Significance
DNA is a stiff polymer that is condensed in vivo by polyamines and cationic proteins, many of which are intrinsically disordered. In eukaryotes, where DNA is packaged as chromatin, condensation of internucleosomal (linker) DNA is effected by the C-terminal tail of histone H1. Its structure and DNA binding are poorly understood, as is the link between H1 phosphorylation and chromatin structural changes during the cell cycle. Here we present a model system, with which we show that H1 binds tightly to DNA but retains a high level of disorder. Phase-separated condensates form, containing higher-order structures that are highly sensitive to the phosphorylation state of H1, suggesting a mechanism by which condensation of the chromatin fiber and other assemblies might be regulated.
Keywords: intrinsic disorder, histone H1, chromatin, phosphorylation, phase separation
Abstract
Disordered proteins play an essential role in a wide variety of biological processes, and are often posttranslationally modified. One such protein is histone H1; its highly disordered C-terminal tail (CH1) condenses internucleosomal linker DNA in chromatin in a way that is still poorly understood. Moreover, CH1 is phosphorylated in a cell cycle-dependent manner that correlates with changes in the chromatin condensation level. Here we present a model system that recapitulates key aspects of the in vivo process, and also allows a detailed structural and biophysical analysis of the stages before and after condensation. CH1 remains disordered in the DNA-bound state, despite its nanomolar affinity. Phase-separated droplets (coacervates) form, containing higher-order assemblies of CH1/DNA complexes. Phosphorylation at three serine residues, spaced along the length of the tail, has little effect on the local properties of the condensate. However, it dramatically alters higher-order structure in the coacervate and reduces partitioning to the coacervate phase. These observations show that disordered proteins can bind tightly to DNA without a disorder-to-order transition. Importantly, they also provide mechanistic insights into how higher-order structures can be exquisitely sensitive to perturbation by posttranslational modifications, thus broadening the repertoire of mechanisms that might regulate chromatin and other macromolecular assemblies.
Molecular recognition of disordered proteins by their partners has long been assumed to involve a disorder-to-order transition (1). However, recently, there have been several examples of complexes in which the proteins retain a high level of disorder (2–5), some of very high affinity (6). Many of these proteins have low sequence complexity (7), consisting of, or containing stretches of, charged amino acid residues, and behave as flexible polyelectrolytes. Binding of oppositely charged species causes ion displacement and entropically driven binding (8). Nucleic acids also behave as polyelectrolytes by virtue of their spine of negatively charged phosphates, and, in solutions of cationic polymers, DNA condenses into a compact, solvent-excluded state (9, 10). The formation of DNA−protein condensates plays an essential role in a wide variety of biological processes, as a means of overcoming the inherent rigidity of DNA, thereby facilitating its packing (10). The compact state and the conformational transitions between condensed forms are of interest for their relevance to chromosome structure and the packing of DNA in viruses. The DNA condensate in model systems that is “polymer- and salt-induced,” originally described by the acronym “ψ” (psi) (11), is an ordered assembly of B-DNA, arranged in parallel twisted helical segments with a well-defined spacing (12). Ψ-DNA produces an anomalously large scattering signal in circular dichroism (CD) spectra (13). More recently, RNA−protein binding has been shown to drive formation of membraneless liquid organelles capable of compartmentalizing biomolecules through charge-mediated liquid−liquid phase separation or “complex coacervation” (14). Phase separation has been postulated to be key to gene silencing by heterochromatin protein 1 (HP1) (15, 16). Of particular relevance to our study is the assembly and maintenance of the condensed chromatin fiber by linker histones (17).
Many cellular processes are regulated by protein phosphorylation. Phosphorylation has a regulatory role in the assembly of RNA granules (18), complex coacervation of model organelles (19), and droplet formation by HP1 (15). Phosphorylation of H1 occurs reversibly during the cell cycle (20) and results in an increased mobility of H1 in the cell nucleus, presumably due to lower affinity for DNA (21). In S phase, phosphorylation correlates with a more open chromatin structure that would facilitate replication and transcription and, in M phase, probably enables rearrangement of the condensed chromatin structure (e.g., to allow entry of condensins) (22). The chromatin-condensing properties of H1 are mainly conferred by its ca. 100-residue-long polycationic C-terminal tail (23), CH1. In chromatin, the tail, which contains several phosphorylation sites, interacts with and condenses internucleosomal linker DNA (24–26).
We have identified a model system (CH1 and an optimized dsDNA) that allows us to study separately the stages before and after complex coacervation. We use the model further to study the effect of phosphorylation. We find that the complexes contain highly disordered CH1, and form phase-separated droplets, which contain higher-order structures. These structures do not form when the protein is phosphorylated. Firstly, our findings demonstrate that tight binding and a high level of disorder are not mutually exclusive; secondly, they suggest how phosphorylation may affect condensation and higher-order structuring in macromolecular assemblies including chromatin.
Results
CH1 Is Highly Disordered, both Free and When Bound to DNA.
