTBP1.4-transfected KO cells compared with WT (Fig. 3B). Importantly, reintroduction of PTBP1.4 into the KO cells sharply reduced miR-124 by 40%, but had little effect on miR-294 or miR-9. From these data we conclude that PTBP1 inhibits expression of mature miR-124 in mESCs.
The induction of PTBP2 in the Ptbp1 KO mESCs, and the coexpression of miR-124 and PTBP2 in neurons indicate that PTBP2 does not repress miR-124 to the same degree as PTBP1. To examine whether miR-124 continues to be responsive to PTBP1 after neuronal differentiation, we transduced cultured cortical neurons with FLAG-PTBP1.4 using an adenoassociated virus (AAV). In these cells, miR-124 is expressed at high levels from both the miR-124-1 and miR-124-2 genes, while endogenous PTBP1 expression is repressed and replaced with PTBP2 (Fig. 3C, Bottom and Table 1). We found that reexpression of PTBP1 repressed miR-124 expression by 40%, corresponding to the proportion of miRNA arising from the pri-miR-124-1 gene (Table 1). The magnitude of the reduction in miR-124 also correlated with the titer of the transducing AAV (SI Appendix, Fig. S5). Altogether, these data confirm that PTBP1 represses mature miR-124 expression in vivo.
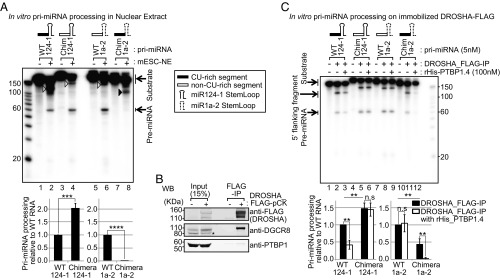
The repression of miR-124 could occur by PTBP1 blocking DROSHA cleavage. However, most regulators of DROSHA processing have been found to bind in the stem–loop of the target RNA (51–53), while PTBP1 binds upstream of the stem. To test the effect of PTBP1 on DROSHA action, we used an in vitro pri-miRNA processing assay. We transcribed two pri-miRNA substrates: pri-miR-124-1 and pri-miR-1a-2, each including ∼107 nt upstream and ∼10 nt downstream of the miRNA stem–loop. We also designed two chimeric RNAs where the upstream regions of pri-miR-124-1 and pri-miR-1a-2 were switched. These RNAs were incubated in mESC nuclear extract active for DROSHA processing and their cleavage products characterized by gel electrophoresis. With incubation in nuclear extract, the synthetic pri-miR-124-1 was converted to the ∼60-nt pre-miRNA with moderate efficiency, and a 130-nt species the size of the detached 5′ flanking fragment from upstream of the pre-miR-124-1 was also produced (Fig. 4A, lane 2, open arrowhead). Using the chimeric RNA, where the PTBP1 binding site was replaced with the upstream sequence from miR-1a-2, production of the pre-miRNA increased twofold (Fig. 4A, lane 4). Note that the 5′ flanking fragment from the chimeric RNA yields a lower-intensity band than the WT RNA due to its lower U content (Fig. 4A). Interestingly, while the synthetic pri-miR-1a-2 was efficiently processed in the extract, this DROSHA cleavage was completely abolished by replacing its upstream segment with the PTBP1 binding sequence of miR-124-1 (Fig. 4A, lanes 6 and 8, with bar graph below). Instead, we observed an aberrant product of ∼100 nt that does not correspond in size to any of the expected DROSHA cleavage products (Fig. 4A, lane 8, black arrowhead). These data indicate that the sequence upstream of the miR-124-1 stem–loop is inhibitory for cleavage at the expected DROSHA processing sites and may induce aberrant products from DROSHA or other activities.
Fig. 4.
