Significance
Organisms frequently exchange costly resources with other species. Theory suggests that this paradoxical cooperation between species might have its origins in waste consumption. When a species benefits from the waste of another, the recipient can evolve to aid the waste producer. The waste producer could then be selected to provide costly resources in return. We previously demonstrated the first step of this theorized process: Salmonella enterica evolved to secrete a costly amino acid to increase access to a byproduct generated by Escherichia coli. Here, we provide demonstration of a waste producer switching to costly cooperation. E. coli repeatedly evolved novel secretion of sugar to feed S. enterica. The results validate long-standing theory about the evolutionary origins of costly mutualism.
Keywords: mutualism, cross-feeding, genome-scale metabolic modeling
Abstract
Mutualisms are essential for life, yet it is unclear how they arise. A two-stage process has been proposed for the evolution of mutualisms that involve exchanges of two costly resources. First, costly provisioning by one species may be selected for if that species gains a benefit from costless byproducts generated by a second species, and cooperators get disproportionate access to byproducts. Selection could then drive the second species to provide costly resources in return. Previously, a synthetic consortium evolved the first stage of this scenario: Salmonella enterica evolved costly production of methionine in exchange for costless carbon byproducts generated by an auxotrophic Escherichia coli. Growth on agar plates localized the benefits of cooperation around methionine-secreting S. enterica. Here, we report that further evolution of these partners on plates led to hypercooperative E. coli that secrete the sugar galactose. Sugar secretion arose repeatedly across replicate communities and is costly to E. coli producers, but enhances the growth of S. enterica. The tradeoff between individual costs and group benefits led to maintenance of both cooperative and efficient E. coli genotypes in this spatially structured environment. This study provides an experimental example of de novo, bidirectional costly mutualism evolving from byproduct consumption. The results validate the plausibility of costly cooperation emerging from initially costless exchange, a scenario widely used to explain the origin of the mutualistic species interactions that are central to life on Earth.
Species frequently exchange costly resources with other species (1, 2). Plants exchange carbon for nutrients from rhizobia and mycorrhizal fungi, and insects frequently exchange nutrients for essential amino acids from bacterial symbionts (1, 2). Providing costly resources to another species can be advantageous if the benefit received in return outweighs the cost of provisioning. However, this generates a “chicken and egg” problem. How do mutualisms initially arise if cooperation is only favored in a species if its partner is already cooperating?
It has been proposed that costless byproducts may provide the foundation from which costly mutualisms evolve (2–4). Organisms frequently produce waste products that can be utilized by other species. For example, the feces generated by ungulates provides resources for a host of insects and microbes. This unidirectional provisioning of costless benefits could select consumers to provide a costly resource in return, thereby increasing production of byproducts. This selection requires cooperative consumers to get disproportionate access to the increased byproducts and the benefit of the differential access to outweigh the cost of cooperating (2, 5). Once the byproduct producer receives a benefit, it too may experience selection to provide costly resources to its partner. Indeed, it has been suggested that once species obtain a degree of benefit from each other, selection may drive an “orgy of mutual benefaction” (6), favoring the evolution of ever more costly cooperation between species.
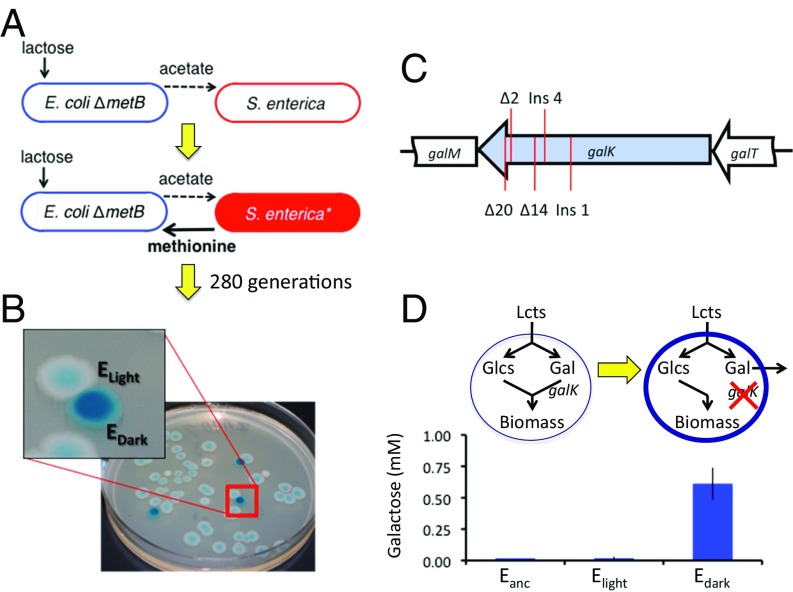
Costly cooperation in a byproduct consumer has been experimentally evolved (4), validating the first step proposed by theory (2, 3). It was previously shown that Salmonella enterica serovar Typhimurium evolved to secrete methionine in exchange for acetate byproducts from an Escherichia coli strain that was an auxotroph for methionine (Fig. 1A) (4). Secretion of the costly amino acid, methionine, was only selected when the coculture was grown on a resource that forced S. enterica to rely on byproducts from E. coli for a source of carbon and energy. Furthermore, spatial structure was critical to ensure that cooperative S. enterica isolates received more byproducts than the wild type.
