Significance
The involvement of biotransformation enzymes in drug metabolism is one of the most crucial objectives during preclinical research, since they ultimately determine the bioavailability of medicinal drugs. The mARC N-reductive enzyme system was found to be a highly effective counterpart to one of the most prominent biotransformation enzymes, CYP450, and is involved in activation of amidoxime prodrugs as well as inactivation of other drugs containing N-hydroxylated functional groups. Owing to its potent N-reductive capacity toward a broad range of compounds, including mutagenic N-oxygenated nucleobase analogs, mARC plays a crucial role in pharmacology. Our crystal structure of human mARC forms the basis for predictions on the metabolism of drug candidates and structure–activity relationships. Moreover, it indicates the evolutionary development of different molybdoenzyme families.
Keywords: biotransformation, detoxification, drug metabolism, molybdenum cofactor, MOSC
Abstract
Biotransformation enzymes ensure a viable homeostasis by regulating reversible cycles of oxidative and reductive reactions. The metabolism of nitrogen-containing compounds is of high pharmaceutical and toxicological relevance because N-oxygenated metabolites derived from reactions mediated by cytochrome P450 enzymes or flavin-dependent monooxygenases are in some cases highly toxic or mutagenic. The molybdenum-dependent mitochondrial amidoxime-reducing component (mARC) was found to be an extremely efficient counterpart, which is able to reduce the full range of N-oxygenated compounds and thereby mediates detoxification reactions. However, the 3D structure of this enzyme was unknown. Here we present the high-resolution crystal structure of human mARC. We give detailed insight into the coordination of its molybdenum cofactor (Moco), the catalytic mechanism, and its ability to reduce a wide range of N-oxygenated compounds. The identification of two key residues will allow future discrimination between mARC paralogs and ensure correct annotation. Since our structural findings contradict in silico predictions that are currently made by online databases, we propose domain definitions for members of the superfamily of Moco sulfurase C-terminal (MOSC) domain-containing proteins. Furthermore, we present evidence for an evolutionary role of mARC for the emergence of the xanthine oxidase protein superfamily. We anticipate the hereby presented crystal structure to be a starting point for future descriptions of MOSC proteins, which are currently poorly structurally characterized.
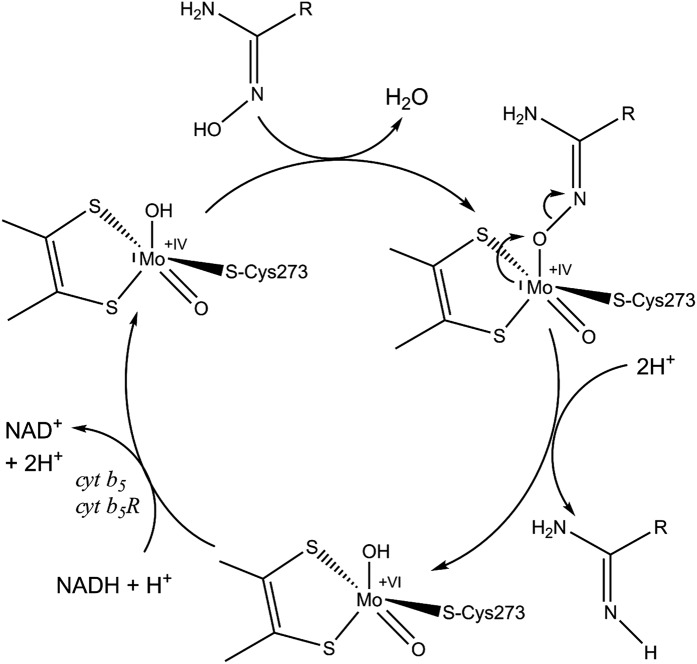
To enable the body to create a viable environment homeostasis, complex biochemical transformations such as reversible metabolic cycles of oxidative and reductive reactions are required. In this respect, metabolisms involving nitrogen are of high pharmaceutical and toxicological relevance, since a number of nitrogen-containing functionalities can undergo N-oxygenations to N-oxides or N-hydroxylated compounds (NHCs) by cytochrome P450 (CYP)- or flavin-dependent monooxygenase (FMO)-catalyzed xenobiotic metabolism. The resulting metabolites have different pharmacological properties and, in some cases, even highly toxic, mutagenic, or carcinogenic N-hydroxylated metabolites are produced (1, 2). Thus, retroreduction of such first-generation metabolites to their parent compounds can be regarded as a detoxification reaction. In this context, the mitochondrial amidoxime-reducing component (mARC) was discovered in our laboratory in 2006 as a thus-far unknown molybdenum-containing protein (3). It was identified as being an extremely effective reductase for a multitude of N-oxygenated molecules such as hydroxylamines (4), N4-hydroxycytosine, and N6-hydroxyadenine, including their corresponding nucleosides (5), hydroxyamidines (6), amidoxime prodrugs and hydroxyguanidines (7), oximes (8), N-oxides (8, 9), hydroxamic acids (10), and sulfohydroxamic acids (11). mARC therefore plays a pivotal role as a counterpart to CYP- and FMO-mediated oxygenation reactions in metabolic cycles. Furthermore, recent studies suggest that mARC is important for organisms to ensure reductive detoxification strategies, for example of toxic hydroxylamines (4) or mutagenic N-hydroxylated nucleobases (5, 12). After its discovery, subsequent studies have shown that the enzyme is able to reduce the full range of N-oxygenated compounds, including the capacity to reduce inorganic nitrite to nitric oxide (13) and Nω-hydroxy-l-arginine to arginine (14). All annotated genomes of mammals appear to possess two copies of mARC genes, with both copies showing strong similarities on nucleotide and amino acid levels, thus making a discrimination difficult, but defining them as paralogous proteins. Nevertheless, different substrate preferences, especially for N-oxides (8, 9) and hydroxamic acids (10), as well as different tissue-specific expression levels have been shown for the two paralogs (5). Besides sulfite oxidase, aldehyde oxidase, and xanthine oxidoreductase, mARC is only the fourth molybdenum-containing enzyme found in humans and other mammals (15). With a molecular mass of around 35 kDa, it represents the simplest form among them, only binding the molybdenum cofactor (pyranopterin as a prosthetic group coordinated to molybdenum; further referred to as Moco) and its substrates. In the presence of NADH, mARC proteins exert N-reductive activity toward NHCs in concert with the two electron transport proteins cytochrome b5 and NADH cytochrome b5 reductase. Even though mARC proteins alone have been found to be associated not only with mitochondria but also with peroxisomes, the entire three-component system is located on the outer mitochondrial membrane and is expressed in every tissue studied so far (16). The catalytic cycle was proposed to be analogous to the described mechanism of nitrate reduction by nitrate reductase (17, 18) (Fig. 1).