PTBP1 binding inhibits pri-miR-124-1 processing in vitro. (A) The CU-rich segment inhibits pri-miRNA processing in vitro. Pri-miR-124-1, pri-miR-1a-2, and chimeric substrates (∼3 nM) were incubated in mESC nuclear extract (NE) and DROSHA-cleaved products were resolved by urea-PAGE. For the chimeric RNAs the upstream sequences of miR-124-1 (black box, containing the PTBP1 binding CU-rich segment) and miR-1a-2 (open box) were switched. The stem–loops of miR-124-1 and miR-1a-2 are indicated by solid and dashed lines, respectively. The pri-miRNA substrates were uniformly labeled with [α-32P]UTP. Unprocessed RNAs and the pre-miRNA products are indicated by arrows. Open arrowheads in lanes 2, 4, and 6 indicate the 5′ processing products (5′ flanking fragments) of pri-miR-124-1, chimera-miR-124-1, and pri-miR-1a-2 by DROSHA. The black arrowhead in lane 8 indicates an unidentified product of chimeric pri-miR-1a-2, whose size (∼100 nt) does not match any of the expected DROSHA cleavage products. Representative gel images are from three replicates (Top). Pri-miRNA processing efficiency was calculated by dividing pre-miRNA signal by unprocessed substrate (incubated with buffer only), normalized by the number of labeled U residues in each substrate, and then to the WT construct in each replicate (Bottom) (n = 3 biological replicates; Student’s t test; ***P ≤ 0.001, ****P ≤ 0.0001; error bars are SEM). (B) PTBP1 is not pulled down with the DROSHA-FLAG-IP beads in mESC (E14). DROSHA-FLAG immunoprecipitates were analyzed by Western blotting. DGCR8 immunoprecipitated with DROSHA-FLAG efficiently, while PTBP1 did not. Asterisk marks nonspecific band in DGCR8 blot. (C) Recombinant His-PTBP1 inhibits processing of pri-miRNAs that have an upstream CU-rich segment. The same pri-miRNA substrates analyzed in A were subjected to in vitro pri-miRNA processing on immunoprecipitated DROSHA-FLAG (B) with or without added rHis-PTBP1.4. Unprocessed substrates and the DROSHA processing products (5′ flanking fragments and pre-miRNAs) are indicated by arrows. A longer exposure of the gel image is in SI Appendix, Fig. S7. Representative gel images from three replicates (Top). Pri-miRNA processing efficiency measured as in C is presented in the bar graph (Bottom) (n = 3 biological replicates; Student’s t test; **P ≤ 0.01; error bars are SEM). n.s, not significant.
We also observed faint bands of correct size for the 3′ fragments produced by DROSHA cleavage (Fig. 4A). To confirm their identity and to further quantify the extent of DROSHA cleavage on each RNA, we used 3′-end-labeled pri-miRNA substrates (SI Appendix, Fig. S6). These experiments gave equivalent results. The short cleavage products were indeed derived from the 3′ ends of the RNAs. Removing the PTBP1 binding site from pri-miR-124-1 again led to a twofold increase in DROSHA cleavage. Adding the PTBP1 binding site to pri-miR-1a-2 led to a 10-fold decrease in DROSHA cleavage—essentially identical results to those with uniformly labeled RNA.
To demonstrate that PTBP1 mediates the inhibition of DROSHA processing by the upstream CU-rich segment, we tested processing of the same four pri-miRNAs on immobilized DROSHA. We expressed Flag-tagged DROSHA in mESCs and pulled the protein down with anti-Flag antibodies. These IPs did not contain PTBP1 detectible by Western blot (Fig. 4B), allowing simple addition of exogenous protein. This system exhibited greater DROSHA activity and allowed clearer identification of the processing products than crude nuclear extract. When incubated with the DROSHA-bound beads, all four pri-miRNAs were cleaved to produce the pre-miRNA, the 5′ flanking fragment and the 3′ flanking fragment (Fig. 4C, lanes 2, 5, 8, and 11). The 3′ flanking fragments can be observed with longer exposure (SI Appendix, Fig. S7). Processing of the two RNAs that lack a PTBP1 binding site (WT pri-miR-1a-2 and chimeric pri-miR-124-1) was not affected by added recombinant PTBP1.4 (Fig. 4C, lanes 5, 6, 8, and 9, with bar graph below). In contrast, processing of both WT pri-miR-124-1 and the chimeric pri-miR-1a-2 containing the upstream PTBP1 binding site were strongly inhibited by the addition of PTBP1 (Fig. 4C, lanes 2, 3, 11, and 12, with bar graph below). Taken together, these results demonstrate that PTBP1 inhibits DROSHA–DGCR8 processing of pri-miR-124-1 by binding to a CU-rich segment upstream of the stem–loop.
The Host Gene for miR-124-1 Is Not Expressed as an mRNA.