Fig. 1.
A novel bidirectional costly mutualism evolved from byproduct secretion. (A) Initially, the ancestral E. coli excreted acetate as a waste product, which S. enterica consumed. Then, S. enterica evolved costly secretion of methionine, the amino acid needed by the E. coli auxotroph. This unidirectional mutualism was evolved for an additional ∼280 generations. (B) After evolving in the coculture, two E. coli phenotypes arose in five of six replicate communities. A novel dark blue colony phenotype was apparent when the community was diluted on rich media plates with X-gal. Light blue E. coli colonies retained the ancestral phenotype. S. enterica remained white on these plates. (C) Dark blue E. coli acquired different galK mutations in each replicate community. (D) The frameshift mutations in galK lead to an inability to metabolize the galactose generated during metabolism, and as a result, abundant galactose was measured in spent media for dark blue E. coli. Bars are the mean excretion of isolates from each community, and error bars represent SE. The abbreviations lcts, glcs, and gal refer to lactose, glucose, and galactose, respectively.
Here we test whether reciprocal benefits and spatial structure are sufficient to drive a system to bidirectional costly mutualism. Despite theoretical predictions (2, 3), de novo evolution of systems with two costly resources has not previously been observed. It has also been noted that local competition between partners can favor competitive strategies (7, 8), and therefore alternative mechanisms may be necessary for the evolution of costly reciprocity (9).
We experimentally evolved six replicate communities of the mutualism described above on lactose minimal media plates (auxotrophic E. coli and methionine-secreting S. enterica are henceforth referred to as the ancestral strains). These cocultures grew as dense lawns, which were repeatedly transferred with mixing and dilution for ∼280 generations. Evolutionary changes in communities were subsequently analyzed. E. coli repeatedly evolved to supplement its secretion of waste with a highly costly sugar, although this phenotype never swept to fixation. We analyzed the mechanistic and selective basis of this evolution and found support for the long-standing theory that mutualisms with two costly resources can evolve from byproduct consumption.
Results
Plating replicate evolved E. coli–S. enterica communities revealed E. coli isolates with a distinct phenotype on indicator agar medium. Quantification of the population sizes of E. coli and S. enterica in the evolved communities was performed by plating the mixture on permissive, rich medium with X-gal, a beta-galactosidase indicator that turns E. coli colonies blue while leaving S. enterica white. In five of six replicate communities, dark blue E. coli colonies were observed in addition to the ancestral light blue colonies (Fig. 1B). The frequency of dark blue colonies varied from <1–32% of the E. coli population in different communities. As the dark blue colonies represented a substantial divergence from the ancestral phenotype, we investigated these isolates further.
The repeated evolution of dark blue E. coli was driven by parallel genetic changes. Genome resequencing of a dark blue E. coli isolate from community 1 revealed a mutation in galactokinase (galK). Subsequent sequencing of galK in isolates from each community revealed that dark blue isolates all had distinct frameshift mutations in the gene (Fig. 1C). The different mutations in each replicate confirm that the dark blue isolates evolved in parallel from independent origins.