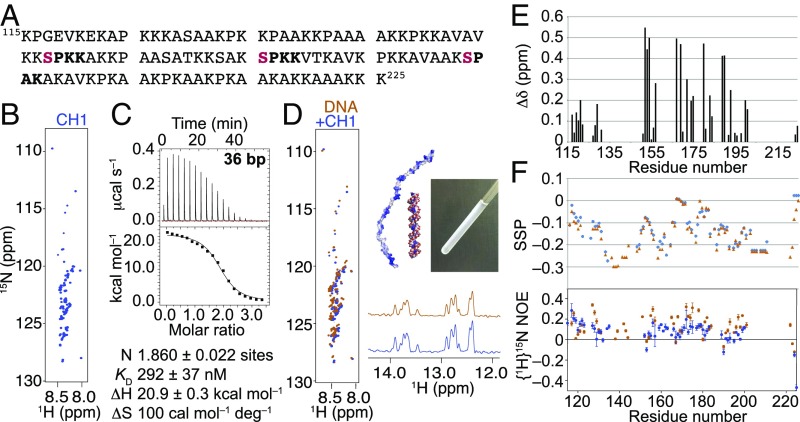
The free polypeptide (Fig. 1A) was characterized extensively by NMR spectroscopy and, as has long been known (27), shows an extremely high level of disorder, displaying low chemical shift dispersion and narrow line widths in 15N-heteronuclear single-quantum coherence (HSQC) spectra (Fig. 1B), a distinguishing feature of disordered proteins (28). The lack of dispersion is further exacerbated by the very low sequence complexity (41% lysine, 31% alanine, 13% proline) and the number of repeated sequence motifs (e.g., PKAAK occurs three times, KSPKK occurs twice, and PKKAV occurs twice).
Fig. 1.
CH1 is a highly disordered polypeptide, both free and complexed with DNA. (A) CH1 amino acid sequence, with consensus CDK2/Cyclin A sites in bold, and phosphorylatable serines in red. (B) The 15N-HSQC spectrum of free CH1 (blue) displays the sharp line widths and poor chemical shift dispersion characteristic of a highly disordered protein. (C) Isotherm from titration of CH1 into 36-bp DNA (I = 0.16 M) and thermodynamic parameters from fitting to a one-site model. (D) Titration of 15N CH1 into 36-bp DNA produces a turbid solution (Inset), but, in the complex (0.5:1 CH1:DNA; orange), the polypeptide is still detectable by 15N HSQC (Left) and the DNA is detectable by 1H imino resonances (Bottom Right). (E) Chemical shift differences compared with free CH1 are quantified. (F) SSP and HNOE values.
Previous studies of the binding of the related mouse subtypes H1.1, H10, and H1.4 to short dsDNA using isothermal titration calorimetry (ITC) reported a Kd of ca. 100 nM, and a DNA site size of 32 bp to 36 bp for the full-length proteins and 28 bp for the C-terminal tail of H10 (29, 30). [A similar value of 24 bp was reported for the average spacing of H1 molecules in complexes of two duplexes bridged by a cooperatively bound array of H1 molecules (31).] These lengths are similar to the length of internucleosomal linker DNA (ca. 34 bp) in “canonical” chromatin. As a starting point, we therefore measured the binding affinity of CH1 for 36-bp DNA by ITC (Fig. 1C), at approximately “physiological” ionic strength (I ca. 0.16 M; 10 mM sodium phosphate and 150 mM NaCl). The fitted Kd was 292 nM, and the molar stoichiometry (CH1:DNA) was 1.86:1, indicating that the DNA was long enough to bind more than one CH1. We then conducted a titration by NMR. The 15N CH1 was added to a solution of 36-bp DNA, which became increasingly cloudy following each addition (Fig. 1D). A viscous liquid layer appeared at the bottom of the NMR tube on standing—evidence of phase separation. Despite this, there were only minor differences between the CH1/DNA 15N-HSQC spectrum and that of CH1, namely, small but widespread chemical shift changes and slightly increased chemical shift dispersion and line widths. However, the peak intensities did not increase by the expected amount following each CH1 addition, indicating that some CH1 had partitioned into an NMR-invisible state. The polypeptide that was “visible” retained all of the striking features of a highly disordered protein despite the evidence from ITC that the majority would be bound at the concentrations present in the NMR tube (100 μM; Kd = 292 nM). The DNA also appeared essentially unchanged in structure, as judged by the presence of 1H imino resonances, indicating base pairing (Fig. 1D). The only visible change was a reduction in intensity.
Sequence-specific assignment of CH1 using a conventional triple-resonance approach was challenging due to the near-perfect overlap of the 13C nuclei of the 45 lysine and 34 alanine residues. However, reasonable dispersion in the 15N dimension allowed most of the backbone nuclei in both the free and DNA-bound bulk-phase states to be assigned with confidence using the HNN/HN(C)N approach (32). Three PKAAK repeats near the C terminus and a run of four alanines following proline (141PAAAA145) gave rise to heavily overlapped peaks, many of which could not be assigned sequence-specifically; the sequences were nevertheless clearly disordered, as judged by chemical shifts close to random coil values. With the assignments that could be obtained, the chemical shift changes associated with DNA binding were mapped (Fig. 1E). The largest changes were observed for V152 and V154 (Δδ 0.55 and 0.50 ppm, respectively), closely followed by various serine and threonine residues distributed through the central portion of the sequence. Two sensitive measures of secondary structure were used to look for subtle differences between the free and DNA-bound CH1: secondary structure propensity (SSP) scores and {1H}15N heteronuclear NOE (HNOE) to probe motions on a timescale faster than overall tumbling (Fig. 1F). SSP scores are ca. +1 or −1 in well-formed α- and β-structures, respectively (33). Our data for free CH1 indicate a disordered polypeptide with a slight propensity for β-type structures (mean SSP −0.12 ± 0.07), as expected for a highly expanded polyelectrolyte chain (6). There were only slight changes to the SSP scores on binding DNA (shift of the mean to −0.16 ± 0.09). For free CH1, the HNOE had an average value of 0.07 ± 0.11, with no single residue exhibiting a value of >0.3. On binding DNA, the average value increased slightly to 0.12 ± 0.16, and to slightly over 0.3 for K127, S172, and S175. The overall picture of the DNA-bound polypeptide is one of a highly dynamic chain.