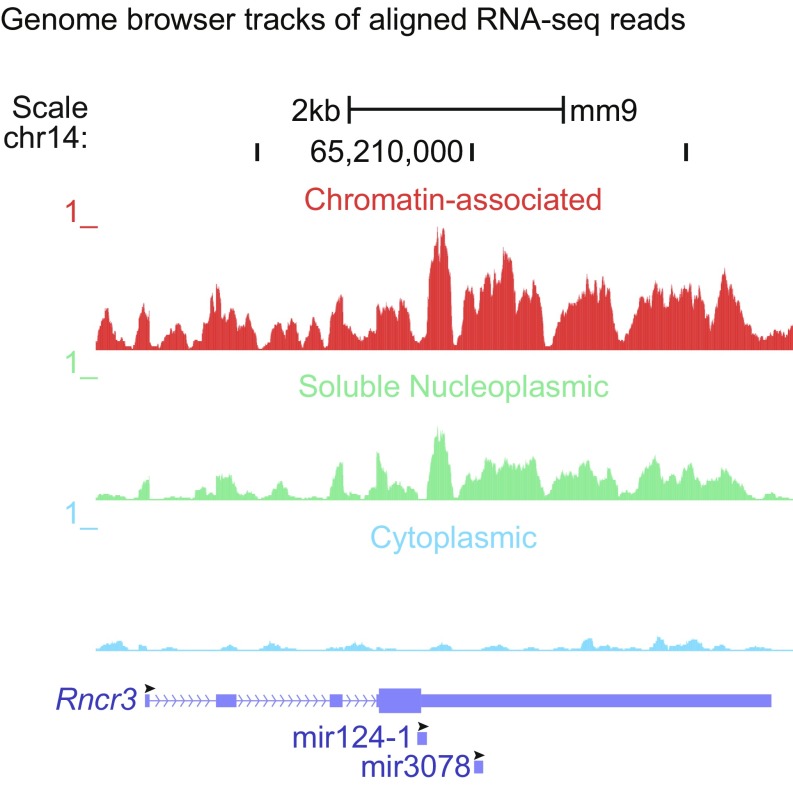
The expression of pri-miR-124-1 in mESCs raises questions regarding the function of the transcript in these cells. The host gene for miR-124-1 (Rncr3) is annotated as a potential protein coding gene with an ORF that terminates within the miR-124 stem–loop, making the expression of the putative mRNA and the mature miRNA mutually exclusive. Thus, PTBP1 repression of DROSHA processing could allow expression of the host transcript as an mRNA. To examine this, we assessed the host gene transcripts that are spliced and exported to the cytoplasm in mESCs. As described previously, we isolated RNA from three subcellular fractions: nascent RNA that is associated with chromatin in the nucleus, RNA from the soluble nucleoplasm, and RNA from the cytoplasm where an mRNA will be translated (54, 55). We generated RNA sequencing datasets from each fraction and aligned the sequences to the genome to specifically examine reads mapping to the pri-miR-124-1 locus from these three compartments. A more complete analysis of these data will be presented elsewhere. A typical protein coding gene exhibits abundant exon reads with limited intronic reads in the cytoplasm derived from the spliced mRNA, as well as some reads derived from both the exons and introns of the nascent RNA in the nuclear fractions. This pattern was not observed for the pri-miR-124-1 transcript, which expressed abundant reads for nascent RNA in the chromatin-associated fraction, similar to our observations by RT-qPCR (Fig. 1A and Table 1). RNA from the locus was also present at lower levels in the soluble nucleoplasm (Fig. 5). However, reads from this gene were present at very low levels in the cytoplasm and these reads were sporadically distributed without the typical exon peaks of a spliced mRNA. The lack of spliced RNA in the cytoplasm indicates that the host gene is not being expressed as an mRNA in mESC, although it could function as an mRNA in other cells.
Fig. 5.
Pri-miR-124-1 is expressed in mESCs as chromatin-associated RNA but not as cytoplasmic RNA. Genome browser tracks showing poly-A+ RNA sequencing reads mapping to the pri-miR-124-1 host gene (Rncr3, AK044422) from three subcellular fractions: the chromatin-associated pellet (Top), soluble nucleoplasm (Middle), and cytoplasm (Bottom). Fractionation followed the method of Wuarin and Schibler to enrich for nascent RNA in the chromatin fraction (54, 87, 88) and was assessed by Western blot for separation of diagnostic proteins (SI Appendix, Fig. S8). Read number scale (RPM) is set to 1 for all three fractions, the maximum peak height for the chromatin fraction. The positions of the two encoded miRNA stem–loops miR-124-1 and miR-3078 are indicated. Black arrowheads indicate the 5′ to 3′ direction of each of RNA. Tracks shown are the combined reads from three biological replicates.
The Regulation of miR-124 by PTBP1 Enforces Commitment to Neuronal Differentiation.
We find that PTBP1 inhibits miR-124 expression in mESCs, while previous studies showed that miR-124 inhibits PTBP1 expression in neurons (21). One role for the early nonproductive expression of miR-124 could be to alter the dynamics of the neuronal differentiation program. Since both miR-124 expression and PTBP1 depletion are known to induce neuronal differentiation, it was difficult to experimentally define a role for the PTBP1/miR-124-1 interaction through perturbation of the concentrations of each component. To examine how the antagonistic regulatory loops connecting PTBP1 and miR-124 affect their expression profiles over development, we constructed a kinetic computational model of miR-124 and PTBP1 production (Fig. 6A). The model incorporated the known regulatory loops affecting PTBP and miR-124 expression and was assessed with or without the newly discovered PTBP1 inhibition of miR-124-1 processing (Fig. 6A, red line).
Fig. 6.