The galK mutations found in all dark blue E. coli led to incomplete substrate utilization and sugar secretion. E. coli cleave lactose into glucose and galactose during carbon metabolism. In dark blue E. coli mutants, mutations in galK made cells incapable of metabolizing galactose, and as a result the sugar was secreted into the medium (Fig. 1D). Analysis of spent media from ancestral and light blue evolved E. coli showed little to no release of galactose (Fig. 1D). Secreting one galactose per lactose consumed means that dark blue E. coli lose half of all of the energy and carbon available in each lactose molecule they consume.
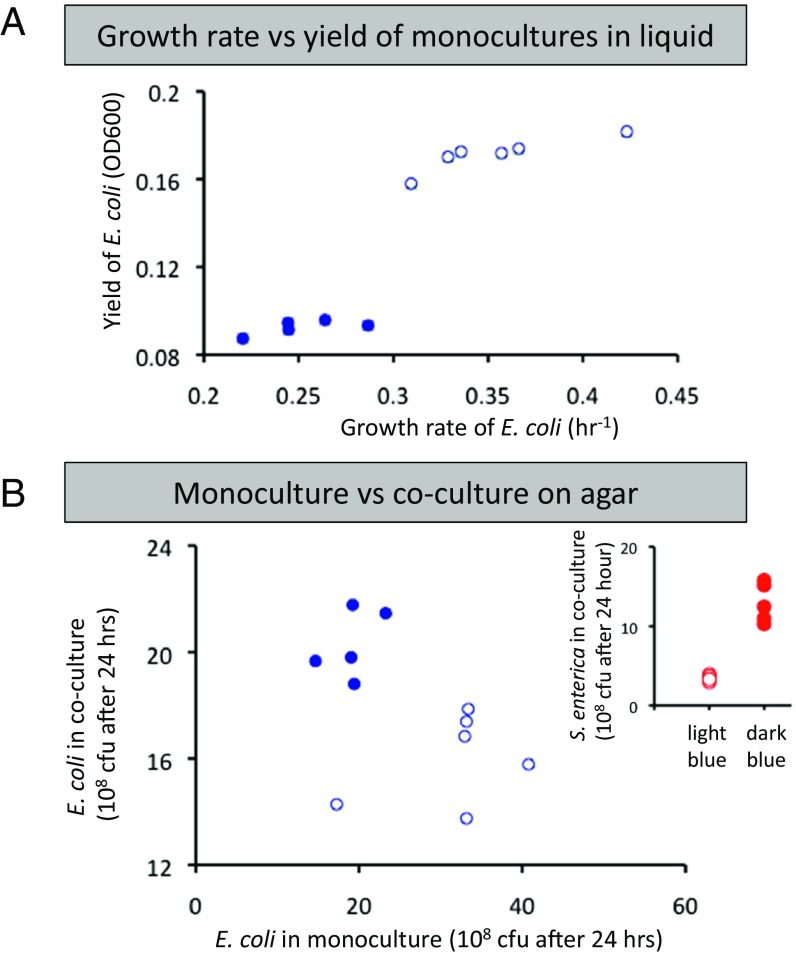
Why would incomplete substrate utilization and sugar excretion rapidly and repeatedly evolve? In previous work, metabolic strategies that reduce yield have evolved because they increased growth rate and therefore provided a competitive advantage (10–13). In our system in liquid media with methionine supplied in excess, dark blue E. coli grew to a significantly lower final yield than light blue E. coli and also grew significantly slower (Fig. 2A, t test, P value < 0.001 for each). This indicates that sugar excretion did not evolve to maximize growth rate of E. coli in isolation. Thus, the evolution of E. coli with inefficient lactose metabolism cannot be explained without considering the partnership with S. enterica.
Fig. 2.
Dark blue E. coli had lower growth rate and yield in monoculture but grew faster in coculture with S. enterica. (A) Dark blue E. coli isolates (solid circles) grew slower and to a lower yield than light blue E. coli (open circles) in liquid minimal media. (B) On agar plates, dark blue E. coli grew worse in monoculture, but better in coculture than light blue isolates. Growth was measured as cfu/mL after 24 h of growth; this measurement time represents midlog. (Inset) S. enterica also grew better in coculture when paired with dark blue E. coli isolates (solid circles) than when paired with light blue E. coli isolates (open circles).