Identification of the Optimal DNA Substrate.
The stoichiometry of 1.86:1 CH1:DNA measured by ITC indicated that the 36-bp DNA is long enough to bind more than one CH1 molecule. In an attempt to generate well-defined 1:1 complexes, we screened three shorter dsDNAs (32, 26, and 20 bp) at the same I (0.16 M). We found that stoichiometry corresponds closely to DNA length (Fig. 2A), and that 20 bp provides the best match, with a stoichiometry of 0.996:1 (Fig. 2B). Do these stoichiometries represent complexes in which all charge is neutralized? CH1 has a net charge of +43; the total charge of synthetic dsDNA with n bp is 2(n − 1), i.e., −70 and −38 for the 36- and 20-bp DNAs, respectively. The stoichiometries of 1.86:1 and 0.996:1 for the 36- and 20-bp complexes therefore lead to a residual net charge of +10 and +5, respectively, which, presumably, either is stabilized by the phosphate and chloride ions in the buffer or the overall charge is modified by charge regulation effects (34).
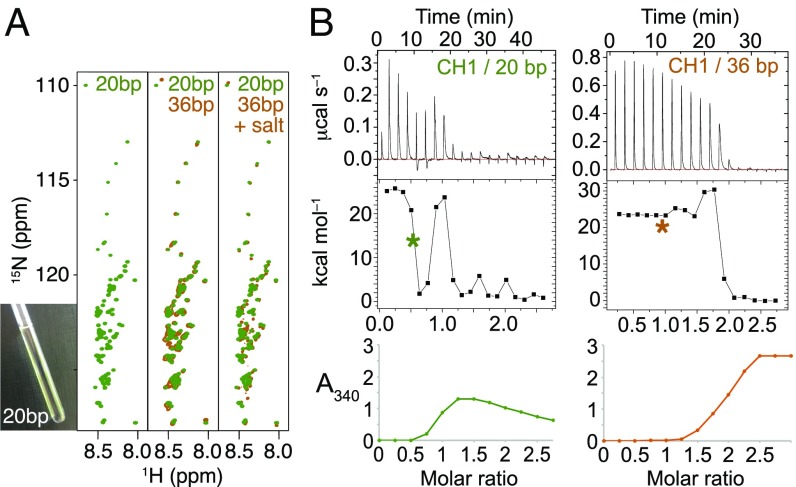
Fig. 2.
Optimization of DNA length. (A) Binding stoichiometry from ITC vs. DNA length. (B) Isotherm from titration of CH1 into 20 bp DNA (I = 0.16 M), and thermodynamic parameters from fitting to a one-site model.
The charge dependence of the interaction was tested with both the 36- and 20-bp DNA at a much lower I (ca. 10 mM), which would be expected to provide less charge screening. There was little difference in the 15N-HSQC spectra with DNA of different lengths (Fig. 3A). Although the 15N-HSQC spectra looked similar to the 36-mer at I = 0.16 M, the low I solutions remained clear during titration until after a 0.5:1 CH1:DNA stoichiometry was reached. The binding isotherms measured by ITC acquired bimodal features indicating a two-stage process; the second process correlated with increasing turbidity (Fig. 3B). Similar bimodal isotherms have been reported previously for other polyelectrolyte pairs (35, 36), and have been interpreted as a primary ion-pairing event followed by complex coacervation, consistent with turbidity measurements and observation of a coalesced liquid layer after centrifugation, as for our complexes. The two stages were unusually distinct for the 20-bp DNA, where they were baseline-separated, and had defined molar stoichiometries of ∼0.5 and 1.0, respectively. A range of DNA lengths (18 bp to 36 bp) was screened by ITC and turbidity, but the baseline-separated stages were only observed for the 20-bp/CH1 complex (SI Appendix, Fig. S1). This is likely to be due to a match between molar stoichiometry and charge neutralization at this particular DNA length. When binding to the full-length protein was explored, it was clear that this is dominated by the C-terminal tail (SI Appendix, Fig. S1). The 20-bp DNA with CH1 at low I therefore constitutes a good model system of defined stoichiometry, in which the distinct formation of a noncoacervating (1:2) and coacervating (1:1) CH1:DNA complex permits their separate study.
Fig. 3.
Distinct stages of binding of CH1 to 20 bp DNA at low I (ca. 10 mM). (A) Titration of 15N CH1 into 20- or 36-bp DNA at low I followed by 15N HSQC produces a clear solution at a molar ratio of 0.5:1 CH1:DNA (20-bp complex shown in Inset);15N HSQC of CH1:20 bp DNA complex at low I (green, Left) and superimposed on the CH1:36 bp complex (orange) at low I (Middle) and I = 0.16 M (Right) for comparison. (B) Isotherms (Top) and turbidity measurements (Bottom) obtained at low I. The isotherms show bimodal features indicative of two stages of complex formation, which are most dramatic for the 20-bp DNA (see also SI Appendix, Fig. S1). Asterisks mark the stoichiometry at which the 15N-HSQC spectra in A were obtained.