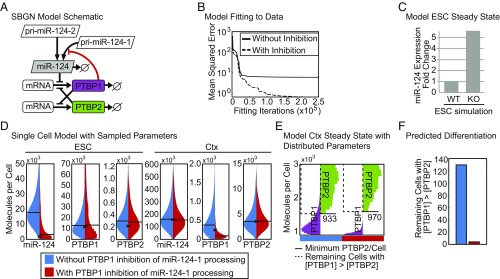
PTBP1 inhibition of miR-124-1 maturation mitigates cell-to-cell variability during neuronal differentiation. (A) Schematic of the ordinary differential equation model of miR-124 expression presented in Systems Biology Graphical Notation [SBGN (89)]. Known regulatory interactions are indicated with black lines and the inhibition of miR-124 processing by PTBP1 identified here is indicated by a red line. (B) Mean squared error between best simulation fits and the molecule numbers measured experimentally (Table 1). Parameter estimation was performed by particle swarm optimization in either the model with the inhibition of pri-miR-124-1 processing by PTBP1 (dashed line) or without the addition of this mechanism (solid line). (C) Prediction of miR-124 levels in the WT and PTBP1 KO ESC from simulations maintaining or removing the synthesis of PTBP1. (D) Distribution of mature miR-124, PTBP1, and PTBP2 levels in ESCs and cortical neurons (Ctx) in 1,000 single-cell simulations run with (red) and without (blue) PTBP1 inhibition of pri-miR-124-1 processing. The experimentally measured value is indicated with an x and the median molecule numbers are indicated with horizontal lines. (E) PTBP1 and PTBP2 molecules in 1,000 simulated single cells of the Ctx stage at the end of the modeled time course, with (red bar) and without (blue bar) PTBP1 inhibition of pri-miR-124-1 processing (shown in SI Appendix, Fig. S9). The minimum PTBP2 molecule number is indicated with a solid line and those cells with more PTBP1 than this number are indicated in the region marked with a dashed line. (F) Number of cells from 1,000 single-cell simulations in which the concentration of PTBP1 in the neuronal population was above the minimum PTBP2 concentration at this stage with (red) and without (blue) PTBP1 inhibition of pri-miR-124-1 processing (number of cells within the region marked with a dashed line in E).
To provide input parameters for the model, we used the above-measured per-cell levels of each RNA species in each of three cell types: mESC, mNPC before the induction of miR-124, and mCtx (Table 1). We also performed quantitative fluorescent Western blots of PTBP1 and PTBP2, compared with a standard curve of recombinant protein, to measure protein molecules per cell (Table 1 and SI Appendix, Table S1). These concentrations were used to parameterize a system of ordinary differential equations describing the concentrations of all proteins and miRNAs determined by their rates of synthesis and decay, and the inhibition of these processes. The entire space of kinetic constants affecting miR-124 and PTBP1 concentrations was then scanned over 12 orders of magnitude to capture all potential parameter combinations (Fig. 6B). Parameters were iteratively fit to the models to identify combinations that minimized the distance to experimentally observed concentrations.
We found that incorporating the inhibition of pri-miR-124-1 processing by PTBP1 into the model enabled a substantially better fit to the experimental data than was possible with a model lacking this inhibition (Fig. 6B, dashed line). In fact, the model without PTBP1 repression of pri-miR-124-1 was unable to recapitulate the experimentally determined levels of protein and RNA with any parameterization, suggesting the experimental data cannot be quantitatively explained without this mechanism (Fig. 6B, solid line). To assess how well the model with PTBP1 repression of miR-124-1 recapitulated the underlying conditions, we removed expression of PTBP1 to predict the effect of knocking out PTBP1 (a condition not used to parameterize the model). This simulation produced an approximately fivefold increase in mature miR-124 upon loss of PTBP1—notably close to the approximately fivefold increase we observed in Ptbp1 KO mESCs (Figs. 3A and 6C). We conclude that the inhibition of miR-124-1 processing by PTBP1 is required to explain the quantitative experimental data and plays a substantial role in determining the balance of these molecules in vivo.
Feedback loops involving miRNAs and transcription factors add robustness to differentiation pathways (56–59). By reducing the impact of variation in environment and genotype, these regulatory circuits make developmental transitions more reliable. We wanted to examine whether similar properties would result from regulatory loops involving miRNAs and RBPs. To test the effects of miR-124-1 regulation by PTBP1 on the path of neuronal differentiation under conditions of heterogeneous environments or intrinsic variability, we added stochastic parameter sampling to the kinetic models. Parameter sampling has been widely used to model cellular heterogeneity (60–62). Simulations of 1,000 individual cells were run for each model with each parameter sampled from a fourfold range distribution centered on the optimal parameter identified by the original fitting to experimental data (Fig. 6D). We found that the addition of feedback not only reduced the overall level of mature miR-124 in ESCs but it also reduced cell-to-cell heterogeneity in this level (Fig. 6D, leftmost panel; miR-124 level in ESC, red vs. blue distribution). Interestingly, we found that heterogeneity in PTBP1 levels was also greatly reduced in cortical neurons when miR-124-1 was repressed by PTBP1 (Fig. 6D, fifth panel; PTBP1 levels in Ctx, red vs. blue distribution). The tighter regulation of PTBP1 in the model affects the transition to PTBP2 expression in the presence of biological variability, with fewer cells reaching a state of incompletely down-regulated PTBP1, and ensuring that a neuronal, PTBP2-only state is reached (Fig. 6 E and F). Thus, the computational modeling indicates that PTBP1-meditated inhibition of miR-124-1 can act to mitigate biological variability and ensure that robust differentiation occurs in all cells.