Could secretion instead be an adaptation for mutualism? The benefit of galactose secretion became apparent when E. coli relied on methionine from S. enterica. Dark blue isolates grew significantly faster than light blue isolates when each was paired with S. enterica on agar plates (Fig. 2B, t test, P value = 0.004). Additionally, S. enterica grew significantly faster when paired with dark blue E. coli than when paired with light blue (Fig. 2B, Inset, t test, P value < 0.001). This suggests that galactose secretion is adaptive because it increases the growth of a mutualistic partner. Further, dark blue E. coli had slower growth than light blue in monocultures grown on agar (Fig. 2B, t test, P value < 0.002), indicating that galactose secretion is costly on agar as well as liquid media. Despite these clear trends, substantial variation in growth was observed between isolates from different replicates (Fig. 2), likely due to additional mutations present in these evolved E. coli isolates. However, introducing the galK frameshift mutation from community 1 or a complete deletion of galK in ancestral E. coli is sufficient to recapitulate the observed reduction in monoculture growth and improved growth of the consortia on agar plates (SI Appendix, Fig. S1). These data demonstrate that evolution of galactose excretion in E. coli represents the evolution of costly cooperation and indicate that the system has now evolved into a mutualism with bidirectional provisioning of costly resources.
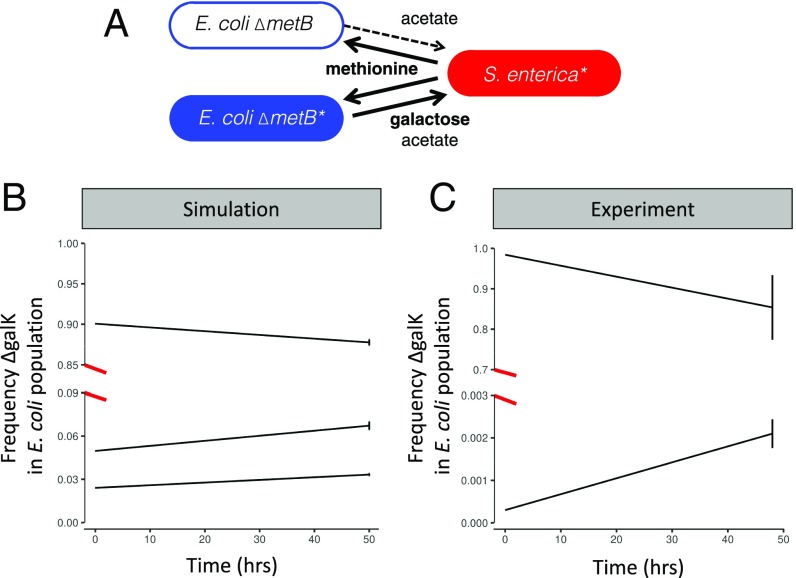
Finally, given that dark blue E. coli have a growth rate and yield advantage when grown with their partner, why were they not observed at higher frequencies? One explanation could be that the dark blue mutants would ultimately sweep to fixation if the experiment were carried out longer. An alternative possibility is that the balance of individual costs and frequency-dependent collective benefits of galactose secretion stabilize dark blue genotypes at an intermediate frequency in the E. coli population. We used genome-scale metabolic models with explicit spatial structure to examine the potential for negative frequency dependence to emerge between the two E. coli strategies. A metabolic model representing our ancestral E. coli was generated by removing the metabolic reaction associated with metB. A model for the dark blue genotype was then generated by additionally knocking out the metabolic reaction associated with galK. We simulated competition between these genotypes in the computational platform COMETS (14), which uses dynamic flux balance analysis and diffusion across a 2D grid to predict microbial growth and interactions based on optimal intracellular metabolism operating in each genotype or species. Consistent with expectation, the dark blue genotype rapidly decreased in frequency when competed against the ancestral E. coli genotype in the absence of cross-feeding (i.e., if methionine was provided) or the absence of spatial structure (SI Appendix, Fig. S2). However, the dark blue genotype was able to invade the E. coli population in spatially structured simulations with S. enterica (Fig. 3B and SI Appendix, Fig. S3). This is consistent with previous genome-scale analysis that identified galK as one of only six E. coli metabolic reactions whose loss would lead to a significant increase in S. enterica growth during coculturing (15). Simulations also suggested that selection for dark blue E. coli was frequency-dependent, as cooperation was only favored when the dark blue genotype started at <40% of the E. coli population. Frequency-dependent selection for ∆galK E. coli in coculture was further supported by experimental invasion assays (Fig. 3C). In the coculture, ∆galK E. coli increased in frequency relative to ancestral E. coli only when the mutant was initially rare in the E. coli population. These results suggest that the intermediate frequencies of dark blue isolates in experimental populations represent stable maintenance of diversity rather than a transient point in the midst of a selective sweep. The data highlight that mutualism can generate negative frequency-dependent selection that drives divergence of metabolic strategies within a population.