Fig. 1.
Catalytic cycle of mARC enzymes. Reducing equivalents supplied by NADH are passed to cytochrome b5 reductase, then to cytochrome b5, before being relayed to mARC to prime the MoIV active site for substrate reduction. It is assumed that N-hydroxylated substrates are reduced by cleavage of the N–O bond, in analogy to the described mechanism of nitrate reduction by nitrate reductase (17). This is accomplished by protonation of the hydroxyl group and subsequent leaving of one water molecule (18).
Based on in silico analysis, Anantharaman and Aravind (19) proposed that the C-terminal domain of molybdenum cofactor sulfurases represents a common feature among various enzymes in eukaryotes and prokaryotes. Reflecting this, proteins carrying this domain were referred to as molybdenum cofactor sulfurase C-terminal (MOSC) domain-containing proteins. In humans, only the Moco sulfurase itself and mARC belong to this quite diverse protein family (15). Structural knowledge of MOSC proteins is so far limited to the bacterial Moco-dependent proteins YuaD and the recently published YiiM (20). However, these crystal structures are lacking the molybdenum cofactor and therefore only allow speculation about the nature of the active site and substrate binding mode.
Here we present the high-resolution crystal structure of human mARC, which reveals detailed insights into MOSC proteins and their coordination of the Mo-molybdopterin cofactor. In addition, the structure provides evidence for the evolutionary link between the sulfite oxidase and xanthine oxidase families of molybdoenzymes. This structure is crucial for a deeper understanding of the observed substrate spectrum in drug metabolism.
Results
Crystal Structure of Human mARC1.
The crystal structure of the fusion protein comprising T4 lysozyme (T4L) and N-terminally truncated human mARC1 (hmARC1) was determined by molecular replacement (SI Appendix, Fig. S1). The 1.78-Å dataset was refined to a final R factor of 16.9% (Rfree 20.8%). The final model consists of 444 residues (with 283 belonging to hmARC1 and 161 to T4L), one Moco, four molybdate ions, one phosphate ion, one bis-Tris propane molecule, and 439 water molecules in the asymmetric unit. The ions and small molecules are derived from the purification and crystallization buffers, respectively. The C-terminal His6 tag is disordered, and could not be modeled due to the absence of appropriate electron density. Residues D301 to K310 are poorly defined and display high B factors, yet their positions could be traced at low σ-contouring. The electron density maps for the rest of the model were of high quality and could be modeled with high confidence.
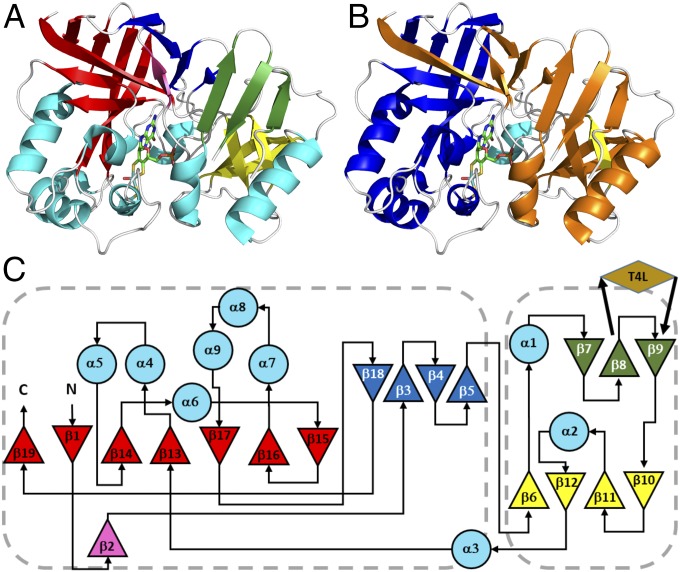
hmARC1 comprises two structural domains which consist of nonsuccessive secondary-structure elements. Most parts of the protein are dominated by β-strands which form three- and four-stranded antiparallel β-sheets (Fig. 2 A and C; green and dark blue, respectively), a small four-stranded antiparallel β-barrel (yellow), and a large, seven-stranded mostly antiparallel β-barrel (red), which is slightly deformed. Nine α-helices of different lengths complete the crystal structure. Two structural domains can be distinguished: One is composed of β-strands 7 to 12 as well as helices α1 and α2, and the other encloses the large β-barrel, a four-stranded β-sheet forming a “lid” of the barrel and helices α4 to α9. Buried within the cleft between the two domains lies the Moco, accompanied by helix α3.
Fig. 2.
Crystal structure and topology of hmARC1. (A) Cartoon representation of hmARC1 colored by secondary-structure elements. (B) Cartoon representation colored by in silico predictions of MOSC_N (orange) and MOSC domains (blue). (C) Topology diagram of hmARC1. Secondary-structure elements are colored as in A. Triangles represent β-strands and large circles represent α-helices. The insertion site of the crystallization-facilitating fusion partner T4 lysozyme is indicated by a brown rhombus. Domains are highlighted in orange (MOSC_N) and blue (MOSC) according to current database predictions. Dashed boxes encompass the definitions of MOSC and MOSC_N domains as deduced from the 3D structure of hmARC1.
Structural Insight into MOSC Proteins.