Discussion
A Posttranscriptional Feedback Loop Affecting Neuronal Differentiation.
Neuronal differentiation from progenitor cells involves a complex regulatory program affecting all levels of the gene expression pathway. Notch signaling through its receptor Jagged maintains the neuronal progenitor cell population and must be turned off to initiate differentiation (63). The transcriptional repressor REST and the C-terminal phosphatase SCP1 are down-regulated to allow induction of neuron-specific genes (64, 65), and the transcription factors Sox9 and histone methyltransferase Ezh2 that regulate glial cell fate are also repressed (27). As neuronal progenitors exit mitosis and begin to form early neurons, chromatin remodeling complexes acquire new subunits, including BAF53b in place of BAF53a to form the nBAF complex (66). At the same time PTBP1 is down-regulated, inducing expression of the neuronal paralog PTBP2 and shifting the cells to a neuronal program of alternative splicing (21, 39–41). In addition to these general regulators, there are a large number of localized signaling pathways and specialized transcription factors that drive development of specific neuronal subtypes and lineages (67–69). As a master regulator of neuronal cell fate, miR-124 is induced at the beginning of NPC differentiation into neurons and directly represses expression from Jagged, REST, SCP1, BAF53a, Sox9, PTBP1, and many other mRNAs.
Of the three miR-124 loci, only pri-miR-124-1 has been characterized genetically and was found to be essential for brain and retinal development (25, 26). MiR-124 expression is modulated by feedback loops where particular target molecules can alter miR-124 function or expression. For example, all three loci contain REST binding sites, which may limit their transcription in nonneuronal cells (24). In other studies, 3′UTR binding by PTBP1 was found to block miR-124 action on its targets RCOR1 (CoREST) and SCP1 (37). These additional regulatory loops can repress miR-124 activity before neuronal differentiation.
We identify a feature of the complex regulatory circuit controlling miR-124 that acts on its biogenesis rather than its transcription or functional activity. PTBP1 binding to pri-miR-124-1 blocks DROSHA cleavage and prevents formation of the pre-miRNA. PTBP1 has long been known to be repressed by miR-124 (21), and knockdown of PTBP1 is sufficient to induce neuronal differentiation in nonneuronal cell lines (37). This action was attributed to the loss of PTBP1, allowing miR-124 to then target CoREST and SCP1 (37). However, our results indicate that an additional effect of PTBP1 depletion is the direct up-regulation of mature miR-124 itself. This may be the primary driver of the neuronal differentiation seen with PTBP1 knockdown. Previously miR-124 was observed to be induced upon PTBP1 depletion in colorectal cancer cells (33). Our findings indicate that this effect on miRNA expression may result from the direct regulation of miR-124 biogenesis by PTBP1 (Figs. 3 and 4).
The block to miR-124 biogenesis by PTBP1 reinforces the antagonistic regulatory interactions of these two molecules. Upon neuronal differentiation, the loci for both pri-miR-124-1 and pri-miR-124-2 are transcriptionally induced. Since pri-miR-124-2 is not affected by PTBP1 the level of mature-miR-124 increases. This miR-124 will down-regulate PTBP1, with the consequent increase in pri-miR-124-1 processing. The system thus constitutes a positive feedback loop that drives cells toward the neuronal state of high miR-124 and low PTBP1. Feedback loops between transcription factors that drive development of particular cell lineages and miRNAs that regulate mRNAs in that lineage have been described (57, 59). In these systems, it is found that the feedback improves efficiency of commitment to differentiation and reduces noise. We find that the PTBP1/miR-124-1 feedback loop alters the dynamics of neuronal differentiation, enforcing PTBP1 expression before differentiation and miR-124 afterward, and making the cell fate decision more reliable in the presence of biological variability. This is similar to what is proposed for transcriptional feedback loops but here involves posttranscriptional steps of gene expression (56, 57, 70).
The Regulation of miRNA Biogenesis.