Phosphorylated CH1 Is Disordered, both Free and in DNA-Bound Complexes.
Given the dependence of coacervation events on phosphorylation in other systems (15, 18, 19), we tested the effect of phosphorylation on the noncoacervating and coacervating CH1/DNA complexes. The 15N CH1 was enzymatically phosphorylated to produce CH1-P, in which S157, S175, and S193 in the three SPxK CDK2/Cyclin A consensus sites were fully phosphorylated, as evidenced by their pronounced downfield 1H shifts (37, 38) in 15N-HSQC spectra (Fig. 4A). With the exception of these residues, backbone chemical shift changes are relatively small and localized to residues i − 1 to i + 4 (i is pSer). SSP and HNOE values (Fig. 4B) revealed a similar picture of small, localized changes. The average SSP score was unchanged after phosphorylation (−0.12). HNOE values were all <0.3, consistent with a highly dynamic chain that samples multiple conformations on a rapid timescale, with a slight stiffening of the chain around the pSer, consistent with an intraresidue hydrogen bond between the phosphate and backbone amide (37). CD spectroscopy also showed both CH1 and CH1-P to be disordered (Fig. 4C).
Fig. 4.
Secondary structure and dynamics of CH1 and CH1-P. (A) (Top) Phosphorylation of the three serine residues in SPxK sites shown by their pronounced downfield shift in CH1-P (red) compared with CH1 (blue; see dotted ellipse) (37). (Bottom) Chemical shift changes are quantified. (B) SSP scores and HNOE values. (C) CD signatures consistent with disorder.
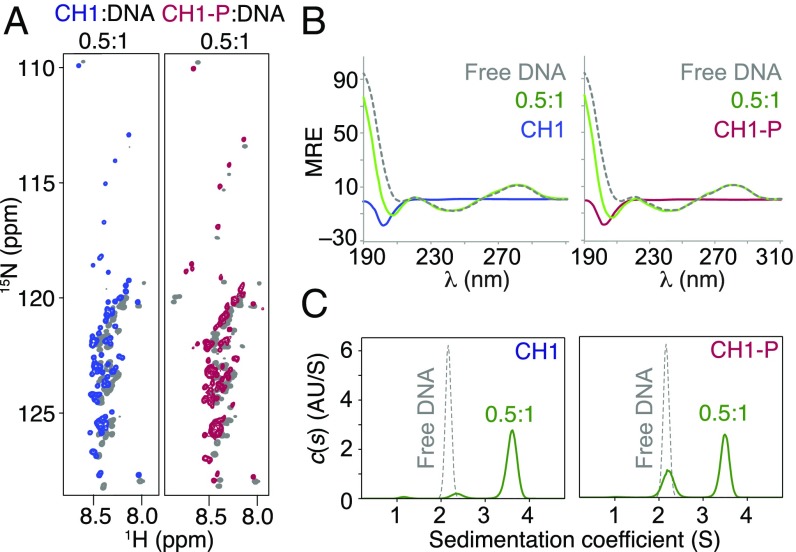
In the complex formed with 20-bp DNA at low I, CH1-P displayed small but widespread chemical shift changes and slightly increased chemical shift dispersion, as observed for CH1 (Fig. 5A). However, the complex solution was slightly turbid (see below). No gain of structure was seen by CD; the profile obtained for the complexes was the sum of the spectra of the free polypeptide and DNA (Fig. 5B), implying that they consist of a highly disordered polypeptide and unchanged B-form DNA. The size and composition of the complexes was investigated using analytical ultracentrifugation (AUC). CH1 and the 20-bp DNA have similar masses (11.3 and 12.2 kDa, respectively) but very different shapes, CH1 being a flexible extended chain and the DNA a short rigid rod. Free CH1 and CH1-P did not sediment, due to their high frictional ratio (f/f0), but the 20-bp DNA sedimented with s20,w of ca. 2.2 (Fig. 5C) and a fitted f/f0 of 1.55. The CH1/20-bp complex was well defined, sedimenting with s20,w of ca. 3.5 and a fitted molecular mass of ca. 35 kDa. The fitted f/f0 (1.65) was only slightly higher than that for free DNA, implying a high degree of compaction of the protein in the complex. The composition of the complex could be calculated by comparison of the integrals obtained from the UV detector and interferometer; the complex was 66% DNA by mass, or 61% DNA in molar terms, close to the expected 2:1 ratio. Therefore, the initial complex is a discrete, high-affinity complex in solution, containing one CH1 bound to two 20-bp DNA molecules. For CH1-P, a similarly sedimenting complex formed, but the peak was reduced in intensity and a significant amount of DNA remained in the free state, implying that the CH1-P complex was of lower affinity. This was investigated further by ITC.
Fig. 5.
CH1-P retains disorder in the DNA complex. The polypeptides in the 0.5:1 complexes at low I retain a high degree of disorder by (A) 15N-HSQC (free proteins in gray) and (B) CD, in which the resultant spectra (green) are the sum of the B-form DNA (gray, dotted) and disordered polypeptide (CH1 in blue, CH1-P in red) components. (C) Sedimentation velocity (SV) continuous c(s) distributions show that in each case the major species has an s20,w 3.5 (cf. s20,w 2.2 for free DNA). (The 20-bp DNA and CH1 are 12.2 and 11.3 kDa, respectively.)
Thermodynamics of the Two-Stage Process.