Fig. 3.
Simulations with genome-scale metabolic models and experiments suggest that dark blue and light blue E. coli should coexist in coculture. (A) A model of the light blue E. coli genotype was competed against the dark blue genotype in the presence of S. enterica in spatially explicit simulations. (B) In simulations the dark blue genotype showed negative frequency-dependent selection. Dark blue E. coli increased in frequency in the E. coli population when initially rare, and decreased when initially common. Four replicates with randomized biomass positions were run for each starting frequency. (C) Negative frequency dependence was also observed experimentally. The ∆galK genotype decreased relative to the ancestor when the mutant started at 98% of the population. However, when ∆galK started at 0.03% it increased in frequency. The points denote the mean of three replicates, and bars represent SEs. Note that the scale of the y axis changes at the break.
Discussion
Our results demonstrate that bidirectional costly mutualism can evolve from byproduct consumption, a transition which had been theorized (2, 3), but not experimentally observed. We show that the challenge of initiating bidirectional cooperation was overcome in a stepwise progression from generation of a useful byproduct (acetate), to mutualism with a single costly resource (methionine), to exchange of two costly resources (methionine and galactose). The evolution of bidirectional costly mutualism was highly parallel and arose in many independent replicates. However, there were also constraints on evolution of mutualism with galactose-secreting isolates only rising to low frequency in the E. coli population. The benefits of mutualism can drive rapid and repeated evolution of novel bidirectional cooperation between species.
The observed galactose secretion evolved due to selection for mutualistic benefits, although inefficient metabolism can also emerge due to selfish benefits. Incomplete metabolism of a resource can allow cells to require fewer enzymes, driving fast but wasteful growth (10–13). Such inefficient use of resources can generate “tragedies of the common” that reduce the total yield of a population, especially in well-mixed environments (11, 12). Dark blue E. coli engage in inefficient metabolism, failing to metabolize half of the carbon they consume; however, this strategy does not provide a boost in maximum monoculture growth rate. Galactose secretion was selected because it enhances cooperation with a mutualistic partner. By secreting sugar, E. coli increased the abundance of S. enterica, thereby increasing the production of methionine. The spatial structure of the agar plate afforded dark blue E. coli preferential access to the additional methionine, allowing the benefits of cooperation to outweigh the costs. Sugar secretion by E. coli represents a de novo transition to mutualism with exchange of two costly resources.
Mutualism drove a divergence in metabolic strategies. The E. coli population split into a stable polymorphism of cooperative and efficient genotypes. Although providing galactose to S. enterica is adaptive, it did not sweep to fixation. Rather, efficient E. coli excreting just acetate became favored as the strongly cooperative E. coli excreting galactose became more common. As cooperators became common, increasing amounts of methionine diffused to noncooperative E. coli, allowing noncooperators to gain the benefits of cooperation without paying the cost. Such maintenance of both cooperative and noncooperative strategies in structured environments is likely to often occur due to the private nature of costs, versus the semipublic nature of returned benefits. Indeed, it was also seen in the first evolutionary step of this mutualism. When S. enterica initially evolved to secrete methionine in exchange for acetate byproducts from auxotrophic E. coli, the cooperative genotype rose to ∼80% of the S. enterica population (4). The frequency at which cooperators stabilized during the first and second step of evolution is a function of the balance of costs and benefits of mutualism. Galactose secretion is both more costly and less beneficial than methionine secretion; galactose enhanced growth rate of the mutualism, while methionine secretion was absolutely required for growth of the coculture. Reductions in the relative benefit of additional cooperation are likely to frequently constrain selection for an “orgy of mutual benefaction” (6).