There are multiple examples of RBPs regulating miRNA processing, and recent studies identify widespread interactions between RBPs and stem–loop RNAs (71, 72). The classic example of this is in early development, where the let-7 stem–loop is bound by LIN28A protein that blocks DROSHA processing in the nucleus (73, 74) and DICER processing in the cytoplasm (10, 11, 75). In later somatic tissues, the terminal loop of let-7 is bound by KSRP and hnRNPA1 proteins, which enhance or repress miRNA processing, respectively. HnRNPA1 also binds the terminal stem–loop of miR-18a, where it stimulates DROSHA cleavage (52, 76–78). Like let-7, DROSHA processing of miR-7 is regulated by multiple RBPs. Mature miR-7 is primarily expressed in brain and pancreas, but the miR-7-1 stem–loop is within an intron of the widely expressed hnRNPK transcript (79, 80). Proteins implicated in repressing pri-miR-7-1 processing in nonneuronal cells include HuR and Musashi-2, again by binding to the terminal loop (81). The terminal loop of miR-9 is also bound by LIN28A protein. In this case the interaction destabilizes the pre-miRNA, leading to low expression of mature miR-9 early in neuronal differentiation (82). MiR-9 has other interesting parallels to miR-124 in being brain-enriched and being transcriptionally repressed by the REST complex and posttranscriptionally repressed by RBPs in early development.
The regulation of miR-124-1 by PTBP1 differs from the above systems in that an extended inhibitory binding site is upstream from the stem–loop structure. A recent study identified the Quaking 5 protein (QKI5) as a positive regulator of pri-miR-124-1 processing during human erythropoiesis (83). The QKI5 binding site is upstream of the PTBP1 sites identified here and the role of PTBP1 in erythropoiesis has not been explored.
The CU-rich PTBP1 binding segment that inhibits DROSHA–DGCR8 processing extends nearly to the base of the stem where DROSHA cleavage occurs (Fig. 2A). Thus, PTBP1 may block access of DROSHA–DGCR8 to its cleavage site. Alternatively, PTBP1 binding may change the folded structure of pri-miR-124-1 to disrupt DROSHA–DGCR8 recognition. Interestingly, we observe reduced DGCR8 binding to the pri-miRNA in the presence of the upstream PTBP1 binding site. However, PTBP2, which is strongly induced in the Ptbp1 KO cell line and is seen to bind pri-miR-124-1 in vitro and in iCLIP analyses of brain tissue (SI Appendix, Fig. S10), does not inhibit miR-124 expression (44). PTBP1 and PTBP2 have very similar RNA binding properties yet have different activities in repressing the splicing of certain exons (35, 39, 50). Thus, it appears that the repression of DROSHA cleavage by PTBP1 involves more than simple binding and occlusion of the cleavage site. It will be interesting to assess how the protein interactions of the two PTB proteins differ when they are bound to pri-miR-124-1.
We identified PTBP1 binding sites in other primary miRNA transcripts, including those of the miR-7-1 allele regulated by HuR and Musashi-2. Unlike the miR-7-1 host transcript, the repression of pri-miR-124-1 processing does not lead to the expression of the host RNA as an mRNA, at least in mESCs. It is possible that in some cells the pri-miR-124-1 transcript does serve a functional role and that PTBP1 acts to allow its expression. It will be interesting to look at this in the retina, where expression of pri-miR-124-1 RNA is observed earlier than the mature miRNA, possibly due to PTBP1 repression (26).
Materials and Methods
Transfection and Protein/RNA Extraction.
The plasmids pAAV-nEF-tRFP and pAAV-nEF-tRFP-p2A-FLAG-PTBP1.4 were transfected into mESC (Ptbp1 WT and KO cell lines) using Lipofectamine 2000 (Thermo Fisher Scientific). Forty-eight hours posttransfection, protein was extracted with RIPA buffer (50 mM Tris⋅Cl, pH 8.0, 150 mM NaCl, 1% Igepal CA-C630, 0.5% sodium dexolycholate, 0.1% SDS, 1× phosphatase inhibitor, and 1× protease inhibitor) and RNA was extracted using TRIzol (Thermo Fisher Scientific).
Immunoprecipitation.
mESCs (E14) were harvested and sonicated in cold buffer D-200K (20 mM Tris⋅Cl, pH 8.0, 200 mM KCl, and 0.2 mM EDTA). After centrifugation at 20,000 × g for 15 min at 4 °C, the supernatant was incubated with anti-PTBP1 (PTB_NT) or rabbit IgG isotype control (Thermo Fisher Scientific) at 4 °C. After 1 h, proteinG Sepharose beads (GE Healthcare) were added to the reaction and further incubated for 1 h. The beads were washed four times in BufferD-200K, and RNA was extracted with phenol.
RNA Isolation, RT-PCR, RT-qPCR, and Quantification.