MOSC domains were revealed by computational analysis as a novel, yet ancient, superfamily of β-strand–rich domains, which occur either as stand-alone forms or fused to other domains. They were predicted to be sulfur-carrier domains, which receive formerly enzymatically abstracted sulfur on a highly conserved cysteine residue and further deliver it for the formation of diverse metal–sulfur clusters (19). In eukaryotes, this domain superfamily solely comprises the two mARC proteins and the Moco sulfurase (15), while in prokaryotes the Moco-dependent enzymes YcbX and YiiM, among others, can also be found (19). In addition to the MOSC domain, larger family members also share a distinct N-terminal domain, which is referred to as the MOSC_N domain. This domain was nowhere detectable as a stand-alone form but was predicted to adopt a β-barrel–like structure and to be involved in substrate recognition and binding, while the MOSC domain contains a conserved cysteine needed for sulfur transition. In hmARC1, the MOSC_N and MOSC domains are predicted to be formed by residues 56 to 175 and 187 to 335, respectively (Fig. 2 B and C). Residues of the MOSC domain, which enclose the β-strands 13 to 19 and helices α4 to α9, bind the Moco and contain the conserved cysteine residue coordinating the central molybdenum ion. However, in contrast to predictions and current annotations in databases, the MOSC_N domain, which is currently defined as comprising β-strands 1 to 11, does not form a β-barrel–like structure. In fact, only β1 is part of the large β-barrel, which is present within the hmARC1 crystal structure, but mainly comprises residues of the MOSC domain. Still, the four β-strands 6 and 10 to 12 from the MOSC_N domain form a small β-barrel. While there are two domains distinguishable within the crystal structure of hmARC1, these do not correlate with the two computationally predicted MOSC and MOSC_N domains. Tertiary-structure elements like the large β-barrel or a four-stranded β-sheet (β3 to β5 and β18) rather comprise residues from both predicted domains.
Using the Dali server, the structure-based alignment of hmARC1 with the recently published crystal structure of Geobacillus stearothermophilus (gs)YiiM (20) enabled us to identify conserved structural elements within these two proteins. It reveals our domain definition to be in concert with other MOSC proteins (SI Appendix, Fig. S2). YiiM is a smaller representative of the MOSC protein superfamily, solely comprising the MOSC domain and an additional C-terminal helix bundle. Despite the low sequence identity between hmARC and gsYiiM (SI Appendix, Fig. S2A), the secondary-structure elements and overall fold of the MOSC domain are conserved within the proteins’ 3D structures (SI Appendix, Fig. S2B). The root-mean-square distance for 129 aligned Cα atoms is 2.3 Å with a sequence identity of 13%. The crystal structure of gsYiiM [Protein Data Bank (PDB) ID code 5YHH] does not contain the essential molybdenum cofactor. The proposed coordination of the Moco indicates a weak binding between the β-barrel and the C-terminal helix bundle as well as a different relative orientation compared with hmARC1. In hmARC1, the Moco is very well buried between the β-barrel and the MOSC_N domain.
Based on this comparison, we propose a clarification for the definition of the MOSC_N as well as MOSC domain and therefore for the MOSC protein family, which takes into account not only predictions based on sequence analysis but also the structural arrangement and composition of conserved domains. In the future, definitions for the MOSC_N domain should contain β-strands 6 to 12 as well as helices α1 and α2 (residues 93 to 183), while the MOSC domain should comprise β-strands 1 to 5 and 13 to 19 as well as helices α4 to α9 (residues 52 to 92 and 210 to 335).
Molybdenum Cofactor Coordination.
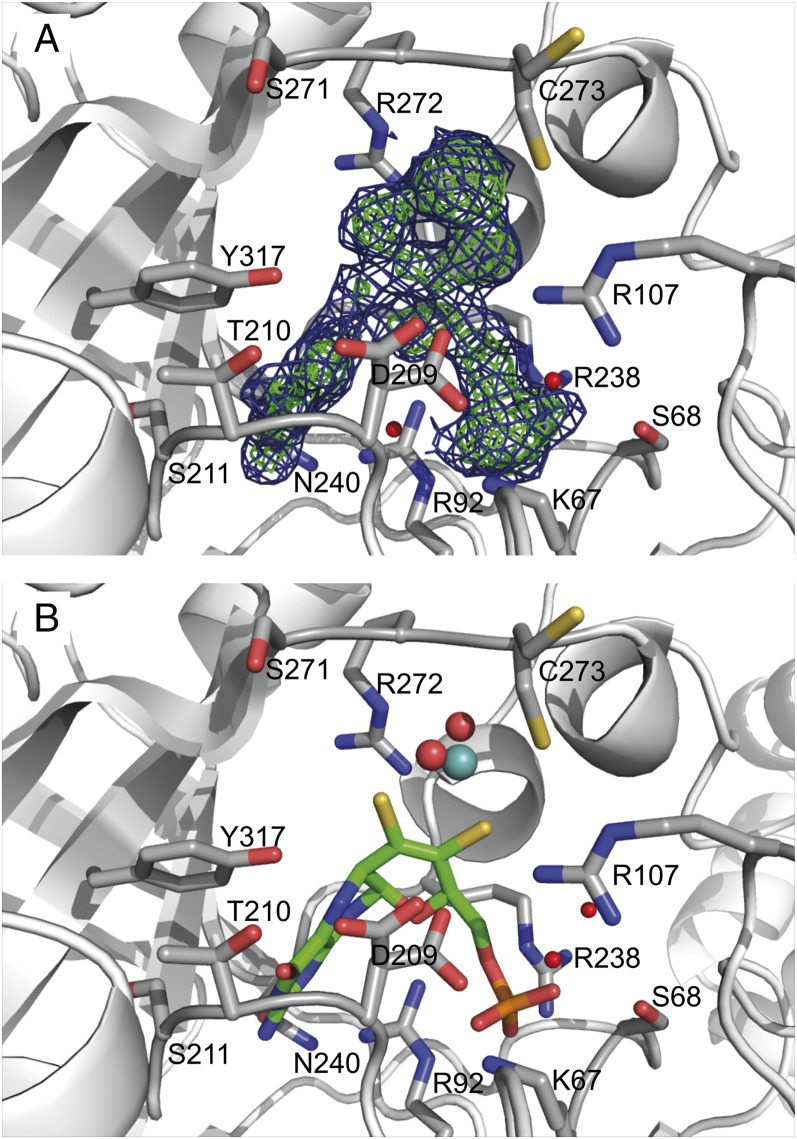
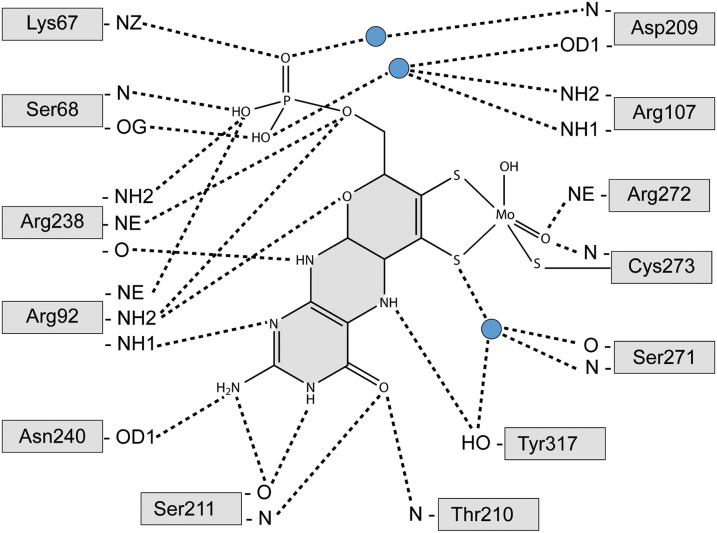