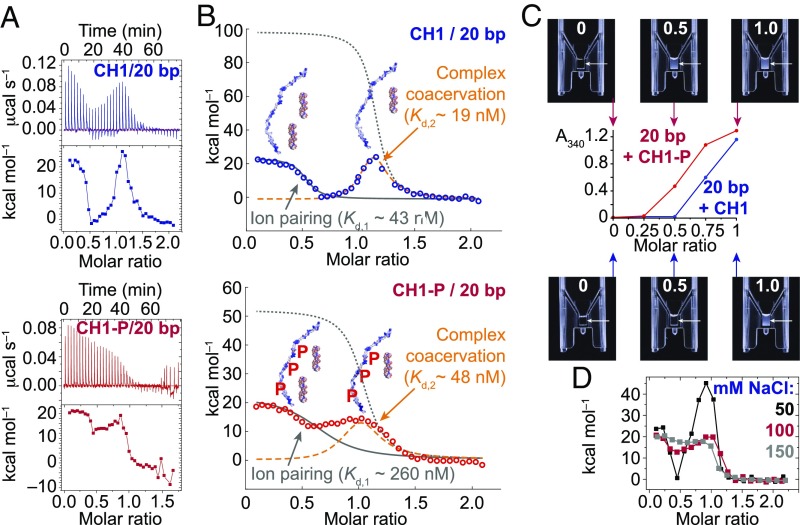
High-resolution isotherms were obtained in triplicate to fit the more complex profiles obtained for CH1 and CH1-P titrated into 20-bp DNA at low I (Fig. 6A). The isotherms fit a model with two independent sites, modified for sequential binding with apparent positive cooperativity due to coacervation (see Materials and Methods) (Fig. 6B). CH1-P showed a similar stoichiometry to CH1, but a less pronounced dip in the isotherm between the two stages. This appears to be due primarily to a reduced affinity of CH1-P for DNA in the first binding event (Kd,1 for CH1-P is ca. 260 nM vs. 43 nM for CH1), such that the steps partially merge. This implies that the onset of complex coacervation is earlier for CH1-P, consistent with the slight turbidity seen in the NMR tube, and with turbidity measurements (Fig. 6C).
Fig. 6.
ITC and two-step model fitting. (A) High-resolution isotherms from titration of CH1 (blue) and CH1-P (red) into 20-bp DNA at low I. (B) Fitting to a two-step model (see Materials and Methods) yields approximate affinities for ion pairing to form an initial complex at a protein:DNA molar ratio of ca. 0.5, followed by formation of a second complex at a molar ratio of ca. 1.0, which undergoes coacervation. (C) Turbidity assessed visually in microvolume cuvettes and by A340 measurements. (D) ITC profiles of CH1 titrated into 20-bp DNA at a range of I (50, 100, and 150 mM NaCl).
The steps also merge with increasing ionic strength. A gradual change in profile with increasing I (from biphasic to merged) is clear (Fig. 6D and SI Appendix, Fig. S1), and, at I of ca. 0.11 M (10 mM sodium phosphate and 100 mM NaCl), the ITC profile closely resembles that for CH1-P (Fig. 6A). Again, merging reflects a reduced affinity (Kd for CH1 at I = 0.16 M is 292 nM for 36 bp and 101 nM for 20 bp; Figs. 1B and 2B). The origin of the reduced affinity on increasing I or on phosphorylation appears to be entropic, since the enthalpy is essentially constant (ca. +20 kcal⋅mol−1; y intercept of the isotherms). The entropy gain for counterions released from polyelectrolyte chains is reduced when there are already plenty of free counterions present in the solution, as has been shown by simulations (39). At low I, the second event—formation of a complex of 1:1 stoichiometry accompanied by coacervation—was distinct and fitted a sequential binding model (Fig. 6B). ΔH2 is not well determined by such a complex fit, but is ca. twofold lower for CH1-P than for CH1 (+50 kcal⋅mol−1 vs. +100 kcal⋅mol−1), implying ΔS2 is also reduced upon phosphorylation, since the interaction of CH1-P is both lower in affinity and less endothermic.
Phosphorylation Reduces Partitioning from the Bulk to the Coacervate Phase.
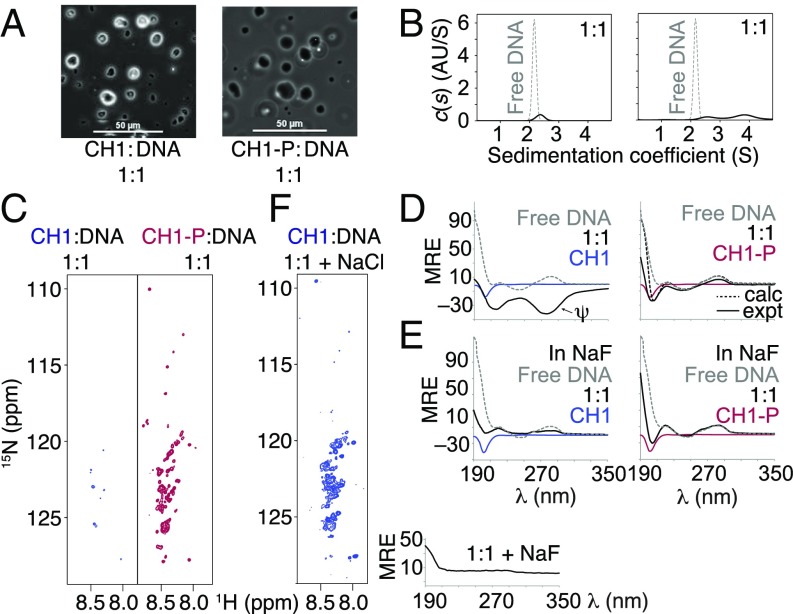
Solutions of complexes containing a 1:1 molar ratio of CH1-P or CH1:20 bp DNA were visibly cloudy. Optical microscopy revealed the presence of micrometer-scale droplets (Fig. 7A). Only a small quantity of free DNA was detectable in the bulk phase by AUC, the majority sedimenting too quickly to be detected (Fig. 7B). However, for CH1-P, a small peak was present at ca. 3.8 S, indicating that some CH1-P:DNA complex was present in the bulk phase. The 15N-HSQC spectra showed a loss of signal intensity (Fig. 7C), which was severe for the CH1:DNA complex, indicating that most of the material had entered the separated phase, whereas, for CH1-P, consistent with the centrifugation results, a greater proportion of the material remained in the solution phase.