Total RNA was collected from cell cultures using TRIzol (Thermo Fisher Scientific). RNA was treated with DNase I (Takara) followed by phenol extraction; 0.8–1 µg of total RNA was used for random priming in a 10-µL reaction with or without 100 units of SuperScript III RT (Thermo Fisher Scientific) for the plus or minus RT reactions, respectively. PCR was performed using Phusion DNA polymerase (Fisher Scientific). PCR conditions are described in SI Appendix, SI Materials and Methods. The miR- 294 and miR-295 stem–loops are located 48 nt apart within the same precursor RNA. This precursor is denoted as miR-294/295. RT-PCR products were run on 1.5% agarose gels with ethidium bromide staining and visualized under UV light, and the band intensities were measured using ImageJ. RT-qPCR was performed using the SensiFAST SYBR Lo-ROX Kit (Bioline) on a QuantStudio 6 Real-Time PCR System (Thermo Fisher Scientific). One microgram of total RNA was used to generate cDNA for quantification with gene-specific primer. Absolute RNA levels were determined using standard curves generated with known amounts of synthesized cDNAs (84). Primer pairs for the RT-qPCR of pri-miRNAs were designed to amplify a ∼65-nt region upstream of the stem–loop (SI Appendix, Fig. S2B). Mature miRNA reverse transcription and qPCR were performed using TaqMan MicroRNA Reverse Transcription kit and TaqMan Universal PCR Master Mix (Thermo Fisher Scientific; SI Appendix, Table S3). Synthesized small RNA was used to make a standard curve for absolute quantification. A list of primer sequences is presented in SI Appendix, Table S2.
In Vitro RNA Pull-Down Assays.
The RNA pull-down assay was modified from Heo et al. (11). WT and chimeric pri-miR-124-1s were prepared by in vitro transcription from PCR product templates (SI Appendix, SI Materials and Methods). The 5′-biotinylated adapter DNAs complementary to the 3′ extensions of the transcripts were used to immobilize the WT and chimeric pri-miR-124-1. Adapter DNAs were first incubated with streptavidin-conjugated Sepharose beads (GE Healthcare) in buffer I-200K (20 mM Tris⋅Cl, pH 8.0, 200 mM KCl, and 2 mM EDTA, pH 8.0) for 1 h at 4 °C. Empty streptavidin binding sites were then blocked with free biotin for 10 min. The beads were washed twice with buffer D-300K (20 mM Tris⋅Cl, pH 8.0, 300 mM KCl, and 0.2 mM EDTA) and then incubated with pri-miRNA in buffer D-200K with 100 units/mL of RNase inhibitor (Thermo Fisher Scientific) for 2 h at room temperature. The beads were washed twice with buffer D-300K and once with buffer D-200K. mESCs were collected and sonicated in buffer D-200K. After clearing cell debris by centrifugation, total cell extract was added to the immobilized pri-miRNAs and incubated with constant rotation for 2–3 h at 4 °C. After washing with buffer D-200K twice, proteins were isolated in SDS buffer (50 mM Tris⋅Cl, pH 6.8, 0.05% bromophenol blue, 10% glycerol, 2% SDS, and 0.1 M DTT) and separated by 8% SDS/PAGE for Western blotting. Western blot images were taken on a Typhoon imager. RNA was extracted from the beads in phenol, run on standard urea gels, stained with SYBR gold (Thermo Fisher Scientific), and imaged on a Typhoon Imager (GE Healthcare). Fluorescent band intensities of Western blots and RNA gels were quantified with ImageQuant software (GE Healthcare).
Computational Modeling.
All computational simulations were performed in COPASI (85). Ordinary differential equation-based models were constructed as depicted in Fig. 6A as described previously (86). The full system of differential equations with PTBP1 meditated inhibition of miR-124 processing is presented in SI Appendix, SI Materials and Methods.
Details of all other procedures are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Feng Guo (University of California, Los Angeles) for the DGCR8 antibody; V. Narry Kim (Seoul National University) for the DROSHA-FLAG-pCK plasmid; Grigori Enikolopov (Cold Spring Harbor Laboratory) for the Nestin-GFP mouse line; and Areum Han, Celine Vuong, and Manuel Ares Jr. (University of California, Santa Cruz) for critical reading of the manuscript and helpful discussion. This work was supported by National Institutes of Health Grant R01 GM049662 (to D.L.B.) and U01HG007912 (to A.H. and D.L.B.). K.-H.Y. was supported by a postdoctoral fellowship from Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at University of California, Los Angeles. A.J.L. was supported by the University of California, Los Angeles Medical Scientist Training Program and the training programs in neural repair (T32 NS07449) and neurobehavioral genetics (T32 NS048004) at University of California, Los Angeles. S.Z. was supported by National Institutes of Health Grant R00MH096807. We also acknowledge support from a Quantitative and Computational Biosciences Collaboratory Postdoctoral Fellowship (to S.M.) and the Quantitative and Computational Biosciences Collaboratory community, directed by Matteo Pellegrini.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809609115/-/DCSupplemental.