The Moco is tightly bound within the core region of hmARC1 by mostly positively charged amino acids and residues carrying a hydroxyl group in their side chain (Fig. 3). Besides the two dithiolene sulfurs from the molybdopterin backbone and two oxygen ligands, the central molybdenum is coordinated by C273, forming a slightly distorted coordination geometry (neither ideal square pyramidal nor trigonal bipyramidal). This cysteine residue is highly conserved among all mARC proteins throughout all species and is part of a common CxxC motif. The most prominent Moco-coordinating side chain is R92, which interacts with several atoms of the cofactor via polar and ionic interactions, keeping it strictly in place. The pterin ring system is further bound by residues T210, S211, P212, R238, N240, and Y317, while the phosphate moiety is coordinated by K67, S68, R92, and R238 (Fig. 4). This multitude of specific interactions between the cofactor and surrounding protein residues allows for a tight coordination of the molybdopterin backbone within the core of the enzyme. However, the reactive site comprising the molybdenum and its ligands remains exposed to the surface of the protein, where NHCs are being recruited and subsequently reduced.
Fig. 3.
Molybdenum cofactor binding site. The hmARC1 protein backbone is depicted in cartoon representation. Residues interacting with the Moco are shown as sticks and colored according to different atom types (gray, carbon; red, oxygen; blue, nitrogen; yellow, sulfur; orange, phosphorus). (A) Composite omit map of the Moco. Blue, 2Fo − Fc map, contoured at 1.0σ; green, Fo − Fc map, contoured at 2.5σ. (B) Representation of the cofactor as it was modeled into the electron density map. The molybdopterin backbone is depicted in stick representation, and the molybdenum ion and its oxygen ligands are shown as spheres.
Fig. 4.
Coordination of the molybdenum cofactor. Residues interacting with the Moco are depicted as gray boxes. Dashed lines represent hydrogen bonds between polar atoms. Blue circles indicate water molecules.
Assignment to the Xanthine Oxidase or Sulfite Oxidase Family.
Eukaryotic molybdenum enzymes are currently classified into two different families, which are distinguishable from each other by the composition of the five-coordinate ligand sphere of the molybdenum center. In both families, the molybdenum is coordinated by the dithiolene sulfurs of the molybdopterin as well as two oxygen (either oxo or hydroxyl) ligands. Members of the sulfite oxidase (SO) family contain a protein-derived cysteine sulfur as the fifth ligand, while molybdenum enzymes of the xanthine oxidase (XO) family carry an inorganic “terminal sulfur” ligand, which is essential for catalytic activity (15). Abstracting this terminal sulfur ligand from members of the XO family by cyanide treatment leads to inactivation of the respective enzyme (21, 22). Human mARC proteins, however, did not release any sulfur in the form of thiocyanate after cyanide treatment, excluding their belonging to the XO family (23). In contrast, mARC homologs from different source organisms were assigned to the SO family of molybdenum enzymes by pulsed electron paramagnetic resonance (24), X-ray absorption near-edge structure, and extended X-ray absorption fine structure at the molybdenum K edge (25), as well as biochemical studies and activity assays performed with mARC variants (26). Taken together, these studies clearly identified a protein-derived cysteine as the fifth ligand, which is indeed confirmed by our hmARC1 crystal structure.
Apart from physical and biochemical investigations, the pyranopterin conformations of currently available protein structures of mononuclear molybdenum and tungsten enzymes were analyzed (27). By deriving a distortion coordinate based on dihedral angles within the prosthetic group, the authors could show that the pyranopterin conformation can be correlated with their former biochemical assignment to either the XO or SO enzyme family. Interestingly, despite being well-characterized as a member of the SO family, the hmARC1 crystal structure reveals a pyranopterin conformation with dihedral angles of α = −43.3° and β = 73.4° (SI Appendix, Fig. S3). Remarkably, this angle combination would assign hmARC to the XO family based on the analysis of Rothery et al. (27).
Therefore, we suggest that mARC proteins should not be directly assigned to either of the two currently proposed and distinct mononuclear molybdenum enzyme families, since they (at least hmARC1) exhibit combined characteristics of both the SO and XO families.
Composition of the Active Site and Substrate Binding Area.