Fig. 7.
Partitioning and structure in the CH1 and CH1-P:20 bp DNA coacervate phases. (A) Phase-separated droplets are micrometer-scale in diameter. (Scale bar: 50 μm.) (B) Sedimentation velocity (SV) continuous c(s) of the CH1 (Left) and CH1-P (Right) complexes. (C) The 15N-HSQC spectra show dramatic loss of signal for CH1, and partial loss for CH1-P. (D) CH1-P/DNA CD spectrum resembles the sum of the spectra of the components (compare solid and dotted black lines); CH1/DNA gains negative ellipticity characteristic of ψ-DNA. (E) Complexes prepared in 150-mM NaF do not form higher-order ψ-type structures. (F) Addition of 150-mM salt destroys preformed higher-order ψ-structures (loss of negative ellipticity) and causes some complexes to repartition into the solution phase (HSQC spectrum).
Long-Range Order in the Coacervate Phase Is Highly Phosphorylation- and Salt-Dependent.
Despite the similarity in the phase separation behavior of the CH1 and CH1-P complexes, differences were revealed by CD (Fig. 7D). The CH1:DNA complex gave a pronounced negative ellipticity characteristic of long-range DNA order with a long helical periodicity typical of ψ-DNA (11, 13), in which side-by-side twisted packing of B-form DNA has been inferred. In striking contrast, the CD spectrum for the CH1-P/DNA complex could be approximated to an appropriately weighted sum of the individual components. CD spectra of both CH1 and CH1-P complexes acquired in the presence of 150 mM NaF (I of ca. 0.16 M) show the protein to be disordered and the DNA to be simple B form (Fig. 7E). Addition of salt to preformed ψ-DNA structures destroyed the long-range order irreversibly, and the 15N-HSQC spectrum showed some recovery of solution-phase signals (Fig. 7F). The translational diffusion coefficient of these recovered signals was measured (40) as ca. 8 × 10−7 cm2⋅s−1, consistent with soluble species in the bulk phase, and the peaks were also still observable after the coacervate had been spun down. Taken together, this demonstrates that addition of salt results in the release of disordered complexes within the droplets and a degree of repartitioning into the solution phase. Thus, as indicated by ITC, addition of salt appears to recapitulate the effects of phosphorylation.
Discussion
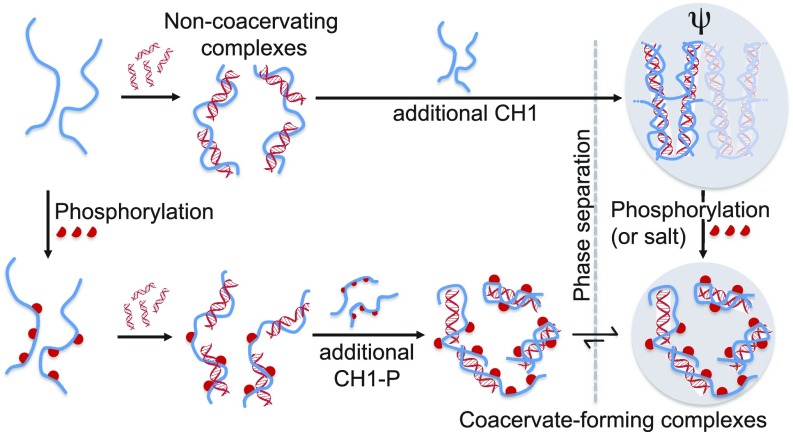
Together, these data demonstrate that the long C-terminal tail of linker histone H1 forms high-affinity complexes with DNA in which the polypeptide component is, as measured by several different methods, highly disordered. Phosphorylation of CH1 acts to modulate the thermodynamics of binding, lowering the affinity. This is achieved by phosphorylation of just three residues, spaced along the tail and bringing down the net charge by only 10%. Similar effects are recapitulated at ionic strengths approaching physiological; triphosphorylation and an increase in I of 0.1 M produce a similar reduction in the affinity. Binding is a two-step process in which the steps are fully separable only for the 20-bp DNA at low I, when the protein is unphosphorylated (the tightest binding conditions). The second step involves an increase in the protein:DNA stoichiometry accompanied by complex coacervation. The events are summarized in Fig. 8. Reduced coacervation of the CH1-P/DNA complexes is in line with a study of RNA/protein liquid organelles, in which diphosphorylation of a model peptide containing four arginine residues effectively neutralized the charge (at pH 7.4) and dissolved the coacervate phase completely (19).