Evolution was both repeatable and predictable even in a spatially structured microbial community. E. coli repeatedly evolved sugar secretion through mutations in the same gene, reminiscent of the parallel adaptations through which S. enterica previously evolved methionine secretion (16, 17). In addition to being repeatable, genome-scale metabolic models also made evolution predictable in this system. Models accurately identified that galactokinase-deficient E. coli stabilize at an intermediate frequency. Further, genome-scale models previously identified galactokinase (15) as one of only six reactions in E. coli whose loss would dramatically increase S. enterica density when grown in this mutualistic scenario. Despite the complexity of evolution in structured microbial communities, our results highlight that evolutionary outcomes can be not only repeatable, but predictable.
We now have experimental support for the theory that mutualisms with exchange of two costly resources can evolve from byproduct consumption. In addition to enhancing theoretical understanding, this knowledge is important for management and engineering of microbial systems. E. coli rapidly evolved to secrete half of its resources. This work suggests that evolving bacteria in structured environments with mutualistic partners may be a useful tool for generating a range of novel microbial excretions.
Materials and Methods
Strains and Media.
The ancestral strains used were a methionine auxotrophic Escherichia coli K12 and a methionine-excreting Salmonella enterica serovar Typhimurium LT2 (3). The E. coli auxotrophy was generated by a ∆metB mutation (4, 14). The secretion of methionine by S. enterica was driven by a base pair change in metA (16) and an IS element inserted in front of metJ (17). In lactose minimal medium, E. coli relies on methionine from S. enterica, and S. enterica relies on carbon in the form of acetate generated by E. coli’s metabolism of lactose. Additional variants of the ancestral E. coli were constructed with either a galK deletion or the galK replaced with the mutated gene from evolved replicate 1.
The strains were evolved on hypho minimal medium plates with lactose [2.92 mM lactose, 7.26 mM K2HPO4, 9.38 mM NaH2PO4, 1.89 mM (NH4)2SO4, 0.41 mM MgSO4, 0.6 μM ZnSO4, 9.98 μM CaCl2, 0.5 μM MnCl2, 1 μM (NH4)6Mo7, 0.5 μM CuSO4, 1 μM CoCl2, 0.169 μM Na2WO4, 8.88 μM FeSO4; based on ref. 18). Monoculture growth assays were done on hypho media that were supplemented with methionine or glucose to allow single species growth. Nutrient broth plates with X-gal (5-bromo–4-chloro–3-indolyl–β–d-galactopyranoside) were also used to distinguish S. enterica (white colonies) and the two phenotypes of E. coli colonies (light and dark blue). Note that there was not a lac inducer added, such as IPTG (isopropyl β–d-1-thiogalactopyanoside), which is why the ancestral (and some evolved) phenotype was a light shade of blue.
Evolution Conditions.
Cocultures were evolved in lawns on lactose minimal media plates at 30 °C. Cocultures were grown for 48 h and then scrubbed with a spreader from the plate surface using 1 mL of minimal media. Cells were then vortexed and transferred at a 1/128 dilution and spread on a fresh plate. Cocultures were evolved for 40 transfers or ∼280 generations.
Genomic Analysis.
E. coli genotypes were analyzed through a mix of whole-genome and Sanger sequencing. For whole-genome sequencing, DNA was extracted from lysed cells via phenol chloroform CTAB extraction (19) and prepared for Illumina sequencing using a TruSeq kit (Illumina). Samples were sequenced on an Illumina HiSeq 2000 and analyzed using breseq (20). Subsequent investigation of galK sequence was carried out through Sanger sequencing of PCR products.
Spent Media Analysis.