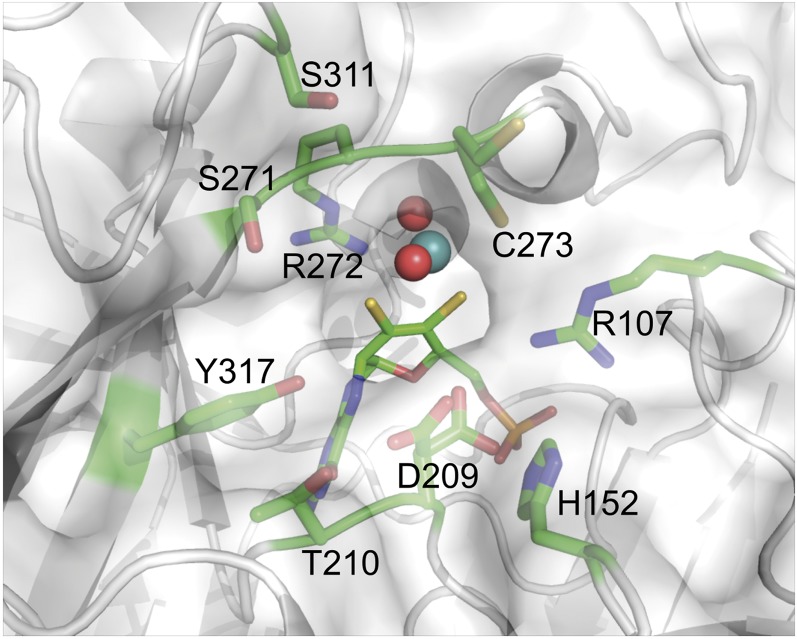
While the molybdopterin moiety itself is tightly anchored within the enzyme, the reactive center is well-accessible from the solvent area. There are few spatial limitations to the active site and substrate binding area, which is mainly composed of residues C273, D209, R272, S271, R107, Y317, T210, H152, and S311 (Fig. 5). With the exception of T210, H152, and S271, these residues are highly conserved throughout mARC proteins from different organisms, suggesting quite similar substrate spectra among them. However, we were able to identify two potential key residues close to the active site which are indicative for either mARC1 or mARC2. While H152 did not appear to be conserved among all analyzed mARC enzymes, it is indeed highly conserved among the mARC1 orthologs, whereas in mARC2 there is a likewise conserved phenylalanine in this position. The second discriminator between the two paralogs is S271. Among mARC1 sequences, mostly serine or threonine residues can be found in this position, whereas mARC2 proteins predominantly display a proline (SI Appendix, Fig. S4). To examine the impact of these paralog-specific amino acids, we performed preliminary reductase activity assays with a potential marker substrate of hmARC1 and the hmARC2_P270S variant. The hydroxamic acid derivative CP544439, a matrix metalloproteinase inhibitor, is almost exclusively reduced by hmARC1 (10). In contrast to the almost inactive wild-type hmARC2, its P270S variant shows a remarkably higher activity toward this substrate, even though reductase activity levels are not comparable to those of hmARC1 (SI Appendix, Fig. S5).
Fig. 5.
Representation of the hmARC1 active site. The enzyme is shown in cartoon as well as semitransparent surface representation. The Moco is depicted in stick (molybdopterin backbone) and sphere representation (molybdenum ion and oxygen ligands). Residues in close proximity to the reactive molybdenum center are shown in stick representation and colored according to different atom types (green, carbon; red, oxygen; blue, nitrogen; yellow, sulfur; orange, phosphorus).
The solvent-exposed reactive center as well as the absence of any highly specific substrate binding site clearly account for the broad substrate spectrum of mARC enzymes. Also, given the presence of some residues with alternative conformations (C273 and D209), the active site allows for some plasticity, making it even more adjustable toward different kinds of substrates. hmARC1 displays positively charged residues surrounding the active site (SI Appendix, Fig. S6). Since a variety of substrates contain nitrogen atoms with at least a partial positive charge, this was unexpected. However, there is one negatively charged, highly conserved, residue (D209) in direct proximity to the molybdenum center, which is crucial for catalytic activity (discussed below) and is most likely involved in the binding of the hydroxylated nitrogen of any given substrate. The surrounding positively charged patch might instead be crucial for the interaction between mARC1 and its electron-delivering redox-partner protein cytochrome b5, which displays negatively charged residues on its surface surrounding the heme cofactor [information derived from PDB ID code 3NER (28)].
Discussion
The high-resolution crystal structure of hmARC1 allows detailed insight into the fourth mammalian molybdenum-dependent enzyme. Previous biochemical characterizations of mARC proteins can be directly correlated with the 3D architecture of the enzyme. Recently, the mARC homolog from the green alga Chlamydomonas reinhardtii (crARC) was intensively investigated with regard to highly conserved amino acid residues among all mARC enzymes (29). The authors identified three residues which are essential for reduction activity toward NHCs. These residues correspond to hmARC1 residues D209, F237, and R298 (SI Appendix, Fig. S7). Another two residues (corresponding to hmARC1 R298 and F237) were shown to have an impact on Moco coordination, while some others (corresponding to hmARC1 L180, R238, E251, and E289) were identified to lead to inactivation of the enzyme when exchanged for alanine. Our crystal structure of hmARC1 strongly supports these findings. The aspartic acid residue D209 seems to have a Moco-independent impact on mARC enzymatic activity. D209 is situated in direct proximity to the molybdenum ion and its hydroxo ligand (SI Appendix, Fig. S8A). We propose that this residue is responsible for binding the hydroxylated nitrogen atom of any NHC and coordinating it in such a way that allows for the hydroxo ligand of the Moco to be exchanged for the NHC, resulting in a reaction intermediate (Fig. 1). Furthermore, D209 is the only negatively charged residue within a mostly alkaline substrate binding area, which further supports its role in recruiting hydroxylated nitrogen compounds. Other identified amino acid residues are essential for the structural integrity of the mARC enzyme. F237 is the central amino acid of a hydrophobic core between the large β-barrel and helices α4, α7, and α8, securing the 3D arrangement of the Moco binding site (SI Appendix, Fig. S8B). R298 is the central residue which connects the large β-barrel and helices α6 and α8 via polar and ionic interactions as well as cation–π stacking (SI Appendix, Fig. S8C). Replacement of either of these residues probably results in a partial collapse of these domains and leads to the observed decrease or even loss of Moco-binding capacity and ultimately the loss of NHC reductive activity. Other residues proposed to be involved in the interaction between the redox-partner proteins and/or the electron transfer between them (equal to hmARC1 L180, R238, N240, E251, D252, and L295) are essential for structural integrity, as can be deduced from the presented hmARC1 structure (SI Appendix, Fig. S8 D–H).