Fig. 8.
Summary of findings. Our model system permits isolation of a tight-binding CH1:DNA (1:2) complex at low I that is noncoacervating, requiring additional CH1 to phase separate. Higher-order structures (ψ-DNA) are formed, which are dispersed by addition of salt. Accompanying the release of higher-order structure is a partial repartitioning to the solution phase. We propose that a similar function may be achieved in chromatin by H1 phosphorylation, since salt and phosphorylation have similar effects on the thermodynamics of H1/DNA complex formation, higher-order structure formation, and bulk/droplet partitioning.
In all but the conditions of tightest binding (low I and no phosphorylation, where analysis of CH1 is made difficult due to the swamping effect of the ψ-DNA signal), it is clear that CH1 is disordered even in the condensed, phase-separated state. In the tightest-binding conditions (ψ-DNA), there was no obvious indication from CD that CH1 adopts defined secondary structures, including the cross β-structure observed for Sup35 (41), and FUS (42). Thus, we conclude that CH1 is highly disordered, both free and in various DNA-bound states. This most probably explains the difficulty in directly visualizing the H1 tail in chromatin by cryo-EM (43) and is consistent with a previous suggestion that the “stem” containing linker DNA emerging from nucleosomes (44)—which is the binding site for the H1 C-tail—is, in fact, an ensemble of thermally fluctuating, partially protected structures (45). Retention of disorder may represent a more entropically favorable way of achieving charge neutralization than a disorder-to-order transition.
Disordered protein/protein complexes have previously been observed between the H1 tail and the long uniformly acidic tail of the architectural transcription factor, HMGB1 (4), and recently for H1 in complex with the linker histone chaperone protein ProTα (46), where the binding is even tighter (picomolar) (6). From a structural viewpoint, there are clear differences between the dynamic snake-like association of two disordered polymers in the latter case and a highly flexible extended chain condensing on to the surface of a structured rod-like partner in the CH1/20-bp DNA complex. If HMGB1 and ProTα facilitate the rapid exchange of H1 between chromatin sites in vivo, it seems likely that this role is facilitated by the disorder itself; the tight binding is the sum of multiple noninteracting weaker-binding sites that can be “peeled away” one by one by either HMGB1 or ProTα, thus facilitating the exchange.
It remains to be seen whether the higher-order structure we detect in our model system in any way reflects the condensation of linker DNA stem structures in chromatin compaction, and this will be the subject of further study. However, it is probably relevant that the higher-order structure in the low-I coacervate is only metastable, being easily dispersed by salt, which results in some repartitioning to the bulk phase. In vivo, where the ionic conditions are relatively constant, the dispersal and repartitioning mechanism could be phosphorylation.
In the different but not entirely unrelated situation of chromatin compaction by HP1, it has recently been proposed that the formation of phase-separated droplets is promoted by either DNA binding or by phosphorylation of the HP1 N-terminal extension (15, 16). When phosphorylated HP1 phase separates in the cell nucleus, chromatin permeates the droplets and condenses, allowing formation of heterochromatin regions. We did not observe phase separation of CH1 or CH1-P under any of the conditions we studied, but a mechanism of phase separation before DNA binding cannot be excluded in vivo, as this could be achieved by binding to other partners.
Conclusion
CH1 binds tightly to DNA, retaining its high level of disorder. The weakened binding of the phosphorylated protein causes reduced partitioning to the coacervate phase. The higher-order structures formed by CH1 and the 20-bp DNA under the conditions of tightest binding (here, low ionic strength) might reflect the situation inside the condensed chromatin domain where linkers are juxtaposed, and might even drive the condensation. Whether or not the phase transitions we observe in vitro occur in vivo, the environment is likely to resemble the solvent-excluded conditions that occur in the chromatin fiber. Within a highly condensed phase, phosphorylation could be the primary mechanism for dispersing long-range order, acting as a driver of dramatic changes in structural plasticity.
Materials and Methods
Recombinant chicken histone H1 subtype H1.11L and CH1 (residues 115 to 225) were purified (4) and phosphorylated (47). Oligonucleotides were based on 36-mer dsDNAs (30). ITC data were fit to a one-site model in Origin, or to a two-step sequential process based on the Multiple Non-Interacting Sites model developed to fit two-site processes where Kd,2 < Kd,1 (48). Full details are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Tim Hunt for the GST-CDK2 and cyclin A3 constructs, Rebecca Michael for expression and purification of the CDK2/cyclin complex, Ian Hu for assistance with microscopy, and Chris Green for assistance with photography. This work was supported by the Biotechnology and Biological Sciences Research Council, initially through the award of Grant BB/D002257/1 (to J.O.T.) and a Research Studentship (to L.C.), and then through the award of Grant BB/N022181/1 (to K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Chemical shift assignments have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 27633).
See Commentary on page 11868.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805943115/-/DCSupplemental.