E. coli excretion profiles were determined by analyzing spent media with gas chromotography/mass spectrometry (GC-MS). E. coli were grown to saturation in lactose minimal media supplemented with methionine, and then the cells were filtered out with a 0.2-µm filter. Three milliliters of spent media were acidified with 100 μL of 4% HCl, and 3 μL of 10% U-13C glucose were added as an internal standard. Media were passed through solid phase extraction Chromaband C18 columns per manufacturer directions (Macherey-Nagel) and eluted in 500 μL methanol. After removal of methanol in a vacuum centrifuge, samples were resuspended in 50 μL methoxamine (MOX) reagent and incubated for 3 h at 85 °C. Then, 50 μL of N-(tertbutyldimethylsilyl) –N-methyltrifluoroacetamide (MSTFA) were added, and the sample was incubated for an additional hour. Derivatized samples were injected into a Shimadzu QP2010 GC-MS. The injection source was 230 °C. The oven was held at 80 °C for 3 min, increased to 280 °C at a rate of 5 °C per minute, and held at 280 °C for 2 min. Column flow rate was 1 mL/min, and the split ratio was 0. The column was a 30-m DB column (Restek). Results were analyzed in GC-MS Postrun Analysis (Version 2.70; Shimadzu).
Growth Assays.
Growth rates were analyzed both in liquid and on plates. Liquid assays were carried out in 48-well plates with shaking at 30 °C. Optical densities were obtained every 30 min to 1 h on a Wallac Victor 2 plate reader (Perkin-Elmer) until cultures reached saturation, using a previously described automated measurement system (21). Agar assays involved spotting 0.5 μL of media containing cells with an OD600 = 10−3 onto a minimal media plate (∼200 cells per spot). Cocultures were plated at the same total cell density, with even species ratios in terms of OD600. Petri dishes were incubated at 30 °C on a Canon Perfection V600 scanner, and a 600-dpi image was taken of the plate, agar side down, every hour. Tracking colony area over time was performed using custom software in Matlab (26). We measured the growth rate of colonies on Petri dishes via the diameter of a hypothetical circular colony with same area (22). The growth rate was calculated by regressing diameter over time for the first 12 h (12 frames) once initial spots began spreading radially.
Genome-Scale Metabolic Modeling.
To determine the metabolic mechanism underlying observed evolutionary patterns we used constraint-based metabolic modeling. Genome-scale metabolic networks were obtained for E. coli (iJO_1366) (23) and S. enterica (iRR_1083) (24). Methionine excretion in the mutualist S. enterica was modeled by connecting excretion of the amino acid to the biomass equation that serves as the objective function (14, 15, 25). In the E. coli models, flux through metB was blocked, and dark blue models were generated by additionally blocking flux through galK. COMETS v. 2.2.11 was used to simulate the metabolic interactions and growth in a spatially structured community (14). Spatial simulations used a square lattice of 50 × 50 “boxes,” mimicking a Petri dish environment with 2.5 cm per side (i.e., box length = 0.5 mm per side). The simulation environment was homogenous and contained excess trace metals, excess ammonia, and 1.8 × 10−6 mmol lactose per box. Fifty percent of boxes were randomly chosen and initiated with 1 × 10−10 g (dry weight) of bacterial biomass. Of the occupied boxes, half received the iRR_1083 model. The remaining half of occupied boxes received E. coli models, either ancestor or dark blue, at the various frequencies shown in Fig. 3. Only one type of biomass was allowed per box. At least three simulations were run for each E. coli dark blue frequency, with locations of occupied boxes randomized each time. Each simulation was run for 50 simulated hours. The time step was 1/100 of an hour for biomass growth and 1/1,000 of an hour for metabolite diffusion. Each biomass growth time step, dFBA calculated changes in biomass and metabolites, including excretion of methionine from iRR_1083 and excretion of acetate and galactose from E. coli. Metabolite uptake was calculated using Monod kinetics with a default Vmax of 10 mmol g−1 h−1 and Km of 5 × 10−6 mM. Metabolites diffused to adjacent boxes with a diffusion rate of 5 × 10−6 cm2 s−1. Biomass did not diffuse. Changes in dark blue frequency were calculated as: [final dark blue/(final dark blue + final Ancestor)]/[initial dark blue/(initial dark blue + initial Ancestor)].
Frequency-Dependent Fitness Assays.