Furthermore, the presented structure of hmARC1 will allow structural interpretation of prospectively discovered MOSC proteins and their computationally predicted MOSC and MOSC_N domains. While there are two domains distinguishable within the hmARC1 crystal structure, these do not correlate with the in silico domain definitions. In contrast to former hypotheses, the MOSC_N domain does not represent a β-barrel–like fold. Although there is a large β-barrel, which contributes to a major part of one structural domain, this one mainly comprises β-strands from the predicted MOSC domain in concert with the very N-terminal β-strand of the MOSC_N domain. Since both predicted domains of MOSC proteins are intertwined on a structural level, we propose a definition of MOSC domains to be deposited in databases which takes into account sequence motifs as well as the now-available structural information. Of note, the recently published crystal structures of the bacterial enzyme YiiM (20) (PDB ID codes 5YHH and 5YHI) also contribute to the MOSC superfamily description. However, they belong to another subfamily and are lacking their essential molybdopterin prosthetic group, and the MOSC_N/MOSC-specific topology is not discussed.
The hmARC1 crystal structure revealed an unexpected conformation of the bound Moco, which is usually exclusively observed in molybdenum-dependent enzymes belonging to the XO family. However, mARC proteins from different organisms have been clearly identified as members of the SO family based on biochemical and biophysical features. Therefore, in contrast to previous assumptions, XO and SO families of Moco-containing enzymes might not be strictly separated from each other—at least not in all cases. On the other hand, mARC proteins might represent an evolutionary link between the two enzyme families. This hypothesis is supported by the high sequence similarity between mARC and the C terminus of Moco sulfurase. The latter protein is composed of two functional domains and catalyzes the final maturation step, the sulfuration of the Moco as it is found within enzymes of the XO family (22, 30). In a multistep reaction, this enzyme abstracts sulfur from a free l-cysteine with its cysteine desulfurase domain, transfers it to a cysteine residue within the MOSC domain, and subsequently sulfurates recruited SO Moco. Finally, the sulfurated Moco is released and inserted into enzymes of the XO family (31). Two things can be concluded from this reaction: (i) members of the XO family are likely to have emerged later than the SO enzymes, and (ii) Moco sulfurase needs to be able to bind both, SO-type as well as XO-type Moco, via its MOSC domain. It directly links both families of molybdenum enzymes to each other. There are indications that Moco sulfurases evolved from MOSC proteins like mARC by domain fusion with the aforementioned cysteine desulfurase-like domains (19). Conclusively, mARC enzymes could be the ancestors of Moco sulfurase proteins and represent an evolutionary link between different families of Moco-dependent enzymes while still retaining their ability to bind the SO type of Moco, though in a conformation usually observed within members of the XO family.
The active site of hmARC1 is revealed to be almost completely solvent-exposed. Like many other enzymes involved in biotransformation, mARC proteins are generalists rather than specialists, which provide a fast and efficient detoxification system for a variety of NHCs. This necessitates a free access of any substrate to the active site which is not buried inside a restricting binding pocket. The limited substrate specificity of mARC enzymes is realized by only a few residues surrounding the reactive molybdenum center, which are strictly conserved throughout different organisms. This explains why it is very difficult to derive structure–activity relationships (6). Thus, functional groups with N-hydroxylated components are reduced irrespective of the rest of the molecule. For example, ximelagatran, a thrombin inhibitor and a large molecule with many other functional groups, is reduced to a similar extent as a simple N-hydroxylated benzamidine (benzamidoxime) (7).
By comparison of mARC sequences from different eukaryotes, we could identify two residues close to the active site which appear to be paralog-specific and can therefore be used to discriminate between mARC1 and mARC2. The impact of one of these residues was supported by preliminary results of an hmARC2 protein variant which gained hmARC1-specific reductase features. These findings might explain the few differences in their substrate preferences and specific enzymatic activity, despite their mostly overlapping substrate spectrum. Analysis of the importance of these residues will be the subject of future research on a variety of mARC variants. Furthermore, these paralog-specific residues might be used for future annotations of mARC enzymes which are not yet deposited in databases.
Methods
Protein Expression, Purification, and Crystallization.
The design, protein expression, purification, functional characterization, and crystallization of the hmARC1–T4L fusion construct have been described in detail before (32). Briefly, the N-terminally truncated fusion protein was expressed in Escherichia coli TP1000 cells and purified by sequential affinity and cation-exchange chromatography. Purified protein was analyzed for N-reductive activity and subjected to hanging-drop vapor-diffusion crystallization setups, yielding hmARC1-T4L crystals of the orthorhombic space group P212121, which diffracted to a resolution of 1.65 to 3.5 Å.
NADH cytochrome b5 reductase and cytochrome b5 were expressed and purified as described before (23). Expression vectors for the hmARC2_P270S variant were generated according to the QuikChange site-directed mutagenesis protocol (Stratagene) by using the Phusion high-fidelity DNA polymerase (Fermentas) with the pQE80-hmARC2 plasmid as template. Expression and purification were performed in analogy to the wild-type enzyme (23).
Reductase Activity Assay.
N-reductive activity toward CP544439 was performed according to Ginsel et al. (10) with slight modifications: The amount of used protein was doubled, incubation volume was reduced to 75 µL, and injection volume for HPLC analysis was set to 20 µL. Activity of used enzyme sources was verified by incubation with benzamidoxime, showing that all mARC proteins have comparable activity toward the model substrate.
Data Collection, Phasing, Model Building, and Refinement.
Diffraction data were collected at beamline P14 (DESY PETRA III; EMBL) by using a Pilatus 2M detector. Data were collected at 100 K, detector distance of 136.2 mm, wavelength of 0.9789 Å, oscillation range of 0.1°, and exposure time of 0.00146 s per frame. Due to needle-shaped crystal morphology and to limit radiation damage, a helical data collection strategy along the longitudinal axis of the crystals was chosen. The best dataset derived from a single crystal which diffracted to a resolution of 1.78 Å.