References
- 1.Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tat TT, Maroney PA, Chamnongpol S, Coller J, Nilsen TW. Cotranslational microRNA mediated messenger RNA destabilization. eLife. 2016;5:e12880. doi: 10.7554/eLife.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 7.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 11.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 16.Kawahara H, Imai T, Okano H. MicroRNAs in neural stem cells and neurogenesis. Front Neurosci. 2012;6:30. doi: 10.3389/fnins.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun AX, Crabtree GR, Yoo AS. MicroRNAs: Regulators of neuronal fate. Curr Opin Cell Biol. 2013;25:215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staahl BT, Crabtree GR. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr Opin Neurobiol. 2013;23:903–913. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe Y, et al. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004;18:889–900. doi: 10.1101/gad.1179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanuki R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 27.Neo WH, et al. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J Biol Chem. 2014;289:20788–20801. doi: 10.1074/jbc.M113.525493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deo M, Yu J-Y, Chung K-H, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuta M, et al. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 31.Xia H, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi K, et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi K, et al. Positive feedback of DDX6/c-Myc/PTB1 regulated by miR-124 contributes to maintenance of the Warburg effect in colon cancer cells. Biochim Biophys Acta. 2015;1852:1971–1980. doi: 10.1016/j.bbadis.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 34.Shibayama M, et al. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J. 2009;276:6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 35.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng S, et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388, S1. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linares AJ, et al. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Licatalosi DD, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gueroussov S, et al. An alternative splicing event amplifies evolutionary differences between vertebrates. Science. 2015;349:868–873. doi: 10.1126/science.aaa8381. [DOI] [PubMed] [Google Scholar]
- 43.Xue Y, et al. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci. 2016;19:807–815. doi: 10.1038/nn.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vuong JK, et al. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal pre-mRNA splicing. Cell Reports. 2016;17:2766–2775. doi: 10.1016/j.celrep.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124-1–131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 47.Oberstrass FC, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 48.Han A, et al. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLOS Comput Biol. 2014;10:e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spellman R, et al. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 50.Markovtsov V, et al. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 52.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 54.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vera J, Lai X, Schmitz U, Wolkenhauer O. MicroRNA-regulated networks: The perfect storm for classical molecular biology, the ideal scenario for systems biology. Adv Exp Med Biol. 2013;774:55–76. doi: 10.1007/978-94-007-5590-1_4. [DOI] [PubMed] [Google Scholar]
- 59.Posadas DM, Carthew RW. MicroRNAs and their roles in developmental canalization. Curr Opin Genet Dev. 2014;27:1–6. doi: 10.1016/j.gde.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shokhirev MN, et al. A multi-scale approach reveals that NF-κB cRel enforces a B-cell decision to divide. Mol Syst Biol. 2015;11:783. doi: 10.15252/msb.20145554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao J, Pilko A, Wollman R. Distinct cellular states determine calcium signaling response. Mol Syst Biol. 2016;12:894–905. doi: 10.15252/msb.20167137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell S, Hoffmann A. Identifying noise sources governing cell-to-cell variability. Curr Opin Syst Biol. 2018;8:39–45. doi: 10.1016/j.coisb.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 64.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Yeo M, et al. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 66.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kessaris N, Magno L, Rubin AN, Oliveira MG. Genetic programs controlling cortical interneuron fate. Curr Opin Neurobiol. 2014;26:79–87. doi: 10.1016/j.conb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo J, Anton ES. Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol. 2014;24:342–351. doi: 10.1016/j.tcb.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandler RC, Mayer C, Fishell G. Cortical interneuron specification: The juncture of genes, time and geometry. Curr Opin Neurobiol. 2017;42:17–24. doi: 10.1016/j.conb.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Treiber T, et al. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol Cell. 2017;66:270–284.e13. doi: 10.1016/j.molcel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 72.Nussbacher JK, Yeo GW. Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol Cell. 2018;69:1005–1016.e7. doi: 10.1016/j.molcel.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 76.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 77.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsu SD, et al. miRNAMap 2.0: Genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choudhury NR, et al. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013;27:24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nowak JS, Choudhury NR, de Lima Alves F, Rappsilber J, Michlewski G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat Commun. 2014;5:3687. doi: 10.1038/ncomms4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang F, et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017;27:416–439. doi: 10.1038/cr.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 85.Hoops S, et al. COPASI–A COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 86.Mitchell S, Mendes P. A computational model of liver iron metabolism. PLOS Comput Biol. 2013;9:e1003299. doi: 10.1371/journal.pcbi.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeom K-H, Damianov A. Methods for extraction of RNA, proteins, or protein complexes from subcellular compartments of eukaryotic cells. Methods Mol Biol. 2017;1648:155–167. doi: 10.1007/978-1-4939-7204-3_12. [DOI] [PubMed] [Google Scholar]
- 89.Le Novère N, et al. The systems biology graphical notation. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.