References
- 1.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Mittag T, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Watson M, Stott K, Fischl H, Cato L, Thomas JO. Characterization of the interaction between HMGB1 and H3-a possible means of positioning HMGB1 in chromatin. Nucleic Acids Res. 2014;42:848–859. doi: 10.1093/nar/gkt950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgia A, et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Insights into protein structure and function from disorder-complexity space. Proteins. 2007;66:16–28. doi: 10.1002/prot.21055. [DOI] [PubMed] [Google Scholar]
- 8.Mascotti DP, Lohman TM. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc Natl Acad Sci USA. 1990;87:3142–3146. doi: 10.1073/pnas.87.8.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro JT, Leng M, Felsenfeld G. Deoxyribonucleic acid-polylysine complexes. Structure and nucleotide specificity. Biochemistry. 1969;8:3219–3232. doi: 10.1021/bi00836a014. [DOI] [PubMed] [Google Scholar]
- 10.Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T, Venable JH, Jr, Lerman LS. The structure of psi DNA. J Mol Biol. 1974;84:37–64. doi: 10.1016/0022-2836(74)90211-3. [DOI] [PubMed] [Google Scholar]
- 12.Maestre MF, Reich C. Contribution of light scattering to the circular dichroism of deoxyribonucleic acid films, deoxyribonucleic acid-polylysine complexes, and deoxyribonucleic acid particles in ethanolic buffers. Biochemistry. 1980;19:5214–5223. doi: 10.1021/bi00564a010. [DOI] [PubMed] [Google Scholar]
- 13.Shin YA, Eichhorn GL. Formation of ψ (+) and ψ (-) DNA. Biopolymers. 1984;23:325–335. doi: 10.1002/bip.360230211. [DOI] [PubMed] [Google Scholar]
- 14.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 15.Larson AG, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednar J, Hamiche A, Dimitrov S. H1-nucleosome interactions and their functional implications. Biochim Biophys Acta. 2016;1859:436–443. doi: 10.1016/j.bbagrm.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Wang JT, et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aumiller WM, Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 20.Gurley LR, D’Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 21.Contreras A, et al. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haering CH, Nasmyth K. Building and breaking bridges between sister chromatids. BioEssays. 2003;25:1178–1191. doi: 10.1002/bies.10361. [DOI] [PubMed] [Google Scholar]
- 23.Hendzel MJ, Lever MA, Crawford E, Th’ng JPH. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 24.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 25.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas JO. Histone H1: Location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 27.Bradbury EM, et al. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The conformation of histone H1. Eur J Biochem. 1975;52:605–613. doi: 10.1111/j.1432-1033.1975.tb04032.x. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 29.Machha VR, et al. Calorimetric studies of the interactions of linker histone H1(0) and its carboxyl (H1(0)-C) and globular (H1(0)-G) domains with calf-thymus DNA. Biophys Chem. 2013;184:22–28. doi: 10.1016/j.bpc.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Machha VR, et al. Exploring the energetics of histone H1.1 and H1.4 duplex DNA interactions. Biophys Chem. 2014;185:32–38. doi: 10.1016/j.bpc.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Clark DJ, Thomas JO. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986;187:569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- 32.Panchal SC, Bhavesh NS, Hosur RV. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: Application to unfolded proteins. J Biomol NMR. 2001;20:135–147. doi: 10.1023/a:1011239023422. [DOI] [PubMed] [Google Scholar]
- 33.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lund M, Jönsson B. On the charge regulation of proteins. Biochemistry. 2005;44:5722–5727. doi: 10.1021/bi047630o. [DOI] [PubMed] [Google Scholar]
- 35.Priftis D, Megley K, Laugel N, Tirrell M. Complex coacervation of poly(ethylene-imine)/polypeptide aqueous solutions: Thermodynamic and rheological characterization. J Colloid Interface Sci. 2013;398:39–50. doi: 10.1016/j.jcis.2013.01.055. [DOI] [PubMed] [Google Scholar]
- 36.Vitorazi L, et al. Evidence of a two-step process and pathway dependency in the thermodynamics of poly(diallyldimethylammonium chloride)/poly(sodium acrylate) complexation. Soft Matter. 2014;10:9496–9505. doi: 10.1039/c4sm01461h. [DOI] [PubMed] [Google Scholar]
- 37.Du JT, et al. Low-barrier hydrogen bond between phosphate and the amide group in phosphopeptide. J Am Chem Soc. 2005;127:16350–16351. doi: 10.1021/ja054568p. [DOI] [PubMed] [Google Scholar]
- 38.Theillet FX, et al. Cell signaling, post-translational protein modifications and NMR spectroscopy. J Biomol NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou Z, Muthukumar M. Entropy and enthalpy of polyelectrolyte complexation: Langevin dynamics simulations. J Chem Phys. 2006;124:154902. doi: 10.1063/1.2178803. [DOI] [PubMed] [Google Scholar]
- 40.Wu DH, Chen A, Johnson CS. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J Magn Reson A. 1995;115:260–264. [Google Scholar]
- 41.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bednar J, et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol Cell. 2017;66:384–397.e8. doi: 10.1016/j.molcel.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 45.Meyer S, et al. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: Nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011;39:9139–9154. doi: 10.1093/nar/gkr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.George EM, Brown DT. Prothymosin α is a component of a linker histone chaperone. FEBS Lett. 2010;584:2833–2836. doi: 10.1016/j.febslet.2010.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welburn J, Endicott J. Methods for preparation of proteins and protein complexes that regulate the eukaryotic cell cycle for structural studies. Methods Mol Biol. 2005;296:219–235. doi: 10.1385/1-59259-857-9:219. [DOI] [PubMed] [Google Scholar]
- 48.Kim W, Yamasaki Y, Kataoka K. Development of a fitting model suitable for the isothermal titration calorimetric curve of DNA with cationic ligands. J Phys Chem B. 2006;110:10919–10925. doi: 10.1021/jp057554e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.