Phasing was performed by molecular replacement (MR) using MOLREP (33). Due to the lack of structurally characterized mARC homologs, MR had to be performed in sequential steps using partial models of the fusion protein. First, T4L (PDB ID code 206L) was used as a search template. A convincing solution with just one molecule within the asymmetric unit was found. After rigid-body refinement performed by REFMAC5 (34), reliable electron density distribution was found for the lysozyme molecule. Some additional electron density was visible, which had to account for hmARC1, yet it was poorly defined and did not allow modeling the molecule. To improve phases, additional molecular replacement runs were performed with T4L as a fixed input model and additional structure elements of truncated hmARC1 models. These incomplete homology models were generated using MODELLER within the HHpred server (35, 36) and the crystal structure of the bacterial Moco-dependent enzyme YuaD (PDB ID code 1ORU; sequence identity to hmARC1: 25.38%). MR solutions were found using a model containing several β-strands as well as an α-helix (residues W94 to N98 and S116 to F183) and, subsequently, a model containing the predicted β-barrel. The position of the β-barrel was found, yet the orientation was inverted and residues were incorrectly assigned. However, due to improved phases, the electron density distribution was sufficiently well defined to allow for manual inspection in Coot (37) and correctly building the hmARC1 Cα chain step by step. Iterative refinement cycles were performed using Coot and REFMAC to complete the model; 97.9% of all modeled residues lie within the favored region, and 2.1% lie within the allowed region of the Ramachandran plot. There are no outliers. Refinement statistics are summarized in SI Appendix, Table S1.
Visualization.
The pH-dependent electrostatic potential maps at pH 7.4 were calculated by using the software APBS with the PARSE force field (38). Input files in PQR format for APBS were generated from files containing hmARC1 atom coordinates in PDB format with the program PDB2PQR to calculate pKa values and set protonation states for titratable groups accordingly (39). Composite omit maps were generated using the program PHENIX (40). Simulated annealing cycles were performed to erase model bias. The representation of residues involved in binding the Moco was derived from an analysis with LigPlot+ (41). The WebLogo representation of conserved mARC paralog discriminators was created using the online tool at weblogo.berkeley.edu/logo.cgi (42). The multiple sequence alignment of 58 mARC1 and 65 mARC2 proteins from different mammalian organisms as well as the alignment of hmARC1 with crARC were performed with Clustal Omega (43). The mARC sequences were derived from the Ensemble website (www.ensemble.org/), GeneTree StableID ENSGT00530000063150, and node_id 20095130 (mARC1 orthologs) and 20094778 (mARC2 orthologs) as of March 17, 2018. Incomplete sequences were sorted out before analysis. The final figure for the hmARC1–crARC alignment was prepared using the program ESPript (44). The secondary structure of hmARC1 was derived by DSSP (45). The sequence alignment of hmARC1 with gsYiiM is deduced from a structure-based alignment using the pairwise structure comparison option within the Dali server (46). All visualization and preparation of 3D structural images were performed using the program PyMOL (47).
Data and Material Availability.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.wwpdb.org/) under ID code 6FW2.
Supplementary Material
Acknowledgments
We thank Tracy Palmer and Grant Buchanan (University of Dundee) for sharing E. coli strain TP1000. We gratefully acknowledge access to the core facilities of the BiMo/LMB of Kiel University. Diffraction data were collected on beamline P14 operated by the EMBL at the PETRA III storage ring. Our research received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStructX (Grant Agreement 283570). Additionally, we are grateful for access to the HTX crystallization facility and beamtime at P14 by means of a grant from BioStructX at the EMBL Outstation Hamburg.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6FW2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808576115/-/DCSupplemental.
References
- 1.Foti RS, Dalvie DK. Cytochrome P450 and non-cytochrome P450 oxidative metabolism: Contributions to the pharmacokinetics, safety, and efficacy of xenobiotics. Drug Metab Dispos. 2016;44:1229–1245. doi: 10.1124/dmd.116.071753. [DOI] [PubMed] [Google Scholar]
- 2.Hlavica P. N-oxidative transformation of free and N-substituted amine functions by cytochrome P450 as means of bioactivation and detoxication. Drug Metab Rev. 2002;34:451–477. doi: 10.1081/dmr-120005646. [DOI] [PubMed] [Google Scholar]
- 3.Havemeyer A, et al. Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J Biol Chem. 2006;281:34796–34802. doi: 10.1074/jbc.M607697200. [DOI] [PubMed] [Google Scholar]
- 4.Ott G, et al. Reduction of sulfamethoxazole hydroxylamine (SMX-HA) by the mitochondrial amidoxime reducing component (mARC) Chem Res Toxicol. 2014;27:1687–1695. doi: 10.1021/tx500174u. [DOI] [PubMed] [Google Scholar]
- 5.Krompholz N, et al. The mitochondrial amidoxime reducing component (mARC) is involved in detoxification of N-hydroxylated base analogues. Chem Res Toxicol. 2012;25:2443–2450. doi: 10.1021/tx300298m. [DOI] [PubMed] [Google Scholar]
- 6.Bauch E, et al. Electrochemical and mARC-catalyzed enzymatic reduction of para-substituted benzamidoximes: Consequences for the prodrug concept “amidoximes instead of amidines.”. ChemMedChem. 2015;10:360–367. doi: 10.1002/cmdc.201402437. [DOI] [PubMed] [Google Scholar]
- 7.Gruenewald S, et al. The fourth molybdenum containing enzyme mARC: Cloning and involvement in the activation of N-hydroxylated prodrugs. J Med Chem. 2008;51:8173–8177. doi: 10.1021/jm8010417. [DOI] [PubMed] [Google Scholar]