The ΔgalK E. coli was competed against the ancestor in coculture. Each strain was streaked onto Nutrient Broth medium and grown at 30 °C for 48 h. A single colony of each strain was then inoculated into 5 mL of species-specific hypho medium and grown with shaking until early log phase, ∼6 h at 30 °C. The OD600 was then measured for each strain, which was used to calculate an approximate cfu/mL for each. A total of 106 cells of each species were plated onto hypho plates. The E. coli population started with either ∼98 or ∼0.03% ΔgalK mutants. Plates were incubated at 30 °C for 48 h. Plates were then scraped using 1,600 μL E. coli-specific hypho, serially diluted, and plated for cfu onto E. coli hypho (to count total E. coli), or E. coli hypho containing 50 μg/mL kanamycin (to select for only ΔgalK mutants). Plates were incubated at 30 °C for 48 h, and cfu were enumerated. Percent frequency of ΔgalK mutants was determined by dividing the cfu/mL of cells on kanamycin plates by the cfu/mL of cells on antibiotic-free plates.
Supplementary Material
Acknowledgments
We thank Mike Travisano, Lisa Fazzino, and Sarah Hammarlund for insightful comments. This work was funded by a Department of Energy Award (DE-SC0006731, to C.J.M.) and a National Institutes of Health Award (1R01-GM121498, to W.R.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11874.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810949115/-/DCSupplemental.
References
- 1.Bronstein JL, editor. Mutualism. Oxford Univ Press; New York: 2015. [Google Scholar]
- 2.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation. Q Rev Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 3.Connor RC. Pseudoreciprocity: Investing in mutualism. Anim Behav. 1986;34:1562–1584. [Google Scholar]
- 4.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 5.Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J Evol Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- 6.May RM. Theoretical Ecology: Principles and Applications. 2nd Ed Oxford Univ Press; Oxford: 1981. [Google Scholar]
- 7.Wilson DS, Pollock GB, Dugatkin LA. Can altruism evolve in purely viscous populations? Evol Ecol. 1992;6:331–341. [Google Scholar]
- 8.Bull JJ, Harcombe WR. Population dynamics constrain the cooperative evolution of cross-feeding. PLoS One. 2009;4:e4115. doi: 10.1371/journal.pone.0004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivers RL. The evolution of reciprocal altruism. Q Rev Biol. 1971;46:35–57. [Google Scholar]
- 10.Pfeiffer T, Bonhoeffer S. Evolution of cross-feeding in microbial populations. Am Nat. 2004;163:E126–E135. doi: 10.1086/383593. [DOI] [PubMed] [Google Scholar]
- 11.MacLean RC, Gudelj I. Resource competition and social conflict in experimental populations of yeast. Nature. 2006;441:498–501. doi: 10.1038/nature04624. [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 13.Carlson RP, et al. Competitive resource allocation to metabolic pathways contributes to overflow metabolisms and emergent properties in cross-feeding microbial consortia. Biochem Soc Trans. 2018;46:269–284. doi: 10.1042/BST20170242. [DOI] [PubMed] [Google Scholar]
- 14.Harcombe WR, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chubiz LM, Granger BR, Segrè D, Harcombe WR. Species interactions differ in their genetic robustness. Front Microbiol. 2015;6:271. doi: 10.3389/fmicb.2015.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas SM, Chubiz LM, Harcombe WR, Ytreberg FM, Marx CJ. Parallel mutations result in a wide range of cooperation and community consequences in a two-species bacterial consortium. PLoS One. 2016;11:e0161837. doi: 10.1371/journal.pone.0161837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas SM, Chubiz LM, Harcombe WR, Marx CJ. Identification of the potentiating mutations and synergistic epistasis that enabled the evolution of inter-species cooperation. PLoS One. 2017;12:e0174345. doi: 10.1371/journal.pone.0174345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaney NF, et al. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One. 2013;8:e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 2001;56:2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 20.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delaney NF, Rojas Echenique JI, Marx CJ. Clarity: An open-source manager for laboratory automation. J Lab Autom. 2013;18:171–177. doi: 10.1177/2211068212460237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wimpenny JW. The growth and form of bacterial colonies. J Gen Microbiol. 1979;114:483–486. doi: 10.1099/00221287-114-2-483. [DOI] [PubMed] [Google Scholar]
- 23.Orth JD, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism–2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harcombe WR, Betts A, Shapiro JW, Marx CJ. Adding biotic complexity alters the metabolic benefits of mutualism. Evolution. 2016;70:1871–1881. doi: 10.1111/evo.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chacón JM, Möbius W, Harcombe WR. The spatial and metabolic basis of colony size variation. ISME. 2018;12:669–680. doi: 10.1038/s41396-017-0038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.