- 8.Jakobs HH, et al. The mitochondrial amidoxime reducing component (mARC): Involvement in metabolic reduction of N-oxides, oximes and N-hydroxyamidinohydrazones. ChemMedChem. 2014;9:2381–2387. doi: 10.1002/cmdc.201402127. [DOI] [PubMed] [Google Scholar]
- 9.Schneider J, Girreser U, Havemeyer A, Bittner F, Clement B. Detoxification of trimethylamine N-oxide by the mitochondrial amidoxime reducing component mARC. Chem Res Toxicol. 2018;31:447–453. doi: 10.1021/acs.chemrestox.7b00329. [DOI] [PubMed] [Google Scholar]
- 10.Ginsel C, et al. The involvement of the mitochondrial amidoxime reducing component (mARC) in the reductive metabolism of hydroxamic acids. Drug Metab Dispos. 2018;46:1396–1402. doi: 10.1124/dmd.118.082453. [DOI] [PubMed] [Google Scholar]
- 11.Havemeyer A, et al. Reduction of N-hydroxy-sulfonamides, including N-hydroxy-valdecoxib, by the molybdenum-containing enzyme mARC. Drug Metab Dispos. 2010;38:1917–1921. doi: 10.1124/dmd.110.032813. [DOI] [PubMed] [Google Scholar]
- 12.Plitzko B, Havemeyer A, Kunze T, Clement B. The pivotal role of the mitochondrial amidoxime reducing component 2 in protecting human cells against apoptotic effects of the base analog N6-hydroxylaminopurine. J Biol Chem. 2015;290:10126–10135. doi: 10.1074/jbc.M115.640052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llamas A, Chamizo-Ampudia A, Tejada-Jimenez M, Galvan A, Fernandez E. The molybdenum cofactor enzyme mARC: Moonlighting or promiscuous enzyme? Biofactors. 2017;43:486–494. doi: 10.1002/biof.1362. [DOI] [PubMed] [Google Scholar]
- 14.Kotthaus J, et al. Reduction of N(ω)-hydroxy-L-arginine by the mitochondrial amidoxime reducing component (mARC) Biochem J. 2011;433:383–391. doi: 10.1042/BJ20100960. [DOI] [PubMed] [Google Scholar]
- 15.Hille R, Nishino T, Bittner F. Molybdenum enzymes in higher organisms. Coord Chem Rev. 2011;255:1179–1205. doi: 10.1016/j.ccr.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott G, Havemeyer A, Clement B. The mammalian molybdenum enzymes of mARC. J Biol Inorg Chem. 2015;20:265–275. doi: 10.1007/s00775-014-1216-4. [DOI] [PubMed] [Google Scholar]
- 17.Fischer K, et al. Structural basis of eukaryotic nitrate reduction: Crystal structures of the nitrate reductase active site. Plant Cell. 2005;17:1167–1179. doi: 10.1105/tpc.104.029694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havemeyer A, Lang J, Clement B. The fourth mammalian molybdenum enzyme mARC: Current state of research. Drug Metab Rev. 2011;43:524–539. doi: 10.3109/03602532.2011.608682. [DOI] [PubMed] [Google Scholar]
- 19.Anantharaman V, Aravind L. MOSC domains: Ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including molybdenum cofactor sulfurases. FEMS Microbiol Lett. 2002;207:55–61. doi: 10.1111/j.1574-6968.2002.tb11028.x. [DOI] [PubMed] [Google Scholar]
- 20.Namgung B, Kim JH, Song WS, Yoon SI. Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site. Sci Rep. 2018;8:3304. doi: 10.1038/s41598-018-21660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massey V, Edmondson D. On the mechanism of inactivation of xanthine oxidase by cyanide. J Biol Chem. 1970;245:6595–6598. [PubMed] [Google Scholar]
- 22.Wahl RC, Warner CK, Finnerty V, Rajagopalan KV. Drosophila melanogaster ma-l mutants are defective in the sulfuration of desulfo Mo hydroxylases. J Biol Chem. 1982;257:3958–3962. [PubMed] [Google Scholar]
- 23.Wahl B, et al. Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J Biol Chem. 2010;285:37847–37859. doi: 10.1074/jbc.M110.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajapakshe A, et al. Structural studies of the molybdenum center of mitochondrial amidoxime reducing component (mARC) by pulsed EPR spectroscopy and 17O-labeling. Biochemistry. 2011;50:8813–8822. doi: 10.1021/bi2005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giles LJ, et al. Molybdenum site structure of MOSC family proteins. Inorg Chem. 2014;53:9460–9462. doi: 10.1021/ic5015863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamizo-Ampudia A, Galvan A, Fernandez E, Llamas A. The Chlamydomonas reinhardtii molybdenum cofactor enzyme crARC has a Zn-dependent activity and protein partners similar to those of its human homologue. Eukaryot Cell. 2011;10:1270–1282. doi: 10.1128/EC.05096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothery RA, Stein B, Solomonson M, Kirk ML, Weiner JH. Pyranopterin conformation defines the function of molybdenum and tungsten enzymes. Proc Natl Acad Sci USA. 2012;109:14773–14778. doi: 10.1073/pnas.1200671109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parthasarathy S, et al. Accommodating a nonconservative internal mutation by water-mediated hydrogen bonding between β-sheet strands: A comparison of human and rat type B (mitochondrial) cytochrome b5. Biochemistry. 2011;50:5544–5554. doi: 10.1021/bi2004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamizo-Ampudia A, Galvan A, Fernandez E, Llamas A. Study of different variants of Mo enzyme crARC and the interaction with its partners crCytb5-R and crCytb5-1. Int J Mol Sci. 2017;18:E670. doi: 10.3390/ijms18030670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amrani L, et al. Comparison of the sequences of the Aspergillus nidulans hxB and Drosophila melanogaster ma-l genes with nifS from Azotobacter vinelandii suggests a mechanism for the insertion of the terminal sulphur atom in the molybdopterin cofactor. Mol Microbiol. 2000;38:114–125. doi: 10.1046/j.1365-2958.2000.02119.x. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- 32.Kubitza C, et al. T4 lysozyme-facilitated crystallization of the human molybdenum cofactor-dependent enzyme mARC. Acta Crystallogr F Struct Biol Commun. 2018;74:337–344. doi: 10.1107/S2053230X18006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Jurrus E, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 42.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrödinger LLC 2015. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.